Abstract
The transcriptional repressor REST/NRSF (RE-1 silencing transcription factor/neuron-restrictive silencer factor) and the transcriptional regulator REST4 share an N-terminal zinc finger domain structure involved in nuclear targeting. Using this domain as bait in a yeast two-hybrid screen, a novel protein that contains three LIM domains, putative nuclear localization sequences, protein kinase A phosphorylation sites, and a CAAX prenylation motif was isolated. This protein, which is localized around the nucleus, is involved in determining the nuclear localization of REST4 and REST/NRSF. We propose the name RILP, for REST/NRSF-interacting LIM domain protein, to label this novel protein. RILP appears to serve as a nuclear receptor for REST/NRSF, REST4, and possibly other transcription factors.
One of the major transcription factors regulating the expression of neuronal genes is the repressor element 1 (RE-1) silencing transcription factor (REST) (7), also called the neuron-restrictive silencer factor (NRSF) (24), which binds to a 21-bp DNA regulatory element, RE-1, also known as the neuron-restrictive silencer element (NRSE). REST/NRSF is a modular protein containing an N-terminal repression domain, a DNA binding domain composed of eight consecutive Cys2-His2 zinc fingers, a highly basic region, and a C-terminal repression domain containing a single Cys2-His2 zinc finger motif. The mechanism whereby REST/NRSF represses gene transcription has not been fully elucidated. It has been reported that the silencing activity of REST/NRSF correlates with the recruitment of the corepressor mSin3 to a site near the N terminus, which then forms a complex with histone deacetylase (6, 13, 17). This results in the deacetylation of nearby histones, compaction of the DNA, and loss of transcriptional activity. In addition, a novel protein called Co-REST (2) binds to the C-terminal region of REST/NRSF and helps to repress gene transcription by an unknown mechanism. Co-REST appears to act independently of the action of histone deacetylase. REST/NRSF thus has a bipartite mechanism for repression of gene expression.
Several variants of REST/NRSF mRNA, derived by alternative splicing of REST/NRSF pre-mRNA, are expressed in mature neurons of the adult rat brain, albeit at low levels (19). They encode protein isoforms with four or five zinc finger motifs. Two of these splice variants have an insertion of either 16 nucleotides (REST4) or 28 nucleotides (REST5) in the region of the gene encoding a spacer between zinc fingers 5 and 6 and produce truncated proteins containing only five of the nine zinc finger domains found in full-length REST/NRSF.
We have previously reported that REST4 acts as a dominant negative and blocks the ability of REST/NRSF to bind to DNA (25). We reported that REST4 could derepress choline acetyltransferase gene expression in a model PC12 cell line (A126.1B2), presumably by blocking the repressor activity of REST/NRSF. We proposed that REST4 acts as a modulator or “antisilencer” of REST/NRSF transcriptional repression. Recently, Tabuchi et al. (27) confirmed the competitive interaction between REST/NRSF and REST4 in primary rat cortical neurons, where REST4 was shown to reverse the silencing activity of REST/NRSF.
REST4 was shown to localize to the nucleus, although the nuclear localization signal found in REST/NRSF (11) is absent from REST4 (26). Deletion mutagenesis and generation of point mutations suggested the presence of three different signals within zinc fingers 2 to 5 of REST4: a signal for nuclear targeting, a signal for nuclear entry, and a signal for release from the nuclear translocation machinery (26). In the present study we have cloned a novel protein which interacts with REST4 zinc finger domains. This protein has been named RILP (for REST/NRSF interacting LIM domain protein). We present evidence that RILP is required for the nuclear translocation of REST/NRSF and REST4.
MATERIALS AND METHODS
Yeast two-hybrid system.
A yeast two-hybrid screen was conducted with the MATCHMAKER yeast two-hybrid system 3 (Clontech), using mouse REST/NRSF (REST4) zinc finger domains 2 to 5 (amino acids 213 to 321) as bait. A DNA fragment encoding amino acids 213 to 321 of mouse REST/NRSF was amplified from its cDNA by using PfuTurbo Hotstart DNA polymerase (Stratagene) and the primers CCC ATC CGC TGT GAC CGC TGT and TCA CCC AAC TAG ATC ACA CTC TGA GTG AGT ACG CAT GTG. The fragment was first inserted into the SmaI site in pBluescript (confirmed by DNA sequencing) and then inserted in frame into the bait vector, pGBKT7. A rat brain MATCHMAKER cDNA library constructed in the pGAD10 vector (Clontech) was used for screening. Positive clones were identified by nutritional selection (SD−His−Leu−Trp) and α-galactosidase staining as specified by the manufacturer.
Isolation of rat and human RILP cDNAs.
Full-length rat and human cDNAs were obtained using 3′ and 5′ rapid amplification of cDNA ends (RACE) employing the Marathon-Ready cDNA library (Clontech), which is a pre-made library of adaptor-ligated double-stranded cDNAs ready for use as templates. For cloning the human RILP cDNA, primers TCA TGA TCA GCC CAT TCT TCA GG and TGT GAA ACC TGT GGG GAA CAT AT were used. The RACE products were linked, cloned into pBluescript, and sequenced. In parallel, a full-length cDNA of RILP was cloned from a human brain cDNA library (Clontech) by screening with partial fragments of rat RILP obtained from the yeast-two hybrid screen. A full-length clone was obtained using 5′ and 3′ RACE.
Northern blot analysis.
