Abstract
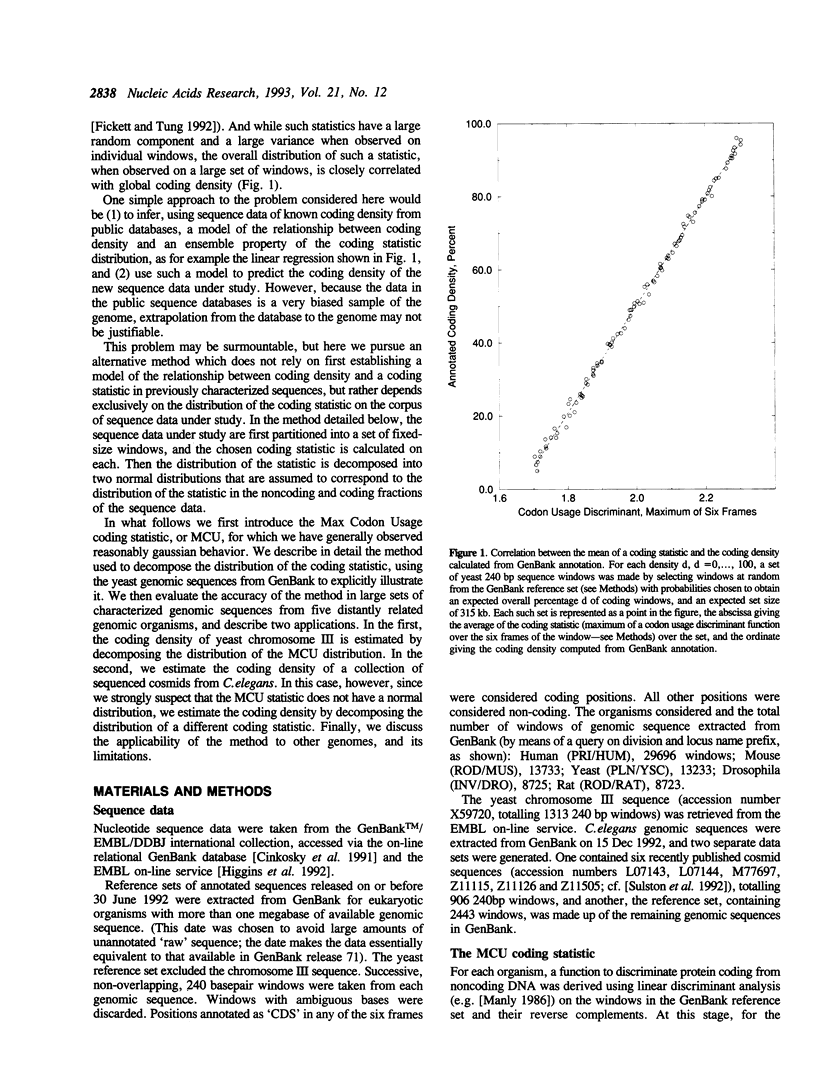
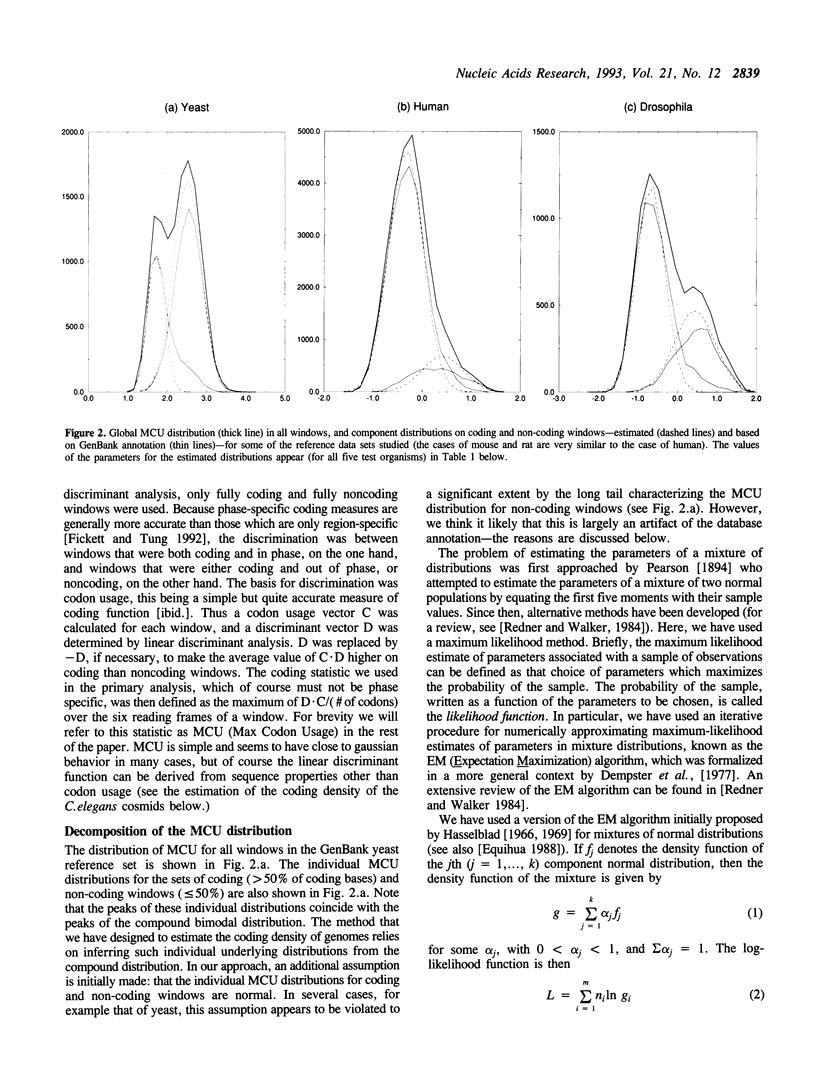
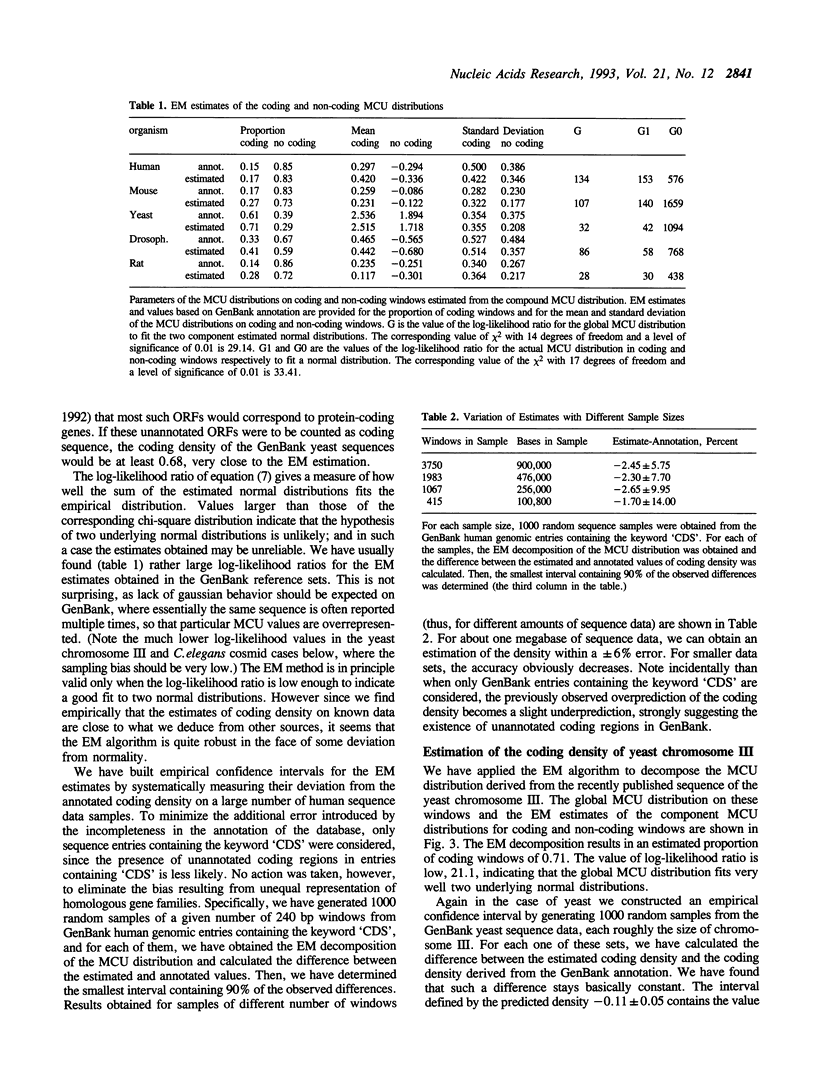
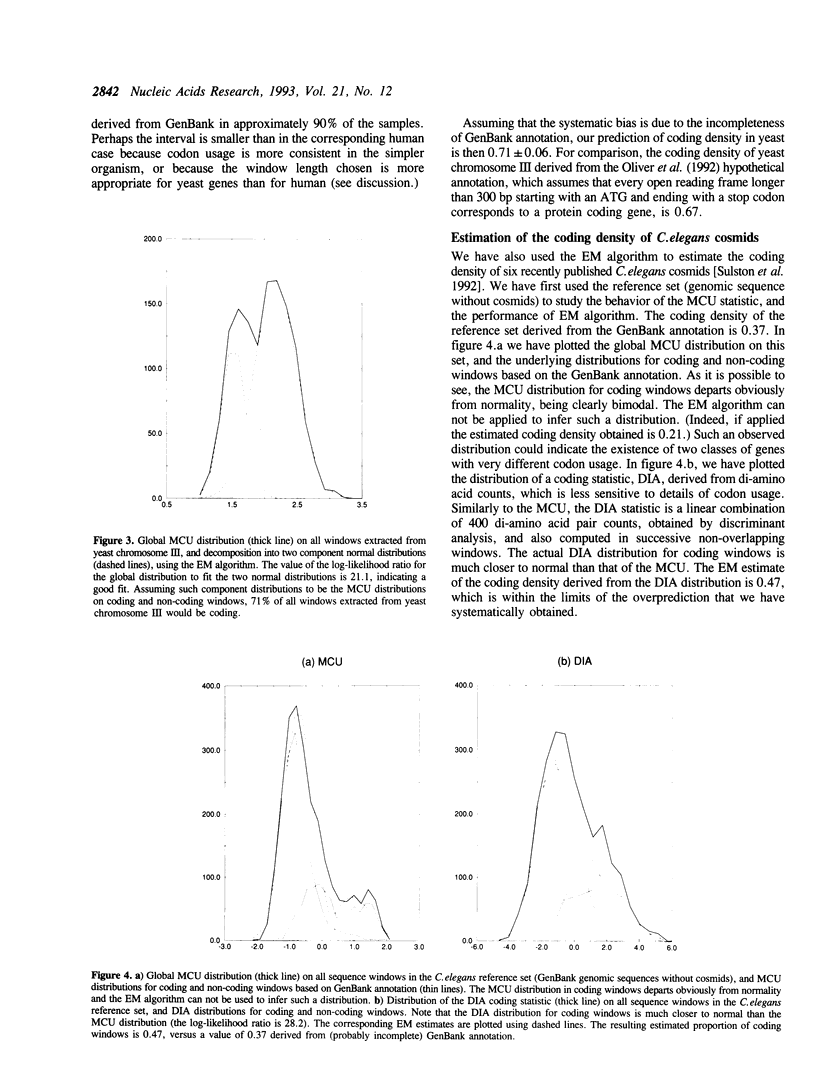
A number of experimental methods have been reported for estimating the number of genes in a genome, or the closely related coding density of a genome, defined as the fraction of base pairs in codons. Recently, DNA sequence data representative of the genome as a whole have become available for several organisms, making the problem of estimating coding density amenable to sequence analytic methods. Estimates of coding density for a single genome vary widely, so that methods with characterized error bounds have become increasingly desirable. We present a method to estimate the protein coding density in a corpus of DNA sequence data, in which a 'coding statistic' is calculated for a large number of windows of the sequence under study, and the distribution of the statistic is decomposed into two normal distributions, assumed to be the distributions of the coding statistic in the coding and noncoding fractions of the sequence windows. The accuracy of the method is evaluated using known data and application is made to the yeast chromosome III sequence and to C. elegans cosmid sequences. It can also be applied to fragmentary data, for example a collection of short sequences determined in the course of STS mapping.