Abstract
Although the human papillomavirus (HPV) E7 oncogene is known to contribute to the development of human cervical cancer, the mechanisms of its carcinogenesis are poorly understood. The first identified and most recognized function of E7 is its binding to and inactivation of the retinoblastoma tumor suppressor (pRb), but at least 18 other biological activities have also been reported for E7. Thus, it remains unclear which of these many activities contribute to the oncogenic potential of E7. We used a Cre-lox system to abolish pRb expression in the epidermis of transgenic mice and compared the outcome with the effects of E7 expression in the same tissue at early ages. Mice lacking pRb in epidermis showed epithelial hyperplasia, aberrant DNA synthesis, and improper differentiation. In addition, Rb-deleted epidermis (i.e., epidermis composed of cells with Rb deleted) exhibited centrosomal abnormalities and failed to arrest the cell cycle in response to ionizing radiation. Transgenic mice expressing E7 in skin display the same range of phenotypes. In sum, few differences were detected between Rb-deleted epidermis and E7-expressing epidermis in young mice. However, when both E7 was expressed and Rb was deleted in the same tissue, increased hyperplasia and dysplasia were observed. These findings indicate that inactivation of the Rb pathway can largely account for E7's phenotypes at an early age, but that pRb-independent activities of E7 are detectable in vivo.
Human papillomaviruses (HPVs) are small DNA viruses that infect various epithelial tissues, causing the formation of warts. A subset of HPVs that infect the anogenital tract, the high-risk HPVs, including HPV type 16 (HPV-16), are associated with almost all cases of cervical cancer, a leading cause of cancer mortality in women worldwide (54). In these cancers, the HPV genome is often found integrated into the host genome (59), and this integration results in increased expression of two viral genes, E6 and E7 (20). These data suggest that E6 and E7 expression is necessary for the development of the vast majority of cervical cancers.
HPV-16 E7 is a small nuclear phosphoprotein with potent transforming and oncogenic properties. Coexpression of E6 and E7 is necessary and sufficient to transform primary human keratinocytes (31), and E7 is strongly positive in a number of other in vitro transformation assays (2, 27, 36, 46, 49, 50). In culture, E7-expressing cells exhibit genomic instability (10, 39, 51, 52) and lack normal responses to DNA damage (7, 44). In addition, expression of E7 in primary human keratinocytes results in abnormal centrosome synthesis, with resulting multipolar mitoses and aneuploidy (8-10). Previously, our lab generated mice transgenic for HPV-16 E7 under the control of the keratin 14 (K14) promoter, targeting E7 expression to the basal layer of stratified squamous epithelia such as the skin and cervical epithelium (16). These mice, which express E7 at levels similar to the levels seen in human cervical carcinoma cell lines (I. Frazier, unpublished data), have a broad spectrum of phenotypes. These include epithelial hyperplasia, increased cell cycle progression in all epithelial cell layers, disrupted epithelial differentiation, loss of DNA damage-induced cell cycle arrest, spontaneous skin tumors, and cervical cancers in estrogen-treated mice (16, 40, 45).
The first described function of E7 was binding and inactivation of the retinoblastoma tumor susceptibility gene product, pRb (11). Interaction between E7 and pRb results in proteasomal degradation of pRb in cultured cells (4, 12, 22) and disrupts the ability of pRb to bind to and inactivate the transactivation activity of cellular E2F transcription factors (5, 35). Disruption of the Rb regulatory pathway is frequent in most human cancers and pRb represents a target common to many highly tumorigenic or transforming viruses. E7 has also been reported to bind and inactivate other known cell cycle regulators in vitro, including the pRb family members p107 and p130, and the cyclin-dependent kinase (cdk) inhibitors p21 and p27 (12, 21, 22, 53). Remarkably, at least 14 additional cellular binding partners of the 98-amino-acid E7 protein have now been reported from in vitro studies (6, 30). Thus, E7 is a multifunctional protein in vitro with pleiotropic effects in vivo.
It is unclear which of E7's reported binding interactions is responsible for its phenotypes observed in vivo. pRb inactivation is likely to account for many of E7's effects, since pRb is known to affect cell cycle regulation, differentiation, and DNA damage responses in many cell types in vitro and in vivo (19, 28, 44). Consistent with this, E7 mutants that fail to bind or inactivate pRb fail to induce any phenotypes in vivo (13). These mutants, however, are also deficient for binding multiple other cellular targets, so they do not isolate the importance of the E7-pRb interaction. In addition, the effects of pRb inactivation cannot be studied in adult animals by using traditional knockout technology, since the Rb−/− status is lethal to embryos(19).
In the present study, we analyzed the effect of somatic pRb inactivation in murine skin by using a Cre-lox based system. This system allows comparison of pRb-deleted tissue to E7-expressing tissue in adult mice to determine which effects of E7 expression may be attributable to pRb inactivation. We demonstrate that pRb inactivation recapitulates all known phenotypes associated with E7 expression in murine skin. However, pRb-independent activities of E7 are detectable when E7 is expressed and Rb inactivated in the same tissue. Possible interpretations of these results are discussed.
MATERIALS AND METHODS
Transgenic mice.
K14E7 mice have been previously described (16). K14Cre mice were obtained from Anton Berns at The Netherlands Cancer Institute; details on their generation and characterization will be described elsewhere. For our studies, we verified the tissue-specific expression of Cre in stratified squamous epithelia by using the Z/AP Cre recombinase reporter strain (26) (data not shown). The Rb floxed mice have been described previously (41). Briefly, this mutant strain contains lox P sites flanking Rb exon 3, with no other additional sequences; a Puro cassette incorporated during the generation of the mutant allele was removed from this mouse strain to eliminate any potential hypomorphic effects. All mice were bred and maintained in the American Association for Accreditation of Laboratory Animal Care-approved McArdle Laboratory Cancer Center Animal Care Facility. All studies were performed on a mixed 129-FVB-C57BL/6 background, with all genotypes bred to contain the same levels of genetic heterogeneity. All mice were genotyped by PCR for K14E7, K14Cre, and Rb by using the following primers: for E7, oligonucleotides 709-1 (5′-GGCGGATCCTTTTATGCACCAAAAGAGAACTG-3′) and 709-4 (5′-CCCGGATCCTACCTGCAGGATCAGCCATG-3′); for Cre, oligonucleotides Cre-5 (5′-GCACGTTCACCGGCATCAAC-3′) and Cre-3 (5′-CGATGCAACGAGTGATGAGGTTC-3′); and for Rb, oligonucleotides 5′-lox (5′-CTCTAGATCCTCTCATTCTTC-3′) and 3′-lox (5′-CCTTGACCATAGCCCAGCAC-3′). One hour prior to sacrifice, all mice were intraperitoneally injected with bromodeoxyuridine (BrdU; 10 μl per g of body weight of a 12.5-mg/ml solution). For the irradiation study, mice were exposed to 0 or 5 Gy of ionizing radiation from a 137Cs source 24 h prior to BrdU administration.
