Abstract
The acquisition of the metastatic melanoma phenotype is associated with increased expression of the melanoma cell adhesion molecule MCAM/MUC18 (CD146). However, the mechanism by which MUC18 contributes to melanoma metastasis remains unclear. Herein, we stably silenced MUC18 expression in two metastatic melanoma cell lines, A375SM and C8161, and conducted cDNA microarray analysis. We identified and validated that the transcriptional regulator, Inhibitor of DNA Binding-1 (Id-1), previously shown to function as an oncogene in several malignancies including melanoma, was downregulated by 5.6-fold following MUC18 silencing. Additionally, we found that MUC18 regulated Id-1 expression at the transcriptional level via ATF-3, which itself was upregulated by 6.9 fold in our cDNA microarray analysis. ChIP analysis showed increased binding of ATF-3 to the Id-1 promoter after MUC18 silencing. To complement these studies, we rescued the expression of MUC18, which reversed the expression patterns of Id-1 and ATF-3. Moreover, we showed that MUC18 promotes melanoma invasion through Id-1, as overexpression of Id-1 in MUC18-silenced cells resulted in increased MMP-2 expression and activity. To our knowledge, this is the first demonstration that MUC18 is involved in cell signaling regulating the expression of Id-1 and ATF-3 in contributing to melanoma metastasis.
Keywords: Melanoma, Metastasis, MCAM, MUC18, Id-1, MMP-2
Introduction
Melanoma develops in a multistep, sequential process by which normal melanocytes are transformed to nevi, continuing to the radial growth phase (RGP) and subsequently to the vertical growth phase (VGP) and the metastatic phenotype (1). During the transition from the non-metastatic to the metastatic phenotype, the interactions between the transformed melanoma cells, the microenvironment, and the extracellular matrix (ECM) are altered (2, 3).
One adhesion molecule which has been shown to play an important role during melanoma progression from the RGP to the VGP is MCAM/MUC18 (4, 5). MUC18, also known as CD146, is a 113-kDa transmembrane glycoprotein (5). MUC18 contains an extracellular domain composed of five immunoglobulin–like domains, a single transmembrane domain, and a short cytoplasmic domain consisting of several protein kinase-like recognition motifs, suggesting its potential involvement in cell signaling (6). MUC18 was first identified as a human melanoma antigen, rarely expressed on normal epidermal melanocytes, but abundantly expressed in advanced primary and metastatic melanoma (5). Furthermore, MUC18 expression correlates with poor patient prognosis (7).
We have previously demonstrated that ectopic expression of MUC18 in the non-metastatic melanoma cell line, SB-2 (MUC18 negative), increased their tumorigenic and metastatic potential in vivo (8, 9). The role of MUC18 in melanoma tumor growth and metastasis was further studied utilizing a fully human anti-MCAM/MUC18 monoclonal Ab (ABX-MA1; produced by Abgenix). Treatment with ABX-MA1 decreased tumor growth and experimental lung metastases in metastatic melanoma cell lines in nude mice (10). Inhibition of MUC18 by ABX-MA1 disrupted the adhesive function of MUC18 and decreased angiogenesis and cell invasion both in vitro and in vivo via regulation of MMP-2 (10).
Nevertheless, the role of MUC18 in melanoma intracellular signaling has not yet been fully elucidated. To further define the mechanism of MUC18 as a signaling molecule and its role in melanoma metastasis, we conducted a cDNA microarray analysis, following MUC18 silencing, in two metastatic melanoma cell lines and identified Id-1 as a downstream target of MUC18. Id-1 is part of the Id protein family (‘Inhibitor of differentiation’ or ‘Inhibitor of DNA binding’) of helix-loop-helix (HLH) proteins that hetero-dimerize to basic HLH (bHLH) transcription factors and inhibit their activity (11–13). It has been suggested that Id proteins, particularly Id-1, function as oncogenes in several malignancies. Upregulation of Id-1 expression has been demonstrated in primary human cancers including melanoma (14, 15).
Herein, we establish that signaling through MUC18 regulates Id-1 expression via modulation of ATF-3 expression and binding to the Id-1 promoter. Id-1, in turn, regulates the expression of MMP-2. Collectively, our studies have identified a novel mechanism by which MUC18 contributes to the acquisition of the malignant phenotype of melanoma.
Material and Methods
Cell Lines
The generation and maintenance of A375SM, SB-2 and C8161-c9 cell lines were previously described (16,17). Briefly, A375SM and SB-2 cell lines were maintained in cell culture as monolayers in Eagle’s minimal essential medium supplemented with 10% fetal bovine serum. C8161-c9 cells were maintained in Dulbecco’s modified Eagle’s medium-F12 (DMEM-F12) supplemented with 5% fetal bovine serum (17). The 293FT cells (Invitrogen) used to produce the lentiviral shRNA were maintained as previously described (16). All cell lines used in our studies were tested prior to their usage for authentication by DNA fingerprinting using the STR method.
Western Blot Analysis
To detect the expression of ATF-3, Id-1, MUC18, MMP-2, Ets-1 and Sp-1, 20 μg of protein lysates were loaded on SDS-PAGE as previously described (16). Blots were incubated with primary antibodies (1:1000, anti-ATF-3 or anti-Id-1; 1:10000, anti-Ets-1, anti-Sp1, Santa Cruz Biotechnology) (1:1000 anti-MMP-2, Cell Signaling Technology) (1:1000 anti-MUC18, BD Bioscience). For detecting Ets-1, nuclear extracts were prepared using a nuclear extraction kit (Panomics). Densitometry was carried out by usingImageJ software (NIH).
Chromatin Immunoprecipitation Assay
ChIP Assay was performed utilizing ChIP-IT Express Kit from Active Motif as described previously according to the manufacturer’s protocol (16). Protein-DNA complexes were pulled down with anti-ATF-3 antibody (sc-188, Santa Cruz), anti-Sp1 antibody (sc-14027X, Santa Cruz), anti-Ets-1 antibody (sc-55581X, Santa Cruz), anti-AP-2α antibody (sc-184X, Santa Cruz), anti-CREB antibody anti-p53 antibody (sc-126X, Santa Cruz). See Supplemental Materials and Methods for primer sequences.
siRNA/shRNA
MUC18 targeting shRNA 5′-TGATATCGCTGCTGAGTGA-3′ and a nontargeting (NT) shRNA 5′-TTCTCCGAACGTGTCACGT-3′ were cloned into the lentiviral vector, pLVTHM, and transfected into 293FT (human embryonic kidney cell) to generate lentiviral particles. The lentivirus system and cell transduction were generated as described previously (18, 19) and were kindly provided by Didier Trono (Ecole Polytechnique Fédérale de Lausanne). Id-1 siRNA (Hs 01_00246328) was purchased from (Sigma) and transfected into SB-2 cells overexpressing MUC18 by using HiPerFect Transfection Reagent (QIAGEN) per manufacturer’s instructions.
