Abstract
Trypanosoma brucei lacks mitochondrial genes encoding tRNAs and must import nuclearly encoded tRNAs from the cytosol. The mechanism and specificity of this process remain unclear. We have identified a unique sequence motif, YGG(C/A)RRC, upstream of the genes encoding mitochondrially localized tRNAs in T. brucei. Both in vitro import studies and in vivo transfection studies indicate that deletion of the YGG(C/A)RRC sequence alters mitochondrial localization of tRNALeu, and in vivo studies also show a decrease in the cellular abundance of tRNALeu. These studies provide direct evidence for cis-acting RNA motifs within precursor tRNAs that facilitate the selection of tRNAs for mitochondrial import in trypanosomes. Furthermore, we found that mutations to the YGG(C/A)RRC sequence also altered the intracellular distribution of other endogenous tRNAs, suggesting a general role for this sequence in tRNA trafficking in trypanosomes.
Mitochondria typically encode all of the rRNAs and tRNAs necessary for autonomous protein synthesis. However, import of nuclearly encoded tRNAs has been documented in evolutionarily diverse species, including yeast, protozoa, plants (reviewed in reference 25), and marsupials (6). The numbers and identities of imported tRNAs are highly variable, but the only organisms identified to date that must import all of their mitochondrial tRNAs are the kinetoplastid protozoa Trypanosoma and Leishmania (8, 28).
tRNAs contain highly conserved intragenic RNA polymerase III promoter elements and a nearly ubiquitous tertiary structure (7). The sequence and structure conservation of tRNAs complicates how they might be specifically targeted to mitochondria. Most of the tRNAs in kinetoplastids are shared between the cytosol and mitochondrion but are localized to the mitochondrion with differing efficiencies, ranging from 1 to 7.5% of the total cellular tRNA in Trypanosoma brucei (30). tRNA structure is important for mitochondrial localization in T. brucei; however, this alone may not explain the differences in the abundance of imported tRNAs (11). It has been proposed that mitochondrially localized tRNAs in Leishmania may contain sequences that positively influence targeting and transport into the mitochondrion (3, 4, 16, 18, 22). A positive import determinant was discovered in the D arms of Leishmania tRNAIle(16) and tRNATyr (18). Swapping the D arms of imported tRNAIle and a cytosolically localized tRNAGln conferred mitochondrial import to the hybrid tRNAGln but did not eliminate mitochondrial localization of the hybrid tRNAIle, suggesting that there are multiple signals for import (16). In vitro studies with Leishmania mitochondria suggested that there might be different sequence or structural requirements for crossing the outer and inner membranes of mitochondria, indicating that import may involve two distinct steps (3). Furthermore, a SELEX (systematic evolution of ligands by exponential enrichment) procedure was used to isolate sequence aptamers that were imported into mitochondria with high efficiencies. One set of the import-competent aptamers contained the motif YGGYAGAGY, which is present in the anticodon or D arms of many tRNAs, whereas another set contained the motif UG3-4U, found in the V-T region of other tRNAs. These aptamers were able to interact with the inner membranes of isolated Leishmania mitochondria. Interestingly, the first motif is found in the D arm of the imported tRNATyr and the second is found in the imported tRNAIle (4). Although the previous studies suggest the presence of positive import determinants, an alternative view proposes that the mitochondrial import machinery may not discriminate between different tRNAs but may be negatively regulated by sequences or nucleotide modifications that inhibit import (12).
Although the role of 5′ flanking sequences in localization of tRNAs to the mitochondria of trypanosomes is highly debated, previous studies have shown the presence of tRNA precursors in the mitochondria of T. brucei (15). Transcription of one precursor tRNA was shown to initiate 14 nucleotides upstream of the tRNASer coding sequence and extend through a 59-nucleotide intergenic sequence and a downstream tRNALeu (15). These findings suggest that at least some tRNAs may be imported as precursors rather than as processed, mature tRNAs (15). Consistent with this result is the presence of RNase P activity in T. brucei mitochondria, although the ability of this activity to process in vitro imported precursor tRNAs to a mature size has not been demonstrated (9, 23).
In this paper, we present evidence that sequences upstream of tRNA coding regions, within the 5′ leader sequence, influence localization of tRNAs in T. brucei. Sequence analysis of the immediate 5′ flanking sequences of T. brucei tRNAs revealed the presence of a highly conserved dinucleotide GG within a conserved sequence motif, YGG(C/A)RRC. By 5′ rapid amplification of cDNA ends (RACE), we have determined that precursor tRNALeu, including this motif within the 14-nucleotide 5′ leader, is localized in both the cytosol and the mitochondrion. Interestingly, this sequence is similar to the previously published motif YGGYAGAGY found in import-competent tRNAs and aptamers in Leishmania (3, 4). Using an in vitro import system, we tested 5′ deletions of a tRNA precursor for the ability to be imported into mitochondria. A significant decrease in import was observed when the YGG(C/A)RRC motif was removed from the precursor tRNALeu. We also developed an in vivo system to further characterize the influence of flanking sequences on import. Mutations to the YGG(C/A)RRC sequence indicate that this 5′ flanking sequence is involved in maintaining both the abundance and cellular distribution of tRNALeu. Finally, not only does mutation of this sequence motif affect the localization of the tagged tRNALeu, it also has a global effect on localization of other endogenous tRNAs to the mitochondrion.
MATERIALS AND METHODS
Trypanosome growth, purification of mitochondria, and isolation of RNA.
T. brucei procyclic cells (TREU 667) were grown at 27°C in semidefined medium (5) containing 10% heat-inactivated fetal bovine serum (Sigma) and 20 μg of gentamicin sulfate (Life Technologies, Inc.) per ml. Mitochondria were isolated from cells at a density of 1 × 107 to 1.5 × 107/ml by use of a nitrogen cavitation bomb (minibomb cell disruption chamber; Kontes, Vineland, N.J.), as described previously for in vitro import assays (21, 32). Mitochondria were isolated from 4 to 8 liters of culture at a cell density of 1 × 107 to 1.5 × 107 cells/ml as described previously for in vivo import studies (10). Briefly, cells were suspended in hypoosmotic buffer and then lysed by passage through a 26-gauge needle. Mitochondrial vessels were isolated from a 20 to 35% Percoll gradient. Mitochondrial vessels were treated with 0.6 U of micrococcal nuclease (USB Corporation) per 3.5 × 106 cells in 1 ml of 10% glycerol-10 mM Tris-HCl (pH 8.0)-1 mM CaCl2 for 20 min at room temperature. The reaction was stopped by the addition of 0.5 M EDTA, pH 8.0, to a final concentration of 10 mM. The vesicles were recovered by centrifugation at 32,500 × g for 15 min (adapted from reference 8). The mitochondrial RNA was extracted from the vesicles with TriPure isolation reagent (Boehringer Mannheim) according to the manufacturer's instructions. Cytosolic RNA was isolated from procyclic T. brucei (TREU 667) as described previously (8), except that the RNA was extracted with Tripure isolation reagent (Boehringer Mannheim) after treatment with DNase I (Roche).
In vitro import of 5′ deleted tRNAs.
The 5′ deletions of the dicistronic tRNASer (CGA)-tRNALeu (CAA) were constructed from the previously described D1 genomic clone for in vitro import (15). The 5′ oligonucleotides for PCR amplification were generated by mixing 2 μM primer 19 (5′-GTTTTTGGCATTGCATAAATA-3′), primer 4 (5′-GTGTCTAAAACAAATCTGTTGTGG-3′), primer 5′ (5′-GGCAAGATGGCCGAGTGGTCTAAG-3′), or primer 1 (5′-GGTGGCGGGGCGGTGTCACC-3′) in 1× kinase buffer (New England Biolabs), 2 μM ATP, and 20 U of T4 polynucleotide kinase at 37°C for 2 h. After being subjected to the kinase, 100 pmol of the oligonucleotide was ligated with 2 μM primer 002 (T7 promoter is indicated in bold) (5′-GGCGCGCCTAATACGACTCACTATAG-3′). The ligation of primer 002 to primers 19, 4, and 5′ was carried out in a reaction containing 1× RNA ligation buffer, 5 μl of dimethyl sulfoxide, and 80 U of T4 RNA ligase (New England Biolabs) at 37°C for 2 h. The 5′ deletions were generated by PCR amplification with oligonucleotides as follows: Δ1-95, primers 002 and 19; Δ1-121, primers 002 and 4; and Δ1-154, primers 002 and 5′. The PCRs were denatured at 94°C for 1 min, annealed at 53°C for 1 min, and extended at 72°C for 2 min for 30 cycles. The PCR products were gel purified and T7 transcribed as described previously (32). The in vitro import assays were carried out as described previously (32).
Cloning of transfection vectors.