Immunohistochemical and immunofluorescence analysis of epidermis.
Skin and ear epidermal samples were fixed in 10% phosphate-buffered formalin, embedded in paraffin, and cut into 5-μm sections. Serial sections were used for hematoxylin and eosin staining, immunohistochemical staining for pRb and BrdU, and immunofluorescence staining for K14, K10, and gamma-tubulin.
For immunohistochemical stains, sections were deparaffinized in xylenes and rehydrated through a graded series of ethanol-water solutions. Endogenous peroxidase activity was quenched by treatment in 3% H2O2 in methanol for 10 to 20 min. Slides were washed in phosphate-buffered saline (PBS) and heated in boiling 10 mM sodium citrate (pH 6.0) for 20 min. Further unmasking was achieved with 20 min of immersion in 2 N HCl. Samples were blocked for 30 min at ambient temperature in 5% horse serum in PBS. Primary antibodies were diluted in blocking buffer and applied as follows: 1:50 anti-pRb (clone G3-245, Pharmingen catalog no. 554136) or 1:40 anti-BrdU (Oncogene catalog no. NA20-100UG) for 2.5 h at room temperature. After washes in PBS, biotinylated secondary antibody and streptavidin-peroxidase conjugate were applied according to the Vectastain ABC kit instructions (Vector Labs catalog no. PK-6200). Staining was developed in 3,3′-diaminobenzidine (DAB) solution (Vector Labs catalog no. SK-4100) for 1 to 2 min and then quenched in H2O. Slides were counterstained with hematoxylin, dehydrated through a series of ethanols and xylenes, and covered with a coverslip.
For immunofluorescent stains, sections were deparaffinized and rehydrated as described above. For cytokeratin stains, slides were blocked with 10% horse serum-3% bovine serum albumin for 30 min. A 1:1,000 dilution of anti-mouse K14 (Covance catalog no. PRB-155P) in blocking solution was applied overnight at 4°C. Slides were then washed in PBS, and biotinylated universal secondary antibody (Vector Labs catalog no. PK-6200) at 1:250 was applied for 30 min at room temperature. After washes in PBS, streptavidin-Texas red (Vector Labs catalog no. SA-5006) at 1:150 was applied for 30 min at room temperature, followed by additional washes. Slides were then incubated overnight with fluorescein isothiocyanate-conjugated anti-keratin 10 (Covance catalog no. FITC-159L) at 1:200 in blocking buffer. Slides were rinsed in PBS, mounted in Vectashield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole; Vector Labs catalog no. H-1200), and visualized by using a Zeiss Axiophot fluorescence microscope. For centrosome stains, slides were deparaffinized as described above and boiled in 10 mM sodium citrate buffer (pH 6.0) for 30 min, followed by pepsin treatment (Digest-All 3; Zymed, South San Francisco, Calif.) for 10 min at 37°C. Sections were blocked with 10% donkey serum in destilled H2O for 15 min at room temperature, followed by incubation with an anti-gamma-tubulin monoclonal antibody (GTU-88; Sigma, St. Louis, Mo.) at a 1:1,000 dilution in PBS overnight at 4°C. After a wash in PBS, cells were incubated with a rhodamine red-conjugated donkey anti-mouse secondary antibody (Jackson Immunoresearch, West Grove, Pa.) at a 1:100 dilution in PBS for at least 2 h at 37°C. After a final wash in PBS, sections were counterstained with DAPI and then analyzed by using a Leica DMLB epifluorescence microscope.
Quantitation of pRb loss and cell proliferation.
To quantify the loss of pRb expression, sections of ear epidermis were stained for pRb and counted. Similarly, BrdU incorporation into newly synthesized DNA was used as a measure of keratinocyte proliferation by counting BrdU-stained skin and ear sections. All keratinocytes in 10 visual fields were scored as either positive (brown) or negative (blue) for pRb expression or BrdU incorporation in both basal and suprabasal layers of the epidermis. For counting purposes, even slightly brown cells were counted as positive. Three to six mice were counted per genotype. Statistical analyses of results were performed by using the two-sided Wilcoxon rank sum test.
Quantitation of centrosome abnormalities.
On each tissue section, cells showing a dot-like staining for gamma-tubulin were identified, and the number of centrosomes was determined. Since normal cells contain up to two centrosomes, cells with more than two centrosomes were considered abnormal. Due to tissue sectioning, centrosomes were detectable only in a proportion of cells on each section. On average, 164 cells were assessed per section, and the mean ± standard error of the percentage of cells with more than two centrosomes was calculated. Statistical significance was assessed by using Student two-tailed t test for independent samples.
RESULTS
Somatic inactivation of Rb in murine skin.
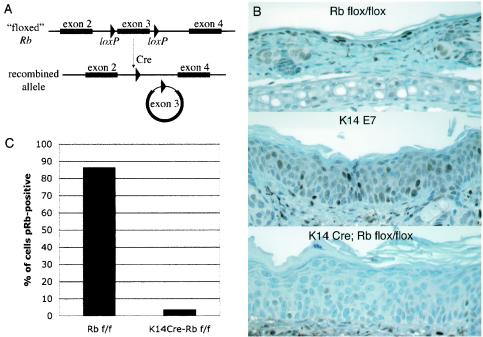
To eliminate pRb expression in murine skin, mice transgenic for Cre under the control of the K14 promoter were crossed to mice carrying a “floxed” Rb allele (Rbf ). This allele is described further elsewhere (41). Briefly, the Rbf allele contains two loxP sites in the introns flanking exon 3, with no other inserted sequences. In the presence of Cre recombinase, recombination excises the regions between the loxP sites (Fig. 1A). Excision of exon 3 from the Rb gene results in a frameshift, so that only the extreme N terminus of pRb is produced from the recombined allele. The recombined allele thus resembles the previously characterized null allele of Rb, which contains two termination codons in exon 3 (19). Mouse embryonic stem cells containing the recombined floxed allele lack detectable pRb protein and have no detectable pRb-E2F complexes in electrophoretic mobility shift assays (42). Use of the K14 promoter-driven Cre transgene limits Cre expression to the basal layer of stratified squamous epithelia, leaving pRb expression intact in other tissues (data not shown). Since epidermal stem cells are believed to be K14 positive, this transgene should cause permanent inactivation of pRb in murine epidermis (3, 47).