The human multiple-tissue Northern blot (MTN) (Clontech) was hybridized with a RILP RNA probe derived from a PstI-BamHI fragment of human RILP as the template. The single-stranded RNA probe was made with Strip-EZ RNA and labeled with the Bright-Star Psoralen-Biotin nonisotopic detection system (Ambion). Hybridization and detection were performed as specified by the manufacturer. β-Actin mRNA was detected using a single-stranded DNA probe as a positive control.
Construction of plasmids.
A cDNA was constructed in which a FLAG epitope tag (DYKDDDDK) was added to the N terminus of RILP (FLAG-RILP). The fragment containing the FLAG epitope was amplified by PCR using PfuTurbo Hotstart DNA polymerase, subcloned into pBluescript SK, and sequenced. The FLAG-human RILP cDNA was cloned into the pcDNA3.1 mammalian expression vector (Invitrogen) and sequenced. Myc-tagged REST4 and Myc-tagged REST/NRSF were prepared as previously described (25). The primers used were as follows: MShFLAG-ATG261-278, ATG GAC TAC AAG GAC GAC GAC GAC AAG ATG CCT TTG GAG ATG GAG; MSh1205-1188, CAG TTA AGA GGC ATG GAC GTC TTC; MSh2762-2742, ACT TGG TTA AGA AAT AAT ACA; and MSh2741-2724, CCG TTA ATT TTT GCC CTT GTG TCC.
Coimmunoprecipitation of RILP and REST4.
FLAG-human RILP and Myc-REST/NRSF or FLAG-human RILP and Myc-REST4 were cotransfected in HEK293 cells. A cell lysate, prepared in mild lysis buffer (Immunocatcher; Cytosignal), was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis with either anti-FLAG or anti-Myc antibodies. Lysates in which FLAG-RILP and Myc-REST/NRSF or FLAG-RILP and Myc-REST4 were coexpressed were immunoprecipitated with agarose-conjugated anti-FLAG or anti-Myc antibodies. Immunoprecipitates were subjected to SDS-PAGE followed by Western blot analysis with anti-FLAG or anti-Myc antibodies and visualized with the ECL Plus detection system (Amersham).
Cell fractionation.
HeLa cells were cotransfected with Myc-REST4 (or Myc-REST/NRSF) and FLAG epitope-tagged full-length RILP cDNA or with FLAG-RILP(ΔCIIS) in which the C-terminal CIIS sequence was deleted. Transfection was performed with the Effectene transfection reagent (Qiagen) as specified by the manufacturer. Cellular localization was determined by analysis of nuclear and cytosolic extracts as well as by whole-cell fluorescence. Nuclear and cytosolic extracts were prepared as described previously (25) with modification. Briefly, the cells were washed with phosphate-buffered saline and collected. The cell pellet was resuspended in 5 packed-cell volumes of buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride [PMSF]), maintained on ice for 10 min, and centrifuged at 1,000 × g for 5 min. The cells were then homogenized in 2 packed-cell volumes of buffer A by using a Wheaton Dounce homogenizer. The cytosol and nuclei were separated by centrifugation (12,000 × g for 30 min) and used for further analysis.
Proteinase K digestion of nuclei from HeLa cells.
Nuclei from HeLa cells, prepared as described above and suspended in 200 μl of buffer A without PMSF, were incubated with 10 μl (10 mU) of proteinase K-agarose (Sigma) at room temperature for 10 min. A protease inhibitor cocktail (5 μl [P-2714; Sigma]) and 2.5 mM PMSF were added to stop the reaction, and the immobilized protease was removed by gentle centrifugation. The nuclei were then sonicated and centrifuged at 100,000 × g for 30 min, yielding the nuclear extract. Buffer A (200 μl) containing Triton X-100 was added to the pellet, which was sonicated and then centrifuged at 100,000 × g for 30 min to yield a solubilized pellet. Alternatively, nuclei were disrupted by sonication prior to their treatment with proteinase K-agarose. Aliquots (10 μl) of each supernatant were subjected to SDS-PAGE (10% polyacrylamide gel), followed by Western blot analysis with anti-RILP and anti-Myc antibodies.
Intracellular localization of RILP by immunohistochemistry.
HeLa cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 37°C in 5% CO2. For transfection, the cells were plated on glass coverslips (22 by 22 mm) in a six-well plate and transfected the next-day. The transfected cells were then grown for 24 h, fixed in dry ice-methanol for 5 min, washed three times with PBS, and blocked in TBS-T-M (20 mM Tris-HCl [pH 7.6], 137 mM NaCl, 0.1% Tween 20, 5% skim milk). Fixed cells were incubated with antibodies (anti-RILP antibody at 10 μg/ml, anti-FLAG antibody at 35 μg/ml, and anti-NRSF antibody at 1:100) in TBS-T-M at room temperature for 30 min. The cells were washed three times in TBS-T-M and then incubated with AlexaFluorR594- or AlexaFluor488-linked immunoglobulin G (IgG) (Molecular Probes) in TBS-T-M. The cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in PBS (1:1,000) for 5 min and then washed with PBS. Coverslips were mounted in Vectashield H-1000 (Vector Laboratories, Inc.). Fluorescence was observed using an E600 epifluorescence microscope (Nikon, Melville, N.Y.) or a laser confocal microscope.
Preparation of rabbit anti-RILP polyclonal antibody.
An antiserum against RILP was generated from the C-terminal region of rat RILP expressed in Escherichia coli with the pET-32a(+) system (Novagen). After the C-terminal part of RILP (BamHI-HindIII fragment) was expressed in E. coli BL21 (DE3) as a His tag-fusion protein, it was purified on a nickel-nitrilotriacetic acid column. The antiserum was prepared in rabbits by Bethyl Labs (Montgomery, Tex.) and affinity-purified over a column of the antigen. Preimmune serum was obtained from the rabbits prior to injection of the RILP antigen.