Non-targetable MUC18 Expression Vector
Five silent point mutations were introduced into the MUC18 expression vector at the region targeted by the MUC18 shRNA, utilizing the QuikChange II XL site-directed mutagenesis kit (Staratagene) and the following primers: forward-5′-CAGGGCCTGGACTTGGACACCATGATTTCCCTCCTCAGCGAACCACAGGAACTACTGGTG-3′, reverse-5′-CACCAGTAGTTCCTGTGGTTCGCTGAGGAGGGAAATCATGGTGTCCAAGTCC AGGCCCTG -3′. The amplified mutated sequence was ligated into the pLVX-DsRed-Monomer-C1 vector (Clontech) replacing the red protein coding sequence of DsRed and a final rescue lentiviral vector.
To rescue MUC18 expression in stably transduced MUC18-silenced cells, the cells were transduced with the virus containing either the non-targetable MUC18 expression vector or empty vector control. Cells were selected with growth medium containing 500 μg/ml puromycin.
Luciferase Assay
A375SM, C8161 and SB-2 were plated in a 24-well plate. Transfections of 0.8 μg luciferase reporter plasmids (pGL3-Id-1, pGL3-Id-1 mutant promoter, pGL3-MMP-2 and pGL3-MMP-2 mutant promoter) were performed 48 h after cells were plated utilizing Lipofectamin-2000 (Invitrogen) as described previously (16).
Zymography
To determine MMP-2 activity zymography assay was performed as described previously (9, 10).
Matrigel Invasion Assay
Invasion assay was carried out using BioCoat Matrigel invasion chambers (BD Biosciences) as described previously (16).
Immunohistochemistry
Immunohistochemistry was performed as described previously (16). Slides were incubated with anti-MMP-2 (1:400; Chemicon), anti-Id-1(polyclonal, 1:50; Santa Cruz) or anti-Ki67 (1:100; Abcam). For MUC18 staining, 1X Divi deckloaker (BioCare Medical) was utilized for antigen retrieval. Fragment blocking was used overnight at 4 °C with Affini fragment blocking anti mouse antibody (1:10; Jackson ImmunoResearch) Slides were incubated overnight at 4 °C with anti-MUC18 (1:50; BD Biosciences). Pictures were taken at X20 magnification using a Leica DFC 320 R2. TUNEL and CD31 staining were described previously (16)
Quantitative Real-Time PCR
RNA was harvested using the RNAqueous kit (Ambion) according to the manufacturer’s instructions and as described previously (16). The primers were obtained from Applied Biosystems (MMP-2: Assay ID Hs01548727_m1). Reaction components for reverse transcription-PCR and amplifications were described previously (16).
Animals, Tumor Growth, and Metastasis
Female athymic BALB/c nude mice were purchased from the National Cancer Institute-Frederick Cancer Research and Development Center (Frederick, MD) and housed in specific pathogen-free conditions. All studies were approved and supervised by The University of Texas M.D. Anderson Cancer Center Institutional Animal Care and Use Committee (IACUC). Subcutaneous tumors and experimental lung metastases were produced as described previously (16). Briefly, 5 × 105 A375SM cells or 2.5 × 105 C8161 cells were injected into the right flank of each mouse (n=7/group). Mice were sacrificed (according to guidelines of IACUC). To determine metastatic potential, 5×105 A375SM cells and 3.75×105 C8161 cells were injected into the tail vein of nude mice (n=7/group). Mice were sacrificed 35 days after injections.
cDNA Microarray
Total RNA was isolated from NT A375SM cells and MUC18-silenced A375SM cells utilizing the Clontech Advantage RT-for-PCR kit according to the manufacturer’s instructions. A human genome U133 Plus 2.0 array (Affymetrix) was used for the microarray analysis. The microarrays were produced in the microarray core facility of Codon Bioscience (Houston, TX). Affymetrix software was utilized to analyze the results as described previously (20).
Statistical Analysis
Student’s t test was used to evaluate the statistical significance of differences for the in vitro data. Statistical analysis of the results of the animal studies was performed using the Mann-Whitney U test. Values for tumor growth are given as a mean volume ± S.E.M., and p values < 0.05 were considered statistically significant.
Results
Effect of MUC18 Silencing on Melanoma Tumor Growth and Metastasis
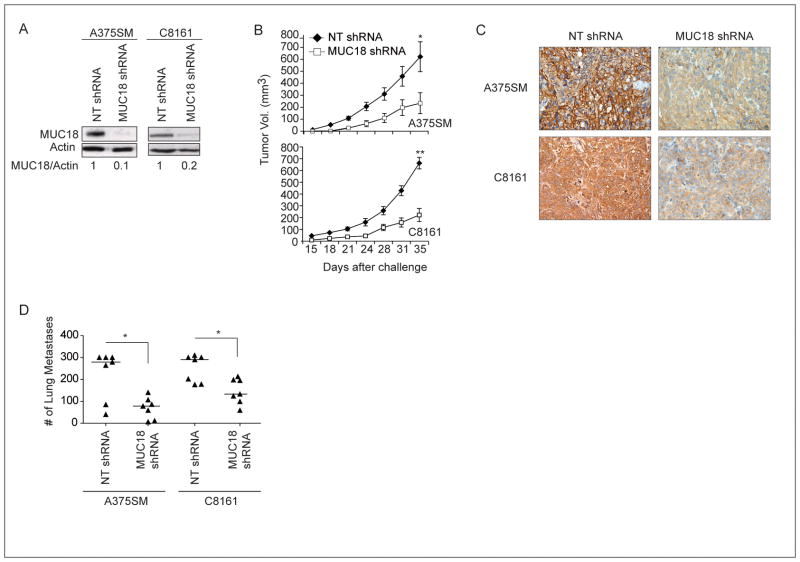
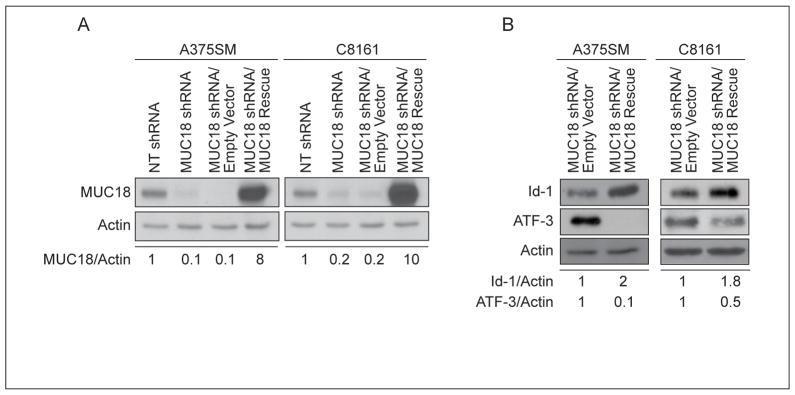
To establish the role of MUC18 in promoting tumor growth and metastasis of human melanoma, we stably silenced MUC18 expression in two metastatic human melanoma cell lines, A375SM and C8161, using a lentiviral vector. MUC18 protein expression decreased by 90% and 80% in MUC18-silenced A375SM and C8161 cells, respectively, as compared to the non-targeting (NT) shRNA transduced cells (Figure 1A).
Figure 1. Effect of MUC18 silencing on tumor growth and metastasis of human melanoma cells in nude mice.
