Abstract
Purpose
To determine the mechanisms by which tumors situated in extra-hepatic sites can cause profound changes in hepatic drug clearance, contributing to altered drug response and chemotherapy resistance.
Experimental Design
We studied in wild type or transgenic CYP3A4 reporter mice implanted with the murine Engelbreth–Holm–Swarm sarcoma, changes in nuclear receptor and hepatic transcription factor expression and/or function, particularly related to CYP3A gene regulation.
Results
Repression of hepatic CYP3A induction was dramatic and associated with reduced levels of C/EBPβ isoforms and impaired PXR and CAR function. Unexpectedly, extra-hepatic tumors strongly reduced nuclear accumulation of RXRα in hepatocytes, providing a potential explanation for impaired function of nuclear receptors that rely on RXRα dimerization. Profiling revealed 38 nuclear receptors were expressed in liver with 14 showing between 1.5 and 4 fold reduction in expression in livers of tumour-bearing animals, including Car, Trβ, Lxrβ, Pparα, Errα/β, Reverbα/β and Shp. Altered Pparα and γ induction of target genes provided additional evidence of perturbed hepatic metabolic control elicited by extra-hepatic tumors.
Conclusions
Extra-hepatic malignancy can affect hepatic drug metabolism by nuclear receptor re-localization and decreased receptor expression and function. These findings could aid the design of intervention strategies to normalize drug clearance and metabolic pathways in cancer patients at risk of chemotherapy-induced toxicity or cancer cachexia.
Keywords: drug metabolism, cancer, CYP3A, nuclear receptors
INTRODUCTION
A major challenge to the effective use of cancer chemotherapy is wide inter-patient variability in clearance, and consequently, induced side effects of cytotoxic drugs. There is accumulating evidence that the presence of malignancy is accompanied by widespread changes in hepatic gene expression. This is clinically relevant as the liver is responsible for an extensive range of metabolic processes. Clinical studies have also demonstrated that cancer patients with elevated inflammatory markers/symptoms induced by their malignancy have reduced hepatic drug clearance leading to worse toxicity from anticancer drugs (1–3). In advanced cancer patients reduced cytochrome P450 3A4 (CYP3A4)-mediated drug metabolism, as indicated by the erythromycin breath test, had reduced plasma clearance of the anti-cancer drug docetaxel and increased toxicity following weekly injections. In these clinical studies reduced CYP3A4 activity correlated with inflammatory markers, such as CRP and IL-6 (1, 4). The finding of significantly worse myelosuppression in lymphoma patients with inflammatory (B) symptoms compared to those without indicated the clinical relevance of this result (3). CYP3A4 is the major enzyme involved in the metabolic clearance of many commonly used anti-cancer drugs (5). Furthermore, CYP3A4 is also central to the metabolism of an extensive range of endogenous compounds, making a significant contribution to the termination of the action of steroid hormones (6) and bile acid detoxification (7). We have previously demonstrated transcriptional repression of CYP3A-mediated drug metabolism in mouse models of extra-hepatic cancer, including sarcoma, melanoma and breast tumors (8, 9). Repression of the mouse CYP3A4 homologue, Cyp3a11 in livers of these tumor-bearing mice was associated with elevated circulating IL-6 concentrations as well as increased expression of the murine acute phase protein SAP, indicating a tumor-associated inflammatory response.
Such tumor-induced perturbations in hepatic metabolism could also contribute to the development of cancer-related cachexia. The cancer cachexia syndrome (CCS) is generally defined as a hypermetabolic wasting disease, which results in progressive depletion of lipid depots and skeletal muscle, irrespective of nutritional intake (10). Cachexia occurs in approximately 50% of cancer patients. However, the incidence of cachexia varies depending on the tumor type, ranging from 70–80% in patients with carcinomas of the pancreas and stomach to 8% in patients with cancer of the esophagus (11). Cancer cachexia contributes to morbidity and mortality in these patients, directly accounting for 20–30% of all cancer deaths (12). As a consequence, cachexia is considered a late event that once established has no effective treatment (13). The mechanisms of CCS are likely to be complex involving crosstalk between cytokine and endocrine signaling pathways with homeostatic regulation of metabolism and energy balance (10, 14).