Construction and transfection of siRNA.
Double-stranded small interfering RNA (siRNA) was made using the Silencer siRNA construction kit (Ambion). Briefly, target mRNA was chosen at the 5′ sequence of human RILP. The oligonucleotides for the sense (MS1) and antisense (MS2) strands of mRNA, which contain additional T7 promoter sequences, were synthesized. After being filled in with Klenow DNA polymerase, each RNA was transcribed with T7 RNA polymerase in an individual tube, mixed, combined, and then digested with RNase to remove the T7 leader sequence. The siRNA double-stranded DNA was analyzed and purified as specified by the manufacturer. The sequence of oligonucleotide MS1 (hRILP22sense) is AAG ATG AGC AAA CTG GCC TTT CCT GTC TC; while that of MS2 (hRILP22antisense) is AAA AAG GCC AGT TTG CTC ATC CCT GTC TC. The following random oligonucleotides were used for negative controls: MS1 (random), AAA GGT GAA CAA TCG GCC TTC TTC TGT CC; MS2 (random), AAA AGA CGA CTG TTC GCT TAC CTC TGT CC.
Nucleotide sequence accession numbers.
The rat and human RILP cDNA sequences have been deposited in the GenBank database (accession numbers RILP-AF399843 and RILP-AF399844; respectively).
RESULTS
Isolation of rat and human RILP cDNAs.
Previous studies (26) showed that REST4 was localized to the nucleus but that REST1, which has only four of the five zinc finger domains at the C terminus, is localized to the cytosol. Point mutations that disrupt the zinc finger structures of REST4 caused mislocalization to the nuclear membrane. To identify proteins that interact with the zinc finger domains of REST4 (and REST/NRSF) a yeast two-hybrid screen was conducted with mouse REST/NRSF zinc finger domains 2 to 5 (amino acids 213 to 321) as bait and a rat brain cDNA library constructed in the pGAD10 vector. As a result of this screen, one positive clone was isolated, verified as a true positive by rescreening, and sequenced, yielding a partial clone containing a unique sequence distinct from any cDNA in the GenBank database.
The initial clone did not contain an in-frame stop codon, and it was unclear whether the complete 5′ sequence was present. A full-length cDNA was thus obtained by using 3′ and 5′ RACE. In parallel, a full-length cDNA was cloned from a human brain cDNA library. The full-length human cDNA was found to be almost identical to the rat cDNA. Both the rat and human sequences have been deposited in the GenBank database.
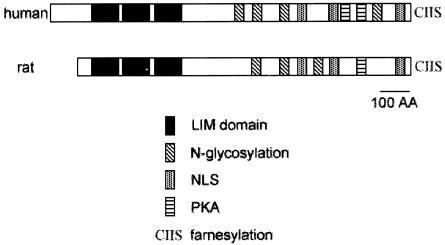
We used the deduced amino acid sequences of the rat and human cDNAs to search for conserved domains. For the human RILP clone, the coding sequence is 3,312 bp and the deduced protein is 831 amino acids. As illustrated in Fig. 1, there are three LIM domains at the N terminus. A LIM domain is a cysteine-histidine-rich, zinc-coordinating domain, consisting of tandemly repeated zinc fingers (8, 14). LIM domains are structurally similar to the GATA-type zinc fingers (20, 21). Also shown in Fig. 1 are the presence of putative N-glycosylation sites (four in human RILP and three in rat RILP), cyclic AMP-dependent protein kinase phosphorylation sites (two in human RILP and one in rat RILP), three nuclear localization signals, and a C-terminal prenylation motif. Since these novel cDNAs contain several LIM domains and interact with REST/NRSF (see below) we have named the protein the REST/NRSF-interacting LIM domain Protein (RILP).
FIG. 1.
Schematic of the structure of RILP. The amino acid sequence of RILP was deduced from its cDNAs. The human and rat RILP show greater than 95% identity, except that the human RILP has a 121-amino-acid (AA) N-terminal extension. The possible three LIM domains, three NLSs, and conserved N-glycosylation sites (four in human RILP and three in rat RILP) are indicated. Putative cAMP-dependent protein kinase A sites (two in human RILP and one in rat RILP) and a putative prenylation motif (CIIS) are also indicated.
Human genome sequence database search analysis.
The sequence of the human cDNA was used as the query sequence for BLAST searches (1) of the human genome database. The result showed that this gene was localized on chromosome 12q12 and that the RILP gene contains at least eight exons which encode the protein. We found a very similar sequence located on human chromosome 3. The gene on chromosome 3 is unknown and seems to be either an isoform or a pseudogene of RILP, since some parts of the RILP sequence are missing or exhibit low sequence identity.
Tissue distribution of RILP mRNA.
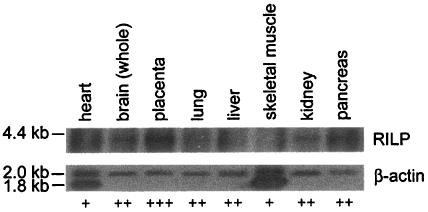
If RILP interacts with REST/NRSF or REST4 in vivo, we would expect it to be expressed in the brain as well as in most other tissues where neuronal gene expression is repressed. We therefore used a RILP RNA as a probe for the human multiple-tissue Northern blot (MTN) analysis (Clontech). As shown in Fig. 2, a single RILP mRNA band of ∼4.4 kb was detected in each of the tissues examined, albeit at low levels. This mRNA is large enough to encompass the cloned 3,312 bp of the human RILP cDNA. An RNA probe was used since initial attempts to detect RILP mRNA with a single-stranded DNA probe were unsuccessful. This finding, coupled with the observation that β-actin mRNA was easily detected using a single-stranded DNA probe (Fig. 2), shows that the expression of RILP mRNA was much lower than that of β-actin. This is in keeping with the low levels of REST/NRSF expressed in tissues (19). RILP mRNA was expressed at its highest levels in the placenta, with lower levels expressed in the brain, lungs, liver, kidneys, and pancreas.