FIG. 1.
Cre-mediated inactivation of pRb in 21-day-old murine epidermis. (A) Schematic of the floxed Rb allele before and after recombination. Cre expression under the K14 promoter limits recombination to stratified squamous epithelia. (B) pRb immunohistochemistry of ear epidermal sections. (C) Quantification of pRb inactivation. Keratinocytes from Rbf/f or K14CreRbf/f sections stained for pRb were counted either as pRb positive (brown) or negative (blue).
Mice expressing Cre and heterozygous for the floxed Rb allele (K14CreRbf/wt mice) had no apparent phenotypes, either overtly or in any of the assays reported in the present study. In addition, no differences were detected between Rbwt/wt and Rbf/f mice or between K14E7Rbwt/wt and K14E7Rbf/f mice. These observations indicate that neither the presence of loxP sites in Rb nor the expression of Cre had any direct effects on murine epidermis. K14Cre mice homozygous for the floxed Rb allele (K14CreRbf/f mice) were generated by mating K14CreRbf/wt mice to Cre-negative, Rbf/f mice. From these crosses, K14CreRbf/f mice were obtained in the expected Mendelian ratios. These pups had several overt phenotypes reminiscent of K14E7 mice, including wrinkled, thickened skin, a scruffy appearance associated with a less dense coat of hair, and a slightly smaller size than littermate controls, although these mice grew to normal weight by about 6 weeks of age (data not shown). Both males and females of this genotype were fertile and bred normally.
To assess the efficiency of Cre-induced inhibition of pRb expression, ear and dorsal skin epidermal sections from mice of different ages were stained for pRb. To minimize variations in staining intensity, all slides were stained simultaneously using the same reagents and developed in DAB for identical times. When the animals were 21 days old, pRb staining was drastically reduced in K14CreRbf/f ear and skin epidermis but still strongly evident in Cre-negative Rbf/f epidermis (Fig. 1B). Since Cre-mediated recombination could occur independently in each keratinocyte, inactivation of pRb was quantified on a per-cell basis by counting cells as positive or negative for pRb staining. This revealed a 96% decrease in the fraction of cells expressing detectable pRb in K14CreRbf/f mice compared to Cre-negative Rbf/f controls, with the remaining positive cells in K14CreRbf/f epidermis staining very weakly (Fig. 1C). Rb inactivation was only slightly less efficient in dorsal skin, where a 91% reduction in pRb-positive cells was seen. Thus, pRb expression is inhibited in the vast majority of keratinocytes in K14CreRbf/f mice by 21 days of age. Interestingly, pRb staining was detected in most keratinocytes in K14E7 mice, although staining intensity in individual cells was often diminished compared to Cre-negative Rbf/f epidermis. This observation supports the hypothesis that E7 not only binds but also degrades pRb in vivo (4, 12, 22). Since nearly complete loss of detectable pRb was achieved in K14CreRbf/f mice by 21 days of age, all further analyses were performed on 21-day-old mice.
Hyperproliferation in Rb-deleted and E7-expressing epidermis.
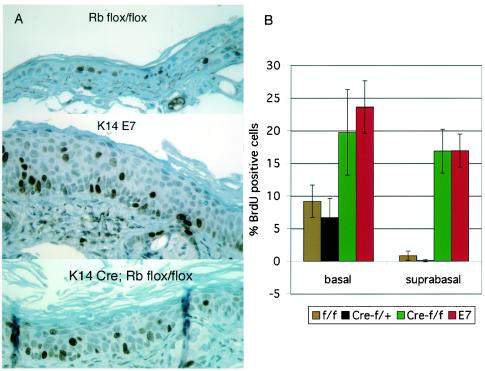
Pronounced hyperplasia was evident in the ear epidermis of both K14E7 and K14CreRbf/f mice (Fig. 1B and 2A; see also Fig. 5). To compare quantitatively the hyperproliferative effects of E7 expression versus loss of pRb, mice were injected with the deoxynucleotide analog BrdU 1 h prior to sacrifice. Sections were stained with anti-BrdU antibody to detect DNA synthesized during the hour prior to sacrifice (Fig. 2A). Counting of these sections revealed several phenotypes in Rb-deleted epidermis. These phenotypes included an ∼2-fold increase in DNA synthesis in the basal layer of keratinocytes, as well as inappropriate DNA synthesis in normally quiescent suprabasal keratinocytes (Fig. 2B). These phenotypes are strikingly similar to those seen in K14E7 mice (Fig. 2). Indeed, the fraction of cells incorporating BrdU in K14CreRbf/f mice was indistinguishable from that in K14E7 mice in both the basal and suprabasal epidermal layers. Since the severity of E7-associated phenotypes is known to vary with the expression level of E7 (16), it is important to note here that the line of K14E7 mice used in these studies expresses E7 in ear epidermis at a level similar to the E7 expression levels in human cervical carcinoma cell lines, as measured by an enzyme-linked immunosorbent capture assay (Frazier, unpublished).
FIG. 2.
Hyperplasia in E7-expressing and Rb-deleted epidermis. (A) BrdU immunohistochemistry of ear epidermal sections from mice injected with BrdU 1 h prior to sacrifice. (B) Quantification of BrdU incorporation. Basal and suprabasal cells were counted as BrdU positive (brown) or BrdU negative (blue), and the percentage of keratinocytes in each layer incorporating BrdU was calculated (basal cells are in contact with the basement membrane, but suprabasal cells are not). Increases in DNA synthesis in K14CreRbf/f (Cre-f/f) and K14E7 (E7) basal and suprabasal layers were statistically significant compared to Rbf/f (f/f) and K14CreRbf/wt (Cre-f/+) mice (P < 0.03). Differences between K14CreRbf/f and K14E7 mice were not statistically significant.