(A) Western blot of MUC18 expression in A375SM and C8161 after stable transduction with MUC18 shRNA or a Non-Targeting (NT) control vector, showing 90% and 80% reduction in MUC18 expression in A375SM and C8161, respectively. Data shown is a representative of three independent experiments. (B) MUC18 silencing resulted in a significant decrease in tumor growth. (C) Immunohistochemical staining demonstrated downregulation of MUC18 expression in MUC18-silenced tumors, 35 days after injections with MUC18-shRNA as compared to NT-shRNA-transduced A375SM or C8161 cells. Images are shown at X20 magnification. (D) Inhibition of experimental lung metastasis following MUC18 silencing in A375SM and C8161 (*p <0.05 **p<0.001)
We have previously shown that blocking MUC18 with the fully human antibody, ABX-MA1, significantly decreases both tumor growth and experimental lung metastasis (10). To corroborate these results and further determine the effect of MUC18 silencing on melanoma tumor growth, we injected both MUC18-silenced melanoma cell lines subcutaneously (s.c.). MUC18 silencing resulted in a significant decrease in tumor growth in both cell lines, as compared to mice injected with the NT shRNA-transduced cells; mean tumor volume at day 35 was 621.4 mm3 in mice injected with NT A375SM cells, whereas in mice injected with MUC18-silenced A375SM cells it was 233.6 mm3 (*p<0.05) (Figure 1B). Similar results were observed with NT shRNA and MUC18-silenced C8161 cells; mean tumor volume was 662.3 mm3 versus 221.9 mm3 (*p<0.001) (Figure 1B).
Immunohistochemical staining of the s.c. tumors demonstrated that MUC18 expression decreased in tumors derived from MUC18-silenced A375SM and C8161 (Figure 1C), as compared to tumors from NT-shRNA-transduced cells. The decreased expression of MUC18 was sustained until the end of the experiment at day 35.
Although silencing of MUC18 resulted in decreased tumor growth in vivo, we did not observe any difference in cell proliferation in vitro for either cell line (data not shown). However, immunohistochemical staining demonstrated reduced tumor cell proliferation (Ki67), accompanied by enhanced apoptosis (TUNEL) and diminshed vessel size and number (CD31) in tumors obtained from MUC18-silenced cells (Supplemental Figure S1), suggesting the importance of the tumor microenvironment in regulation of MUC18-mediated tumor growth in vivo.
To determine the effect of MUC18 silencing on melanoma metastasis, MUC18-silenced A375SM and C8161 cells were injected intravenously (i.v.) into nude mice. Lung metastasis was significantly decreased in mice injected with MUC18-silenced A375SM and C8161 cells, as compared to NT-transduced cells (median= 78 vs 279 for A375SM, median= 133 vs 290 for C8161) (*p< 0.05) (Figure 1D).
MUC18 silencing decreases in vitro cell invasion
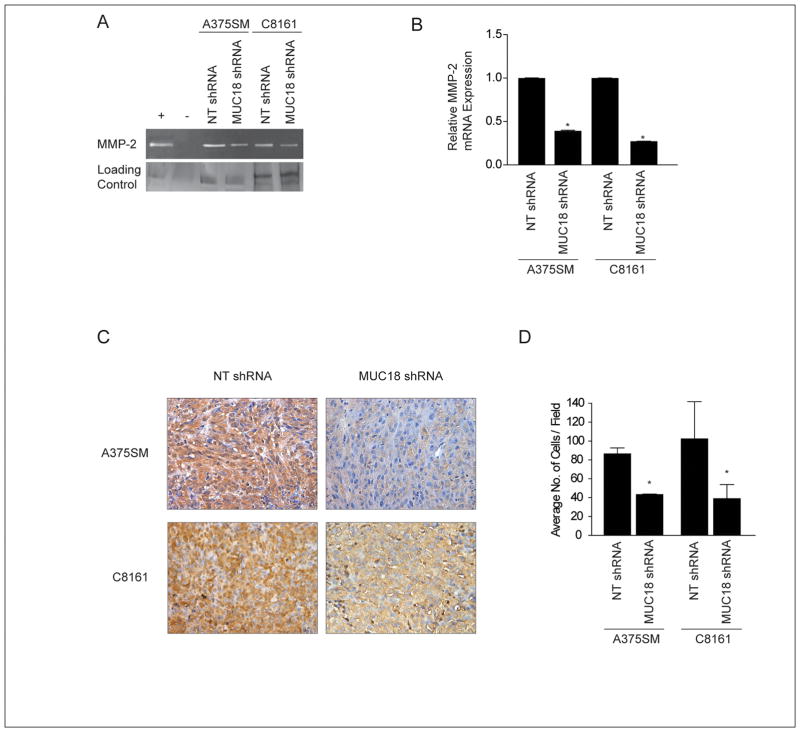
We have previously demonstrated that MUC18 contributes to melanoma cell invasion through regulation of MMP-2 activity (10). In the present study, we also observed decreased MMP-2 activity in MUC18-silenced A375SM and C8161 cells (Figure 2A). Real time PCR demonstrated decreased MMP-2 mRNA expression, following MUC18 silencing, by more than 2-fold in A375SM cells and 3-fold in C8161 cells (Figure 2B). Furthermore, a significant decrease of MMP-2 promoter activity was observed in MUC18-silenced A375SM cells (*p< 0.01) (Supplemental Figure S2). Reduced MMP-2 expression was also confirmed in vivo, by immunohistochemical analysis of tumor samples obtained from mice 35 days after injection with MUC18-silenced cells as compared to NT-transduced cells (Figure 2C). Furthermore, Matrigel invasion assays demonstrated a greater than 2-fold significant decrease in the invasive capability of MUC18-silenced cells (*p<0.05) (Figure 2D).
Figure 2. Effect of MUC18 silencing on melanoma cell invasion and MMP-2 expression.
(A) Zymography shows decreased MMP-2 activity after MUC18 silencing in both A375SM and C8161 compared to NT transduced cells. Control of zymography gel demonstrates equal loading of samples as indicated by silver staining. Images are representative of three experiments performed. (B) Quantitative real-time PCR validates dimished MMP-2 expression following MUC18 silencing in both A375SM and C8161, compared to NT transduced cells(*p <0.001). Expression values are illustrated as fold change of mean values ± SEM of three independent experiments in each cell line, relative to the NT-shRNA cells, after normalization with 18s RNA. (C) Immunohistochemical staining demonstrating downregulation of MMP-2 expression in MUC18-silenced tumors 35 days after injections as compared with NT-transduced A375SM and C8161 cells. Images are shown at X20 magnification. (D) Matrigel invasion assay shows a significant decrease in invasion following MUC18 silencing in both A375SM and C8161 (*p<0.05). Data are expressed as mean ± SEM of three experiments performed.