Nuclear hormone receptors are a superfamily of transcription factors with 48 distinct members identified within the human genome (15). In addition to the classic steroidal hormone receptors, other nuclear receptors act as metabolic sensors that respond to compounds of dietary origin, intermediates in metabolic pathways, drugs and other environmental factors, integrating homeostatic control over many metabolic processes (16–18). For example, aspects of drug metabolism and transport are regulated by pregnane X receptor (PXR) and constitutive androstane receptor (CAR); energy and glucose metabolism through peroxisome proliferator-activated receptor gamma (PPARγ); fatty acid, triglyceride and lipoprotein metabolism via PPAR alpha (α), delta (δ) and γ; reverse cholesterol transport and cholesterol absorption through liver X receptor (LXR) and bile acid metabolism through farnesoid X receptor (FXR) (17–19). Given that nuclear receptors are central to the regulation of these various metabolic pathways, an understanding of their overall function in tumor-induced metabolic disturbances needs to be developed. Such investigations may aid in understanding the mechanisms underlying metabolic changes which impact on drug clearance pathways in cancer patients, as well as the dysregulated energy balance that produces cancer cachexia.
In the following study we employed the Engelbreth-Holm-Swarm sarcoma (EHS) mouse model, a non-metastatic tumor implanted in the quadriceps muscle, to investigate the expression and function of hepatic transcription factors and nuclear receptors, particularly in the regulation of drug metabolism involving CYP3A-mediated pathways. The EHS tumor model has been previously shown to be associated with a tumor-mediated inflammatory response, as indicated by increased plasma levels of acute phase proteins and high circulating cytokine concentrations (5, 8, 9). Herein, we demonstrate an in vivo tumor-mediated inflammatory model exhibiting impaired action of PXR and CAR in the control of CYP3A expression and more importantly, altered sub-cellular distribution of their obligatory heterodimerization partner retinoid X receptor alpha (RXRα). Furthermore, we demonstrate an extensive effect of extra-hepatic tumor on the expression of a number of hepatic nuclear receptors. Thus, the broad perturbations of metabolism observed in cancer patients may be explained by functional impairment of a wide range of hepatic signaling processes mediated by several nuclear receptors and associated with tumor derived inflammatory stimuli.
MATERIALS AND METHODS
Tumor mice
All animal experimentation was conducted in accordance with the guidelines of the Australian Council on Animal Care under protocols approved by the Westmead Hospital Animal Ethics Committee. Eight to ten week old male FVB mice were aseptically inoculated with 0.3 mL suspension of EHS sarcoma into the right quadriceps muscle using a 16-gauge needle. Control animals were inoculated with the vehicle Dubelcco's modified Eagle's medium (DMEM) (GIBCO, Invitrogen, Mulgrave, Vic, Australia) containing penicillin/streptomycin (GIBCO, Invitrogen). At harvest, the tumor mass reached approximately 3 grams or 10% of total body weight after 2–3 weeks. The liver was immediately harvested, snap frozen in liquid nitrogen then stored at −80°C for downstream analysis.
