Summary
The Odd-skipped related 1 (Osr1) gene encodes a zinc finger protein homologous to the Drosophila Odd-skipped transcription factor. During mouse embryogenesis, Osr1 is expressed in multiple tissues, including the developing heart, kidney, limb, lung, and craniofacial structures. While characterization of targeted mutant mice has revealed essential roles for Osr1 in heart and kidney development, the embryonic lethality of the Osr1 null mice has hindered investigation of its role in many other developmental processes. We report here the generation of conditional mutant mice containing two loxP sites flanking Exon 2 of the Osr1 gene. Mice homozygous for this targeted Osr1fneo allele are normal and fertile. Crossing the Osr1fneo/fneo mice with the EIIa-Cre transgenic mice resulted in Cre-mediated deletion of the loxP-flanked Exon2 in the germ line and mice homozygous for this deletion recapitulated the Osr1 null mutant phenotypes. The Osr1fneo conditional mice will be valuable for tissue-specific analysis of the roles of Osr1 in embryonic and postnatal developmental processes.
Keywords: Cre, loxP, frt, conditional inactivation, odd-skipped, Osr1
The Odd-skipped family of zinc finger transcription factors plays essential roles in embryonic development throughout metazoans. The odd-skipped gene was first identified in a large mutagenesis screen of developmental control genes in Drosophila as mutations in this gene caused loss of portions of the odd-numbered segments in the embryo (Nusslein-Volhard and Wieschaus, 1980; Coulter and Wieschaus, 1988). Since the characterization of the coding sequences of the odd-skipped gene, homologous genes have been identified in mice, human, chick, nematode, Xenopus, and zebrafish (Coulter et al., 1990; So and Danielian, 1999; Lan et al., 2001; Katoh, 2002; Buckley et al., 2004; Stricker et al., 2006; Tena et al., 2007; Mudumana et al., 2008). Analysis of gene knockout mice showed that Odd-skipped related-1 (Osr1) is essential for heart and kidney development whereas Osr2 plays critical roles in palate and tooth development (Lan et al., 2004; Wang et al., 2005; James et al., 2006; Mugford et al., 2008; Zhang et al., 2009). Gene knockdown studies in Xenopus and zebrafish showed that both Osr1 and Osr2 play critical roles in embryonic kidney development in these organisms (Tena et al., 2007; Mudumana et al., 2008).
During mouse embryogenesis, Osr1 mRNA expression is first activated in the nascent intermediate mesoderm during gastrulation and subsequently in the developing heart, limb, lung, and craniofacial structures (Wang et al., 2005). While analyses of the Osr1 null mutant mouse strains have revealed essential roles for Osr1 in the early stages of kidney and heart development, the embryonic lethality of the mutant mice has hindered investigations of the roles of Osr1 in many other developmental processes (Wang et al., 2005). To overcome this problem, we have generated mice for conditional inactivation of the Osr1 gene using the Cre-loxP in vivo DNA recombination system.