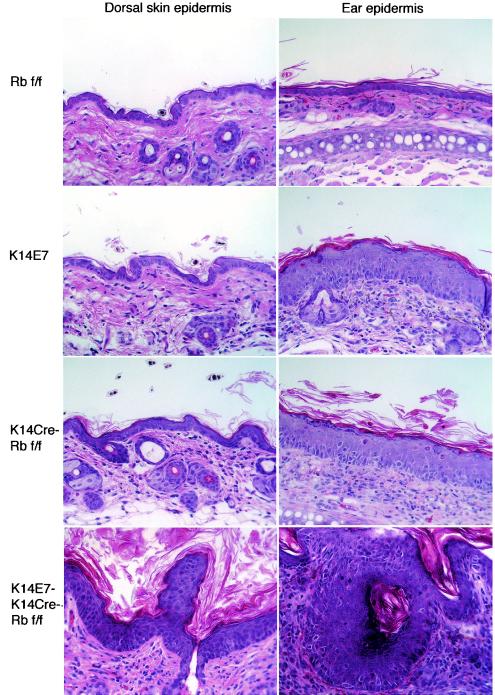
FIG. 5.
Increased phenotype severity in K14E7K14CreRbf/f mice and phenotypic variation between epidermal sites. Hematoxylin-and-eosin-stained sections from dorsal skin and ear of 21-day-old mice are shown. K14E7 and K14CreRbf/f mice have pronounced hyperplasia in ear epidermis but relatively little hyperplasia in dorsal skin epidermis. K14E7K14CreRbf/f mice, however, exhibit pronounced hyperplasia at both epidermal sites. Also shown are examples of dysplastic lesions common in K14E7K14CreRbf/f mice but not seen in other genotypes.
A greater degree of hyperplasia was seen in K14E7 epidermis than in K14CreRbf/f epidermis, as determined by the total number of suprabasal cells per visual field (P < 0.02) (Fig. 2 and data not shown). However, this difference may merely reflect the fact that E7 is expressed in the epidermis of K14E7 mice from day 6 after birth (13), whereas Cre-mediated pRb loss only becomes complete some time between 10 and 21 days of age (see below), thus allowing more time for hyperplasia to develop in K14E7 mice than in K14CreRbf/f mice.
pRb loss inhibits radiation-induced cell cycle arrest.
Having seen similarities between the effects of loss of pRb and the expression of E7 on epithelial cell proliferation, we conducted experiments to determine whether other acute effects of E7 expression in epithelium were reproduced in Rb-deleted tissues.
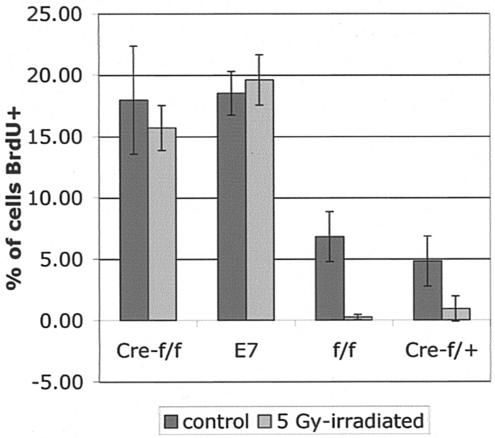
Expression of E7 in stratified squamous epithelia in mice inhibits the ability of these cells to suppress DNA synthesis in response to DNA damage (45). To determine whether loss of pRb also abolishes the normal DNA damage response of epithelia, K14CreRbf/f and K14E7 mice were exposed to 0 or 5 Gy of ionizing radiation at 21 days of age, BrdU was injected 24 h later, animals were sacrificed 1 h after that, and ear epidermal sections were taken. Histological sections were stained for BrdU, and the percentage of epidermal cells staining positively for BrdU in tissues from irradiated versus unirradiated mice was determined (Fig. 3). As expected, the epidermis from control mice (Rbf/f and K14CreRbf/wt mice) displayed a drastic decrease in BrdU incorporation after irradiation compared to that from the unirradiated control mice; in contrast, the epidermis from K14E7 mice failed to display any significant decrease in the frequency of cells supporting DNA synthesis after irradiation. Epidermis of K14CreRbf/f mice also failed to display any decrease in the frequency of cells supporting DNA synthesis after irradiation. Thus, the loss of pRb function is sufficient to inhibit DNA damage-induced responses in the mouse epidermis.
FIG. 3.
Disruption of DNA damage-induced cell cycle arrest by E7 expression or Rb deletion. Mice were exposed to 0 or 5 Gy of ionizing radiation 24 h prior to BrdU injection and sacrificed 1 h after BrdU administration. Epidermal sections were stained for BrdU incorporation and counted as in Fig. 2. Only Rbf/f (f/f) and K14CreRbf/wt (Cre-f/+) mice exhibited significant decreases in BrdU incorporation in response to ionizing radiation (P < 0.05). Cre-f/f, K14CreRbf/f mice; E7, K14E7 mice.
Disrupted differentiation in Rb-deleted epidermis.