FIG. 2.
Northern blot analysis of RILP expression in various human tissues. The human multiple-tissue Northen blot (MTN) (Clontech) was hybridized with a RILP RNA probe. The single-stranded RNA probe was made with Strip-EZ RNA and labeled with the Bright-Star psoralen-biotin nonisotopic detection system (Ambion). The β-actin mRNA was detected using a single-stranded DNA probe as a positive control. In heart and skeletal muscle, a 1.8-kb β-actin band is predominant. The relative RILP/β-actin expression is indicated as + (low) to +++ (high) in the various tissues.
Interaction of RILP with REST4 or REST/NRSF.
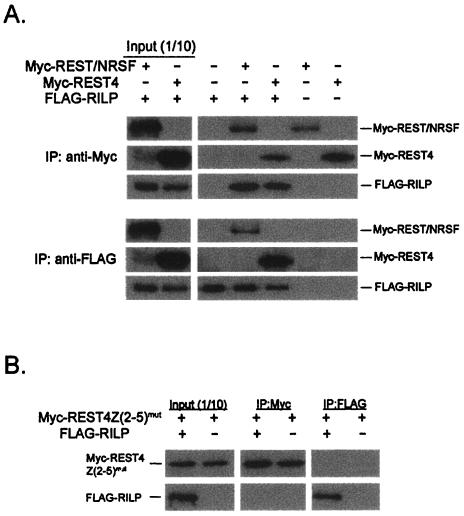
To confirm the interaction between RILP and REST/NRSF or REST4, coexpression followed by coimmunoprecipitation was performed. FLAG-RILP and Myc-REST/NRSF or FLAG-RILP and Myc-REST4 cDNAs were constructed and cotransfected in HEK293 cells. The lysates from cells expressing FLAG-RILP and Myc-REST/NRSF or FLAG-RILP and Myc-REST4 were immunoprecipitated with anti-FLAG or anti-Myc monoclonal antibodies conjugated to agarose (Santa Cruz Biotechnology) and subjected to SDS-PAGE followed by Western blot analysis with anti-Myc or anti-FLAG antibodies. As shown in Fig. 3A, both FLAG-RILP and Myc-REST/NRSF or FLAG-RILP and Myc-REST4 were detected in the immunoprecipitates. Myc-REST4 or Myc-REST/NRSF expressed alone in HEK293 cells were immunoprecipitated with anti-Myc antibody but not with anti-FLAG antibody. Similarly, FLAG-RILP expressed alone in HEK293 cells was immunoprecipitated with anti-FLAG antibody but not with anti-Myc antibody. These experiments show that RILP interacted with both REST4 and REST/NRSF. We further confirmed the specificity of this interaction by showing that FLAG-RILP did not coimmunoprecipitate with Myc-REST4(Z2-5)mut, a previously described (26) REST4 mutant in which the zinc finger structures were disrupted by point mutations (Fig. 3B).
FIG. 3.
Coimmunoprecipitation of FLAG-RILP with Myc-REST/NRSF or with Myc-REST4. (A) Coimmunoprecipitation of FLAG-RILP with Myc-REST/NRSF and FLAG-RILP with Myc-REST4. FLAG-RILP and Myc-REST/NRSF or FLAG-RILP and Myc-REST4 were cotransfected in HEK293 cells. A cell lysate was then prepared in mild lysis buffer (Immunocatcher; Cytosignal). Aliquots of the lysate were subjected to SDS-PAGE followed by Western blot analysis with either anti-FLAG or anti-Myc antibodies to detect the input level of expression of the respective cDNAs. Lysates in which FLAG-RILP and Myc-REST/NRSF or Myc-REST4 were coexpressed were immunoprecipitated with agarose-conjugated anti-FLAG or anti-Myc antibodies. Immunoprecipitates (IP) were subjected to SDS-PAGE followed by Western blot analysis with anti-FLAG or anti-Myc antibodies. The input (1/10) of immunoprecipitation is shown in the presence (+) or absense (−) of plasmid DNA. (B) FLAG-RILP and Myc-REST4Z(2-5)mut are not coimmunoprecipitated. Aliquots of a cell lysate in which FLAG-RILP and a REST4 zinc finger domain mutant [Myc-REST4Z(2-5)mut] were subjected to immunoprecipitation with either agarose-conjugated anti-Myc (IP:Myc) or anti-FLAG (IP:FLAG) antibodies followed by SDS-PAGE and Western blot analysis, as in panel A.
Intracellular localization and endogenous expression of RILP.