The tagged tRNALeu* was made by using the previously described D1 genomic clone (15). The mutant was created by using overlapping PCR. The first round of PCR used primer D15′ (5′-CATAGTTGTTGTGCCAATTTTTTTG-3′) and primer SerMut3′ (5′-CCTACGAATAAAAGAGAACTTG-3′) to create fragment 1 (the single nucleotide change in the variable arm of tRNALeu is indicated in bold). Fragment 2 was made with primer LeuMut5′ (5′-CAAGTTCTCTTTTATTCGTAGG-3′) and primer Leu3′ (5′-CAATCAGCCGATTTGGGTGC-3′). The PCR conditions were as follows: denaturing at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min for 20 cycles, followed by extension for an additional 5 min at 72°C. A second round of PCR was performed on overlapping fragments 1 and 2 with primer KpnD15′ (5′-GGTGGTACCAATTTTTTTGAATTTTCTATAC-3′) and primer LeuXho (5′-GAACTCGAGCAATCAGCCGATTTGGG-3′). This produced a fragment including a 244-nucleotide upstream sequence, tRNASer, the 59-nucleotide intergenic region, sequence-tagged tRNALeu* (C-U change at position 45 in the variable arm), and a 27-nucleotide 3′ sequence with a 5′ KpnI site and a 3′ XhoI site.
All constructs were made in the stable transfection construct pXS2 (generous gift of Jay Bangs) (2). The unique ClaI site in pXS2 was eliminated to make vector pXS2-ClaI. The PCR fragment with the sequence-tagged tRNALeu* and pXS2-ClaI were digested with KpnI and XhoI, and the fragment was cloned into these sites to create vector pSer-Leu*. Mutation of the GGCGG [conserved nucleotides in YGG(C/A)RRC] sequence upstream of tRNALeu* in vector pSer-Leu* was made by overlapping PCR under the previously stated conditions. Fragment 1 was made with primer D15′ and primer Int3′ (5′-GAGCGAACAACAGATTTGTTTTAGACACTTC-3′) by PCR on pSer-Leu*. Fragment 2 was made with primer Int5′ (5′-TCGCTCCGATGGCAAGATGGCCGAGTGGTC-3′) and primer Leu3′ by PCR on pSer-Leu*. The mutation of the sequence GGCGG to TCGCT is shown in bold in the primer sequences. A second round of PCR was performed on fragment 1 and fragment 2 with primer D15′Kpn and primer Leu3′Xho. The resulting mutated fragment was digested with KpnI and XhoI and cloned into the KpnI and XhoI sites of vector pXS2-ClaI. The resulting vector was pSer-Leu* 1. All of the following constructs were created with the ExSite PCR-based site-directed mutagenesis kit (Stratagene). The kit was used according to the manufacturer's instructions. PCR conditions were as follows: denaturing at 94°C for 4 min, annealing at 50°C for 2 min, extension at 68°C for 6 min, continued with 10 cycles of 94°C for 1 min, 56°C for 2 min, and 68°C for 5, and finished with a final cycle of 68°C for 8 min. pSerLeu* 2 was made from pSerLeu* with primer SerMut5′ (5′-GTCACCATACCCAAGTGGTTACGG-3′) and primer LeuMut3′ (5′-P-GAGCGACCGCCACCACCTCTCAAATGCG-3′). pSer-Leu* 2 has the sequence GGCGG directly flanking tRNASer mutated to TCGCT. Vector pLeu*(+) was made from pSer-Leu* by deleting tRNASer and the 59-nucleotide intergenic region upstream of tRNALeu* with primer SerDel5′ (5′-P-GGCAAGATGGCCGAGTGGTCTAAGGCG-3′) and primer SerDel3′ (5′-ACCGCCCCGCCACCTCTCAAATGC-3′). Vector pLeu*(+) has the 5′ flanking sequence of tRNASer directly abutting that of tRNALeu*. Sixteen nucleotides of 5′ flanking sequence were deleted from vector pLeu*(+) to create vector pLeu*(−) with primer SerDel5′ and primer SerUpDel (5′-P-CTCTCAAATGCGTTACCCTTGCGC-3′).
Transfection of trypanosomes.
The following protocol was adapted from the work of Bangs et al. (2). All vectors were Qiagen (Qiagen, Inc.) purified and linearized with ClaI to allow targeted integration in a nontranscribed region upstream of the endogenous tRNASer of the tRNASer-Leu gene cluster (see Fig. 3A). The 5′ flanking sequence for all integrated vectors was retained as the entire 5′ flanking sequence of the endogenous tRNASer because we used this site for integration. Each vector was phenol extracted once, chloroform extracted four times, precipitated with 100% ethanol, washed with 70% ethanol, and resuspended at 100 ng/ml in OptiMEM medium (Life Technologies, Inc.) Mid-log-phase (∼5 × 106 cells/ml) procyclic trypanosomes were washed once in ice-cold phosphate-buffered saline and resuspended at 4 × 107/ml in OptiMEM medium (Life Technologies, Inc.). OptiMEM (0.1 ml) containing linearized vector (total of 1 μg) was added to 0.5 ml of cells (2 × 107) in a 0.4-mm-wide cuvette (Bio-Rad) on ice. The cuvette was electroporated (1.4 kV, 25 Ω) with two pulses, 10 s apart, in a Bio-Rad gene pulser. Cells were transferred to 10 ml of prewarmed (27°C) semidefined medium, and at 48 h G418 (Invitrogen) was added to 50 μg/ml. Stable cell lines typically grew out in 10 to 14 days. Cultures were maintained with 25 μg of G418 per ml.
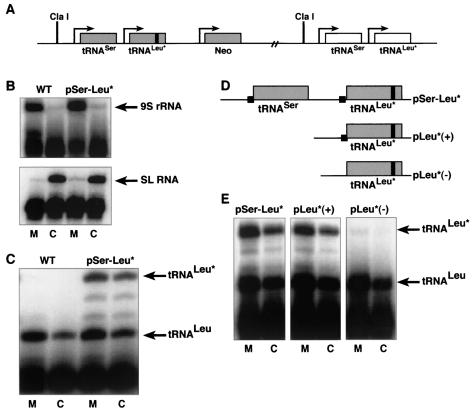
FIG. 3.
Deletion of the upstream motif influences the abundance of tRNALeu* in vivo. (A) T. brucei transfected cell lines containing a sequence-tagged tRNALeu*, with a C-to-U mutation at position 45 upstream of the endogenous tRNASer-tRNALeu gene cluster. (B) A poison primer extension assay was performed on 1 μg of the mitochondrial (M) or cytosolic (C) RNA fraction from wild-type trypanosomes (WT) and the transfectant lines analyzed (only pSer-Leu* is shown). Oligonucleotide 9Sb produces a 22-nucleotide extension product, predominantly in the mitochondrial fraction. Oligonucleotide SL2 also produces a 26-nucleotide extension product, but it is primarily in the cytosol. Mitochondrial or cytosolic fractions with >3% cross-contamination were not used for subsequent experiments. SL RNA, spliced leader RNA. (C) A poison primer extension assay was used to evaluate the abundance and localization of both endogenous tRNALeu and tagged tRNALeu* in mitochondrial (M) and cytosolic (C) RNA fractions with oligonucleotide LE4. Primer extensions were performed on 1 μg of either mitochondrial or cytosolic RNA. This assay produces a 25-nucleotide product for endogenous tRNALeu, present in both wild-type and pSer-Leu* transfected cells, and produces a 28-nucleotide product for the tagged tRNALeu* in the pSer-Leu* transfected cells. (D) Deletion analysis. pSer-Leu*, tagged tRNALeu with wild-type flanking sequences; pLeu*(+), tagged tRNALeu with a YGG(C/A)RRC motif upstream; pLeu*(−), tagged tRNALeu with upstream sequences, lacking both YGG(C/A)RRC motifs. (E) Poison primer extensions on 1 μg of a mitochondrial (M) or cytosolic (C) RNA fraction in transfectants pSer-Leu*, pLeu*(+), and pLeu*(−) detect endogenous tRNALeu, with a 25-nucleotide extension product, and tagged tRNALeu*, with a 28-nucleotide extension product. The extension products were quantitated with a phosphorimager.
Primer extension analysis of tRNA localization in vivo.
