Abstract
Macroautophagy is a cellular process by which cytosolic components and organelles are degraded in double-membrane bound structures upon fusion with lysosomes. A pathway for selective degradation of mitochondria by autophagy, known as mitophagy, has been described, and is of particular importance to neurons, because of the constant need for high levels of energy production in this cell type. Although much remains to be learned about mitophagy, it appears that the regulation of mitophagy shares key steps with the macroautophagy pathway, while exhibiting distinct regulatory steps specific for mitochondrial autophagic turnover. Mitophagy is emerging as an important pathway in neurodegenerative disease, and has been linked to the pathogenesis of Parkinson’s disease through the study of recessively inherited forms of this disorder, involving PINK1 and Parkin. Recent work indicates that PINK1 and Parkin together maintain mitochondrial quality control by regulating mitophagy. In the Purkinje cell degeneration (pcd) mouse, altered mitophagy may contribute to the dramatic neuron cell death observed in the cerebellum, suggesting that over-active mitophagy or insufficient mitophagy can both be deleterious. Finally, mitophagy has been linked to aging, as impaired macroautophagy over time promotes mitochondrial dysfunction associated with the aging process. Understanding the role of mitophagy in neural function, neurodegenerative disease, and aging represents an essential goal for future research in the autophagy field.
Overview of macroautophagy and mitophagy
Although at least three different forms of autophagy exist, the term autophagy is typically used, when referring to the process of macroautophagy, which is a bulk catabolic process that degrades long-lived proteins and recycles organelles. Over the last decade, autophagy has been implicated in many conditions, including cancer, autoimmune disease, neurodegeneration and aging (Cuervo et al., 2005; Deretic, 2005; Mizushima, 2005). Consequently, autophagy has captured the attention of researchers from various fields of study. Autophagy was first observed about four decades ago (Deter and De Duve, 1967). In these studies, cytoplasmic components, including organelles, were engulfed in autophagy-specific vacuoles named autophagosomes, and degradation of vacuolar contents occurred upon fusion of autophagosomes with lysosomes. Observation of a wide range of cytoplasmic components in autophagosomes led to speculation that macroautophagy was a non-specific catabolic process (Deter and De Duve, 1967; Ericsson, 1969a; Ericsson, 1969b). However, directed studies on organelle-specific autophagy, including mitochondrial (Lemasters, 2005), peroxisomal (Sakai et al., 2006) and protein-selective chaperone-mediated autophagy (Cuervo et al., 2004) sparked proposals to the contrary. Today, many forms of selective autophagy are known (Klionsky, 2007). Selective degradation of mitochondria by autophagy, or “mitophagy”, is of particular importance due to the pivotal role that mitochondria play in cellular metabolism. Mitochondria are a source of both energy and highly damaging reactive oxygen species (ROS). In addition, release of cytochrome c from mitochondria is a step in the process of programmed cell death, better known as apoptosis (Liu et al., 1996). Therefore, clearance of damaged and aging mitochondria is a critical process for cell survival (Wallace, 2005).
The presence of mitochondria in lysosomes was first demonstrated in mammalian cells (Ashford and Porter, 1962; Clark, 1957); however, as for macroautophagy, an understanding of mitophagy was pioneered in yeast (Kanki and Klionsky, 2010). In particular, these studies identified genes that are required for mitophagy, but not for other forms of autophagy, highlighting selective regulation of mitophagy (Kanki et al., 2009a). For example, the mitochondrial outer membrane proteins Atg32 and Atg33 are required exclusively for mitophagy (Kanki et al., 2009a; Kanki et al., 2009b; Okamoto et al., 2009). Moreover, Atg32 is a mitochondrial recognition receptor. According to this model, Atg32 is bound by Atg11, which is an adaptor for selective autophagy (Shintani et al., 2002), to mark mitochondria for degradation by mitophagy (Kanki and Klionsky, 2008; Kanki et al., 2009b; Okamoto et al., 2009). Certain genes, however, that are required for autophagy, are dispensable for mitophagy, such as Atg13, which is a critical upstream regulator of macroautophagy (Okamoto et al., 2009). These findings in yeast suggest a model in which mitophagy, like other selective forms of autophagy in yeast, is uniquely regulated, while also utilizing some of the macroautophagy machinery.