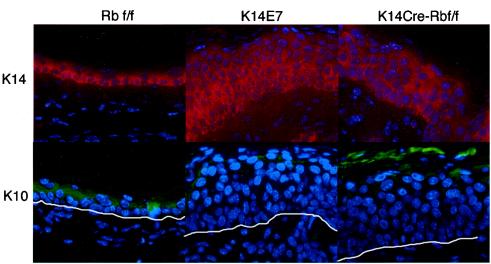
E7 disrupts the differentiation of keratinocytes in vivo, producing an expansion of the poorly differentiated, K14-positive pool of cells from the basal layer into the suprabasal layers where cells normally express the differentiation marker K10 (13, 16). Similarly, pRb also has been shown to affect the differentiation of several cell types, including human keratinocyte cell lines (28, 34). To determine whether E7 expression and loss of pRb function lead to similar effects on keratinocyte differentiation in vivo, K14CreRbf/f and K14E7 mice were stained for K10 and K14 (Fig. 4). In control mice, K14 expression was confined primarily to the poorly differentiated basal layer of the epidermis, with some parabasal cells were also K14 positive. K10 expression in control mice was absent in the basal cell layer but present in nearly every suprabasal cell. In contrast, E7-expressing epidermis was positive for K14 in nearly all cell layers below the squames and was often negative for K10 in at least three cell layers. These data agree with previous observations that E7 delays keratinocyte differentiation in vivo, resulting in an expansion of the poorly differentiated cell population of the epidermis (13, 16, 45). Epidermis from K14CreRbf/f mice typically resembled E7 epidermis, with an expanded K14-positive layer and an increase in K10-negative cell layers. Thus, somatic pRb inactivation can recapitulate the delayed differentiation seen in epidermis of K14E7 mice. However, a subset (two of six) of the 21-day-old K14CreRbf/f mice examined lacked this phenotype. This variability may reflect differences in the timing of Rb gene inactivation. We have noted that Cre-induced recombination of floxed alleles within the epidermis of the K14Cre mice is not complete at day 10, as evidenced by incomplete activation of human placental alkaline phosphatase in crosses with the Z/AP Cre-recombination reporter mice and incomplete loss of pRb expression in K14CreRbf/f mice at this earlier time point (data not shown). Therefore, it is possible that in some K14CreRbf/f mice complete Rb inactivation within the epidermis of 21-day-old mice may have only been reached shortly prior to 21 days of age, and therefore some suprabasal cells may have already undergone their normal differentiation program prior to Rb inactivation.
FIG. 4.
E7 expression or pRb inactivation disrupts epithelial differentiation. Ear epidermal sections of K14E7, K14CreRbf/f, and control mice were stained for the basal layer marker K14 (red) and the suprabasal layer marker K10 (green) and then counterstained with DAPI. For K10-stained sections, the white line represents the basement membrane.
Detection of pRb-independent activities of E7 in pRb-deleted epidermis.
To assess whether E7 confers any acute biological effects on stratified squamous epithelia that are independent of its ability to inactivate pRb, we crossed K14CreRbf/f mice to K14E7Rbf/f mice to generate K14E7K14CreRbf/f mice. K14E7K14CreRbf/f mice were severely stunted in their growth compared to their littermates, with some dying apparently of undernourishment before they reached weaning age. The cause of these deaths is uncertain, but they may have resulted from hyperproliferation in the K14-positive epithelia lining the upper digestive tract, resulting in constriction of the esophagus and forestomach, as seen previously in some lines of K14E7 mice (16). Examination of epidermal sections from surviving K14E7K14CreRbf/f mice revealed dysplastic lesions in both ear and skin epidermis by 21 days of age (Fig. 5). These lesions included unusual projections, enlarged hair follicles, and the formation of keratin pearls (Fig. 5). No such lesions were seen in K14E7 or K14CreRbf/f mice. No increase in the percentage of BrdU-positive cells was seen in ear epidermis of K14E7K14CreRbf/f mice compared to K14E7Rbf/f or K14CreRbf/f mice. In the dorsal skin of K14E7K14CreRbf/f mice, however, the frequency of BrdU-positive epidermal cells was increased compared to that in the K14E7 or K14CreRbf/f mice (P < 0.05), and this reflected an increased thickness of the epidermis (Fig. 5).
Importantly, the dysplasia and hyperplasia seen in K14E7K14CreRbf/f mice is not due to any effect of E7 on the timing or efficiency of Cre-mediated Rb deletion, since the extent of Rb inactivation is the same in K14E7K14CreRbf/f and K14CreRbf/f mice (data not shown). Similarly, since E7-mediated pRb inactivation does not affect the K14Cre transgene in K14E7K14CreRbf/f mice, it is unlikely that Cre-mediated Rb inactivation has any effect on expression of the K14E7 transgene in these mice. Thus, the detection of E7-induced phenotypes in Rb-deleted tissue indicates that E7 posesses pRb-independent effects that lead to either increased dysplasia or increased hyperplasia and dysplasia, depending on the epithelial tissue.
Centrosome abnormalities in K14E7 and K14CreRbf/f mice.
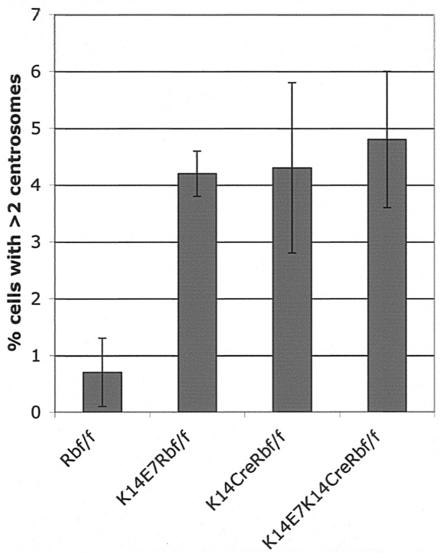
E7 expression has been shown to induce aberrant centrosome duplication and chromosomal instability in keratinocytes in monolayer and organotypic cultures (8-10). To determine whether pRb inactivation is sufficient to deregulate centrosome synthesis, epidermal sections from the ears of mice were stained for the centrosome marker gamma-tubulin, counterstained with DAPI, and the proportion of cells with abnormal centrosome numbers was enumerated (Fig. 6). We detected a statistically significant (P ≤ 0.05) 6.1-fold increase of the proportion of cells with more than two centrosomes when we compared K14CreRbf/f (4.3% ± 1.5%) to Rbf/f controls (0.7% ± 0.6%). Similarly, statistically significant 6- and 6.9-fold increases of cells with numerical centrosome abnormalities were found when we compared K14E7 mice (4.2% ± 0.4%) and K14E7K14CreRbf/f mice (4.8% ± 1.2%), respectively, to controls (P ≤ 0.01). No significant differences were observed between K14E7, K14CreRbf/f, and K14E7K14CreRbf/f (P > 0.05). These data indicate that E7 deregulates centrosome synthesis in vivo and that inactivation of pRb is sufficient to deregulate centrosome synthesis.
FIG. 6.
Centrosome abnormalities in E7-expressing and Rb-deleted epidermis. Ear epidermal sections from 21-day-old animals were stained for gamma-tubulin and counterstained with DAPI. The fraction of cells containing >2 centrosomes was counted for three to seven mice per genotype. K14E7, K14CreRbf/f, and K14E7K14CreRbf/f sections all had increases over Rbf/f sections (P < 0.05), but the former three genotypes did not differ from one another.