Messenger RNA expression
Total RNA was isolated from frozen mouse liver wedges using Trizol reagent (Invitrogen, Mulgrave, Australia). Before cDNA synthesis, RNA was treated with DNAse I (Ambion, Austin, TX) according to the manufacturer's protocol. cDNA was synthesized from 5μg of total RNA with SuperScript III cDNA First-Strand Synthesis System, using random hexamer primers and deoxynucleotides. Taqman or SYBR green protocols were used to amplify cDNAs of interest by real-time quantitative PCR (QPCR) using the Rotor-Gene 3000 and 6000 (Corbett Research, Sydney, NSW, Australia). mRNA levels were initially normalized to GAPDH and 18S ribosomal RNA expression. Normalization to both housekeeping genes gave comparable results and all genes analyzed are shown with GAPDH normalization. Graphs of mRNA levels are shown as expression relative to a standard curve representing 5-fold dilutions of stock cDNA and are not true concentrations of mRNA abundance. Primers used in these studies were C/EBPβ Forward AAGCTGAGCGACGAGTACAAGA, Reverse GTCAGCTCCAGCACCTTGTG; HNF4α Forward CCGGGCTGGCATGAAG, Reverse GACCTCCGCGTGCTGATC; Cyp3a11 Forward TGCTCCTAGCAATCAGCTTGG, Reverse GTGCCTAAAAATGGCAGAGGTT, Probe FAMCCTCTACCGATATGGGACTCGTAAACATGAACTTTAMRA; Gapdh Forward GTCGTGGATCTGACGTGCC, Reverse TGCCTGCTTCACCACCTTCT, Probe VICCCTGGAGAAACCTGCCAAGTATGATGACATTAMRA
Nuclear receptor expression profiling
Total RNA extracted from livers of control and EHS tumor-bearing mice, as described above, were profiled for nuclear receptor expression at the Gene Expression Laboratory, Salk Institute, La Jolla, CA, USA, using a real-time-PCR-based high throughput processing technique. Briefly, cDNA was synthesized from 2 μg of DNase-treated total RNA using Superscript II reverse transcriptase (Invitrogen). Primers and probes were designed using ABI PrimerExpress software for use in the NIH-funded Nuclear Receptor Signaling Atlas Project (NURSA) and were subjected to extensive validation. Sequences of primers and probes are available on the (www.NURSA.org) website. High throughput processing was achieved using a semi-automated Beckman liquid handler, followed by an ABI Prism 7900HT sequence detection system. Relative mRNA levels were calculated using the comparative delta-Ct method and normalized against both GAPDH and U36b4 mRNA levels in the same total RNA samples. Both housekeepers gave comparable results and only GAPDH normalized data are shown.
Western blot analysis
Extraction and preparation of proteins from liver tissue was performed as previously described (20). In brief, 50 μg of liver tissue was homogenized in ERK Buffer (50 mM HEPES, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, 0.1% TritonX-100) containing a mix of protease inhibitors (PMSF, DTT, leupeptin, aprotonin, sodium fluoride and sodium orthovanadate). Protein concentrations for equal loading were determined using the Bio-Rad DC assay kit (Hercules, CA) with BSA as a standard (Sigma-Aldrich, St. Louis, MO). Twenty-fifty μg of extracted protein was loaded and resolved on 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS/-PAGE) under reducing conditions and then transferred to polyvinylidine difluoride membranes. Membranes were blocked with either skim milk or BSA prior to overnight incubation with primary antibodies at 4°C with gentle agitation. Secondary antibodies were incubated for 1 hour at room temperature with gentle agitation. To control for variability in protein loading, membranes were either cut at an appropriate kDa range such that the protein of interest and the normalizing protein, βActin (clone AC15, Sigma-Aldrich) at 42 kDa could be visualized simultaneously, stripped and re-probed for β-Actin or normalized against Coomassie stained protein bands. Proteins detected by specific antibodies were visualized using a SuperSignal West Pico chemiluminescence kit (Pierce Endogen, Rockford, IL) and exposed to autoradiograph film. Protein expression was quantified using densitometric analysis.
Nuclear and cytoplasmic extract preparations
Preparation of nuclear and cytoplasmic protein extracts were made using the ProteoExtract Subcellular Proteome Extraction Kit (Calbiochem, MERCK Darmstadt, Germany, cat no. 539790) as per the manufacturer's instructions. Fifty milligrams (mg) of frozen liver tissue was homogenized by 2–4 passes using a plastic pestle fit for a 1.5 ml microcentrifuge tube. All buffers were provided in the kit and all procedures were performed on ice.