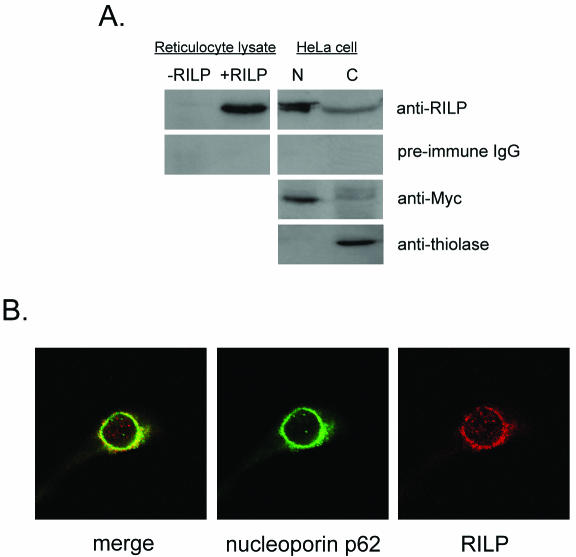
To study endogenous RILP expression and its intracellular localization, an anti-RILP polyclonal antibody was made using the C terminus of RILP as antigen and affinity purified. To test the specificity of the anti-RILP antibody, the human RILP cDNA was transcribed and translated in a rabbit reticulocyte lysate system and subjected to SDS-PAGE followed by Western blot analysis. This affinity-purified antibody detected a single band at ∼100 kDa in sample lysates but not in control lysates. Nuclear and cytosolic extracts were then prepared from HeLa cells and subjected to SDS-PAGE followed by Western blot analysis with the anti-RILP antibody. The quality of each extract was analyzed by using anti-Myc or anti-thiolase antibodies. As shown in Fig. 4A, the nuclear fraction (N) contained a band which is slightly larger than the in vitro-generated RILP. We interpret this to suggest that the nuclear RILP is posttranslationally modified by prenylation (see below), phosphorylation, or both. Interestingly, a smaller amount of RILP was detected in the cytosolic extract (C), corresponding to the same size as in vitro-generated RILP. We next used the anti-RILP antibody for immunostaining of endogenous RILP in HeLa cells. HeLa cells were fixed on a coverslip and treated with anti-RILP antibody followed by fluorescein isothiocyanate-conjugated anti-rabbit antibody. As shown in Fig. 4B, RILP (red fluorescence) was observed mainly around the nucleus, with a faint signal detected in the cytosol. This signal was blocked by adding the recombinant C-terminal domain of RILP prepared in E. coli as a His tag fusion protein. The RILP nuclear signal was detected mainly around the nucleus and localized with nucleoporin p62, suggesting that RILP localized to the nuclear membrane.
FIG. 4.
Intracellular localization of endogenous RILP in HeLa cells. (A) RILP cDNA was transcribed and translated in the reticulocyte lysate system (Promega). Nuclear (N) and cytosolic (C) extracts from HeLa cells were prepared and subjected to SDS-PAGE followed by Western blot analysis with anti-RILP antibody or preimmune serum. Anti-Myc antibody (for nuclear staining) and anti-thiolase antibody (for cytosol staining) were used as controls. (B) HeLa cells were grown on a coverslip, fixed with methanol, and then incubated with rabbit anti-RILP antibody and mouse anti-nucleoporin p62 antibody followed by AlexaFluor594-conjugated goat anti-rabbit IgG and AlexaFluor488-conjugated goat anti-mouse IgG. The images were visualized by confocal microscopy. Also shown are the same cells stained with DAPI (right) and a merged image of the RILP and nucleoporin p62 staining (left).
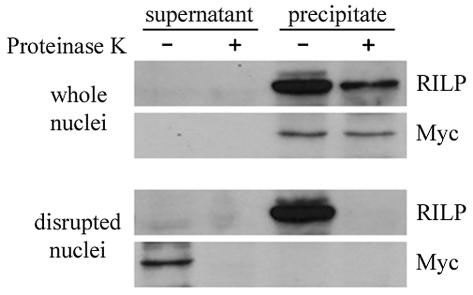
Proteinase K digestion of nuclei from HeLa cells.
To analyze the localization of RILP to the outer or inner membrane of the nuclear envelope, intact HeLa cell nuclei were purified, digested for 10 min at room temperature with proteinase K immobilized on agarose, and centrifuged. The resultant nuclear pellet was subjected to SDS-PAGE and Western blot analysis with anti-RILP antisera. As a positive control for RILP susceptibility to proteinase K, nuclei were first disrupted by sonication and then treated with proteinase K-agarose. As a control for the integrity of the nuclei, Myc protein digestion by proteinase K was also performed. Figure 5 shows that in disrupued nuclei, Myc protein and RILP were both completely digested. With intact nuclei, Myc protein was not digested by proteinase K; however, RILP was significantly proteolyzed. This suggests that at least a portion of RILP is localized to the outer nuclear membrane and that either a fraction is localized to the inner membrane or the orientation of RILP in the outer membrane makes it somewhat resistant to proteinase K.
FIG. 5.
HeLa nuclear digestion with proteinase K. Nuclei (200 μl) from HeLa cells, prepared as described in Materials and Methods, were incubated with 10 μl of proteinase K-agarose (Sigma) at room temperature for 10 min. A protease inhibitor cocktail (P-2714; Sigma) and PMSF were added to stop the reaction, and the protease was removed by centrifugation. After sonication, the mixture was centrifuged (100,000 × g for 30 min) and the supernatant was used as the nuclear extract. The pellet was resuspended in buffer containing Triton X-100, sonicated, and then centrifuged to yield solubilized membrane proteins. Alternatively, nuclei were treated in the same way, except that sonication was done before the incubation with proteinase K. An aliquot (10 μl) of each supernatant was subjected to SDS-PAGE (10% polyacrylamide gel) followed by Western blot analysis with anti-RILP and anti-Myc antibodies.
Intracellular localization of RILP.