DISCUSSION
We report here the first genetic analysis comparing the phenotypes of E7 to that of Rb deficiency in stratified squamous epithelia, the natural site of expression of E7 in infected hosts. We make two important observations. First, the inactivation of pRb largely mimics the acute biological properties of E7. Second, E7 possesses other activities than its inactivation of pRb that contribute to its biological effects.
Consequences of somatic inactivation of pRb in murine epithelia.
We achieved nearly complete (>90%) inhibition of pRb expression in murine stratified squamous epithelia by 21 days of age (Fig. 1). Loss of pRb expression in keratinocytes resulted in multiple phenotypes, including epithelial hyperplasia with increased and deregulated cell cycle progression (Fig. 2). This was accompanied by a wrinkled skin phenotype with visible thickening of the ears and a scruffy appearance. These results were anticipated given the well-established role of pRb in cell cycle regulation (33). Interestingly, however, the extent of hyperplasia induced by Rb deletion varied between different epidermal sites, with pronounced hyperplasia in ear epidermis and only mild hyperplasia in dorsal skin (Fig. 5). This observation indicates different roles for pRb in epidermal tissue from different anatomic locations. A similar difference has long been noted with our K14E7 mice (16) (Fig. 5). There, however, we could not discriminate between a difference in the level of expression of E7 versus a difference in the biology of the different tissues. Inactivation of pRb also disrupted the normal differentiation program of the stratified squamous epitheium, demonstrating that pRb plays an important role in the differentiation of this tissue in vivo (Fig. 4). We observed delayed differentiation in Rb-deleted epidermis, with an expansion of the poorly differentiated, K14-positive cell compartment, and a delay in the onset of expression of the differentiation marker K10. These observations are consistent with studies in which a role for pRb in the differentiation of other cell types has been discerned in vitro and in vivo (28).
We found that the loss of pRb was sufficient to inhibit DNA damage responses in murine epidermis (Fig. 3). Prior studies have argued that E7's binding to pRb was necessary but not sufficient for E7 to inhibit radiation responses in human keratinocytes in tissue culture (14, 15, 44). In that system, E7's inactivation of p21 was also found to contribute to the ability of E7 to inhibit DNA damage responses. A likely explanation is that inactivation of pRb is sufficient to disrupt DNA damage responses both in vitro and in vivo, but E7 does not inactivate pRb solely by binding and degrading pRb. Rather, E7 must also promote pRb phosphorylation by suppressing the activity of cdk inhibitors such as p21. This is of particular importance in the context of DNA-damaging agents, which lead to the induction of p21.
Loss of pRb also deregulated centrosome synthesis in murine keratinocytes (Fig. 6). Although the molecular mechanism of this effect is not clear, centrosome duplication is normally coupled to the onset of DNA replication at the G1/S transition (17, 18, 23) and is dependent on cyclinE-cdk2 activity in many systems (18). Thus, since cyclin E is an E2F-responsive gene (43), loss of pRb could result in inappropriate cyclin E expression and cdk2 activation, triggering abnormal centrosome synthesis.
Centrosome abnormalities result in multipolar mitoses, with consequent genomic instability and aneuploidy (25, 29, 38). Therefore, the demonstration of a direct role for pRb in the regulation of proper centrosome synthesis indicates yet another important tumor suppressor function of pRb. This finding may have broad implications for tumor biology, since the pRb pathway is disrupted in nearly all human tumors (33). Importantly, however, dysregulation of the pRb pathway in primary human epithelial cells by loss of p16INK4A function and/or ectopic expression of the pRb kinase cdk4 does not cause centrosome abnormalities (37). Thus, the effect of pRb pathway disruption on centrosome control may vary depending on precisely how the pathway is disrupted and/or the affected cell type. Further studies will more clearly elucidate the relationship between pRb function and centrosome regulation.
Both HPV-16 E7 and the adenovirus E1A oncoprotein induce abnormal centrosome synthesis (6, 8-10). This activity of E1A is reportedly mediated by an interaction between E1A and Ran-GTPase (6). In our study, loss of pRb function deregulated centrosome synthesis to the same extent as E7 expression, and E7 expression in pRb-deficient cells had no additional effect on centrosome abnormalities. Thus, our data suggest that E7 deregulates centrosome synthesis primarily via a pRb-dependent mechanism rather than a Ran-dependent mechanism. Importantly, cervical epithelium from K14E7 mice expresses increased levels of cyclin E (T. Brake, J. Connor, D. Petereit, and P. F. Lambert, unpublished data), supporting the possibility that E7 deregulates centrosome synthesis by the pRb-dependent activation of cdk2, as described above. Given the variability inherent in counting centrosomes in histological slides, however, we cannot rule out the possibility that interactions between E7 and other binding partners could have subtle effects on centrosome synthesis that were not detected here.
Evidence for additional targets of E7 in mediating its phenotypes.
There are two important implications to the observation that the K14E7K14CreRbf/f mice display increased hyperplasia and dysplasia compared to the K14E7 and K14CreRbf/f mice. First, other activities besides E7's inactivation of pRb must contribute to E7's biological properties in vivo (since, if inactivation of pRb is the only relevant activity, we would not see a difference in the phenotypes between K14CreRbf/f and K14E7K14CreRbf/f mice). It is possible that E7 targets p107, p130, or other cellular factors more efficiently in pRb-null tissue than in pRb-sufficient tissue. However, because E7 does not appear to fully inactivate pRb (see below) and since multiple in vitro studies suggest that non-pRb interactions are important to E7's function (14, 15, 24), we conclude that the pRb-independent activities observed in the K14E7K14CreRbf/f mice are physiologically relevant and not simply a reflection of E7 altering its targets in the absence of pRb.
It is also interesting that the more severe phenotypes bestowed by the combination of E7 expression and pRb deficiency (compared to the consequences of either effect alone) differed between epidermal sites. In the torso epidermis we saw both increased hyperplasia and onset of dysplasia in the K14E7K14CreRbf/f mice, but in the ear epidermis we only observed onset of gross dysplasia without increased hyperplasia in the K14E7K14CreRbf/f mice. Although this difference may simply reflect different efficiencies with which E7 affects the same cellular factors, it could reflect, alternatively, E7 targeting different cellular proteins in these tissues.