Immunofluorescent detection of RXRα
Paraffin fixed liver wedges from control and tumor-bearing mice were cut on a microtome (Leica RM2121RT), 3 mm thick and mounted onto Superfrost® Plus slides (Menzel-Glaser, Braunschweig, Germany). Following paraffin removal, tissues were permeabilized with PBS/(0.1%) Triton X-100 for 15 min, washed and incubated with the following; Image-iT FX signal enhancer (Invitrogen, cat no. I36933) for 30 min, Background Buster (Innovex Biosciences, CA, USA, cat. no. NB306) for 10 min and Streptavidin and Biotin for 15 min each (Vector Laboratories Inc., cat. no. SP-2002). Tissue was then blocked for 1 hour in 2% goat serum with 0.1% cold fish skin gelatine (Sigma-Aldrich, cat. no. G7765)/PBST before an overnight incubation with a 1:100 dilution of anti-rabbit RXRα antibody (Santa Cruz Biotechnology, Santa Cruz, CA, cat. no. sc-553) in a humidified chamber at 4°C. Following PBST washes, slides were incubated for 30min with anti rabbit secondary antibody (ABCAM, Sapphire Biosciences, Sydney, NSW, Australia cat. no. Ab6012) at 1:800 dilution, then for another 30 min with Streptavidin/AlexaFluor 555 (Invitrogen, Molecular Probes, cat. no. S32355) at 1:1000 dilution, light-protected. Nuclei staining was performed using DAPI (Invitrogen, cat. no. D21490). Slides were coverslipped using Prolong Gold antifade reagent (Invitrogen, Molecular Probes, cat. no. P36934) and visualized with a Leica BMBL upright microscope and Spot Advanced version 4.1 software (Diagnostic Instruments, Sterling Heights, MI). Negative controls followed all outlined procedures except RXRα antibody treatment.
Functional assessment of CAR and PXR
Ten to twelve week old male FVB mice hemizygous for the −13kb CYP3A4/lacZ transgene (21), with or without EHS tumor, were administered single daily i.p. injections of pregnenolone-16α-carbonitrile (PCN) (40 mg/kg/day) and 1,4-Bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) (1 mg/kg/day) over 3 days. Control mice received the ligand vehicle corn oil. Ligand injections were performed after 2–3 weeks of tumor growth and 3 days before the due harvest date. PCN was purchased from MP Biomedicals, Inc (Solon, OH, United States), TCPOBOP from Maybridge Chemical Company (Tintagel, Cornwall, PL34 0HW, UK). CAR- and PXR-induced CYP3A4 transgene expression in liver wedges was macroscopically detected and quantified using X-gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside) staining, (Astral Pty. Ltd, Gymea, Australia) and ONPG (O-nitrophenyl-b-D-galactopyranoside) assays, (Sigma-Aldrich), respectively. These procedures have been previously described (21).
Functional assessment of PPARμ and PPARγ
Tumor-bearing and non-tumor male FVB mice were injected with PPARα and PPARγ receptor specific agonists Wy-14643 (Saphire Biosciences, Cayman Chemicals, cat. no. 190-70820) at 100 mg/kg/day and troglitazone (Cayman Chemical, Ann Arbor, MI, USA cat. no. 71750) at 150 mg/kg/day, for 3 days before harvest. Ligand doses were chosen based on existing literature (22–24). Control animals were administered with 100 microlitres (μL) of the vehicle consisting of 1.5% carboxymethylcellulose (CMC) and 0.2% Tween 20 in sterile water. Hepatic PPARα and PPARγ activities in presence of tumor were assessed by analyzing the mRNA level of target gene induction following ligand activation using real time QPCR. Target genes assessed for PPARα included Cyp4a14 Forward GACGCTCCATACCCA, Reverse GCCAGAAACGTGGGT, Hmg-CoA reductase Forward CTTGTGGAATGCCTT, Reverse AGCCGAAGCAGCACATGAT and Cpt1α, Forward CTTCAATACTTCCCGCATCC, Reverse CTGCTGTCCTTGACGTGTTG and for PPARγ included, Lpl Forward GCTGGTGGGAAATGATGTG, Reverse TGGACGTTGTCTAGGGGGTA and Cd36 Forward TTGTACCTATACTGTGGCTAAATGAGA, Reverse CTTGTGTTTTGAACATTTCTGCTT.
Data analysis and statistics
Quantitative data were expressed as mean ± standard error of the mean (SEM). Statistical analyses between control and tumor groups were performed using Student's t-test. Significance was established at p ≤ 0.05.