In mammals, mitophagy has been carefully investigated during the process of erythroid differentiation. Erythroid differentiation involves the programmed clearance of organelles, including the nucleus, to eventually form the nascent reticulocyte (Geminard et al., 2002). A considerable body of evidence suggests that mitochondrial degradation during this process is achieved by mitophagy (Kundu et al., 2008; Schweers et al., 2007). However, this process is not completely understood. Ulk1, the mammalian orthologue of yeast Atg1, is a critical upstream regulator of macroautophagy, and is required for mitophagy during erythroid differentiation (Kundu et al., 2008). Ulk1 knock-out mice develop red blood cells that retain mitochondria. Intriguingly, unlike other macroautophagy gene-deficient mice (Hara et al., 2006; Komatsu et al., 2006; Yue et al., 2003), Ulk1 knock-out mice were viable, and were not defective in starvation-induced macroautophagy (Kundu et al., 2008). In addition to ULK1, a Bcl-2 family member protein, NIX, is required for mitochondrial clearance during erythroid differentiation (Schweers et al., 2007), as Nix knock-out mice fail to target mitochondria to autophagosomes (Sandoval et al., 2008). This is independent of other Bcl-2 family proteins and the macroautophagy pathway, although the mechanistic basis of NIX action upon mitochondria remains unclear (Zhang et al., 2009). Additional studies of mitochondria and organelle turnover in nascent reticulocytes also indicate that mitophagy, at least in this cell type, can occur through an autophagy-independent pathway, as mitochondrial clearance is not eliminated in Atg5 or Atg7 null erthryocytes (Matsui et al., 2006; Zhang et al., 2009). Although such “autophagy-independent” pathways remain to be defined, one recent study proposed that autophagosomes could be formed in a Rab9-dependent manner through the fusion of nascent isolation membranes with vesicles from the Golgi apparatus or endosomes (Nishida et al., 2009). This alternative form of macroautophagy, occurring independently of Atg5 and Atg7, was further shown to promote mitochondrial clearance in reticulocytes. Nonetheless, a previous study had found that mitophagy is markedly diminished in Atg7 null reticulocytes (Zhang et al., 2009).
In summary, these studies imply that, as in yeast, regulation of mitophagy has distinct components in mammalian erythroid differentiation, but may also require factors essential to the macroautophagy pathway. Further evidence in support of a role for ‘canonical’ autophagy pathway factors in mitophagy comes from studies of hypoxia-induced organelle turnover. Hypoxia-induced mitophagy, which is activated to maintain proper oxygen homeostasis, was documented to depend upon autophagy components, including Beclin-1 and Atg5 (Zhang et al., 2008). Regulation of mitophagy in mammalian systems thus requires further clarification. In particular, it remains unclear whether mitophagy, induced by different cues, utilizes specific regulators in certain cases (i.e. Ulk1, NIX, PINK1), or if additional regulators exist. Furthermore, whether all regulators converge upon a central core mitophagy complex, or achieve mitophagy induction through different pathways, remains unknown.
Introduction to Autophagy and Neurodegenerative disease
In recent years, autophagy has attracted considerable attention as a possible therapeutic target for neurodegenerative disease. Studies of autophagy in the nervous system have been difficult due to an inability to observe autophagic structures under physiological conditions (Mizushima et al., 2004). This has been attributed to a very efficient rate of autophagy in neurons, such that autophagic structures are not normally detected (Boland et al., 2008; Cuervo, 2006). Consequently, several studies have reported autophagic structures in the neurons of neurodegenerative disease mouse models and human patient samples (Boland et al., 2008; Kegel et al., 2000; Nixon et al., 2005). It is not clear whether these autophagic structures indicate an increase in autophagy, or a defect in the progression of the autophagy pathway (or both!). Several studies proposed that autophagy helps to relieve the proteotoxic stress of misfolded proteins by degrading toxic oligomers in the cytosol (Levine and Kroemer, 2008; Pandey et al., 2007; Ravikumar et al., 2004). This hypothesis supports a protective role for autophagy; however, several groups have found that autophagy can drive the degeneration of axons and dendrites (Komatsu et al., 2007; Plowey et al., 2008; Yang et al., 2008b). Chaperone-mediated autophagy, a form of selective autophagy, was shown to specifically degrade α-synuclein (Cuervo et al., 2004). Mutated forms of α-synuclein, which cannot be degraded, are found in Lewy bodies in Parkinson’s disease patients. These studies imply that autophagic degradation of misfolded proteins could protect neurons from proteotoxicity. As the focus of this review is restricted to mitophagy, a comprehensive discussion of the role of macroautophagy in neurodegenerative disease can be found in other recent publications (Cherra et al., 2010; Wong and Cuervo, 2010).