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cinkosky M. J., Fickett J. W., Gilna P., Burks C. Electronic data publishing and GenBank. Science. 1991 May 31;252(5010):1273–1277. doi: 10.1126/science.1925538. [DOI] [PubMed] [Google Scholar]
- Clark D. V., Rogalski T. M., Donati L. M., Baillie D. L. The unc-22(IV) region of Caenorhabditis elegans: genetic analysis of lethal mutations. Genetics. 1988 Jun;119(2):345–353. doi: 10.1093/genetics/119.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett J. W., Tung C. S. Assessment of protein coding measures. Nucleic Acids Res. 1992 Dec 25;20(24):6441–6450. doi: 10.1093/nar/20.24.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Fuchs R., Stoehr P. J., Cameron G. N. The EMBL Data Library. Nucleic Acids Res. 1992 May 11;20 (Suppl):2071–2074. doi: 10.1093/nar/20.suppl.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback D. B., Angerer L. M., Davidson N. Improved methods for the formation and stabilization of R-loops. Nucleic Acids Res. 1979 Jun 11;6(7):2499–2317. doi: 10.1093/nar/6.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Olson M., Hood L., Cantor C., Botstein D. A common language for physical mapping of the human genome. Science. 1989 Sep 29;245(4925):1434–1435. doi: 10.1126/science.2781285. [DOI] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. Selfish DNA: the ultimate parasite. Nature. 1980 Apr 17;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Park E. C., Horvitz H. R. Mutations with dominant effects on the behavior and morphology of the nematode Caenorhabditis elegans. Genetics. 1986 Aug;113(4):821–852. doi: 10.1093/genetics/113.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J., Du Z., Thomas K., Wilson R., Hillier L., Staden R., Halloran N., Green P., Thierry-Mieg J., Qiu L. The C. elegans genome sequencing project: a beginning. Nature. 1992 Mar 5;356(6364):37–41. doi: 10.1038/356037a0. [DOI] [PubMed] [Google Scholar]
- Waterston R., Martin C., Craxton M., Huynh C., Coulson A., Hillier L., Durbin R., Green P., Shownkeen R., Halloran N. A survey of expressed genes in Caenorhabditis elegans. Nat Genet. 1992 May;1(2):114–123. doi: 10.1038/ng0592-114. [DOI] [PubMed] [Google Scholar]
- Yoshikawa A., Isono K. Chromosome III of Saccharomyces cerevisiae: an ordered clone bank, a detailed restriction map and analysis of transcripts suggest the presence of 160 genes. Yeast. 1990 Sep-Oct;6(5):383–401. doi: 10.1002/yea.320060504. [DOI] [PubMed] [Google Scholar]