RESULTS
The EHS tumor mouse model
The EHS tumor is a transplantable mouse xenograft tumor that spontaneously arose in a ST/Eh strain mouse (25). The expression profile of EHS using cDNA microarrays has identified the tumor as derived from the parietal endoderm (26) and the tumor itself has been used widely as a cell culture substrate that mimics an extracellular matrix. Once implanted, the EHS tumors were grown for 2–3 weeks such that excessive tumor burden injurious to general animal health was avoided. These tumor-bearing mice have been previously reported to exhibit reduced drug metabolism with decreased CYP3A-mediated enzyme activity. Furthermore, decreased CYP3A enzyme activity was shown to correlate with reduced hepatic Cyp3a protein and mRNA expression, encompassing both the humanized CYP3A4 reporter transgene and its endogenous mouse homologue, Cyp3a11 (8, 9). Thus, Cyp3a mRNA levels are a suitable surrogate of CYP3A-mediated metabolism. In the present study, the EHS mice exhibited similar decreased Cyp3a expression.
Impact of cancer on constitutive regulators of CYP3A
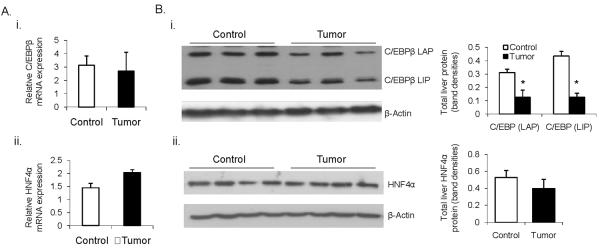
Decreased mRNA expression of CYP3A suggested the EHS tumor affects transcription factors responsible for their regulation in the liver. The impact of malignancy on major constitutive CYP3A regulators showed no statistically significant changes in mRNA for hepatocyte nuclear factor (HNF)4α, CCAAT-enhancer-binding protein (C/EBP)β (Fig 1A); or C/EBPα, HNF3γ and albumin D-site binding protein (DBP) (Data not shown) in tumor-bearing mice as compared to controls. Western blot analysis showed changes in total C/EBPβ protein, whilst HNF4α protein levels were not altered between the control and tumor groups (Fig. 1B). C/EBPβ has a number of isoforms including C/EBPβ Liver Activating Protein (LAP) and C/EBPβ Liver Inhibitory Protein (LIP). These isoforms have different roles in the regulation of CYP3A genes (27) and changes in the LIP:LAP ratio have been shown to be responsible for the IL-6-mediated repression of CYP3A4 in hepatic and non-hepatic cultured cells (28). In our in vivo tumor model there was no difference in the LIP:LAP ratio to explain a similar mechanism of basal CYP3A repression. As determined by densitometric analysis of western blots, both isoforms were decreased equally in the presence of tumor (Fig 1Bi). Nonetheless, significant repression of both C/EBPβ isoforms in tumor-bearing mouse livers may potentially impact on CYP3A basal levels.
Fig. 1.
Expression of constitutive transcriptional regulators of CYP3A genes in livers of EHS tumor-bearing mice. A. Relative mRNA levels of C/EBPβ (i) and HNF4α (ii) showing no significant difference in their expression between tumor-bearing and control animals (n = 8). B. Western blot analysis of total hepatic C/EBPβ (i) and HNF4α (ii) protein. Western blots are normalized against β-actin and protein changes are quantified by densitometric assessment of protein bands. * p < 0.05.
Tumor bearing mice exhibit impaired PXR and CAR function
The predominant inductive transcriptional regulators of CYP3A genes are the nuclear receptors PXR and CAR. Once activated by their ligands, these receptors heterodimerize with RXRα and bind to cis-acting elements in CYP3A genes to enhance transcription. Using real time PCR analysis tumor-bearing animals showed a significant decrease in CAR expression and a trend towards PXR and RXRα repression that did not attain statistical significance (data not shown, see Fig 5 for a summary of profiled NR expression). To investigate the impact of tumor growth on hepatic PXR and CAR, their functional activity in the presence of the EHS sarcoma was examined. Activation of PXR and CAR was achieved by administration of PCN and TCPOBOP, respectively and these agonists were used to determine the integrity of PXR and CAR-mediated CYP3A induction. In addition, mice incorporating a −13 kb CYP3A4/lacZ regulatory transgene were employed providing a direct readout of the function of the human CYP3A4 gene promoter in vivo.