Mitophagy, mitochondrial dynamics, and Parkinson’s Disease
In recent years, mitochondrial physiology in the central nervous system has captured the attention of researchers, due to new insights in Parkinson’s disease (PD) pathogenesis. PD was first defined in 1817, and is the second most common neurodegenerative malady in patients over the age of 50 (Polymeropoulos et al., 1996). Only a small fraction of PD cases have been linked to genetic causes (Feany, 2004). The first causal PD mutation identified was a dominant mutation in α-synuclein (Polymeropoulos et al., 1997). Incorporation of α-synuclein into Lewy bodies thus led to hypotheses linking PD pathogenesis with cytoplasmic protein toxicity. On the other hand, recessive mutations, found in PINK1 (Valente et al., 2004) and Parkin (Kitada et al., 1998), have revealed a novel pathogenic mechanism involving mitochondrial physiology. PINK1 is a serine/threonine kinase, which localizes to mitochondria, whereas Parkin is a cytoplasmic E3 ubiquitin ligase. Two landmark studies in Drosophila established that PINK1 and Parkin are on the same pathway leading to PD pathogenesis, and that their loss causes mitochondrial defects (Clark et al., 2006; Park et al., 2006). In these studies, PINK1 mutations led to dopaminergic neuron and muscle degeneration likely due to mitochondrial defects. Remarkably, PINK1 and Parkin appear to physically interact in vivo, and Parkin expression can ameliorate PINK1 phenotypes, but not vice versa, suggesting that Parkin acts downstream of PINK1. Studies of PINK1 knock-out mice have also identified dysfunctional mitochondria. In these mice, mitochondrial respiration deteriorated in an age-dependent manner, which may account for the development of PD most commonly in the elderly. It should also be noted that these mice did not develop dopaminergic neuron degeneration, but had defects in dopamine release (Gautier et al., 2008; Kitada et al., 2007). Involvement of mitochondria in PD is supported by an extensive literature that has documented mitochondrial oxidative phosphorylation pathway dysfunction in PD patients and mammalian cell lines (Moore et al., 2005; Schapira et al., 1989; Swerdlow et al., 1996). Moreover, recessive mutations in DJ-1, a protein believed to protect cells against oxidative damage, can also cause PD (Bonifati et al., 2003). This further supports the importance of mitochondrial physiology to the nervous system, since dysfunctional mitochondria are the primary source of oxidative damage (Kahle et al., 2009).