Id-1 is a downstream target of MUC18 in melanoma
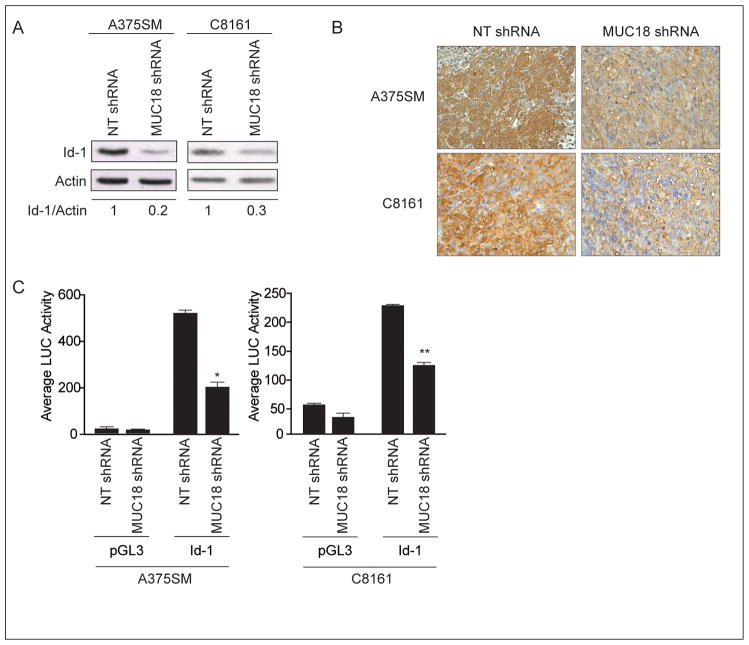
To determine the mechanism by which MUC18 contributes to melanoma tumor growth and metastasis, we performed cDNA microarray analysis comparing the gene expression pattern of NT-transduced cells and MUC18-silenced cells. Our analysis revealed a great number of genes differentially expressed following MUC18 silencing (partial list in Supplemental Table-1). We decided to focus our studies on the differential expression of transcriptional regulators and found that Id-1 was downregulated by 5.6-fold after MUC18 silencing. Western blot validated the cDNA microarray results, illustrating that Id-1 was down-regulated by 80% in MUC18-silenced A375SM cells and by 70% in MUC18-silenced C8161 cells, as compared to NT-shRNA-transduced cells (Figure 3A). These results were obtained when cells were grown in 10% FBS. In serum free medium, following a 48 hour incubation, lower levels of Id-1 were observed (data not shown). Moreover, immunohistochemical analysis of tumor samples after 35 days demonstrated decreased Id-1 expression in MUC18-silenced tumors (Figure 3B).
Figure 3. Regulation of Id-1 expression after MUC18 silencing.
(A) Western blot of Id-1 expression after MUC18 silencing in A375SM and C8161 shows decreased Id-1 expression by 80% and 70% respectively. Data are the representative of three independent experiments. (B) Immunohistochemical staining for Id-1 demonstrating downregulation of Id-1 expression in MUC18-silenced tumors 35 days after injection of mice with MUC18-shRNA as compared to NT-shRNA-transduced A375SM or C8161 cells. Images are shown at X20 magnification. (C) MUC18 silencing significantly decreases Id-1 luciferase driven-promoter activity in A375SM and C8161 (*p<0.01, **p<0.001). Data are the representative of three independent experiments. Lysates were analyzed in triplicate for luciferase activity and expressed as mean ± SEM.
To further analyze the mechanism of Id-1 regulation by MUC18, we cloned the Id-1 promoter in front of a luciferase reporter gene and evaluated Id-1 promoter activity in both A375SM and C8161 MUC18-silenced cells. We found that Id-1 promoter activity decreased by more than 2.5-fold in MUC18-silenced A375SM cells (*p<0.01) and by more than 2-fold in MUC18-silenced C8161 cells (**p<0.001), as compared to NT transduced cells (Figure 3C). These experiments revealed that MUC18 transcriptionally regulates Id-1 expression.
ATF-3 represses Id-1 promoter activity in MUC18- silenced cells
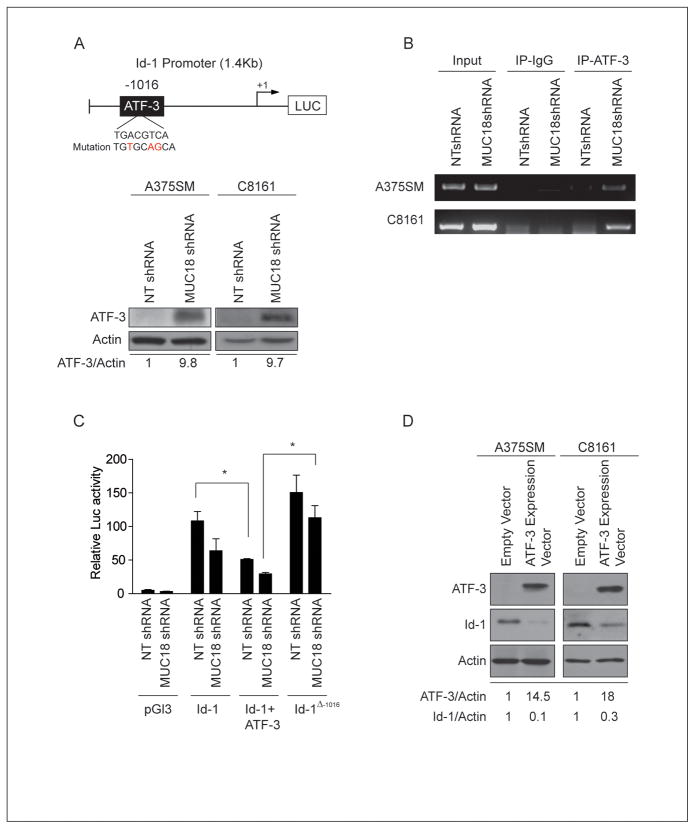
Having shown that MUC18 regulates Id-1 at the transcriptional level, we next wanted to determine the mechanism of Id-1 regulation by MUC18 by analyzing the Id-1 promoter for potential transcription factor binding sites. We found an ATF/CREB family transcription factor binding site located 1016 bp upstream of the transcription initiation site (Figure 4A, upper panel). Interestingly, ATF-3 was up-regulated by nearly 7-fold, after MUC18 silencing in our cDNA microarray analysis (Supplemental Table-1). Previous studies have demonstrated that ATF-3 can act as a repressor of Id-1 expression, through increased binding to the Id-1 promoter (21, 22). Western blot analysis corroborated the cDNA microarray results and demonstrated that, as compared to NT-transduced cells, ATF-3 was upregulated by more than 9-fold in both MUC18-silenced A375SM and C8161 (Figure 4A, lower panel).
Figure 4. MUC18 regulates Id-1 expression at the transcriptional level in metastatic melanoma cells.
(A) Upper panel, Schematic of Id-1 promoter shows an ATF-3 binding site located -1016 bp from the transcription start site. Lower panel, western blot of ATF-3 expression in MUC18-silenced A375SM and C8161 cells demonstrating increased expression of ATF-3 by 9.8-fold and 9.7-fold in A375SM and C8161, respectively. (B) ChIP shows increased binding of ATF-3 to the Id-1 promoter in MUC18-silenced cell lines (A375SM and C8161). (C) Id-1 promoter activity in NT transduced cells is significantly decreased after transfection with an ATF-3 expression vector (*p<0.05). Mutation of the ATF-3 binding site on the Id-1 promoter (Δ-1016) is significantly increased Id-1 promoter activity in MUC18-silenced cells (*p<0.05). Data are the representative of three independent experiments. Lysates were analyzed in triplicate for luciferase activity and expressed as mean ± SEM. (D) Western blot depicting decreased expression of endogenous Id-1 protein by 90% and 70% in A75SM and C8161, respectively, after ATF-3 overexpression. The data shown in A, B, and D correspond to a representative experiment out of three performed.