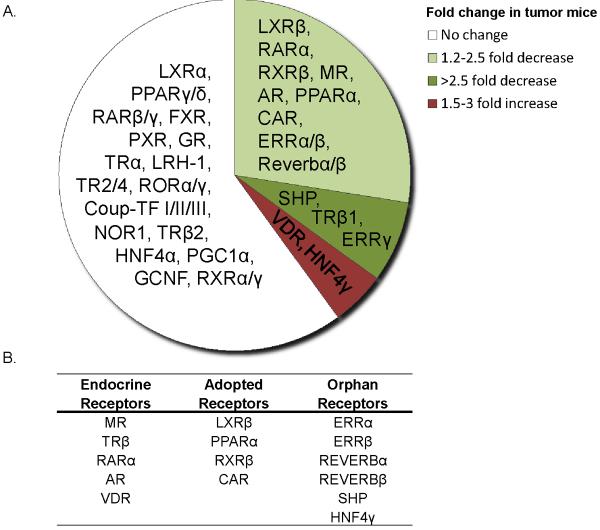
Fig. 5.
Differential expression of nuclear receptors in livers of mice bearing extra-hepatic EHS tumor. A. Pie chart showing the expression fold change of all detected nuclear receptors expressed in livers of tumor-bearing mice as compared to control mice. Varying fold changes are denoted as different colored segments with green representing down-regulation and red, up-regulation. B. A tabular listing of altered nuclear receptor expression under the broadly classified physiological/functional subgroups. Five endocrine, 4 adopted orphan and 7 true orphan receptors are seen with altered mRNA expression. Fold changes with p-value ≤ 0.05 were considered significant (n = 5 per group).
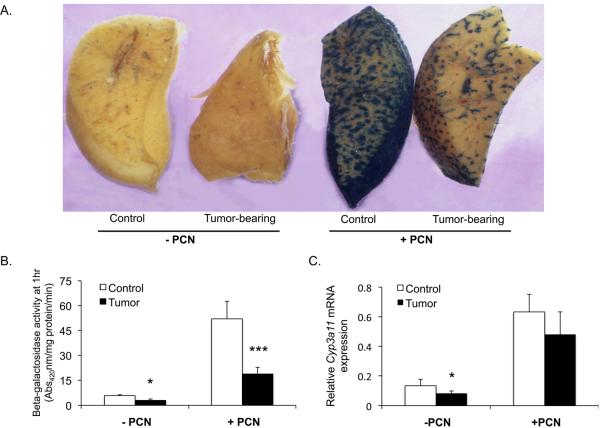
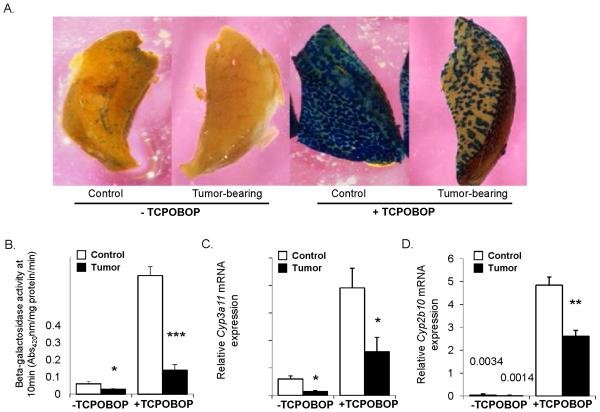
Confirming our previous findings, X-Gal staining of liver wedges without ligand treatment showed reduced basal transcription of the CYP3A4 transgene in tumor-bearing mice (Fig. 2A and 3A) (8). Following PXR and CAR activation by PCN and TCPOBOP respectively, control mice exhibited substantial CYP3A4 induction as determined by both the X-Gal staining and the ONPG assays, while induction by both PCN and TCPOBOP was significantly abrogated in the tumor-bearing cohort (Fig. 2A, 2B and 3A, 3B). Similarly, endogenous mouse hepatic Cyp3a11 and Cyp2b10 mRNA levels were induced by TCPOBOP in the controls with a significantly lower induction in tumor-bearing mice (Fig. 3C and 3D). Following PCN treatment the apparent induction of the endogenous mouse Cyp3a11gene exhibited a trend toward a decreased degree of induction in the tumor mice. However, no statistical significance was reached when compared to the induction potential of activated PXR in the control animals (Fig. 2C).