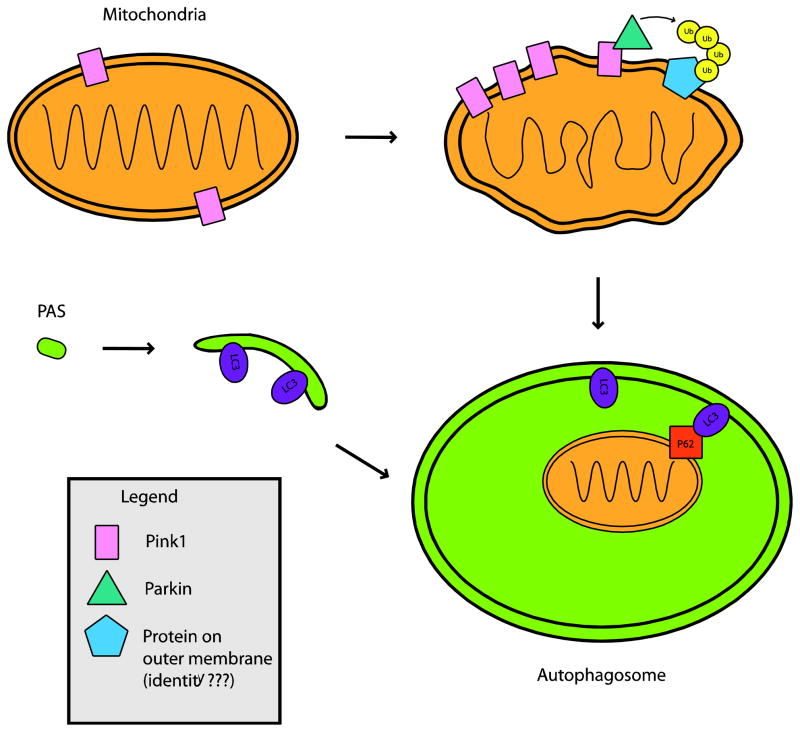
In mammalian systems, an emerging literature has suggested that PINK1 resides on the outer mitochondrial membrane and serves as a sensor for mitochondrial membrane polarization status, such that in normal healthy mitochondria, PINK1 undergoes proteolytic cleavage (Abeliovich, 2010). While PINK1 is a mitochondrial transmembrane protein with its kinase domain exposed to the cytosol (Zhou et al., 2008), Parkin usually resides in the cytosol. Parkin is recruited to dysfunctional mitochondria with low membrane potential, based upon co-localization with mitochondrial TOM20, and may interact with PINK1, which selectively accumulates on these mitochondria (Narendra et al., 2008; Narendra et al., 2010; Vives-Bauza et al., 2010). Studies performed in PINK1 null mouse embryonic fibroblast cell lines and by PINK1 knock-down have found that PINK1 is required for Parkin-induced mitophagy by tracking the loss of mitochondrial fluorescence (Narendra et al., 2010). Thus, upon mitochondrial dysfunction, PINK1 is selectively stabilized on the outer mitochondrial membrane and may recruit Parkin to promote autophagic degradation of impaired mitochondria. This is dependent on PINK1 kinase activity, and results in enhanced ubiquitin ligase activity of Parkin (Kawajiri et al., 2010; Matsuda et al., 2010). Once recruited by PINK1, activated Parkin leads to the formation of poly-ubiquitin chains on certain protein(s), believed to reside on the mitochondrial outer membrane (Figure 1). This triggers recruitment of p62 to mitochondria (Geisler et al., 2010). p62 has a LC3 binding domain, and is an adaptor protein for autophagic degradation of poly-ubiquinated proteins (Pankiv et al., 2007). VDAC1 is one of many proteins shown to be poly-ubiquitinated by Parkin, and its specific role in the recruitment of p62 has not been established and remains speculative; however, Parkin poly-ubiquitination of VDAC1, or more likely some other protein, initiates a process that culminates in mitochondrial degradation by mitophagy (Wild and Dikic, 2010). It is thus plausible that recessive mutations in PINK1 and Parkin, that cause PD, lead to mitophagy failure, promoting accumulation of damaged mitochondria, which contributes to disease pathogenesis; alternatively, it is possible that PINK1 dysfunction promotes excessive turnover of healthy mitochondria, as independent work from another group has shown that stable shRNA knock-down of PINK1 yields mitochondrial fragmentation and mitophagy in SH-SY5Y cells(Dagda et al., 2009). Furthermore, it is not clear whether PINK1 and Parkin interaction is the only path to mitophagy. Several studies in mammalian systems have found that consequent mitochondrial defects in PINK1 mutants can activate mitophagy by mechanisms other than Parkin interaction, such as through increased ROS production, or activation of mitochondrial fission (Dagda et al., 2009; Wood-Kaczmar et al., 2008). Indeed, many of the studies, upon which the PINK1-Parkin mitophagy model is based, have been performed in non-neuronal cells, and rely upon chemical uncoupling agents, which can induce alterations in mitochondrial fluorescence (Chu, 2010). Hence, much more work needs to be done to clarify the mechanistic basis of mitophagy in the central nervous system and its role in PD pathogenesis.
Figure 1. PINK1 and Parkin interact to recruit mitochondria to autophagosomes, and thereby promote mitophagy.