We then sought to determine whether there is differential binding of ATF-3 to the Id-1 promoter. Chromatin immunoprecipitation (ChIP) revealed increased binding of ATF-3 to the Id-1 promoter in both MUC18-silenced A375SM and C8161 cells (Figure 4B). Together, these data suggest that MUC18 regulates ATF-3 expression and its differential binding to the Id-1 promoter.
To further delineate whether ATF-3 acts as a repressor of Id-1 expression, we exogenously expressed ATF-3, along with the Id-1 promoter, in MUC18-silenced and NT-transduced A375SM cells and then measured the promoter-driven luciferase activity. ATF-3 overexpression resulted in a significant decrease of Id-1 promoter activity (*p<0.05) (Figure 4C). In contrast, mutation of Id-1 promoter at the ATF-3 binding site (depicted in Figure 4A, upper panel) significantly increased Id-1 promoter activity (*p<0.05). Furthermore, stable overexpression of ATF-3 in parental A375SM and C8161 cells resulted in decreased expression of endogenous Id-1 protein by 90% and 70%, respectively (Figure 4D). Collectively, MUC18 silencing resulted in enhanced ATF-3 expression and increased binding of ATF-3 to the Id-1 promoter. Accordingly, we conclude that ATF-3 acts to repress Id-1 transcription and protein expression.
Rescue of MUC18 in MUC18-silenced cells reverts the expression of Id-1 and ATF-3
To ascertain that the differential expression of Id-1 and ATF-3 are not off-target effects of MUC18, we utilized a non-targetable MUC18 expression vector to rescue MUC18 expression in MUC18-silenced cells. Figure 5A demonstrates that MUC18 expression was rescued in both MUC18-silenced cells. MUC18 rescue subsequently resulted in increased Id-1 expression and decreased ATF-3 expression (Figure 5B), thereby demonstrating that both Id-1 and ATF-3 were indeed specifically regulated by MUC18.
Figure 5. Rescue of MUC18 expression in MUC18-silenced cells reverts the expression of Id-1 and ATF-3.
(A) MUC18-silenced cells were stably transduced with a non-targetable MUC18 expression vector or empty vector control. MUC18 expression was restored utilizing the rescue vector in both cell lines. (B) Rescue of MUC18 expression in MUC18-silenced cells resulted in increased Id-1 and decreased ATF-3 expression compared to control, in both cell lines. Data are the representative of three independent experiments.
Rescue of Id-1 expression in MUC18-silenced cells increases the invasive phenotype
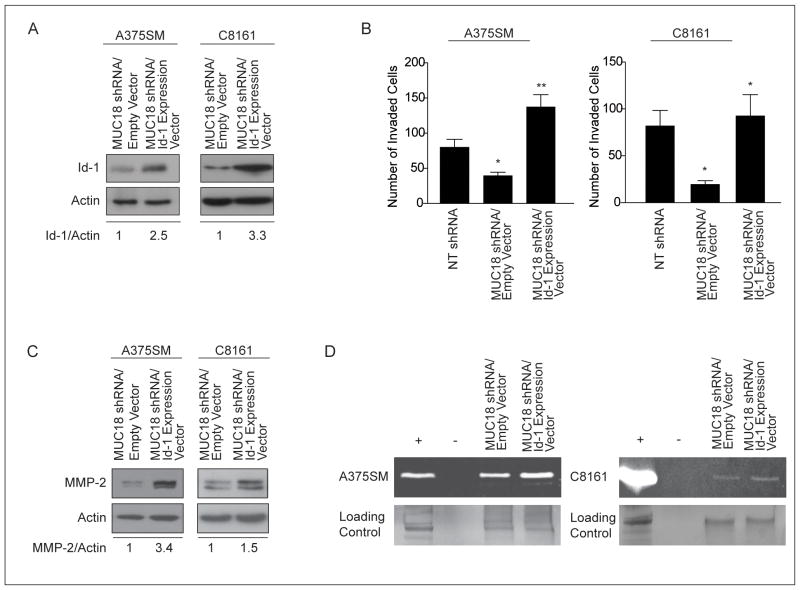
Recent studies have demonstrated that Id-1 plays a role in tumor invasion (23–25). Having established that MUC18 expression promotes melanoma cell invasion and enhanced MMP-2 expression and activity (Figure 2A, B) (8, 10), we assessed whether MUC18 contributes to melanoma cell invasion through Id-1. We therefore overexpressed Id-1 in MUC18-silenced A375SM and C8161 cells and subsequently observed an upregulation of Id-1 expression by 2.5- and 3.3-fold, respectively, as compared to control cells (Figure 6A).
Figure 6. Rescue of Id-1 expression in MUC18-silenced cells restores the invasive phenotype.
(A) Id-1 expression was restored by stable transduction with Id-1 expression vector or empty vector control in MUC18-silenced cells. (B) Matrigel invasion assay shows Id-1 overexpression in MUC18-silenced A375SM and C8161 cells rescues their invasive capacity (*p<0.005, **p<0.001). Data are expressed as mean ± SEM of three independent experiments. (C) Western blot demonstrates increased MMP-2 expression after Id-1 rescue in MUC18-silenced cells. (D) Zymography assay shows increased MMP-2 activity following rescue of Id-1 in MUC18-silenced cells. Control of zymography gel demonstrates equal loading of samples as indicated by silver staining. The data shown in A, C and D correspond to a representative experiment out of three performed.
The invasive capacity of MUC18-silenced cells overexpressing Id-1 was then compared to that of MUC18-silenced cells expressing an empty vector control. As seen in figure 6B, ectopic expression of Id-1 promoted invasiveness of MUC18-silenced A375SM cells by more than 3-fold and of MUC18-silenced C8161 cells by nearly 4-fold (*p<0.005, **p<0.001, respectively). These results suggest that Id-1 rescued the invasive activity of MUC18-silenced cells.
To examine the mechanism by which Id-1 contributes to increased invasiveness of MUC18-silenced cells, we analyzed the effect of Id-1 overexperession on MMP-2 expression and activity. Id-1 overexpression in both MUC18-silenced cells resulted in increased MMP-2 expression and activity, compared to MUC18-silenced cells expressing empty vector control (Figures 6C and 6D respectively). These data confirm that MUC18 modulates MMP-2 expression and activity via Id-1.