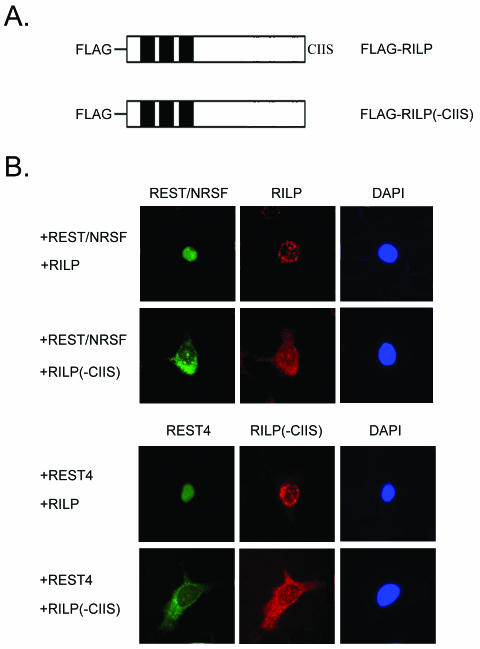
To study the effect of the prenylation motif for intracellular localization of RILP, we made a construct in which the C-terminal prenylation motif (CIIS) was deleted (Fig. 6A). GFP-REST4 and FLAG-RILP or GFP-REST4 and FLAG-RILP(-CIIS) were cotransfected in HeLa cells, and their intracellular localization was determined. As shown in Fig. 6B, FLAG-RILP localized primarily around the nuclear membrane region. However, deletion of the C-terminal CIIS sequence caused mislocalization to the cytosol. These results suggest that the prenylation motif of RILP is important for targeting to the nucleus and that RILP was associated with the nuclear membrane through its C-terminal prenylation motif. GFP-REST4 and GFP-REST/NRSF appeared no longer localized to the nucleus but instead appeared to colocalize with FLAG-RILP(-CIIS), suggesting that they interacted within the cell. It would appear that the overexpressed FLAG-RILP(-CIIS) acted as a dominant negative, masking the activity of the endogenous RILP.
FIG. 6.
Intracellular localization of recombinant RILP expressed in HeLa cells. (A) Schematic of constructs used for studying the intracellular localization of RILP. Shown in the schematic representation is full length RILP (RILP-pcDNA) and RILP with the CAAX motif deleted [RILP(ΔCIIS)-pcDNA]. The black bars represent the LIM domains, the dotted boxes represent the putative NLSs, and CIIS represents the CAAX prenylation sequence. (B) HeLa cells cotransfected with RILP-pcDNA constructs and GFP-REST4 or GFP-REST/NRSF. HeLa cells were transfected with GFP-REST4 or GFP-REST/NRSF as well as RILP-pcDNA or RILP(ΔCIIS)-pcDNA. Localization was determined by whole-cell fluorescence. Green fluorescence is due to GFP-REST4 (left), red fluorescence is RILP (middle), and blue DAPI staining (right) shows the nucleus.
RNA interference to suppress the RILP gene in HeLa cells.
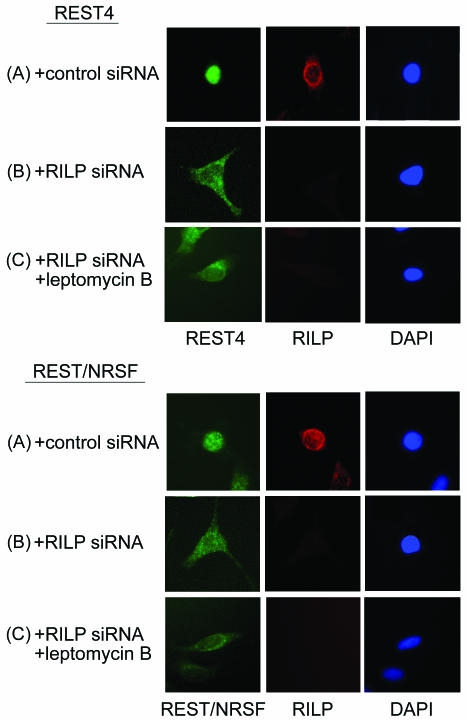
To gain an insight into the function of RILP, RNA interference was performed. HeLa cells were treated with siRNA for 24h to allow for suppression of the RILP gene, and then FLAG-REST4 or FLAG-REST/NRSF was transfected into the cells. The localization of RILP was determined by immunostaining with anti-RILP antibody followed by AlexaFluor594-conjugated goat anti-rabbit IgG, while FLAG-REST4 or FLAG-REST/NRSF was detected by using anti-FLAG antibody. As shown in Fig. 7, panels A, in control HeLa cells transfected with random siRNA (+control siRNA), FLAG-REST4 localized mainly to the nucleus and RILP was detected around the nucleus. On the other hand, with the HeLa cells treated with siRNA Fig. 7, panels B (+siRNA), FLAG-REST4 and FLAG-REST/NRSF were mislocalized to the cytosol. In RILP siRNA-treated cells, but not in random siRNA-treated cells, RILP immunostaining was barely detectable, indicating that the siRNA treatment effectively lowered cellular RILP.
FIG. 7.
Repression of RILP expression by siRNA in HeLa cells. HeLa cells were grown on a coverslip and transfected with RILP siRNA or control siRNA, which has a nonrelated scrambled siRNA sequence. After 24 h of culture, FLAG-REST4 or FLAG-REST/NRSF was transfected; this was followed by culture for an additional 24 h. The cells were then fixed with methanol and incubated with rabbit anti-RILP and mouse anti-FLAG antibodies, followed by fluorescence-labeled IgG. Green fluorescence is due to REST/NRSF or REST4 (left), red fluorescence is due to RILP (middle), and blue is due to DAPI staining (right), which shows the nucleus. (A) Control cells (treated with nonrelated sequence control siRNA). (B) Cells transfected with siRNA. (C) Cells transfected with siRNA and then treated with leptomycin B.