Each of the following oligonucleotides (Invitrogen) was labeled for use in primer extension reactions. Oligonucleotide 9Sb (5′-TATTTGCATATACCTAATGG-3′) is complementary to nucleotides 346 to 364 of the mitochondrial 9S rRNA (29). Oligonucleotide SL2 (5′-GTACAGAAACTGTTCTAATAG-3′) is complementary to the cytosolic 39-nucleotide spliced leader RNA (19). Oligonucleotide LE4 (5′-GGTTCGAACCCACGCCTACGAAT-3′) is complementary to nucleotides 48 to 70 of tRNALeu (CAA). Oligonucleotide Met e (5′-CGGTGAGGCTCGAACTCACGAC-3′) is complementary to nucleotides 46 to 67 of tRNAMet-e (CAU). Oligonucleotide Met i (5′-CCGGTTTTCGATCCAACG-3′) is complementary to nucleotides 47 to 67 of tRNAMet-i (CAU). Each oligonucleotide was 5′ end labeled with T4 polynucleotide kinase (New England Biolabs) under the following conditions: 100 pmol of oligonucleotide, 1× T4 polynucleotide kinase buffer, 20 U of polynucleotide kinase, and 300 μCi of 150-mCi/ml [γ-32P]ATP for 1 h at 37°C. The oligonucleotides were subsequently purified on a 19% polyacrylamide-8 M urea gel. The oligonucleotides were excised from the gel and eluted overnight in 0.5 M ammonium acetate-1 mM EDTA, pH 8.0, at room temperature. After precipitation with 100% ethanol and washing with 70% ethanol, the oligonucleotides were resuspended in double-distilled H2O at 2.25 pmol/μl.
The level of purity of the mitochondrial and cytosolic RNAs was tested by a poison primer extension assay. Both the mitochondrial and cytosolic RNAs extracted from the different transfectant lines were assayed for 9S rRNA and spliced leader RNA with oligonucleotides 9Sb and SL2, respectively. Each primer extension reaction contained the following: 1 μl of primer (2.25 pmol), 4 μl of 5× avian myeloblastosis virus reverse transcriptase (RT) buffer (Promega), 1 μl of 5 mM ddGTP (250 μM final concentration), 1 μl of 10 mM dATP, dTTP, and dCTP mix (500 μM final concentration), 1 μl of RNasin (Promega), 1 μg of RNA, and double-distilled H2O to 18 μl. The primer extension reactions were performed in an MJ Research thermal cycler. The reactions were denatured at 94°C for 3 min and annealed at 55°C for 30 min. Two microliters of avian myeloblastosis virus RT (20 U) (Promega) was added to each reaction, and extension was performed at 48°C for 35 min, followed by a 5-min denaturation step at 85°C. The reactions were analyzed by running on a 19% polyacrylamide-8 M urea gel. Quantitation was performed with a Molecular Dynamics PhosphorImager (model STORM-860). Mitochondrial and cytosolic preparations were used for subsequent analysis if they yielded contamination levels under 3%.
All of the transfectant lines, pSer-Leu*, pSer-Leu* 1, pSer-Leu* 2, pLeu*(+), and pLeu*(−), were assayed for the presence of tagged tRNALeu* and endogenous tRNALeu in both cytosolic and mitochondrial fractions with oligonucleotide LE4, using the assay conditions listed above. Oligonucleotide Met e was used to assay for the presence of tRNAMet-e in mitochondrial and cytosolic fractions for all of the transfectant lines under the same assay conditions. Oligonucleotide Met i was used to assay for tRNAMet-i in both RNA fractions for all of the transfectant lines. The assay conditions for oligonucleotide Met i required a substitution of ddCTP for ddGTP and dATP, dTTP, and dGTP mix for the dATP, dTTP, and dCTP mix listed above. The reactions were analyzed by running on a 19% polyacrylamide-8 M urea gel. Quantitation was performed with a Molecular Dynamics PhosphorImager (model STORM-860). The data were analyzed by calculating the ratio of mitochondrial to cytosolic RNA. This number was normalized to 1 for the results of the tagged transfectant line, pSer-Leu*, and all of the other transfectant lines were compared to that number. An import level of 1 indicates the endogenous steady-state level of each tRNA.
5′ RACE of mitochondrial and cytosolic RNA.
The following protocol was adapted from that of LeBlanc et al. (15). A 27-nt RNA oligonucleotide (5′-CGUACCGCGAUUAUG CUGAGUGAUAUC-3′) (Dharmacon) (500 pmol) was ligated to 10 μg of mitochondrial and cytosolic RNA from pSer-Leu* and pSer-Leu* 1 transfectants. The mixture was heated to 95°C for 2 min, and the volume was adjusted to 100 μl, with final concentrations of 50 mM Tris-HCl (pH 7.8), 20 mM MgCl2, 10 mM dithiothreitol, 10 μg of bovine serum albumin per ml, 10 mM ATP, 10% dimethyl sulfoxide, and 0.4 U of Rnasin per μl. Twenty units of T4 RNA ligase was added and the reactions were incubated at 16°C for 2 h. The RNAs were extracted with phenol and phenol-CHCl3 (1:1) and precipitated with sodium acetate and ethanol. Reverse transcription and PCR were performed according to the manufacturer's instructions for two-step RT-PCR using the enhanced avian HS RT-PCR kit (Sigma). Reverse transcription was performed with 4 μg of ligated RNA and oligonucleotide 2 (LE4) or oligonucleotide 3 (5′-GTGACAAGAGTGGGGTTCGAAC-3′) at 55°C for 1 h. Five microliters of cDNA from each RT reaction was used in the PCR step with RNA oligonucleotide (5′-GCATGGCGCTAATACGACTCACTATAG-3′), which is complementary to the RNA used in the ligation step, paired with oligonucleotide 1 (5′-GACCACTCGGCCATCATTGCC-3′), oligonucleotide 2, or oligonucleotide 3. PCR conditions were as follows: denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min, with a final extension at 72°C for 5 min. The resulting PCR products were cloned into the TA vector (Invitrogen) according to the manufacturer's instructions and transformed into TOP 10F′ competent cells (Invitrogen). One hundred nineteen clones that contained either precursor or mature tRNALeu were sequenced with T7 primer.
RESULTS
Presence of a novel sequence motif upstream of T. brucei tRNAs.
In order to identify sequence elements in mitochondrially localized tRNAs in trypanosomes, we examined 5′ flanking and intragenic sequences of both mitochondrially localized tRNAs and the cytosolically localized tRNAMet-i for conserved motifs. Although 50 putative tRNA genes have been identified in the trypanosome genome, we selected 23 tRNAs for which both expression and cellular localization have been determined (30). Similar analyses were completed with genes for tRNAs from yeast and humans.
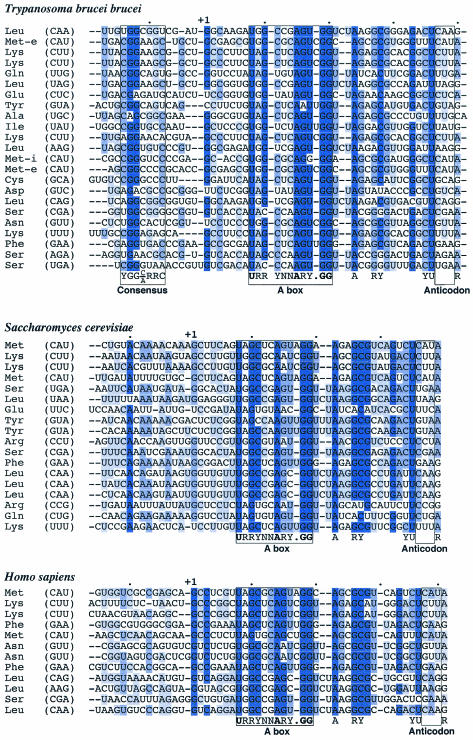
The start site for RNA polymerase III transcription of the tRNALeu and tRNASer genes is 14 nucleotides upstream of the 5′ ends of the mature tRNAs (see Fig. 2C) (15). This led us to examine immediate 5′ flanking sequence as potential leader sequences for the other tRNA genes. Alignments were done with 14 nucleotides of upstream sequence plus tRNA coding sequence through position 37 for tRNAs from T. brucei, Saccharomyces cerevisiae, and humans (Fig. 1). As expected, tRNA genes from all three organisms have highly conserved sequences within the tRNA coding sequence, including the A box promoter element, which overlaps with the D arm. Sequences upstream of the human and yeast tRNAs showed low degrees of sequence conservation, with the exception of a highly conserved A at position −10 in yeast. Alignments of T. brucei tRNAs revealed a highly conserved dinucleotide GG within a consensus sequence, YGG(C/A)RRC, located within 12 nucleotides upstream of the tRNA coding sequences. The dinucleotide GG has a substitution of one of the G residues in 4 of the 23 tRNAs analyzed, but the residue remains a purine in each of these cases. Remarkably, this upstream flanking sequence is similar to a sequence (YGGYAGAGC) identified as an import determinant in the D arm of mitochondrially localized tRNAs in Leishmania (3, 4, 16, 18). Furthermore, the sequence found in the D arm in Leishmania, YGGYAGAGC, overlaps with the conserved A box sequence found in the D arms of all tRNAs, including those in humans and yeast, whereas the motif we have identified upstream of trypanosome tRNAs is not present upstream of human or yeast tRNA genes.