Id-1 regulates MMP-2 transcription
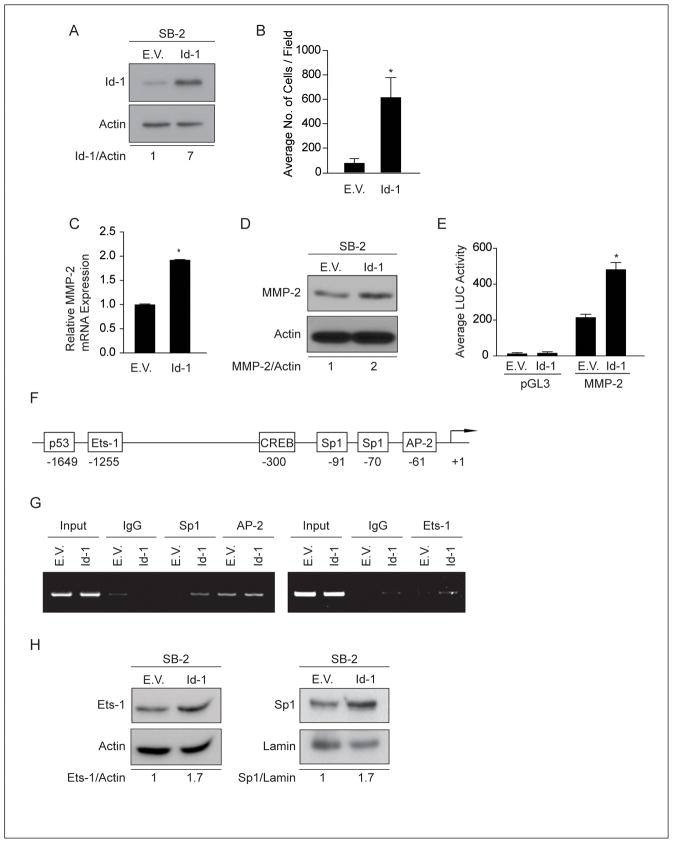
Since melanoma cell invasiveness, as well as MMP-2 expression and activity, were rescued in MUC18-silenced cells overexpressing Id-1, we sought to determine the role of Id-1 in regulating MMP-2 expression and melanoma cell invasion. To that end, Id-1 was stably overexpressed in SB-2 melanoma cells, which do not express endogenous MUC18 and express low levels of Id-1 (Figure 7A).
Figure 7. Regulation of MMP-2 expression by Id-1.
(A) Expression of Id-1 in SB-2 cells stably transduced with Id-1 expression vector. (B) Matrigel invasion assay demonstrates increased SB-2 cell invasion after overexpression of Id-1 (*p<0.01). Data are expressed as mean ± SEM of three independent experiments. (C) Quantitative real-time PCR demonstrates increased MMP-2 mRNA expression after Id-1 overexpression in SB-2 cells, as compared to control cells (*p < 0.001). Expression values are illustrated as fold change of mean values ± SEM of three independent experiments in each cell line, relative to the NT-shRNA cells, after normalization with 18s RNA. (D) Western blot analysis demonstrates that overexpression Id-1 in SB-2 cells resulted in increased MMP-2 expression. (E) MMP-2 luciferase-driven promoter activity is increased by 2.2-fold in Id-1 overexpression SB-2 cells, compared to control cells (*p< 0.001). Data are the representative of three independent experiments. Lysates were analyzed in triplicate for luciferase activity and expressed as mean ± SEM. (F) Schematic of transcription factors binding sites on the MMP-2 promoter. (G) ChIP analyses were performed utilizing Sp1, AP-2 and Ets-1 antibodies. Id-1 overexpression in SB-2 cells increased Sp1 and Ets-1 binding to the MMP-2 promoter as compared to control cells. No differences were observed in AP-2 binding to the MMP-2 promoter. (H) Western blot demonstrates that Id-1 overexpression in SB-2 cells results in increased Ets-1 and Sp1 protein expression. The data shown in A, C, G and H correspond to a representative experiment out of three performed.
We then assessed the function of Id-1 in melanoma cell invasion. SB-2 cells overexpressing Id-1 exhibited a significant increase in cell invasion through Matrigel-coated filters by greater than 3-fold, compared to control cells (Figure 7B). Moreover, overexpression of Id-1 in SB-2 cells resulted in a significant increase in MMP-2 mRNA levels (*p<0.001) and protein expression as compared to empty vector control-transduced cells (Figure 7C and 7D).
To ascertain the mechanism of MMP-2 regulation by Id-1, we cloned the MMP-2 promoter in front of a luciferase reporter gene and evaluated its activity in SB-2 cells overexpressing Id-1. MMP-2 promoter-driven luciferase activity was increased by 2.2-fold in SB-2 cells overexpressing Id-1, compared to control SB-2 cells (*p<0.001) (Figure 7E). These data demonstrate that Id-1 transcriptionally regulates MMP-2 expression.
To determine whether Id-1 regulates MMP-2 transcription via differential binding of transcription factors, we utilized ChIP assay. Previous studies have shown that several transcription factors can positively regulate MMP-2 transcription (26–28). Figure 7F illustrates a schematic of the MMP-2 promoter, depicting potential transcription factor binding sites. Specific primers were designed to amplify the potential transcription binding sites of p53, CREB, Ets-1, Sp1 and AP-2. ChIP assays demonstrated that Id-1 overexpression in SB-2 cells resulted in increased binding of both Ets-1 and Sp1 to the MMP-2 promoter, compared to control cells expressing an empty vector (Figure 7G). However, no effect on AP-2, p53 and CREB binding was observed (data not shown). Additionally, western blot analysis revealed increased expression of Ets-1 and Sp1, after Id-1 overexpression in SB-2 cells (Figure 7H). Moreover, a luciferase assay demonstrated that in SB-2 cells overexpressing Id-1, MMP-2 promoter activity was significantly decreased following deletion of the Ets-1 binding site or mutation of the Sp1 binding site (*p<0.01, **p<0.001) (Supplemental Figures S3A and S3B). These results suggest that Id-1 regulated MMP-2 transcription by affecting both the expression and binding of Ets-1 and Sp1 transcription factors to the MMP-2 promoter.
Finally, to further validate that MUC18 indeed regulates melanoma cell invasion and MMP-2 expression via Id-1, we overexpressed MUC18 and silenced Id-1 expression in SB-2 cells (Supplemental Figures S4A and S4B). Id-1 silencing in these cells resulted in decreased cell invasion as well as MMP-2 mRNA levels (*p< 0.001) (Figure S4C and S4D).
We conclude that MUC18 contributes to melanoma progression by increasing the expression of the transcriptional regulator Id-1 (via ATF-3), which results in positive regulation of MMP-2 expression and activity, thus promoting melanoma cell invasion.
Discussion
The adhesion molecule MUC18 plays an important role in promoting melanoma tumor growth and metastasis (4, 8–10, 29, 30). Previous studies have shown that MUC18 contributes to melanoma progression by promoting both homo- and heterotypic adhesion of melanoma cells either to each other or to endothelial cells.. Remarkably, little is known of what lies downstream of MUC18. We, therefore, further elucidated the downstream molecular events of MUC18 that promote the metastatic phenotype of melanoma.
To that end, we stably silenced MUC18 expression utilizing lentiviral shRNA in two metastatic melanoma cell lines and validated that MUC18 silencing decreased both tumor growth and metastasis of human melanoma cell lines in nude mice. MUC18-silenced melanoma cells were subjected to gene expression profiling to identify potential target genes regulated by MUC18. cDNA microarray analysis identified Id-1 as a novel downstream target gene of MUC18. Id-1 is a regulator of gene transcription that binds to bHLH transcription factors to inhibit their transactivation function (11). The oncogenic role of Id-1 has been suggested since it is widely expressed in human cancers, functioning as a regulator of pleiotropic cellular processes, including cell growth, cellular senescence, apoptosis, angiogenesis and invasion (14). Notably, in melanoma, Id-1 expression has been shown to be associated with inhibition of p16/Ink4a transactivation. Furthermore, increased Id-1 expression was noted in metastatic melanoma cells lines and lesions (31, 32). Utilizing melanoma tissue arrays, Straume et al., have demonstrated that enhanced Id-1 expression is associated with increased melanoma tumor thickness and decreased patient survival (33). This results correlate with previous data demonstrating the association between MUC18 and increased melanoma tumor thickness and metastatic progression (5, 7). We confirmed that Id-1 expression is indeed decreased in MUC18-silenced cells as well as in tumor xenographs, thereby establishing a link between MUC18 and Id-1. Furthermore, utilizing a luciferase driven Id-1 promoter, we demonstrate that MUC18 regulates Id-1 at the transcriptional level. Previous studies have linked ATF-3 to transcriptional inhibition of Id-1 (21, 22). Interestingly, we also identified ATF-3 as a MUC18 downstream target in our cDNA microarray.