To rule out the possibility that FLAG-REST4 and FLAG-REST/NRSF cycle in and out of the nucleus, with RILP shifting the equilibrium to a nuclear localization, we tested the effect of leptomycin in the presence of siRNA. Treatment with leptomycin B (Fig. 7, panels C), which inhibits protein export from the nucleus (29), in conjunction with RILP siRNA did not change the cytosolic localization of FLAG-REST4 and FLAG-REST/NRSF, indicating that RILP has a function in REST/NRSF nuclear entry.
DISCUSSION
Nuclear protein import, including that of transcription factors such as REST4 or REST/NRSF, is a key control point in regulating gene expression. We previously reported that the proper zinc finger structure of the transcriptional regulator REST4 not only functions in DNA recognition but also is involved in protein-protein interaction and in determination of subcellular localization (26). It was suggested that zinc finger domains 2 to 5 are important for nuclear localization. Using these zinc finger domains (amino acids 213 to 321) as bait, a yeast two-hybrid assay screen led to the cloning of a novel protein that we have named RILP.
A conserved-domain search analysis showed that RILP has three LIM domains at its N-terminal region. A LIM domain is a cysteine-histidine-rich, zinc-coordinating domain, consisting of tandemly repeated zinc fingers (8, 14) that are structurally similar to the GATA-type zinc fingers (20, 21). LIM domains have been found to interact specifically with other LIM proteins (9, 23), as well as with a variety of other known proteins including basic helix-loop-helix proteins (10, 28), cytoskeletal components (3, 23), the insulin receptor (30), and POU-HD proteins (5, 15, 31). Since LIM domains are thought to function as protein interaction modules, it is likely that RILP interacts with REST/NRSF and REST4 through its LIM domains. Some proteins that are localized in the nucleus consist primarily of LIM domains, and were therefore named LIM-only proteins (LMO) (22). However, there appear to be no reports that the LIM domains of mammalian LIM domain-containing proteins are involved in DNA binding per se (4).
Coimmunoprecipitation experiments, as well as colocalization of recombinant-tagged RILP and REST4 or REST/NRSF in HEK293 cells, document that RILP interacts with the transcriptional repressor REST/NRSF and the transcriptional regulator REST4. That this interaction is involved in translocation to the nucleus is demonstrated by the finding that reduction of endogenous RILP by siRNA causes the mislocalization of REST4 and REST/NRSF from the nucleus to the cytosol.
The Cys-Ile-Ile-Ser sequence at the C terminus of RILP is possibly a CAAX-type prenylation signal (12, 16, 18), suggesting that RILP may be farnesylated (because of the serine at X) and thus becomes a membrane-associated protein, such as Ras and Lamin. Indeed, in vitro farnesylation analysis suggested that RILP was farnesylated at the C terminus (M. Shimojo and L. Hersh, unpublished data). In the case of lamin B, which is the major constituent of the lamina, its prenylation appears to be involved in membrane targeting, proper nuclear localization, and protein-protein interactions, which are important for signal transduction. That the CAAX motif of RILP is functional is shown by the mislocalization of a mutant RILP in which this motif was deleted. The cytosolic localization of this mutant RILP suggests that prenylation plays a role in RILP associating with the nuclear membrane. The sensitivity of RILP on intact nuclei to protease digestion indicates that some if not all of RILP is localized to the outer nuclear membrane. This suggests that RILP is involved in the translocation of REST/NRSF and REST4 into the nucleus. For nuclear protein import, it is suggested that most nuclear localization signals (NLSs) are recognized by and form a complex with importin/karyopherin. There are three putative NLSs at residues 617 to 623 (PVLRRSK), resdues 673 to 677 (HRRRR), and residues 818 to 821 (KKKK), which were predicted by the PSORT computer program. Deletion and mutation of each NLS caused mislocalization of RILP to the cytosol (unpublished data), suggesting the possibility that importin/karyopherin may be important for RILP nuclear targeting.
Taken together, these data suggest that RILP functions in the trafficking of the transcriptional repressor REST/NRSF and the transcription regulator REST4 into the nucleus. The data support a mechanism in which RILP is bound to the nuclear membrane through its CAAX motif and binds REST/NRSF and REST4, probably through its LIM domains. We therefore suggest that RILP serves as a nuclear receptor for REST/NRSF, REST4, and probably other nuclear proteins, and is intimately involved in their nuclear targeting.