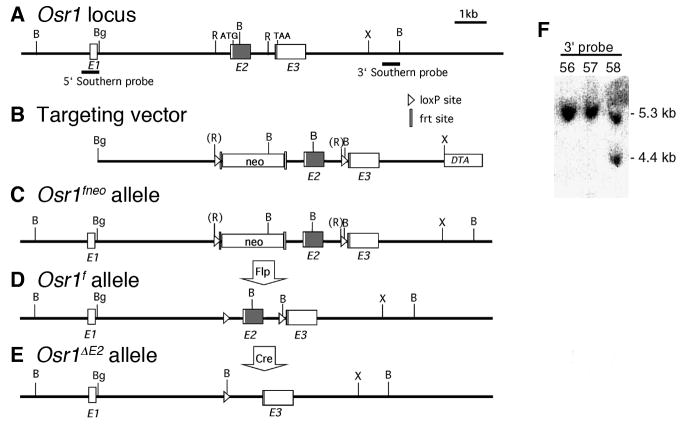
The mouse Osr1 gene contains three exons, spanning approximately 8 kb of genomic DNA sequence (FIG. 1A) (Wang et al., 2005). We isolated an Osr1 genomic DNA fragment containing all three exons from the RPCI-22 129/SvEvTac mouse BAC library (BACPAC Resources, Children’s Hospital of Oakland, Oakland, CA). To construct the targeting vector for generation of Osr1 conditional mice, one loxP sequence was inserted into each of the two EcoRV sites in the first and second introns, respectively (FIG. 1B). An frt-flanked neo expression cassette was inserted immediately 3′ to the loxP sequence in Intron 1 for positive selection. The resultant targeting vector contained the 3.8 kb BglII-EcoRI fragment from Intron 1 as the 5′ homology arm, the 3.2 kb EcoRV-XbaI fragment containing Exon 3 as the 3′ homology arm, and a diphtheria toxin-A (DTA) expression cassette immediately 3′ to the 3′ arm (FIG. 1B). Following sequence confirmation of the construct, the targeting vector was linearized and electroporated into the CJ7 mouse embryonic stem (ES) cells as previously described (Swiatek and Gridley, 1997). G418 resistant ES colonies were screened by Southern hybridization with the 3′ probe to detect integration of the 3′ loxP site in the second intron (FIG. 1F). Southern blots containing DNA samples of positively targeted clones were rehybridized with the 5′ probe (indicated in FIG. 1A) to verify the expected integration of the loxP and neo sequences in the first intron (data not shown). Of 224 colonies screened, three correctly targeted ES clones were identified and two independent clones were used for generation of chimeric mice by microinjection into blastocysts of C57BL/6J mice. Chimeric male mice were bred with C57BL/6J females to test for germ line transmission of the targeted allele. Both ES clones resulted in germ line transmission. F1 mice were genotyped initially by Southern hybridization analysis using tail genomic DNA samples for the presence of the targeted Osr1fneo allele (FIG. 1C). Heterozygous mice were intercrossed and genotyping of F2 progeny at one week after birth detected homozygous Osr1fneo/fneo mice at expected Mendelian ratio. The homozygous Osr1fneo/fneo mice exhibited normal life span and fertility. Some homozygous Osr1fneo/fneo mice were bred to FLPeR mice (Farley et al., 2000) to remove the neo expression cassette in the germ line (FIG. 1D). The resultant Osr1f/+ mice were intercrossed and homozygous Osr1f/f mice were generated and did not exhibit any detectable abnormality.
FIG. 1.
Generation of Osr1 conditional mutant mice. (A) Schematic representation of the Osr1 gene. Boxes represent exons and are labeled E1, E2, and E3, respectively. The protein-coding regions of Exons 2 and 3 are shaded, with the positions of the translation start (ATG) and stop (TAA) codons indicated. Positions corresponding to the Southern hybridization probes used for screening for correct targeting of the Osr1 locus are also indicated. Restriction sites are: B, BamHI; Bg, BglII; R, EcoRV; X, XbaI. (B) The targeting vector consists the BglII/EcoRV fragment form Intron 1 of the Osr1 gene as the 5′ homology arm, followed by an loxP sequences, an frt-flanked neo expression cassette, the EcoRV fragment containing Exon 2 of the Osr1 gene, another loxP sequence, the EcoRV/XbaI fragment containing Exon 3 as the 3′ homology arm, and an DTA expression cassette. (C) Schematic representation of the targeted Osr1fneo allele. (D) Schematic representation of the Osr1f allele, following FLP-mediated deletion of the neo expression cassette from the Osr1fneo allele. (E) Schematic representation of the Osr1ΔE2 allele, following Cre-mediated deletion of the loxP-flanked Exon 2. (F) Southern hybridization band patterns of BamHI-digested DNA samples from two non-targeted clones (Numbers 56 and 57) and one correctly targeted clone (Number 58) using the 3′ Southern probe indicated in A, which hybridizes to the wiltype fragment of 5.3 Kb and a fragment of 4.4 Kb from the correctly targeted Osr1fneo allele.