ATF-3 is a stress-inducible protein and member of the ATF/CREB family of transcription factors induced by the p38 signaling pathway (34). It has been shown to act in a promoter-dependent manner, either as a transcriptional repressor or activator (35). A recent study has described ATF-3 as having a tumor suppressive role in melanoma by negatively regulating IL-6 expression (36). Moreover, Kang et al., have demonstrated that in epithelial cells, ATF-3 binds to the Id-1 promoter and represses its transcription due to a stress response of the cells (21). Our finding that MUC18 silencing increased the expression of ATF-3 further supports the tumor-suppressive role of ATF-3 in melanoma.
These results raised the possibility that MUC18 negatively regulates ATF-3 expression, possibly through inhibition of the p38 signaling pathway, to promote upregulation of Id-1. In this regard, promoter and ChIP analyses showed that indeed, in MUC18-silenced cells, upregulation of ATF-3 results in increased binding of ATF-3 to the Id-1 promoter and repression of Id-1 transcription. Additionally, stable overexpression of ATF-3 in two metastatic melanoma cell lines, A375SM and C8161, decreased endogenous Id-1 expression, thus further establishing the role of ATF-3 in repression of Id-1 expression. The possibility that other transcription factors are involved in Id-1 transcription is currently being investigated.
To further establish the link between MUC18, Id-1 and ATF-3, we rescued MUC18 expression in MUC18-silenced cells. The expression patterns of Id-1 and ATF-3 were reverted after rescue of MUC18 expression in both A375SM and C8161, confirming that the modulation of Id-1 and ATF-3 was not an off-target effect of MUC18 silencing.
Previous studies have demonstrated the role of Id-1 in promoting tumor invasion and MMP-2 expression (14, 23). Herein, we propose that MUC18 promotes melanoma cell invasion through the regulation of MMP-2 expression and activity via Id-1. Our data demonstrate that Id-1 overexpression in MUC18-silenced cells significantly increased the invasive capacity of melanoma cells. Additionally, MMP-2 expression and activity were rescued by Id-1 in MUC18-silenced cells. This suggests that Id-1 is an intermediary in the regulation of melanoma cell invasion by MUC18.
Although Id-1 has been shown to increase MMP-2 expression, the mechanism of regulation of MMP-2 by Id-1 has not been elucidated. To assess the role of Id-1 in the regulation of MMP-2 expression and melanoma cell invasion, we overexpressed Id-1 in a non-metastatic melanoma cell line, SB-2, which lacks expression of MUC18 and expresses low levels of Id-1. Interestingly, we found that Id-1, alone, can drive melanoma cell invasion and positively regulate MMP-2 transcription, as demonstrated by enhanced MMP-2 promoter activity and mRNA levels. Although Id-1 functions as an inhibitor of DNA binding it was recently demonstrated byQian et al., that it can act as a positive regulator of transcription in human breast cancer cells (37). Thus, we further aimed to elucidate the mechanism by which it transcriptionally regulates MMP-2 expression. Studies have demonstrated that transcriptional regulation of MMP-2 is mediated by differential binding of several transcription factors, such as Sp1 and Ets-1, to the MMP-2 promoter (26, 27). Our studies reveal that Id-1 promotes MMP-2 expression through increased expression and binding of both Ets-1 and Sp1 to the MMP-2 promoter. While MMP-2 expression can be modulated by CREB, AP-2 and p53, these transcription factors do not seem to play a role in MMP-2 expression in our system, as no differential binding of these transcription factors was found following Id-1 overexpression in SB-2 cells. Although we demonstrated that Id-1 positively regulated the expression and binding of Ets-1 to the MMP-2 promoter, it should be noted that in other systems Id-1 has been shown to inhibit Ets proteins function as transactivators (32). It is possible that in our system increased expression of Ets-1 promotes its interaction with other transcription factors or co-activators (i.e. c-Jun, c-fos and CBP/p300), rather than being inhibited by Id-1, thus competing with Id-1 and leading to transactivation of MMP-2.
Taken together our data reveal a novel mechanism by which the adhesion molecule MUC18 contributes to melanoma metastasis. The current studies support the notion that MUC18 does not simply function to promote cell adhesion, but rather is involved in cellular signaling events that affect the expression of genes, such as Id-1 and ATF-3. We further demonstrate that MUC18 promotes melanoma cells invasion by upregulating MMP-2 transcription and protein expression through Id-1, thus contributing to the malignant phenotype of melanoma.
Supplementary Material
Acknowledgments
We thank Dr. Pierre-Yves Desprez for providing the Id-1 expression vector and Dr. Douglas D Boyd for providing the ATF-3 expression vector. We thank Walter Pagel and Emily C. Brantley for editing the manuscript, and Donna Reynolds for assistance with immunohistochemical techniques. We thank Didier Trono for kindly providing the lentiviral backbone vectors used to incorporate MUC18 shRNA.
Grant Support
This work was supported by NIH RO1 grant CA76098 (MBE) and the Joanna M. Nicolay Melanoma Foundation Research Scholar Award (MZ).