PINK1 is present on mitochondria normally, but upon mitochondrial depolarization, PINK1 rapidly accumulates on mitochondria. This is followed by recruitment of Parkin, and the subsequent poly-ubiquitination of an unknown target protein by Parkin. Poly-ubiquitinated mitochondria are shuttled to developing autophagosomes through an interaction between the poly-ubiquitin chain, p62, and LC3 on the autophagosome membrane. Once inside an autophagosome, depolarized mitochondria can be degraded by lysosomes, when autophagosome-lysosome fusion occurs (not shown). In Parkinson’s disease caused by recessive PINK1 or Parkin mutations, depolarized mitochondria are no longer efficiently degraded by this pathway, contributing to the hallmark mitochondrial dysfunction observed in this disorder.
PAS = Pre-autophagosomal structure
A fundamentally important advance in our understanding of mitochondrial biology has been the realization that mitochondria are extremely dynamic organelles engaged in a constant process of fission and fusion (Detmer and Chan, 2007). Although the relationship between mitochondrial dynamics and function will require further work, a set of proteins has been shown to mediate this process. Mitofusins 1 and 2 (Mfn1, Mfn2) and OPA1 are required for fusion, while dynamin-related protein 1 (Drp1) and Fis1 are required for fission (Detmer and Chan, 2007). Amazingly, germline mutations in Mfn2 cause Charcot-Marie-Tooth hereditary neuropathy type 2A (CMT2A), while germline mutations in OPA1 produce autosomal dominant optic atrophy (ADOA) and are the most common genetic cause of optic atrophy in humans (Knott and Bossy-Wetzel, 2008). These discoveries underscore the importance of mitochondrial fusion for neuron health. The story for cerebellar Purkinje neurons is even more compelling, as Mfn2 conditional knock-out mice, crossed with a neuron-specific Cre driver line, undergo rapid degeneration and death of Purkinje neurons (Chen et al., 2007). Mitochondrial fission is also important for neuron function, as dominant-negative Drp1 mutation can cause a lethal infantile neurodegenerative phenotype (Waterham et al., 2007). Very recently, Drp1 knock-out mice have been produced, and display embryonic lethality characterized by abnormal brain development and failed synapse formation (Ishihara et al., 2009; Wakabayashi et al., 2009). An important aspect of this work has been the discovery of a link between mitochondrial fission and mitophagy. Inhibition of mitochondrial fission with dominant-negative Drp1 or by Fis1 RNA knock-down prevents mitophagy, resulting in accumulation of damaged mitochondria (Twig et al., 2008). At the same time, mitochondrial fission has been linked to apoptotic cell death of neurons (Knott and Bossy-Wetzel, 2008), indicating that excess fission is also deleterious.
It is not entirely clear how the PINK1/Parkin pathway is involved in mitochondrial physiology, but PINK1 and Parkin function likely intersects with the process of “mitochondrial dynamics”, and vice versa, although further studies in mammals are ongoing to resolve this question (Chu, 2010). Nonetheless, significant advances in our understanding of mitochondrial dynamics have come out of work with Drosophila, and have linked Drp1-mediated fission with the Parkin and PINK1 mutations that cause PD in humans (Poole et al., 2008; Yang et al., 2008a). These studies in Drosophila indicate that PINK1 and Parkin are maintaining mitochondrial quality control, as increased Drp1 expression in Parkin or PINK1 mutant flies can rescue mitochondrial defects, while decreased Drp1 dosage in combination with Parkin or PINK1 mutation is lethal. Although one study of PINK1 in mammalian systems has linked PINK1-dependent mitophagy to mitochondrial fission (Dagda et al., 2009), the role of Parkin and PINK1 in mitochondrial dynamics, and the nature of the cross-talk between mitochondrial dynamics and mitophagy in the mammalian nervous system remain ill-defined.