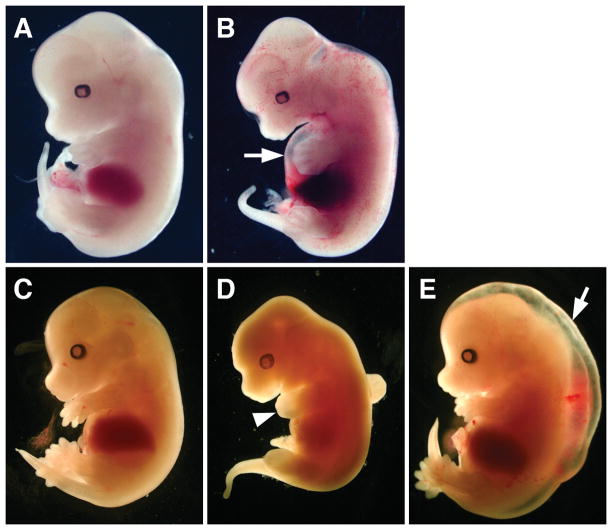
To test for Cre mediated inactivation of the Osr1 conditional allele in vivo, we crossed the Osr1fneo/fneo mice with EIIa-Cre transgenic mice, which express Cre in the early embryo and causes deletion of loxP-flanked sequences in most tissues including the germ line (Lakso et al., 1996). Whereas the resultant Osr1ΔE2/+ mice were normal, analysis of embryos from Osr1ΔE2/+ heterozygous intercrosses revealed that the Osr1ΔE2/ΔE2 mutant embryos died before birth (FIG. 2, FIG. 3). The Osr1ΔE2/ΔE2 embryos phenocopied the phenotypes of the Osr1−/− mutant embryos as previously reported (Wang et al., 2005), with most of them dying at around E12.0 (FIG. 3, B and D). Similar to the Osr1−/− embryos, a small percentage of Osr1ΔE2/ΔE2 embryos survived to late gestation and had generalized edema (FIG. 3E). Histological analysis showed that all Osr1ΔE2/ΔE2 embryos lacked metanephric kidney and had heart defects (data not shown), similar to the Osr1−/− embryos as previously reported (Wang et al., 2005). These data indicate that Cre-mediated deletion of the loxP-flanked Exon-2 abolished Osr1 gene function. Thus, the Osr1fneo conditional mice will be valuable for tissue-specific genetic analysis of the molecular pathways involving Osr1 in many developmental processes.
FIG. 2.
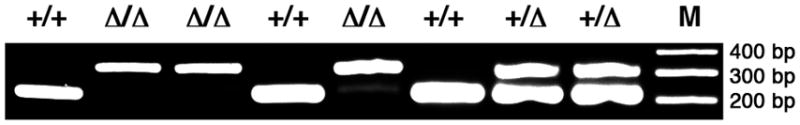
PCR genotyping of a litter of E12 embryos from intercross of a pair of Osr1ΔE2/+ mice. The PCR primers used are: ODDCKO1 (5′-GAGTGTAGCGTCTTGTGGACAG-3′, from the antisense strand in Exon 3), ODDCKO2 (5′-GCACTTTGCTCGATCTTGCTTG-3′, from sense strand upstream of the EcoRV site in Intron II), ODD1CKO3 (5′-GGTTCTGATATTGTTGTAGGACAG-3′, from sense strand upstream of the EcoRV site in Intron I). The ODDCKO1 and ODDCKO2 primers produce a PCR product of 220 bp from the wildtype allele, whereas ODDCKO1 and ODDCKO3 amplify a PCR product of 320 bp from the Osr1ΔE2 allele. +/+, wildtype; +/Δ, Osr1ΔE2/+ heterzygous; Δ/Δ, Osr1ΔE2/ΔE2 homozygous. M, DNA molecular size markers, with the corresponding fragment sizes marked to the right.
FIG. 3.