Acknowledgments
This work was supported in part by grant AG05893 from the National Institute on Aging.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andres, M. E., C. Burger, M. J. Peral-Rubio, E. Battaglioli, M. E. Anderson, J. Grimes, J. Dallman, N. Ballas, and G. Mandel. 1999. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 96:9873-9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arber, S., and P. Caroni. 1996. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev. 10:289-300. [DOI] [PubMed] [Google Scholar]
- 4.Bach, I. 2000. The LIM domain: regulation by association. Mech. Dev. 91:5-17. [DOI] [PubMed] [Google Scholar]
- 5.Bach, I., S. J. Rhodes, R. V. II. Pearse, T. Heinzel, B. Gloss, K. M. Scully, P. E. Sawchenko, and M. G. Rosenfeld. 1995. P-Lim, a LIM homeodomain factor, is expressed during pituitary organ and cell commitment and synergize with Pit-1. Proc. Natl. Acad. Sci. USA 92:2720-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Z.-F., A. J. Paquette, and D. J. Anderson. 1998. REST/NRSF is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20:136-142. [DOI] [PubMed] [Google Scholar]
- 7.Chong, J. A., J. Tapia-Ramirez, S. Kim, J. J. Toledo-Aral, Y. Zheng, M. C. Boutros, Y. M. Altshuller, M. A. Frohman, S. D. Kraner, and G. Mandel. 1995. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80:949-957. [DOI] [PubMed] [Google Scholar]
- 8.Dawid, I. B., J. J. Breen, and R. Toyama. 1998. LIM domains: multiple roles as adaptors and functional modifiers in protein interactions. Trends Genet. 14:156-162. [DOI] [PubMed] [Google Scholar]
- 9.Feuerstein, R., X. Wang, D. Song, N. E. Cooke, and S. A. Liebhaber. 1994. The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc. Natl. Acad. Sci. USA 91:10655-10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.German, M. S., J. Wang, R. B. Chadwick, and W. J. Rutter. 1992. Synergistic activation of the insulin gene by a LIM-homeodomain protein and a basic helix-loop-helix domain protein: building a functional insulin minienhance complex. Genes Dev. 6:2165-2176. [DOI] [PubMed] [Google Scholar]
- 11.Grimes, J. A., S. J. Nielsen, E. Battaglioli, E. A. Miska, J. C. Speh, D. L. Berry, F. Atouf, B. C. Holdener, G. Mandel, and T. Kouzarides. 2000. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem. 275:9461-9467. [DOI] [PubMed] [Google Scholar]
- 12.Holtz, D., R. A. Tanaka, J. Hartwig, and F. McKeon. 1989. The CaaX motif of lamin A functions in conjunction with the nuclear localization signal to target assembly to the nuclear envelope. Cell 59:969-977. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Y., S. J. Myers, and R. Dingledine. 1999. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2:867-872. [DOI] [PubMed] [Google Scholar]
- 14.Jurata, L. W., and G. N. Gill. 1998. Structure and function of LIM domains. Curr. Top. Microbiol. Immunol. 228:75-113. [DOI] [PubMed] [Google Scholar]
- 15.Lichtsteiner, S., and R. Tjian. 1995. Synergistic activation of transcription by UNC-86 and MEC-3 in Caenorhabditis elegans embryo extracts. EMBO J. 14:3937-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moir, R. D., T. P. Spann, and R. D. Goldman. 1995. The dynamic properties and possible functions of nuclear lamins. Int. Rev. Cytol. 162B:141-182. [DOI] [PubMed] [Google Scholar]
- 17.Naruse, Y., T. Aoki, T. Kojima, and N. Mori. 1999. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc. Natl. Acad. Sci. USA 96:13691-13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nigg, E. A., G. T. Kitte, and K. Vorburger. 1992. Targeting lamin proteins to the nuclear envelope: the role of CaaX box modifications. Biochem. Soc. Trans. 20:500-504. [DOI] [PubMed] [Google Scholar]
- 19.Palm, K., N. Belluardo, M. Metsis, and T. Timmusk. 1998. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J. Neurosci. 18:1280-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Alvarado, G. C., C. Miles, J. W. Michelsen, H. A. Louis, D. R. Winge, M. C. Beckerle, and M. F. Summers. 1994. Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nat. Struct. Biol. 1:388-398. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Alvarado, G. C., J. L. Kosa, H. A. Louis, M. C. Beckerle, D. R. Winge, and M. F. Summers. 1996. Structure of the cysteine-rich intestinal protein, CRIP. J. Mol. Biol. 257:153-174. [DOI] [PubMed] [Google Scholar]
- 22.Rabbitts, T. H. 1998. LMO T-cell translocation oncogenesis typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev. 12:2651-2657. [DOI] [PubMed] [Google Scholar]
- 23.Schmeichel, K. L., and M. C. Beckerle. 1994. The LIM domain is a modular protein-binding interface. Cell 79:211-219. [DOI] [PubMed] [Google Scholar]
- 24.Schoenherr, C. J., and D. J. Anderson. 1995. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267:1360-1363. [DOI] [PubMed] [Google Scholar]
- 25.Shimojo, M., A. J. Paquette, D. J. Anderson, and L. B. Hersh. 1999. Protein kinase A regulates cholinergic gene expression in PC12 cells: REST4 silences the silencing activity of neuron-restrictive silencer factor/REST. Mol. Cell. Biol. 19:6788-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimojo, M., J. H. Lee, and L. B. Hersh. 2001. Role of zinc finger domains of the transcription factor neuron-restrictive silencer factor/repressor element-1 silencing transcription factor in DNA binding and nuclear localization. J. Biol. Chem. 276:13121-13126. [DOI] [PubMed] [Google Scholar]
- 27.Tabuchi, A., T. Yamada, S. Sasagawa, Y. Naruse, N. Mori, and M. Tsuda. 2002. REST4-mediated modulation of REST/NRSF-silencing function during BDNF gene promoter activation. Biochem. Biophys. Res. Commun. 290:415-420. [DOI] [PubMed] [Google Scholar]
- 28.Wadman, I., J. Li, R. O. Bash, A. Foster, H. Osada, T. H. Rabbitts, and R. Baer. 1994. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 13:4831-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolff B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type I (HIV-I) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 30.Wu, R.-Y., and G. N. Gill. 1994. LIM domain recognition of a tyrosine-containing tight turn. J. Biol. Chem. 269:25085-25090. [PubMed] [Google Scholar]
- 31.Xue, D., Y. Tu, and M. Chalfie. 1993. Cooperative interactions between the Caenorhabditis elegans homeoproteins UNC-86 and MEC-3. Science 261:1324-1328. [DOI] [PubMed] [Google Scholar]