The second implication of our findings is that E7 does not completely inactivate pRb in this mouse model (since, if E7 were to completely inactivate pRb, there would be no difference in the phenotype of K14E7 and K14E7K14CreRbf/f mice). This conclusion is in agreement with a previous study in which pRb-E2F complexes were detectable in a HPV-18-positive human cervical carcinoma cell line, suggesting that E7's inactivation of pRb is similarly incomplete in human cervical carcinogenesis (48). It is interesting, then, that the K14E7 mice display a phenotype similar to that of the K14CreRbf/f mice. Thus, in addition to partially inactivating pRb, E7 must be targeting other cellular factors, the consequence of which mimics the biological consequences that result from the complete inactivation of pRb. It is unclear which cellular targets of E7 besides pRb contribute to E7's phenotypes in vivo; however, as discussed below, p107 and p130 are the most suspect candidates.
What are the other relevant cellular targets for E7?
E7 is known to bind to at least 18 different cellular proteins in addition to pRb (30). The cellular proteins most likely mediating E7's phenotypes in vivo in addition to pRb are the other pocket proteins, p107 and p130, that are targeted by E7. They, like pRb, bind to and modulate the function of overlapping subsets of E2F family members, transcription factors known to regulate expression of cell cycle regulatory factors and DNA synthesis machinery (28). Interestingly, the combined inactivation of p107 and pRb in the epidermis does lead to a more hyperplastic mouse epidermis (J. Sage and T. Jacks, unpublished data; A. Berns, unpublished data). Also, a mutant version of E7, E7ΔDLYC, encoding a protein defective in binding all three pocket proteins (pRb, p107, and 130) fails to confer any phenotype when directed in its expression to stratified squamous epithelia in mice (13). Thus, p107 and/or p130 likely represent relevant targets for E7 in vivo.
Another potentially important set of cellular factors are the cdk inhibitors targeted by E7, p21 and p27, given their role in cell cycle regulation. Further studies are necessary to discriminate between relevant cellular factors, however, since mutations in E7 often are not sufficiently informative. Particularly, the E7ΔDLYC mutant protein used in the above-cited study (13) is not only defective for binding the pocket proteins (1, 32) but more recently has been found to be defective for binding p21 (21). The experimental mouse shall continue to provide us a unique basis for carrying out these studies by providing the means to disrupt the expression or alter the function of potentially relevant cellular targets in the context of the natural host tissue.
Acknowledgments
We thank Anton Berns for providing the K14Cre mice and sharing unpublished data, and Harlene Edwards, Jane Weeks, and Pamela Wirth for expert histological preparation of samples.
This study was supported by grants CA22443 (P.F.L.), CA84227 (P.F.L.), and CA66980 (K.M.) from the National Institutes of Health. S.B. was supported by training grant T32 GM07215) and the American Cancer Society.
REFERENCES
- 1.Barbosa, M. S., C. Edmonds, C. Fisher, J. T. Schiller, D. R. Lowy, and K. H. Vousden. 1990. The region of the HPV E7 oncoprotein homologous to adenovirus E1A and SV40 large T antigen containts separate domains for Rb binding and casein kinase II phosphorylation. EMBO J. 9:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedell, M. A., K. H. Jones, S. R. Grossman, and L. A. Laimins. 1989. Identification of human papillomavirus type 18 transforming genes in immortalized and primary cells. J. Virol. 63:1247-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bickenbach, J. R., and E. Chism. 1998. Selection and extended growth of murine epidermal stem cells in culture. Exp. Cell Res. 244:184-195. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 5.Chellappan, S., V. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Luca, A., R. Mangiacasale, A. Severino, L. Malquori, A. Baldi, A. Palena, A. M. Mileo, P. Lavia, and M. G. Paggi. 2003. E1A deregulates the centrosome cycle in a Ran GTPase-dependent manner. Cancer Res. 63:1430-1437. [PubMed] [Google Scholar]
- 7.Demers, G. W., S. A. Foster, C. L. Halbert, and D. A. Galloway. 1994. Growth arrest by induction of p53 in DNA damaged keratinocytes is bypassed by human papillomavirus 16 E7. Proc. Natl. Acad. Sci. USA 91:4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duensing, S., A. Duensing, C. P. Crum, and K. Munger. 2001. Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res. 61:2356-2360. [PubMed] [Google Scholar]
- 9.Duensing, S., A. Duensing, E. R. Flores, A. Do, P. F. Lambert, and K. Munger. 2001. Centrosome abnormalities and genomic instability by episomal expression of human papillomavirus type 16 in raft cultures of human keratinoctyes. J. Virol. 75:7712-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duensing, S., L. Y. Lee, A. Duensing, J. Basile, S. Pinoonniyom, S. Gonzalez, C. P. Crum, and K. Munger. 2000. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA 97:10002-10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulliver, G. A., R. L. Herber, A. Liem, and P. F. Lambert. 1997. Both conserved region 1 (CR1) and CR2 of the human papillomavirus type 16 E7 oncogene are required for induction of epidermal hyperplasia and tumor formation in transgenic mice. J. Virol. 71:5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helt, A., J. O. Funk, and D. A. Galloway. 2002. Inactivation of both the retinoblastoma tumor suppressor and p21 by the human papillomavirus type 16 E7 oncoprotein is necessary to inhibit cell cycle arrest in human epithelial cells. J. Virol. 76:10559-10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helt, A., and D. A. Galloway. 2001. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 75:6737-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herber, R., A. Liem, H. Pitot, and P. F. Lambert. 1996. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J. Virol. 70:1873-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinchcliffe, E. H., C. Li, E. A. Thompson, J. L. Maller, and G. Sluder. 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283:851-854. [DOI] [PubMed] [Google Scholar]
- 18.Hinchcliffe, E. H., and G. Sluder. 2001. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 15:1167-1181. [DOI] [PubMed] [Google Scholar]
- 19.Jacks, T., A. Fazeli, E. M. Schmitt, R. T. Bronson, M. A. Goodell, and R. A. Weinberg. 1992. Effects of and Rb mutation in the mouse. Nature 359:295-300. [DOI] [PubMed] [Google Scholar]