Mitophagy in other neurodegenerative diseases
Observation of autophagic vacuoles in the neurons of Alzheimer’s disease (AD) patients suggested that cytoplasmic and organellar turnover might be important during this disease (Boland et al., 2008; Nixon et al., 2005). Moreover, there is evidence that AD patients possess damaged mitochondria; in particular mitochondrial cytochrome oxidase is defective (Maurer et al., 2000; Parker, 1991). Some workers have proposed that the accumulation of β-amyloid fragments within the mitochondria provides a mechanistic insight into mitochondrial dysfunction in AD (Casley et al., 2002a; Casley et al., 2002b). Similarly, Huntington’s disease (HD) has been associated with mitochondrial dysfunction (Gu et al., 1996; Panov et al., 2002). Transcriptional dysregulation of PGC-1α, which has an important role in mitochondria biogenesis, might be a contributing factor to HD pathogenesis (Cui et al., 2006). Importantly, PGC-1α is linked to metabolic and transcriptional defects in HD mice and patients (Weydt et al., 2006). These studies highlight the importance of mitochondrial physiology in HD pathogenesis. It is possible that mitophagy is a defense against neuronal loss in HD by eliminating defective mitochondria, thereby preventing the caspase activation that has been identified in HD and linked to huntingtin protein cleavage (Graham et al., 2006; Sanchez et al., 1999). At least one study suggests that this might also be the case in AD (Moreira et al., 2007), but no direct evidence exists for a role of selective mitophagy activation in ameliorating either disease. Intriguingly, however, a recent study found that HD pathogenesis is associated with autophagic cargo recognition defects that result in the accumulation of dysfunctional mitochondria in the cytoplasm (Martinez-Vicente et al., 2010).
In addition to AD and HD, the Purkinje cell degeneration (pcd) mouse model has provided some insights into mitochondrial physiology in the central nervous system. pcd is a recessive spontaneous mutation that results in the loss of >99% of Purkinje cell neurons by five weeks of age and subsequent ataxia (Mullen et al., 1976). One recent study has shown that Nna1, the protein whose function is lost in pcd, localizes to mitochondria and is required for normal glycolysis and mitochondrial oxidative phosphorylation pathway function (Chakrabarti et al., 2010). As it turns out, mitophagy is up-regulated in pcd mice and this may contribute to the rapid and dramatic neuron cell death (Chakrabarti et al., 2009). Consequently, increased mitophagy may undermine the metabolic needs of the highly electrically active Purkinje cell, causing it to die by reducing mitochondrial number. These findings imply that mitophagy may not always be protective, and highlight the importance of precise regulation of mitochondrial turnover in the nervous system.
Relationship of mitophagy and aging
Some investigators have hypothesized that mitochondria are important in aging in two ways: 1) they are the primary source of ROS production that contribute to cell damage; and 2) their DNA is prone to accumulating mutations, which in turn results in mitochondrial dysfunction (Lemasters, 2005). Early studies of the role of mitophagy in aging again came from yeast. Deletion of the mitochondrial membrane protein Uth1p (“youth 1p”) in yeast leads to a selective defect in mitophagy and decreased lifespan (Kennedy et al., 1995; Kissova et al., 2004). Moreover, an independent screen in yeast identified Uth1p as a protective protein against oxidative damage (Bandara et al., 1998). These studies emphasized the importance of mitochondrial clearance and oxidative damage in cellular aging. Although blocking the autophagy pathway in C. elegans does not by itself lead to shortened lifespan, worms carrying the daf-2 mutation require an intact autophagy pathway for maximum lifespan extension (Melendez et al., 2003). In mammalian cells, aging causes prominent morphological and enzymatic mitochondrial defects (Beregi et al., 1988; Byrne and Dennett, 1992; Ermini, 1976; Navarro and Boveris, 2004). Effects of aging upon energy production or changes in ROS levels could be particularly detrimental in non-proliferating neuronal tissues. Therefore, it is likely that accumulating dysfunctional mitochondria are an important contributor to mammalian aging, and perhaps to age-related disease pathology. Unfortunately, little is known about mechanisms that could reverse the effects of ROS or dysfunctional mitochondria. Dietary restriction is a well-established mechanism of increased longevity across different species, and activates autophagy by inhibiting the Insulin/PI3K/TOR signaling pathway (Yen and Klionsky, 2008). Clearance of damaged mitochondria through mitophagy could be one of the contributing factors of increased longevity by dietary restriction. Studies of normal mitochondrial biology, inherited mitochondrial diseases, and mitophagy together suggest that the ultimate natural demise of every human being on this planet may result from mitochondrial dysfunction. Understanding the role of mitophagy in neural function, neurodegenerative disease, and aging thus represents an essential goal for future research in the autophagy field.