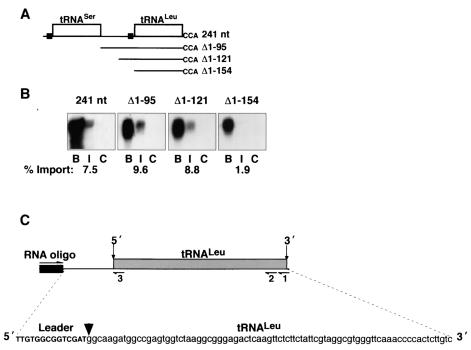
FIG. 2.
RNA import into isolated mitochondria and detection of precursor tRNALeu within trypanosome mitochondria. (A) Schematic of the precursor dicistronic tRNA substrate containing the 14-nucleotide 5′ flanking sequence, tRNASer, the 59-nucleotide intergenic region, tRNALeu, and CCA, with the lengths of each 5′ deletion substrate shown beneath. The 5′ YGG(C/A)RRC motifs are shown upstream of both tRNASer and tRNALeu (black boxes). All substrates were in vitro transcribed with T7 RNA polymerase in the presence of [α-32P]UTP for uniform labeling. (B) Mitochondria were incubated with 5 × 104 cpm of labeled substrate under optimized import conditions. The RNAs were collected by precipitation and treated with 30 μg of proteinase K per ml, which represents binding (lanes B). In the remaining reactions, proteinase K digestion was preceded by the addition of 10 U of micrococcal nuclease, followed by lysis of the mitochondria and extraction of the RNAs, which represents import (lanes I). The controls for resistance to micrococcal nuclease were first treated with 2% CHAPS before proteinase K and micrococcal nuclease treatment (lanes C). The isolated RNAs were run on a 6% polyacrylamide-8 M urea gel. The binding, import, and CHAPS lanes were quantitated with a phosphorimager. The counts per minute for the detergent-treated control were subtracted from those for the binding and import lanes. The percent import was determined as the counts per minute for the import lanes divided by the total counts per minute for import and binding. (C) A 27-nucleotide synthesized RNA oligonucleotide (black box) was ligated to mitochondrial and cytosolic RNA from pSer-Leu* transfectants (see Fig. 3). The RNA was reverse transcribed with oligonucleotides 1 and 2. The resulting cDNAs were amplified with RNA oligonucleotide, complementary to the ligated RNA, and oligonucleotides 1, 2, and 3, separately. The PCR products were ligated into the TA vector and sequenced with T7 primer. The 5′ ends of mature tRNALeu from both mitochondria and cytosol were mapped, as indicated by the arrowhead. The 5′ ends of precursor tRNALeu from both mitochondria and cytosol were mapped and extended 14 nucleotides upstream of the mature 5′ ends.
FIG. 1.
Identification of a conserved sequence upstream of T. brucei tRNAs. The coding and upstream sequences of the tRNAs from T. brucei were collected from the website http://zoosun00.unifr.ch/Trypanos/MITBIO.html (30) and from our own work (15). The S. cerevisiae coding and upstream sequences of the tRNAs were collected from the Saccharomyces Genome Database. Human tRNA sequences were extracted from the UCSC Genome Bioinformatics Database using the human BLAST search for sequences with homology to tRNAs of T. brucei. The resulting sequences were analyzed with tRNAScan-SE to predict the presence of tRNA genes (17). The 14-nucleotide upstream sequences plus tRNA coding sequences through position 37 were aligned for T. brucei, Homo sapiens, and S. cerevisiae with Clustal X. The sequences were adjusted manually with MacClade. The conservation at each residue was determined with Jalview. Conserved residues are highlighted as follows: dark blue, >80%; medium blue, >60%; light blue, >40%. The consensus sequence upstream of T. brucei tRNAs, YGG(C/A)RRC, was determined with MacClade. The highly conserved A box for each alignment is boxed, with the consensus sequence below. Invariant residues are shown in bold. Other universally conserved residues are indicated below the alignments. The anticodons for all tRNAs are boxed.
Deletion of the YGG(C/A)RRC motif causes a decrease in import efficiency in vitro.
Previous work showed the presence of a dicistronic precursor tRNA in the T. brucei mitochondrion (15). Furthermore, we have shown that this precursor RNA is efficiently imported in vitro by an ATP- and proton gradient-dependent pathway (32). In order to evaluate the role of 5′ leader sequences, including the YGG(C/A)RRC motif, in import, we prepared 5′ deletions of the dicistronic tRNASer-tRNALeu substrate. Figure 2A shows a schematic diagram of this RNA import substrate, including the positions of the YGG(C/A)RRC motifs. The 241-nucleotide substrate begins at the start site for tRNASer transcription and consists of a 14-nucleotide 5′ leader sequence, tRNASer, a 59-nucleotide intergenic region, downstream tRNALeu, and a 3′-terminal CCA. The start site for transcription of the tRNASer was determined by 5′ RACE (15). Progressive deletions of this substrate were constructed (Fig. 2A).
Substrate binding to (Fig. 2B, lanes B) and import into (Fig. 2B, lanes I) mitochondria were determined for each of the RNAs described above. Binding represents both RNA bound to the mitochondria and imported RNA. Imported RNA is defined as RNA that is resistant to digestion by micrococcal nuclease treatment of the isolated mitochondrion. To control for the possibility of nonspecific resistance to micrococcal nuclease, mitochondria were lysed with detergent prior to micrococcal nuclease treatment (Fig. 2B, lanes C). Consistent with our previous results, the dicistronic tRNASer-tRNALeu is imported efficiently into the isolated mitochondrion (Fig. 2B) (32). Deletion of tRNASer and its 5′ leader sequence (Δ1-95) and further deletion of an additional 26 nucleotides (Δ1-121) had little effect on the level of import in comparison to the full-length dicistronic tRNASer-tRNALeu, with import levels of 9.6, 8.8, and 7.5%, respectively (Fig. 2B). However, removal of the entire 5′ leader sequence in Δ1-154, including the YGG(C/A)RRC motif, reduced import to 1.9%. These results suggest that import of tRNALeu into the mitochondrion of T. brucei in vitro is affected by the presence of an element within the 5′ leader sequence, possibly acting as a positive determinant for import.
Precursor tRNALeu is found in both the mitochondrion and cytosol.
Although our in vitro studies suggest that the 5′ flanking sequence influences mitochondrial import and a previous study showed that a dicistronic tRNASer-tRNALeu was found in the mitochondrion of T. brucei by both RT-PCR and 5′ RACE, others have reported that tRNAs are processed prior to mitochondrial localization (1, 13, 15, 32). In order to determine whether tRNALeu was transcribed as a precursor and to investigate its cellular localization, we performed 5′ RACE on cytosolic and mitochondrial RNA fractions (Fig. 2C; Fig. 3B). A 27-nucleotide synthetic RNA was ligated to total mitochondrial and cytosolic RNA, followed by reverse transcription with oligonucleotide 1 or 2, which are complementary to the 3′ acceptor stem-D loop and ΤψC loop-variable arm, respectively. The cDNAs were then amplified with RNA oligonucleotide, which is complementary to the ligated synthetic RNA, and oligonucleotide 1, 2, or 3 (Fig. 2C). PCR products were cloned into the TA vector and sequenced with T7 primer. From the mitochondrial fraction, 14 of 40 clones containing tRNALeu had the 14-nucleotide 5′ leader, the sequence of which is shown in Fig. 2C. The remaining 26 clones were mature tRNALeu. From the cytosolic fraction, 8 of 39 clones containing tRNALeu had the 14-nucleotide 5′ leader, with the same sequence seen in the mitochondrial precursor tRNALeu. Twenty-nine clones were mature tRNALeu, and the remaining two clones were dicistronic tRNASer-tRNALeu with a 14-nucleotide 5′ leader and 59 nucleotides of intergenic sequence, which was previously found in the mitochondrial fraction by LeBlanc et al. (15). These results indicate that tRNALeu is synthesized as a precursor that can escape nuclear processing and be imported into the mitochondrion of T. brucei. The unexpectedly high ratio of precursor to mature tRNALeu in mitochondrial and cytosolic fractions is likely to be a consequence of preferential ligation of the RNA oligonucleotide to precursor tRNAs during 5′ RACE.
An in vivo system for analysis of tRNA localization.