Fig. 2.
PXR activity in presence of extra-hepatic EHS tumor. Male mice harboring the −13 kb CYP3A4/lacZ transgene were treated with corn oil vehicle or PCN as described in the experimental procedures. A. Hepatocytes exhibiting transgene expression are visualized as the blue stained areas on the cut liver surface after X-gal histochemical staining. B. Transgene expression as determined by β-galactosidase activity in total liver lysates using the ONPG assay. The units of β-galactosidase activity are given as absorbance at 420nm per milligram of protein per minute. C. Basal and PCN-induced mouse endogenous Cyp3a11 mRNA expression in livers of control and tumor-bearing animals. Graphs express the mean ± SEM for n = 8/9 animals per group. (*** p ⪡ 0.001, * p < 0.05)
Fig. 3.
CAR activity in presence of extra-hepatic EHS tumor. Male mice harboring the −13 kb CYP3A4/lacZ transgene were treated with corn oil vehicle or TCPOBOP as described in the experimental procedures. A. Hepatocytes exhibiting transgene expression are visualized as the blue stained areas on the cut liver surface after incubation with X-gal. Staining intensity reflects the degree of CYP3A4 transgene expression. B. CYP3A4/lacZ transgene expression as determined by β-galactosidase activity in total liver lysates using the ONPG assay. The units of β-galactosidase activity are given as the absorbance at 420nm per milligram of protein per minute. C. Basal and TCPOBOP-induced mouse endogenous Cyp3a11 mRNA expression in livers of control and tumor animals. D. Basal and TCPOBOP-induced mouse endogenous Cyp2b10 mRNA expression in livers of control and tumor animals. Graphs express the mean ± SEM for n = 8/9 animals per group. (*** p ⪡ 0.001, * p < 0.05)
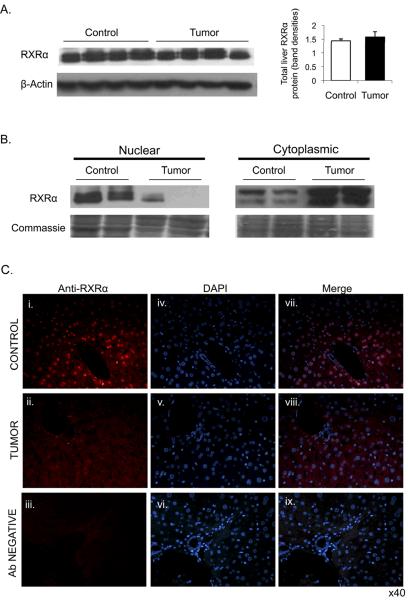
Cytoplasmic accrual of RXRα protein in tumor mice
Following an acute inflammatory response, hepatic RXRα protein has been reported to undergo cytoplasmic re-localization, leading to decreased nuclear RXRα levels (29, 30). To investigate whether the presence of extra-hepatic tumor has similar effects, RXRα localization was examined by Western blot and immunofluorescence in liver sections. Total cellular content of RXRα protein was found to be equivalent between the groups (Fig. 4A). However, nuclear abundance of RXRα in the livers of tumor-bearing mice was substantially decreased, while in the cytoplasmic fraction it was increased relative to controls (Fig. 4B). To confirm the apparent cytoplasmic retention of RXRα in tumor-bearing mice, immunofluorescence staining was carried out on liver tissues (Fig. 4C). In control animals, RXRα was clearly localized predominantly in the nucleus within hepatocytes. In tumor-bearing mice most RXRα was retained in the cytoplasm.
Fig. 4.