Footnotes
This work was supported by NIH RO1 grant CA76098 (MBE) and the Joanna M. Nicolay Melanoma Foundation Research Scholar Award (MZ)
References
- 1.Clark WH, Jr, Elder DE, Guerry Dt, Epstein MN, Greene MH, Van Horn M. A study of tumor progression: the precursor lesions of superficial spreading and nodular melanoma. Hum Pathol. 1984;15:1147–65. doi: 10.1016/s0046-8177(84)80310-x. [DOI] [PubMed] [Google Scholar]
- 2.Hsu M, Andl T, Li G, Meinkoth JL, Herlyn M. Cadherin repertoire determines partner-specific gap junctional communication during melanoma progression. J Cell Sci. 2000;113 (Pt 9):1535–42. doi: 10.1242/jcs.113.9.1535. [DOI] [PubMed] [Google Scholar]
- 3.Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res. 2001;61:3819–25. [PubMed] [Google Scholar]
- 4.Johnson JP. Cell adhesion molecules in the development and progression of malignant melanoma. Cancer Metastasis Rev. 1999;18:345–57. doi: 10.1023/a:1006304806799. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann JM, Holzmann B, Breitbart EW, Schmiegelow P, Riethmuller G, Johnson JP. Discrimination between benign and malignant cells of melanocytic lineage by two novel antigens, a glycoprotein with a molecular weight of 113,000 and a protein with a molecular weight of 76,000. Cancer Res. 1987;47:841–5. [PubMed] [Google Scholar]
- 6.Sers C, Kirsch K, Rothbacher U, Riethmuller G, Johnson JP. Genomic organization of the melanoma-associated glycoprotein MUC18: implications for the evolution of the immunoglobulin domains. Proc Natl Acad Sci U S A. 1993;90:8514–8. doi: 10.1073/pnas.90.18.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann JM, Riethmuller G, Johnson JP. MUC18, a marker of tumor progression in human melanoma, shows sequence similarity to the neural cell adhesion molecules of the immunoglobulin superfamily. Proc Natl Acad Sci U S A. 1989;86:9891–5. doi: 10.1073/pnas.86.24.9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luca M, Hunt B, Bucana CD, Johnson JP, Fidler IJ, Bar-Eli M. Direct correlation between MUC18 expression and metastatic potential of human melanoma cells. Melanoma Res. 1993;3:35–41. doi: 10.1097/00008390-199304000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Xie S, Luca M, Huang S, et al. Expression of MCAM/MUC18 by human melanoma cells leads to increased tumor growth and metastasis. Cancer Res. 1997;57:2295–303. [PubMed] [Google Scholar]
- 10.Mills L, Tellez C, Huang S, et al. Fully human antibodies to MCAM/MUC18 inhibit tumor growth and metastasis of human melanoma. Cancer Res. 2002;62:5106–14. [PubMed] [Google Scholar]
- 11.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 12.Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–11. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh J, Murata K, Itahana Y, Desprez PY. Constitutive expression of the Id-1 promoter in human metastatic breast cancer cells is linked with the loss of NF-1/Rb/HDAC-1 transcription repressor complex. Oncogene. 2002;21:1812–22. doi: 10.1038/sj.onc.1205252. [DOI] [PubMed] [Google Scholar]
- 14.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–14. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 15.Polsky D, Young AZ, Busam KJ, Alani RM. The transcriptional repressor of p16/Ink4a, Id1, is up-regulated in early melanomas. Cancer Res. 2001;61:6008–11. [PubMed] [Google Scholar]
- 16.Dobroff AS, Wang H, Melnikova VO, et al. Silencing cAMP-response element-binding protein (CREB) identifies CYR61 as a tumor suppressor gene in melanoma. J Biol Chem. 2009;284:26194–206. doi: 10.1074/jbc.M109.019836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welch DR, Bisi JE, Miller BE, et al. Characterization of a highly invasive and spontaneously metastatic human malignant melanoma cell line. Int J Cancer. 1991;47:227–37. doi: 10.1002/ijc.2910470211. [DOI] [PubMed] [Google Scholar]
- 18.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–61. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villares GJ, Zigler M, Wang H, et al. Targeting melanoma growth and metastasis with systemic delivery of liposome-incorporated protease-activated receptor-1 small interfering RNA. Cancer Res. 2008;68:9078–86. doi: 10.1158/0008-5472.CAN-08-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mourad-Zeidan AA, Melnikova VO, Wang H, Raz A, Bar-Eli M. Expression profiling of Galectin-3-depleted melanoma cells reveals its major role in melanoma cell plasticity and vasculogenic mimicry. Am J Pathol. 2008;173:1839–52. doi: 10.2353/ajpath.2008.080380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang Y, Chen CR, Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003;11:915–26. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 22.Nemetski SM, Gardner LB. Hypoxic regulation of Id-1 and activation of the unfolded protein response are aberrant in neuroblastoma. J Biol Chem. 2007;282:240–8. doi: 10.1074/jbc.M607275200. [DOI] [PubMed] [Google Scholar]
- 23.Fong S, Itahana Y, Sumida T, et al. Id-1 as a molecular target in therapy for breast cancer cell invasion and metastasis. Proc Natl Acad Sci U S A. 2003;100:13543–8. doi: 10.1073/pnas.2230238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAllister SD, Christian RT, Horowitz MP, Garcia A, Desprez PY. Cannabidiol as a novel inhibitor of Id-1 gene expression in aggressive breast cancer cells. Mol Cancer Ther. 2007;6:2921–7. doi: 10.1158/1535-7163.MCT-07-0371. [DOI] [PubMed] [Google Scholar]
- 25.Ding Y, Wang G, Ling MT, et al. Significance of Id-1 up-regulation and its association with EGFR in bladder cancer cell invasion. Int J Oncol. 2006;28:847–54. [PubMed] [Google Scholar]
- 26.Qin H, Sun Y, Benveniste EN. The transcription factors Sp1, Sp3, and AP-2 are required for constitutive matrix metalloproteinase-2 gene expression in astroglioma cells. J Biol Chem. 1999;274:29130–7. doi: 10.1074/jbc.274.41.29130. [DOI] [PubMed] [Google Scholar]
- 27.Ito H, Duxbury M, Benoit E, et al. Prostaglandin E2 enhances pancreatic cancer invasiveness through an Ets-1-dependent induction of matrix metalloproteinase-2. Cancer Res. 2004;64:7439–46. doi: 10.1158/0008-5472.CAN-04-1177. [DOI] [PubMed] [Google Scholar]
- 28.Park MH, Ahn BH, Hong YK, Min do S. Overexpression of phospholipase D enhances matrix metalloproteinase-2 expression and glioma cell invasion via protein kinase C and protein kinase A/NF-kappaB/Sp1-mediated signaling pathways. Carcinogenesis. 2009;30:356–65. doi: 10.1093/carcin/bgn287. [DOI] [PubMed] [Google Scholar]
- 29.Shih IM, Elder DE, Speicher D, Johnson JP, Herlyn M. Isolation and functional characterization of the A32 melanoma-associated antigen. Cancer Res. 1994;54:2514–20. [PubMed] [Google Scholar]
- 30.Shih IM, Speicher D, Hsu MY, Levine E, Herlyn M. Melanoma cell-cell interactions are mediated through heterophilic Mel-CAM/ligand adhesion. Cancer Res. 1997;57:3835–40. [PubMed] [Google Scholar]
- 31.Ryu B, Kim DS, DeLuca AM, et al. Id1 expression is transcriptionally regulated in radial growth phase melanomas. Int J Cancer. 2007;121:1705–9. doi: 10.1002/ijc.22875. [DOI] [PubMed] [Google Scholar]
- 32.Alani RM, Young AZ, Shifflett CB. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc Natl Acad Sci U S A. 2001;98:7812–6. doi: 10.1073/pnas.141235398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straume O, Akslen LA. Strong expression of ID1 protein is associated with decreased survival, increased expression of ephrin-A1/EPHA2, and reduced thrombospondin-1 in malignant melanoma. Br J Cancer. 2005;93:933–8. doi: 10.1038/sj.bjc.6602792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu D, Chen J, Hai T. The regulation of ATF3 gene expression by mitogen-activated protein kinases. Biochem J. 2007;401:559–67. doi: 10.1042/BJ20061081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7:321–35. [PMC free article] [PubMed] [Google Scholar]
- 36.Karst AM, Gao K, Nelson CC, Li G. Nuclear factor kappa B subunit p50 promotes melanoma angiogenesis by upregulating interleukin-6 expression. Int J Cancer. 2009;124:494–501. doi: 10.1002/ijc.23973. [DOI] [PubMed] [Google Scholar]
- 37.Qian T, Lee JY, Park JH, Kim HJ, Kong G. Id1 enhances RING1b E3 ubiquitin ligase activity through the Mel-18/Bmi-1 polycomb group complex. Oncogene. 2010;29:5818–27. doi: 10.1038/onc.2010.317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.