The presence of precursor tRNALeu in the mitochondrion shown by 5′ RACE, sequence analysis, and in vitro experimental results suggests that trypanosomes may contain elements within 5′ leader sequences that positively influence mitochondrial import. To investigate this possibility, an in vivo transfection system was developed. A tRNALeu that is present in both the mitochondrion and cytosol of T. brucei was mutated by a single C-to-U change at position 45 in the variable arm (designated tRNALeu*) in order to distinguish the transgene from the endogenous tRNALeu. A plasmid construct was made that maintained the genomic organization of tRNALeu. Since this tRNA is part of a gene cluster, the final construct contained 244 nucleotides of 5′ flanking sequence, tRNASer, 59 nucleotides of intergenic sequence, tagged tRNALeu*, and 27 nucleotides of 3′ flanking sequence. The entire plasmid was targeted for integration into a nontranscribed region upstream of the endogenous tRNASer-tRNALeu gene cluster in T. brucei (Fig. 3A). This genomic site was specifically chosen for integration because of its proximity to the endogenous tRNASer-tRNALeu gene cluster. We wanted to ensure that the tagged tRNALeu* localized to a region of the genome that is transcribed by RNA polymerase III. As shown in subsequent experiments, tRNA genes integrated into this site mimic endogenous tRNA transcription levels and cellular localization.
Cell fractionation was used to prepare the mitochondrial and cytosolic RNA fractions for analysis of tRNA abundance and cellular distribution. The purity of mitochondrial and cytosolic fractions was established by a poison primer extension assay to detect abundant markers for mitochondria and cytosol, 9S mitochondrial rRNA (9S rRNA) and spliced leader cytosolic RNA (Fig. 3B), respectively. Quantitation of these data indicated that contamination of mitochondrial and cytosolic RNAs was <3%. A poison primer extension assay detected the endogenous tRNALeu (25-nucleotide product) in both wild-type cells and a cell line transfected with pSer-Leu* (Fig. 3C). The tagged tRNALeu* (28-nucleotide product) was only detectable in the transfected cells (Fig. 3C).
Endogenous tRNALeu was localized in the mitochondrion and cytosol, with mitochondrial/cytosolic RNA ratios of 2.15 and 2.19 in wild-type and pSer-Leu*-transfected cells, respectively, showing that transfection and expression of the tagged tRNALeu* do not affect the cellular localization of endogenous tRNALeu (Table 1). The tagged tRNALeu* had a similar distribution between the mitochondrion and cytosol, with a mitochondrial/cytosolic RNA ratio of 1.70. Also, transfection and expression of the tagged tRNALeu* did not significantly alter the distribution of other endogenous tRNAs, since the mitochondrial/cytosolic RNA ratios of tRNAMet-e were 3.58 and 3.33 in wild-type and pSer-Leu*-transfected cells, respectively (Table 1). Based on these results, it appears that cell fractionation allows for accurate determination of the localization of both tagged tRNAs and endogenous tRNAs within the cell.
TABLE 1.
Expression of tagged tRNALeu* does not alter the mitochondrial/cytosolic ratios of endogenous tRNAs
| Cell line | Ratio of mitochondrial/cytosolic RNAa
|
||
|---|---|---|---|
| tRNALeu | tRNALeu* | tRNAMet-e | |
| Wild type | 2.15 | NAb | 3.58 |
| pSer-Leu* | 2.19 | 1.70 | 3.33 |
Ratios of values generated through phosphorimager analysis of primer extension reactions on 1 μg each of mitochondrial and cytosolic RNA.
NA, not applicable.
5′ flanking sequence influences abundance of tRNALeu.
The role of 5′ leader sequences of tRNALeu in mitochondrial localization was addressed by deletion of upstream sequences in vivo. We made a deletion construct from pSer-Leu* by deleting the upstream tRNASer to the 5′ end of the mature tRNALeu. This deletion replaced the 5′ flanking sequence of tRNALeu* with the 5′ flanking sequence from tRNASer [Fig. 3D, pLeu*(+)]. Although the YGG(C/A)RRC motif (UGGCGGU) directly flanking tRNALeu* was deleted, the same motif (UGGCGGU) flanks tRNASer; therefore, the motif was replaced upstream of tRNALeu*. With pLeu*(+), the cellular localization and abundance of tRNALeu* were unaffected (Fig. 3E), possibly due to retention of the sequence motif in the 5′ flanking sequence of tRNALeu*. In contrast, removal of an additional 16 nucleotides, including the sequence motif, upstream of tRNALeu* reduced the abundance of tagged tRNALeu* by 96% in comparison to pSer-Leu* [Fig. 3E, pLeu*(−)]. Similar results were reported previously when 5′ flanking sequences were removed from tRNATyr and tRNALeu in T. brucei (11, 30). Unfortunately, the low abundance of tRNALeu* observed with pLeu*(−) made it difficult to interpret the import results. However, these results suggest that the YGG(C/A)RRC motif within the 5′ leader sequence of tRNALeu* is necessary for maintaining the cellular abundance of tRNALeu*.
The YGG(C/A)RRC motif is involved in mitochondrial localization in vivo.
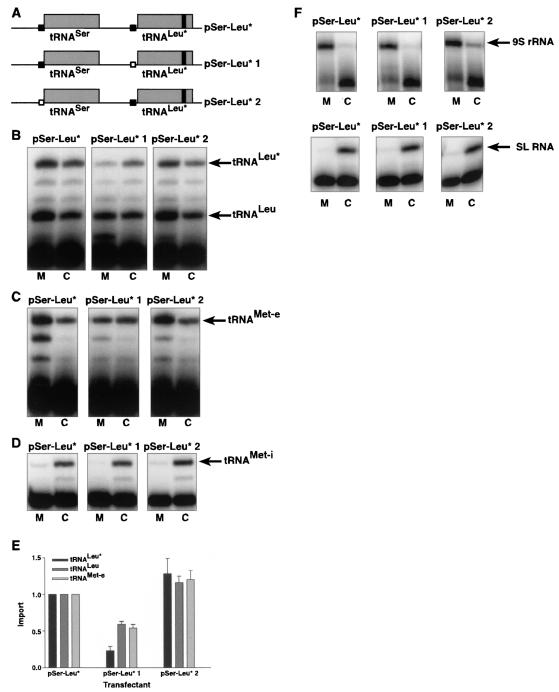
The role of the YGG(C/A)RRC motif in the 5′ leader sequence of tRNALeu* was further analyzed by mutations within the natural genomic context of the tRNASer-tRNALeu gene cluster (Fig. 4A). Due to the severe effects on abundance that were observed when large deletions were made upstream of tRNALeu*, we decided to make more conservative changes to the 5′ flanking sequence of tRNALeu*. Sequence replacement mutations were made in the YGG(C/A)RRC motif upstream of tRNALeu* (pSer-Leu* 1) and upstream of tRNASer (pSer-Leu* 2). We mutated GGCGG to UCGCU, replacing the highly conserved dinucleotide GG. Mutation of the motif upstream of tRNALeu* decreased the mitochondrial localization of tRNALeu* by 77% (Fig. 4B and E, pSer-Leu* 1). This mutation also resulted in a 73% reduction in the abundance of tRNALeu* within the cell (Fig. 4B, pSer-Leu* 1). Mutation of the motif upstream of tRNASer in pSer-Leu* 2 did not negatively affect the localization of tRNALeu* to the mitochondrion (Fig. 4B and E). Mutation of both motifs in the same construct gave the same result as that observed for pSer-Leu* 1 (data not shown). Again, the purity of mitochondrial and cytosolic fractions was established by detection of mitochondrial 9S rRNA and cytosolic spliced leader RNA for the cell lines transfected with pSer-Leu*, pSer-Leu* 1, and pSer-Leu* 2 (Fig. 4F). Quantitation of these data indicated that contamination of mitochondrial and cytosolic RNAs was <3%. These results show that the YGG(C/A)RRC motif in the 5′ flanking sequence directly upstream of tRNALeu* is critical for maintaining normal cellular abundance and mitochondrial localization of tRNALeu*.
FIG. 4.