Acknowledgments
We would like to thank C. Takamatsu for help with the illustration. Autophagy research in the La Spada laboratory is supported by R01 AG33082 and R01 EY14997.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeliovich A. Parkinson’s disease: Mitochondrial damage control. Nature. 2010;463:744–5. doi: 10.1038/463744a. [DOI] [PubMed] [Google Scholar]
- Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara PD, et al. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Curr Genet. 1998;34:259–68. doi: 10.1007/s002940050395. [DOI] [PubMed] [Google Scholar]
- Beregi E, et al. Age-related changes in the skeletal muscle cells. Z Gerontol. 1988;21:83–6. [PubMed] [Google Scholar]
- Boland B, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifati V, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–9. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Byrne E, Dennett X. Respiratory chain failure in adult muscle fibres: relationship with ageing and possible implications for the neuronal pool. Mutat Res. 1992;275:125–31. doi: 10.1016/0921-8734(92)90017-j. [DOI] [PubMed] [Google Scholar]
- Casley CS, et al. Beta-amyloid inhibits integrated mitochondrial respiration and key enzyme activities. J Neurochem. 2002a;80:91–100. doi: 10.1046/j.0022-3042.2001.00681.x. [DOI] [PubMed] [Google Scholar]
- Casley CS, et al. Beta-amyloid fragment 25–35 causes mitochondrial dysfunction in primary cortical neurons. Neurobiol Dis. 2002b;10:258–67. doi: 10.1006/nbdi.2002.0516. [DOI] [PubMed] [Google Scholar]
- Chakrabarti L, et al. Autophagy activation and enhanced mitophagy characterize the Purkinje cells of pcd mice prior to neuronal death. Mol Brain. 2009;2:24. doi: 10.1186/1756-6606-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L, et al. Mitochondrial dysfunction in NnaD mutant flies and Purkinje cell degeneration (pcd) mice reveals a role for Nna proteins in neuronal bioenergetics. Neuron. 2010 doi: 10.1016/j.neuron.2010.05.024. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, et al. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–62. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Cherra SJ, 3rd, et al. Review: autophagy and neurodegeneration: survival at a cost? Neuropathol Appl Neurobiol. 2010;36:125–32. doi: 10.1111/j.1365-2990.2010.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CT. A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Hum Mol Genet. 2010;19:R28–37. doi: 10.1093/hmg/ddq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Clark SL., Jr Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J Biophys Biochem Cytol. 1957;3:349–62. doi: 10.1083/jcb.3.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy in neurons: it is not all about food. Trends Mol Med. 2006;12:461–4. doi: 10.1016/j.molmed.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, et al. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–40. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, et al. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–5. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Dagda RK, et al. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–55. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005;26:523–8. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Deter RL, De Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol. 1967;33:437–49. doi: 10.1083/jcb.33.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–9. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Ericsson JL. Studies on induced cellular autophagy. I. Electron microscopy of cells with in vivo labelled lysosomes. Exp Cell Res. 1969a;55:95–106. doi: 10.1016/0014-4827(69)90462-5. [DOI] [PubMed] [Google Scholar]
- Ericsson JL. Studies on induced cellular autophagy. II. Characterization of the membranes bordering autophagosomes in parenchymal liver cells. Exp Cell Res. 1969b;56:393–405. doi: 10.1016/0014-4827(69)90030-5. [DOI] [PubMed] [Google Scholar]
- Ermini M. Ageing changes in mammalian skeletal muscle: biochemical studies. Gerontology. 1976;22:301–16. doi: 10.1159/000212145. [DOI] [PubMed] [Google Scholar]
- Feany MB. New genetic insights into Parkinson’s disease. N Engl J Med. 2004;351:1937–40. doi: 10.1056/NEJMp048263. [DOI] [PubMed] [Google Scholar]
- Gautier CA, et al. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–9. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Geminard C, et al. Reticulocyte maturation: mitoptosis and exosome release. Biocell. 2002;26:205–15. [PubMed] [Google Scholar]
- Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–91. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Gu M, et al. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann Neurol. 1996;39:385–9. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Ishihara N, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–66. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- Kahle PJ, et al. DJ-1 and prevention of oxidative stress in Parkinson’s disease and other age-related disorders. Free Radic Biol Med. 2009;47:1354–61. doi: 10.1016/j.freeradbiomed.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–93. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. The molecular mechanism of mitochondria autophagy in yeast. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2009.07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009a;20:4730–8. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, et al. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009b;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawajiri S, et al. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584:1073–9. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- Kegel KB, et al. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–78. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, et al. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–96. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kissova I, et al. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–74. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kitada T, et al. Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proc Natl Acad Sci U S A. 2007;104:11441–6. doi: 10.1073/pnas.0702717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Knott AB, Bossy-Wetzel E. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Ann N Y Acad Sci. 2008;1147:283–92. doi: 10.1196/annals.1427.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104:14489–94. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, et al. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–57. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–21. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M, et al. Organelle degradation during the lens and erythroid differentiation is independent of autophagy. Biochem Biophys Res Commun. 2006;339:485–9. doi: 10.1016/j.bbrc.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Maurer I, et al. A selective defect of cytochrome c oxidase is present in brain of Alzheimer disease patients. Neurobiol Aging. 2000;21:455–62. doi: 10.1016/s0197-4580(00)00112-3. [DOI] [PubMed] [Google Scholar]
- Melendez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535–41. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- Mizushima N, et al. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DJ, et al. Molecular pathophysiology of Parkinson’s disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- Moreira PI, et al. Autophagocytosis of mitochondria is prominent in Alzheimer disease. J Neuropathol Exp Neurol. 2007;66:525–32. doi: 10.1097/01.jnen.0000240476.73532.b0. [DOI] [PubMed] [Google Scholar]
- Mullen RJ, et al. Purkinje cell degeneration, a new neurological mutation in the mouse. Proc Natl Acad Sci U S A. 1976;73:208–12. doi: 10.1073/pnas.73.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Boveris A. Rat brain and liver mitochondria develop oxidative stress and lose enzymatic activities on aging. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1244–9. doi: 10.1152/ajpregu.00226.2004. [DOI] [PubMed] [Google Scholar]
- Nishida Y, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Nixon RA, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–22. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- Okamoto K, et al. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–63. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Panov AV, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–6. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Parker WD., Jr Cytochrome oxidase deficiency in Alzheimer’s disease. Ann N Y Acad Sci. 1991;640:59–64. doi: 10.1111/j.1749-6632.1991.tb00191.x. [DOI] [PubMed] [Google Scholar]
- Plowey ED, et al. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–56. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, et al. Mapping of a gene for Parkinson’s disease to chromosome 4q21-q23. Science. 1996;274:1197–9. doi: 10.1126/science.274.5290.1197. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–7. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Poole AC, et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–43. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Sakai Y, et al. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–75. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Sanchez I, et al. Caspase-8 is required for cell death induced by expanded polyglutamine repeats. Neuron. 1999;22:623–33. doi: 10.1016/s0896-6273(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH, et al. Mitochondrial complex I deficiency in Parkinson’s disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–5. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, et al. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH, et al. Origin and functional consequences of the complex I defect in Parkinson’s disease. Ann Neurol. 1996;40:663–71. doi: 10.1002/ana.410400417. [DOI] [PubMed] [Google Scholar]
- Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–46. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–83. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J, et al. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–16. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, et al. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–41. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- Weydt P, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–62. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Wild P, Dikic I. Mitochondria get a Parkin’ ticket. Nat Cell Biol. 2010;12:104–6. doi: 10.1038/ncb0210-104. [DOI] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–11. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood-Kaczmar A, et al. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS One. 2008;3:e2455. doi: 10.1371/journal.pone.0002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008a;105:7070–5. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Transport of autophagosomes in neurites of PC12 cells during serum deprivation. Autophagy. 2008b;4:243–5. doi: 10.4161/auto.5431. [DOI] [PubMed] [Google Scholar]
- Yen WL, Klionsky DJ. How to live long and prosper: autophagy, mitochondria, and aging. Physiology (Bethesda) 2008;23:248–62. doi: 10.1152/physiol.00013.2008. [DOI] [PubMed] [Google Scholar]
- Yue Z, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang J, et al. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–64. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, et al. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008;105:12022–7. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]