- 20.Jeon, S., and P. F. Lambert. 1995. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 92:1654-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, D. L., R. M. Alani, and K. Munger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, D. L., and K. Munger. 1997. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J. Virol. 71:2905-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacey, K. R., P. K. Jackson, and T. Stearns. 1999. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA 96:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam, E., J. Morris, R. Davies, T. Crook, R. Watson, and K. H. Vousden. 1994. HPV-16 E7 oncoprotein deregulates B-myb expression: correlation with targeting of p107/E2F complexes. EMBO J. 13:871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lingle, W. L., and J. L. Salisbury. 2000. The role of the centrosome in the development of malignant tumors. Curr. Top. Dev. Biol. 49:313-329. [DOI] [PubMed] [Google Scholar]
- 26.Lobe, C. G., K. E. Koop, W. Kreppner, H. Lomeli, M. Gertsenstein, and A. Nagy. 1999. Z/AP, a double reporter for Cre-mediated recombination. Dev. Biol. 208:281-292. [DOI] [PubMed] [Google Scholar]
- 27.Matlashewski, G., J. Schneider, L. Banks, N. Jones, A. Murray, and L. Crawford. 1987. Human papillomavirus type 16 DNA cooperates with activated ras in transforming primary cells. EMBO J. 6:1741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulligan, G., and T. Jacks. 1998. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 14:223-229. [DOI] [PubMed] [Google Scholar]
- 29.Munger, K. 2002. Disruption of oncogene/tumor suppressor networks during human carcinogenesis. Cancer Investig. 20:71-81. [DOI] [PubMed] [Google Scholar]
- 30.Munger, K., J. R. Basile, S. Duensing, A. Eichten, S. L. Gonzalez, M. Grace, and V. L. Zacny. 2001. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20:7888-7898. [DOI] [PubMed] [Google Scholar]
- 31.Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinoctyes. J. Virol. 63:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevins, J. R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699-703. [DOI] [PubMed] [Google Scholar]
- 34.Paramio, J. M., S. Lain, C. Segrelles, E. B. Lane, and J. L. Jorcano. 1998. Differential expression and functionally co-operative roles for the retinoblastoma family of proteins in epidermal differentiation. Oncogene 17:949-957. [DOI] [PubMed] [Google Scholar]
- 35.Phelps, W. C., S. Bagchi, J. A. Barnes, P. Raychaudhuri, V. Kraus, K. Munger, P. M. Howley, and J. R. Nevins. 1991. Analysis of trans activation by human papillomavirus type 16 E7 and adenovirus 12S E1A suggests a common mechanism. J. Virol. 65:6922-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phelps, W. C., K. Munger, C. L. Lee, J. A. Barnes, and P. M. Howley. 1988. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 53:539-547. [DOI] [PubMed] [Google Scholar]
- 37.Piboonniyom, S. O., S. Duensing, N. W. Swilling, J. Hasskarl, P. W. Hinds, and K. Munger. 2003. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res. 63:476-483. [PubMed] [Google Scholar]
- 38.Pihan, G. A., and S. J. Doxsey. 1999. The mitotic machinery as a source of genetic instability in cancer. Semin. Cancer Biol. 9:289-302. [DOI] [PubMed] [Google Scholar]
- 39.Reznikoff, C. A., C. Belair, E. Savelieva, Y. Zhai, K. Pfeifer, T. Yeager, K. J. Thompson, S. DeVries, C. Bindley, and M. A. Newton. 1994. Long-term genome stability and minimal genotypic and phenotypic alterations in HPV16 E7-, but not E6-, immortalized human uroepithelial cells. Genes Dev. 8:2227-2240. [DOI] [PubMed] [Google Scholar]
- 40.Riley, R., S. Duensing, T. Brake, K. Münger, P. F. Lambert, and J. Arbeit. 2003. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res., 63:4862-4871. [PubMed] [Google Scholar]
- 41.Sage, J., A. L. Miller, P. A. Pérez-Mancera, J. M. Wysocki, and T. Jacks. 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424:223-228. [DOI] [PubMed] [Google Scholar]
- 42.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 14:3037-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 44.Slebos, R., M. H. Lee, B. S. Plunkett, T. D. Kessis, B. O. Williams, T. Jacks, L. Hedrick, M. B. Kastan, and K. R. Cho. 1994. p53-dependent G1 arrest involves pRb-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc. Natl. Acad. Sci. USA 91:5320-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song, S., G. A. Gulliver, and P. F. Lambert. 1998. Human papillomavirus type 16 E6 and E7 oncogenes abrogate radiation-induced DNA damage responses in vivo through p53-dependent and p53-independent pathways. Proc. Natl. Acad. Sci. USA 95:2290-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka, A., T. Noda, H. Yajima, M. Hatanaka, and Y. Ito. 1989. Identification of a transforming gene of human papillomavirus type 16. J. Virol. 63:1465-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasioukhin, V., L. Degenstein, B. Wise, and E. Fuchs. 1999. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. USA 96:8551-8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Knebel, M., C. Rittmuller, F. Aengeneyndt, P. Jansen-Durr, and D. Spitkovsky. 1994. Reversible repression of papillomavirus oncogene expression in cervical carcinoma cells: consequences for the phenotype and E6-p53 and E7-pRb interactions. J. Virol. 68:2811-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vousden, K. H., J. Doniger, J. A. DiPaolo, and D. R. Lowy. 1988. The E7 open reading frame of human papillomavirus type 16 encodes a transforming gene. Oncogene Res. 3:167-175. [PubMed] [Google Scholar]
- 50.Watanabe, S., and K. Yoshiike. 1988. Transformation of rat 3Y1 cells by human papillomavirus type-18 DNA. Int. J. Cancer 41:896-900. [DOI] [PubMed] [Google Scholar]
- 51.White, A. E., E. M. Livanos, and T. D. Tlsty. 1994. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 8:666-677. [DOI] [PubMed] [Google Scholar]
- 52.Xiong, Y., D. Kuppuswamy, Y. Li., E. M. Livanos, M. Hixon, A. White, D. Beach, and T. D. Tlsty. 1996. Alteration of cell cycle kinase complexes in human papillmavirus E6- and E7-expressing fibroblasts precedes neoplastic tranformation. J. Virol. 70:999-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zerfass-Thome, K., W. Zwerchke, B. Mannhardt, R. Tindle, J. W. Botz, and P. Jansen-Durr. 1996. Inactivation of the cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein. Oncogene 13:2323-2330. [PubMed] [Google Scholar]
- 54.zur Hausen, H. 1996. Papillomavirus infections: a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]