Mutation of the YGG(C/A)RRC motif in the 5′ flanking sequence of tRNALeu* influences localization of tRNALeu* and other mitochondrially localized tRNAs. (A) Black boxes in pSer-Leu* represent the sequence element GGCGG, which is 4 nucleotides upstream of tRNALeu* and directly abuts tRNASer. Open boxes in pSer-Leu* 1 and pSer-Leu* 2 denote mutations of the GGCGG sequence element to UCGCU. (B) Primer extension analysis of pSer-Leu*, pSer-Leu* 1, and pSer-Leu* 2 was performed with oligonucleotide LE4 on 1 μg of mitochondrial (M) or cytosolic (C) RNA. The endogenous tRNALeu is detected by a 25-nucleotide extension product, and the tagged tRNALeu* is detected by a 28-nucleotide product. (C) Localization of endogenous tRNAMet-e was determined by primer extension analysis with oligonucleotide Met e in pSer-Leu*, pSer-Leu* 1, and pSer-Leu* 2. (D) Primer extension analysis with oligonucleotide Met i shows the localization of tRNAMet-i in pSer-Leu*, pSer-Leu* 1, and pSer-Leu* 2. (E) The ratio of mitochondrial/cytosolic RNA for tRNALeu*, endogenous tRNALeu, and tRNAMet-e was analyzed for each transfectant. This ratio was set to an import level of 1 for tRNAs from pSer-Leu*, and the ratios for tRNAs from pSer-Leu* 1 and pSer-Leu* 2 were compared to that from pSer-Leu* in graphical form (n = 6 for tRNALeu* and tRNALeu; n = 4 for tRNAMet-e). (F) A poison primer extension assay was performed on 1 μg of a mitochondrial (M) or cytosolic (C) RNA fraction from the transfectant lines pSer-Leu*, pSer-Leu* 1, and pSer-Leu* 2. Oligonucleotide 9Sb produces a 22-nucleotide extension product, predominantly in the mitochondrial fraction. Oligonucleotide SL2 also produces a 26-nucleotide extension product, but it is primarily in the cytosol. Mitochondrial or cytosolic fractions with >3% cross-contamination were not used for subsequent experiments. SL RNA, spliced leader RNA.
Since the consensus YGG(C/A)RRC motif is not well conserved upstream of the cytosolically localized tRNAMet-i, we replaced tRNALeu* in pSer-Leu* with tRNAMet-i. Although the YGG(C/A)RRC motif within the 5′ leader of tRNALeu* was shown to influence its mitochondrial localization by mutation with pSer-Leu* 1, addition of this sequence motif upstream of tRNAMet-i did not result in mitochondrial localization of tRNAMet-i (Fig. 4B and E; data not shown). This suggests that multiple signals may be required for efficient mitochondrial import.
Mutation of the YGG(C/A)RRC motif affects the presence and localization of precursor tRNALeu*.
We performed 5′ RACE on mitochondrial and cytosolic fractions from pSer-Leu* and pSer-Leu* 1 transfectants, as shown in Fig. 2C, in order to determine whether accurate 5′ processing occurred for the tagged and mutated-tagged tRNALeu*, respectively. Interestingly, the localization of precursors for both the tagged tRNALeu* and endogenous tRNALeu was altered in the pSer-Leu* 1 transfectants. In pSer-Leu* transfectants, 40 clones were analyzed from the mitochondrial fraction that contained tRNALeu; 14 of these clones had tagged tRNALeu*, of which 7 had the 14-nucleotide 5′ leader sequence. The other seven had accurately processed, mature 5′ ends (Fig. 2C). In contrast, 5′ RACE of RNA from the mitochondrial fraction of pSer-Leu* 1 transfectants revealed a single endogenous precursor tRNALeu and no tagged precursor tRNALeu* in a total of 27 tRNALeu clones.
Analysis of the cytosolic fractions also yielded unusual results for the tagged tRNALeu* in pSer-Leu* 1 transfectants. As stated previously, 38 clones resulting from the cytosolic fraction of pSer-Leu* transfected cells contained tRNALeu; of the 8 clones that had the 14-nucleotide 5′ leader, 3 of the precursors were tagged tRNALeu*. Similarly, 24 clones resulting from the cytosolic fraction of pSer-Leu* 1 transfectants contained tRNALeu, of which 12 were mature tRNALeu and 12 were precursor tRNALeu with a 14-nucleotide 5′ leader sequence. None of the 12 precursor tRNALeu was tagged, but there were two mature tagged tRNALeu* clones. Unlike the mitochondrial and cytosolic fractions from pSer-Leu* transfectants, there was no tagged precursor tRNALeu* in either fraction from the pSer-Leu* 1 transfectants. The lack of precursor tagged tRNALeu* in pSer-Leu* 1 transfectants is a sharp contrast to the results obtained with pSer-Leu* transfected cells. The reduced level of mitochondrial localization of tRNALeu* and the corresponding lack of precursor tRNALeu* within the pSer-Leu* 1 transfectants implicate the 5′ leader sequence as an important component of mitochondrial localization (Fig. 4B and C).
Mutation in the YGG(C/A)RRC motif has global effects on cellular tRNA localization.
The data thus far indicate the involvement of cis-acting sequences in the cellular localization of tRNALeu*. Furthermore, closer examination of the data in Fig. 4 also reveals an effect on endogenous tRNALeu localization when the YGG(C/A)RRC motif is mutated (Fig. 4B and E, pSer-Leu* 1). The ratio of endogenous mitochondrial/cytosolic tRNALeu was reduced by 41%, indicating that the distribution of tRNALeu between the mitochondrion and cytosol was altered. The reduction in tRNALeu localization to the mitochondrion is the same effect that was seen on the import level of transfected tRNALeu* (Fig. 4B and E, pSer-Leu* 1) and was dependent on mutation of the YGG(C/A)RRC motif upstream of tRNALeu*. Mutation of the YGG(C/A)RRC motif upstream of tRNASer did not have an effect on the localization of endogenous tRNALeu (Fig. 4B and E, pSer-Leu* 2).
The effect of mutations to the YGG(C/A)RRC motif on endogenous tRNALeu led us to investigate whether other endogenous tRNAs are affected similarly. Poison primer extension assays on endogenous tRNAMet-e, a tRNA shared between both the cytosol and the mitochondrion, revealed that similar effects on mitochondrial localization occur for this endogenous tRNA as well. Mutation of the YGG(C/A)RRC sequence upstream of tRNALeu* reduced the level of mitochondrial import for tRNAMet-e by 46% (Fig. 4C and E, pSer-Leu* 1). Mutation of the YGG(C/A)RRC sequence upstream of tRNASer did not result in a loss of import of tRNALeu* or other endogenous tRNAs (Fig. 4B, to E, pSer-Leu* 2). Other endogenous tRNAs that are shared between the mitochondrion and cytosol, including tRNATyr (GUA) and tRNAArg (CCU), were also tested and gave similar results to those shown for the endogenous tRNALeu and tRNAMet-e (data not shown). Finally, no effect was seen on the cytosolic localization or abundance of the almost exclusively cytosolically localized tRNAMet-i with pSer-Leu* 1 (Fig. 4D), suggesting that the effects caused by the YGG(C/A)RRC mutations are specific for localization of tRNAs to the mitochondrion.
DISCUSSION
We have identified a unique sequence motif, YGG(C/A)RRC, within the 5′ leader sequences of tRNAs in T. brucei. In this paper, we present evidence that the YGG(C/A)RRC sequence, immediately 5′ of the mature tRNALeu gene in T. brucei, influences both the abundance and localization of the resulting tRNA transcript, which contains this motif within its 14-nucleotide 5′ leader. Not only does mutating the 5′ flanking sequence of the tRNALeu* transgene alter the localization of tRNALeu* in the cell, but it also changes the localization of the endogenous tRNALeu and other mitochondrially localized tRNAs. Based on these results, and those of others, we conclude that tRNA import into trypanosome mitochondria is influenced by distinct sequence motifs present within precursor and mature tRNA sequences.
Sequences within a precursor mitochondrial tRNA influence tRNA abundance and cellular localization.
Little is known about RNA polymerase III transcription in trypanosomes; however, the transcriptional start site of tRNASer has been mapped by 5′ RACE to 14 nucleotides upstream of the mature 5′ end (15). We present evidence here from a 5′ RACE experiment that maps the transcriptional start site for tRNALeu to 14 nucleotides upstream of the mature 5′ end (Fig. 2C). Generally, transcription of a tRNA gene initiates at a purine within 10 nucleotides upstream of the mature tRNA coding sequence and continues until the polymerase reaches a cluster of thymidine residues, resulting in a precursor tRNA with short 5′ and 3′ flanking sequences (26, 27). Based on 5′ RACE studies, both tRNASer and tRNALeu from trypanosomes have extended 5′ leader sequences in comparison. Additionally, our 5′ RACE experiments have identified both precursor tRNALeu and tRNASer with 14-nucleotide 5′ leaders within trypanosome mitochondria, suggesting that at least some tRNAs are imported into the mitochondrion as precursors (Fig. 2C) (15). These data appear to be contradictory to previously reported findings (1, 13). In those studies, it was shown that end processing precedes localization of tRNAs to the mitochondria of Leishmania tarentolae and that what appeared to be large precursor tRNAs in T. brucei and L. tarentolae mitochondria were artifacts generated by circularization of mature tRNAs (1, 13). Kapushoc et al. used an RT-PCR assay to show that precursor tRNAs with 29- to 34-nucleotide 5′ and/or 3′ extensions are only localized in the nucleus, but they did not look specifically for precursor tRNAs with a 5′ leader sequence (13). While we have reported the presence of a dicistronic tRNASer-tRNALeu in the mitochondrion, our 5′ RACE data with tRNALeu show that precursor tRNALeu with a 14-nucleotide 5′ leader sequence is much more abundant in the cell. Furthermore, the genomic organization of the trypanosome genome, with singular tRNAs, head-to-head tRNAs, tail-to-tail tRNAs, and head-to-tail tRNAs, would not allow for all tRNAs to be transcribed as multicistronic transcripts (15, 30). We agree that the large tRNA molecules found in mitochondrial fractions are mostly due to ligation of mature tRNAs during our mitochondrial isolation procedure (15).