The Osr1ΔE2/ΔE2 embryos die prenatally. (A, B) A pair of Osr1ΔE2/+heterozygous (A) and Osr1ΔE2/ΔE2 homozygous (B) embryos harvested at E12.0. Note the Osr1ΔE2/ΔE2 homozygous embryo had dilated epicardium (arrow in B) and showed hemorrhaging in the trunk region. (C–E) Osr1ΔE2/+ heterozygous (C) and Osr1ΔE2/ΔE2 homozygous (D and E) embryos harvested at E14.5. The mutant embryo shown in D was dead and arrested development by E12.5 (arrowhead points to the forelimb bud, which shows a characteristic E12 limb morphology, in contrast to the well formed digits in the heterozygous littermate in C). The mutant embryo shown in E was alive but had generalized edema (arrow). These phenotypes are identical to those seen in the Osr1−/−mutant embryos as previously reported (Wang et al., 2005).
Acknowledgments
We thank the University of Rochester Transgenic Mouse Facility for generating chimeric founder mice from our gene-targeted mouse embryonic stem cells. This work was supported by NIH/NIDCR grant R01DE013681.
LITERATURE CITED
- Buckley M, Chau J, Hoppe P. odd-skipped homologs function during gut development in C. elegans. Dev Genes Evol. 2004;214:10–18. doi: 10.1007/s00427-003-0369-x. [DOI] [PubMed] [Google Scholar]
- Coulter DE, Swaykus EA, Beran-Koehn MA, Goldberg D, Wieschaus E, Schedl P. Molecular analysis of odd-skipped, a zinc finger encoding segmentation gene with a novel pair-rule expression pattern. EMBO J. 1990;9:3795–3804. doi: 10.1002/j.1460-2075.1990.tb07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter DE, Wieschaus E. Gene activities and segmental patterning in Drosophila: analysis of odd-skipped and pair-rule double mutants. Genes Dev. 1988;2:1812–1823. doi: 10.1101/gad.2.12b.1812. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (Flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- James RG, Kamei CN, Wang Q, Jiang R, Schultheiss TM. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- Katoh M. Molecular cloning and characterization of OSR1 on human chromosome 2p24. Int J Mol Med. 2002;10:221–225. [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okanoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Kingsley PD, Cho ES, Jiang R. Osr2, a new mouse gene related to Drosophila odd-skipped, exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mech Dev. 2001;107:175–179. doi: 10.1016/s0925-4773(01)00457-9. [DOI] [PubMed] [Google Scholar]
- Lan Y, Ovitt CE, Cho ES, Maltby KM, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004;131:3207–3216. doi: 10.1242/dev.01175. [DOI] [PubMed] [Google Scholar]
- Mudumana SP, Hentschel D, Liu Y, Vasilyev A, Drummond IA. Odd-skipped related 1 reveals a novel role for endoderm in regulating kidney versus vascular cell fate. Development. 2008;135:3355–3367. doi: 10.1242/dev.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford JW, Sipila P, McMahon JA, McMahon AP. Osr1 expression deparcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- So PL, Danielian PS. Cloning and expression analysis of a mouse gene related to Drosphila odd-skipped. Mech Dev. 1999;84:157–160. doi: 10.1016/s0925-4773(99)00058-1. [DOI] [PubMed] [Google Scholar]
- Stricker S, Brieske N, Haupt J, Mundlos S. Comparative expression pattern of Odd-skipped related genes Osr1 and Osr2 in chick embryonic development. Gene Expr Patterns. 2006;6:826–834. doi: 10.1016/j.modgep.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Swiatek PJ, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc-finger gene Krox20. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- Tena JJ, Neto A, de la Calle-Mustienes E, Bras-Pereira C, Casares F, Gomez-Skarmeta JL. Odd-skipped genes encode repressors that control kidney development. Dev Biol. 2007;301:518–531. doi: 10.1016/j.ydbio.2006.08.063. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lan Y, Cho ES, Maltby KM, Jiang R. Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development. Dev Biol. 2005;288:582–594. doi: 10.1016/j.ydbio.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lan Y, Chai Y, Jiang R. Antagonistic actions of Msx1 and Osr2 pattern mammalian teeth into a single row. Science. 2009;323:1232–1234. doi: 10.1126/science.1167418. [DOI] [PMC free article] [PubMed] [Google Scholar]