Based on the mapping of the transcription initiation site to 14 nucleotides upstream of the mature tRNALeu, we performed detailed sequence alignments from −14 nucleotides upstream of the mature tRNAs to position 37 within the coding sequence of 23 T. brucei tRNA genes (Fig. 1). These alignments revealed a conserved sequence motif upstream of the tRNAs with a consensus of YGG(C/A)RRC. We compared the T. brucei upstream sequences to those from human and S. cerevisiae tRNAs. While S. cerevisiae imports a single tRNALys, the mechanism is different from that in kinetoplastids (24, 31). All of the human mitochondrial tRNAs are encoded by the mitochondrial genome, although human mitochondria are able to import tRNALys from S. cerevisiae in the presence of yeast cytosolic factors (14). The alignment of upstream regions of human or S. cerevisiae tRNAs failed to reveal a conserved sequence element, although the regions upstream of S. cerevisiae tRNAs are very A rich. The identification of a conserved motif upstream of T. brucei tRNA genes supported the possibility that sequences within the 5′ leader of precursor tRNAs in T. brucei might be important for tRNA localization.
Initial evidence for the functional significance of the 5′ leader sequence in mitochondrial import came from in vitro import studies. We found that deletion of sequences containing the YGG(C/A)RRC motif, directly upstream of tRNALeu, reduced the efficiency of tRNALeu import (Fig. 2B). In order to evaluate the role of this sequence motif in vivo, we developed a transfection assay for tRNA import and abundance in T. brucei (Fig. 3). A single nucleotide change in the variable arm of tRNALeu allowed us to distinguish the transfected tRNALeu* from endogenous tRNALeu and to determine the steady-state levels of tRNALeu* in the cytosol and mitochondrion. Using this assay, we showed that a deletion of 156 nucleotides of the tRNALeu* 5′ flanking sequence in pSer-Leu* [pLeu*(−)], including the upstream tRNASer and both YGG(C/A)RRC motifs, resulted in a severe decrease in the abundance of tRNALeu* (Fig. 3E). To determine whether the decrease in tRNA abundance was due to deletion of the YGG(C/A)RRC motif, we replaced the motif immediately upstream of tRNALeu* in construct pLeu*(+) (Fig. 3D). This restored the abundance and mitochondrial localization of tRNALeu* to the same level as that seen for pSer-Leu* (Fig. 3E), presumably because the motif located directly 5′ of tRNASer was juxtaposed with the tRNALeu* gene [Fig. 3D and E, pLeu*(+)]. Unfortunately, the low abundance of tRNALeu* observed with pLeu*(−) made it difficult to interpret the import results. The observed decrease in abundance could result from a number of situations, including transcript stability, nuclear export, or transcription. Taken together, these in vitro and in vivo results suggest that the YGG(C/A)RRC motif within the 5′ leader sequence of tRNALeu influences the abundance and localization of tRNALeu in the mitochondrion.
Since large deletions of the 5′ flanking sequence had a severe effect on tRNALeu* abundance, we decided to prepare more limited mutations by scrambling the sequence of each YGG(C/A)RRC motif of tRNASer and tRNALeu* independently (Fig. 4A). We mutated GGCGG in the motif to UCGCU, changing the highly conserved dinucleotide GG to pyrimidines (UC), the central C residue to a purine (G), and the conserved RR residues to pyrimidines (GG to CU). Although mutating the YGG(C/A)RRC motif upstream of tRNASer did not affect the localization of tRNALeu* (pSer-Leu* 2), mutation of the YGG(C/A)RRC motif upstream of tRNALeu* (pSer-Leu* 1) decreased its localization to the mitochondrion significantly (Fig. 4B and E). A striking consequence of this mutation was discovered in 5′ RACE experiments on both pSer-Leu* and pSer-Leu* 1 transfectants. In trying to determine whether the mutation caused an alteration in accurate 5′ processing of tRNALeu*, which it did not, we found a severe reduction in the amount of precursor tRNALeu in the mitochondria of pSer-Leu* 1 transfectants and no precursor tRNALeu* in these cells. While tRNALeu* was accurately 5′ processed, the lack of precursor tRNALeu* with a 5′ leader suggests two possibilities; the mutation may lead to more efficient 5′ processing so that we are unable to detect these precursors in the cell or transcription may initiate at the site of the mature 5′ end of tRNALeu*. If the second possibility occurs, trypanosomes may have the ability to import both mature and precursor tRNAs into the mitochondrion.
At first glance, these data appear to contradict published studies on mitochondrial tRNA import in kinetoplastids. Others have reported that the genomic context of tRNA genes has no influence on their localization to the mitochondrion (16, 30). However, upon reevaluation of published data, it appears that the 5′ flanking sequence influences tRNA localization in experiments with Leishmania. The 5′ flanking sequence of tRNAIle, a tRNA localized in the cytosol and mitochondrion, was interchanged with the 5′ flanking sequence from a cytosolically localized tRNAGln or plasmid sequence. This change decreased the efficiency of localization of tRNAIle to the mitochondrion (16). This is consistent with the results we present here. Studies in T. brucei using tRNALeu showed progressive deletions of the 5′ flanking sequence, with no effect on import (30). Yet, in the experiment in which the YGG(C/A)RRC motif was removed, the abundance of tRNALeu was reduced such that localization of the tRNA into the mitochondrion was difficult to interpret. Our interpretation of the results of Lima and Simpson supports our conclusion that sequences within the 5′ leader sequence of precursor tRNAs influence the efficiency of import, and the results of Tan et al. do not provide sufficient evidence for the conclusion that the 5′ flanking sequence does not influence mitochondrial localization (16, 30).
Multiple sequence motifs influence tRNA import into kinetoplastid mitochondria.
Mutation of the YGG(C/A)RRC sequence within the 5′ leader of tRNALeu* lowers the efficiency of import but does not abolish import of tRNALeu* into T. brucei mitochondria (Fig. 4B). Therefore, other elements within the mature tRNA sequence must influence localization. In Leishmania, exchanging the D loop and stem of cytosolic tRNAGln with those of mitochondrially imported tRNAIle changes the localization of the hybrid tRNAGln so that it imports into mitochondria (16). Yet, exchanging the D loop and stem of the shared tRNAIle with those of cytosolically localized tRNAGln did not exclude the hybrid tRNAIle from mitochondria (16), suggesting the presence of multiple signals within either the mature tRNA sequence or the 5′ leader sequences.
The diversity of sequence and structural motifs that may be involved in tRNA trafficking in trypanosomes was demonstrated by SELEX experiments in which oligoribonucleotide aptamers were selected based on their efficiency of import into Leishmania mitochondria (4). Interestingly, these studies identified the sequence YGGYAGAGY as a positive import signal, and candidate sequences that fit this consensus were identified in the D arm, TψC arm, or variable arm of mitochondrially imported Leishmania tRNAs (4). This sequence is highly homologous to the YGG(C/A)RRC motif that we have identified upstream of tRNA genes for T. brucei.
We replaced the 5′ leader sequence of the cytosolically localized tRNAMet-i with that of tRNALeu, which has a highly conserved YGG(C/A)RRC motif, in order to determine whether this motif is sufficient to direct mitochondrial localization of tRNAMet-i. The motif did not direct mitochondrial localization of tRNAMet-i, possibly because this tRNA lacks the necessary intragenic signals or contains negative regulatory elements specific to an initiator tRNAMet (data not shown). Since most tRNAs are shared between the mitochondria and cytosol and are imported with different efficiencies, it seems likely that multiple signals, both positive and negative, may be required for fine-tuning the distribution and abundance of tRNAs in kinetoplastids. It is also possible that tRNAs contain redundant signals for mitochondrial localization since they are necessary for mitochondrial protein translation.
Mutation of the YGG(C/A)RRC motif has a global effect on tRNA import.
A surprising result from our studies is that not only does mutation of the upstream YGG(C/A)RRC motif alter the localization of tRNALeu*, but it also alters the mitochondrial abundance of other endogenous tRNAs (Fig. 4, pSer-Leu* 1 transfectants). This brings up the question of where mitochondrial and cytosolic localization diverge during tRNA biogenesis. Does sorting take place in the nucleus, in the cytoplasm, or at the mitochondrial membranes? Alternatively, do the multiple signals contained within the precursor tRNAs influence localization at separate points in the pathway? While our studies do not directly address the mechanism of import, the global effect we observed on tRNA localization when the YGG(C/A)RRC motif of tRNALeu* was mutated suggests that this sequence may be involved at a later step in the trafficking of tRNAs to mitochondria.
While previous studies showed the presence of a dicistronic tRNA precursor in the mitochondrion of T. brucei (15), our data with the monocistronic pLeu*(+) cell line conclusively show that this is not the only substrate for tRNA localization to the mitochondrion. Also, the genomic organization of the clustered tRNA genes in T. brucei as head-to-head or tail-to-tail, as well as the presence of nonclustered tRNAs, would not allow for all tRNAs to be transcribed as dicistronic or multicistronic transcripts (20, 30). Although many questions remain concerning the mechanism of tRNA localization to the mitochondrion in T. brucei, we have presented evidence here of a unique sequence motif found within the 5′ leader sequence of a precursor tRNA that influences both the cellular localization and abundance of tRNALeu, suggesting a role for precursor tRNAs in the tRNA localization pathway to the mitochondrion. Furthermore, the motif we have identified upstream of trypanosome tRNAs is unique as an extragenic element that modulates both abundance and localization of tRNAs. In addition, we have shown that mutation of the YGG(C/A)RRC motif, upstream of tRNALeu, causes a global change of tRNA localization to the trypanosome mitochondrion, adding a further level of interest to this unique sequence motif.
Acknowledgments
We thank Jay Bangs for the gift of vector pXS2 and for helpful discussions on transfection of T. brucei. We also thank Jayleen Grams, Dan Golden, Jeff Priest, Bob Sabatini, and other members of the Hajduk lab for their insight and helpful discussions.
This work was supported by the National Institutes of Health grant AI21401 (to S.L.H.).
REFERENCES
- 1.Aphasizhev, R., U. Karmarkar, and L. Simpson. 1998. Are tRNAs imported into the mitochondrion of kinetoplastid protozoa as 5′-extended precursors? Mol. Biochem. Parasitol. 93:73-80. [DOI] [PubMed] [Google Scholar]
- 2.Bangs, J. D., E. M. Brouch, D. M. Ransom, and J. L. Roggy. 1996. A soluble secretory reporter system in Trypanosoma brucei. Studies on endoplasmic reticulum targeting. J. Biol. Chem. 271:18387-18393. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya, S. N., S. Mukherjee, and S. Adhya. 2000. Mutations in a tRNA import signal define distinct receptors at the two membranes of Leishmania mitochondria. Mol. Cell. Biol. 20:7410-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharyya, S. N., S. Chatterjee, and S. Adhya. 2002. Mitochondrial RNA import in Leishmania tropica: aptamers homologous to multiple tRNA domains that interact cooperatively or antagonistically at the inner membrane. Mol. Cell. Biol. 22:4372-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham, I. 1977. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J. Protozool. 24:325-329. [DOI] [PubMed] [Google Scholar]
- 6.Dorner, M., M. Altmann, S. Paabo, and M. Morl. 2001. Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol. Biol. Cell 12:2688-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiduschek, E. P., and G. P. Tocchini-Valentini. 1988. Transcription by RNA polymerase III. Annu. Rev. Biochem. 57:873-914. [DOI] [PubMed] [Google Scholar]
- 8.Hancock, K., and S. L. Hajduk. 1990. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J. Biol. Chem. 265:19208-19258. [PubMed] [Google Scholar]
- 9.Hancock, K., A. J. LeBlanc, D. Donze, and S. L. Hajduk. 1992. Identification of nuclear encoded precursor tRNAs within the mitochondrion of Trypanosoma brucei. J. Biol. Chem. 267:23963-23971. [PubMed] [Google Scholar]
- 10.Harris, M. E., D. R. Moore, and S. L. Hajduk. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 265:11368-11376. [PubMed] [Google Scholar]
- 11.Hauser, R., and A. Schneider. 1995. tRNAs are imported into mitochondria of Trypanosoma brucei independently of their genomic context and genetic origin. EMBO J. 14:4212-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko, T., T. Suzuki, S. T. Kapushoc, M. A. Rubio, J. Ghazvini, K. Watanabe, L. Simpson, and T. Suzuki. 2003. Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism. EMBO J. 3:657-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapushoc, S. T., J. D. Alfonzo, M. A. Rubio, and L. Simpson. 2000. End processing precedes mitochondrial importation and editing of tRNAs in Leishmania tarentolae. J. Biol. Chem. 275:37907-37914. [DOI] [PubMed] [Google Scholar]
- 14.Kolesnikova, O. A., N. S. Entelis, H. Mireau, T. D. Fox, R. P. Martin, and I. A. Tarassov. 2000. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science 289:1931-1933. [DOI] [PubMed] [Google Scholar]
- 15.LeBlanc, A. J., A. E. Yermovsky-Kammerer, and S. L. Hajduk. 1999. A nuclear encoded and mitochondrial imported dicistronic tRNA precursor in Trypanosoma brucei. J. Biol. Chem. 274:21071-21077. [DOI] [PubMed] [Google Scholar]
- 16.Lima, B. D., and L. Simpson. 1996. Sequence-dependent in vivo importation of tRNAs into the mitochondrion of Leishmania tarentolae. RNA 2:429-440. [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahapatra, S., S. Ghosh, S. K. Bera, T. Ghosh, A. Das, and S. Adhya. 1998. The D arm of tRNATyr is necessary and sufficient for import into Leishmania mitochondria in vitro. Nucleic Acids Res. 26:2037-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milhausen, M., R. G. Nelson, S. Sather, M. Selkirk, and N. Agabian. 1984. Identification of a small RNA containing the trypanosome spliced leader: a donor of shared 5′ sequences of trypanosomatid mRNAs? Cell 38:721-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mottram, J. C., S. D. Bell, R. G. Nelson, and J. D. Barry. 1991. tRNAs of Trypanosoma brucei. Unusual gene organization and mitochondrial importation. J. Biol. Chem. 266:18313-18317. [PubMed] [Google Scholar]
- 21.Priest, J. W., and S. L. Hajduk. 1996. In vitro import of the Rieske iron-sulfur protein by trypanosome mitochondria. J. Biol. Chem. 271:20060-20069. [DOI] [PubMed] [Google Scholar]
- 22.Rubio, M. A., X. Liu, H. Yuzawa, J. D. Alfonzo, and L. Simpson. 2000. Selective importation of RNA into isolated mitochondria from Leishmania tarentolae. RNA 6:988-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salavati, R., A. K. Panigrahi, B. A. Morach, S. S. Palazzo, R. P. Igo, and K. Stuart. 2002. Endoribonuclease activities of Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol. 120:23-31. [DOI] [PubMed] [Google Scholar]
- 24.Schneider, A., J. Martin, and N. Agabian. 1994. A nuclear encoded tRNA of Trypanosoma brucei is imported into mitochondria. Mol. Cell. Biol. 14:2317-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider, A., and L. Marechal-Drouard. 2000. Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol. 10:509-513. [DOI] [PubMed] [Google Scholar]
- 26.Schramm, L., and N. Hernandez. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 16:2593-2620. [DOI] [PubMed] [Google Scholar]
- 27.Sharp, S. J., J. Schaack, L. Cooley, D. J. Burke, and D. Soll. 1985. Structure and transcription of eukaryotic tRNA genes. CRC Crit. Rev. Biochem. 19:107-144. [DOI] [PubMed] [Google Scholar]
- 28.Simpson, A. M., Y. Suyama, H. Dewes, D. A. Campbell, and L. Simpson. 1989. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 17:5427-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloof, P., J. Van den Burg, A. Voogd, R. Benne, M. Agostinelli, P. Borst, R. Gutell, and H. Noller. 1985. Further characterization of the extremely small mitochondrial ribosomal RNAs from trypanosomes: a detailed comparison of the 9S and 12S RNAs from Crithidia fasciculata and Trypanosoma brucei with rRNAs from other organisms. Nucleic Acids Res. 13:4171-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan, T. H., R. Pach, A. Crausaz, A. Ivens, and A. Schneider. 2002. tRNAs in Trypanosoma brucei: genomic organization, expression, and mitochondrial import. Mol. Cell. Biol. 22:3707-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarassov, I., N. Entelis, and R. P. Martin. 1995. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 14:3461-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yermovsky-Kammerer, A. E., and S. L. Hajduk. 1999. In vitro import of a nuclearly encoded tRNA into the mitochondria of Trypanosoma brucei. Mol. Cell. Biol. 19:6253-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]