Altered nuclear and cytoplasmic distribution of liver RXRα protein in presence of extra-hepatic tumor. A. Western blot analysis of total hepatic RXRα protein in control (n = 7) and tumor-bearing (n = 8) groups. A representative Western blot of RXRα normalized against β-Actin is shown together with the densitometric measure of protein band quantification. B. Western blot analysis of nuclear and cytoplasmic liver protein extracts for RXRα localization in livers of control verses EHS tumor animals. Representative blots with 50 ug of nuclear or cytoplasmic proteins in each lane are shown with corresponding Coomassie stains of each membrane to confirm equal protein loading. C. Immunofluorescent imaging of RXRα (i –iii), DAPI nuclear staining (iv–vi) and an overlay image of both RXRα fluorescence and DAPI (vii–ix) in hepatocytes of control mice (top panel) and tumor mice (second panel). Bottom panel shows a negative control lacking RXRα antibody. Images are captured at ×40 objective.
Impact of tumor on hepatic nuclear receptor superfamily
Nuclear receptors are pivotal regulators of many metabolic processes, including energy homeostasis and drug metabolism (17–19). Livers from control mice and mice bearing the EHS tumor were profiled at the mRNA level for all 49 murine nuclear receptor superfamily members using high throughput real-time QPCR. Sixteen out of the 40 nuclear receptors expressed in the liver showed significant differential expression in tumor-bearing animals (Figure 5A). Interestingly, with the exception of HNF4γ and VDR, all changes in nuclear receptor levels in the presence of extra-hepatic tumor showed decreased expression. Of these changes, when broadly categorized based on the receptor's physiological ligands and potential functions, 5 nuclear receptors belonged to the endocrine receptor family, 4 belonged to the adopted orphan receptor family and 7 to the true orphan nuclear receptors (Fig. 5B).
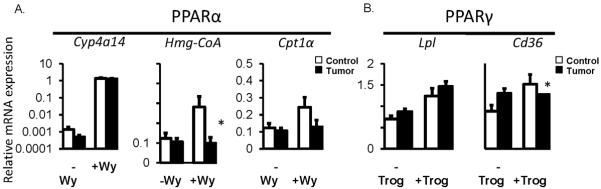
Nuclear receptors, PPARα and PPARγ, which are predominantly involved in lipid and carbohydrate homeostasis were further examined at the functional level. Administration of PPARα and PPARγ receptor specific ligands, Wy-14643 and troglitazone respectively, showed evidence of repressed receptor function in presence of extra-hepatic tumor (Fig 6). Well-characterized PPARα and PPARγ target genes examined all showed significant induction by ligand treatment in control non-tumor bearing mice. The response was reduced for the PPARα target genes, Hmg-CoA and Cpt1α in tumor mice, while Cyp4a14 was robustly induced (Fig. 6A). The induction of PPARγ target genes also showed a mixed response in tumor mice, with Cd36 exhibiting impaired induction while no change in induction of Lpl was observed (Fig. 6B). Evidence of changes in PPARα and PPARγ target gene expression in tumor mice provide supportive evidence of disturbed hepatic function, particularly related to lipid and glucose metabolism.
Fig. 6.
Activity of hepatic PPARα and PPARγ in presence of extra-hepatic EHS tumors. Basal and induced target gene expression for PPARα (A) and PPARγ (B). Induced gene expression in non-tumor controls and tumor-bearing mice as achieved by treatment with Wy-14643 (+Wy) for PPARα activation and troglitazone (+Trog) for PPARγ activation (n = 6–9 per group). * p < 0.05 denotes a significant change in gene expression in control versus tumor groups following ligand activation.
DISCUSSION
These studies show that profound changes in hepatic drug clearance in tumor bearing mice can be due to broad suppression of the transcriptional regulators of genes encoding drug clearance proteins, such as PXR and CAR. This is linked to a reduction in their expression, impaired function and perhaps more importantly, to a concomitant cytoplasmic accumulation of RXRα. Because RXRα interacts with 13 other nuclear receptors, the resulting cumulative changes may underlie more general hepatic perturbations in metabolic pathways and energy balance that are associated with the cancer cachexia syndrome (CCS).
It has been recognized that inflammatory mediators associated with a broad range of disease states can repress hepatic transcription factors such as C/EBPα and HNF4α (31) as well as the major regulators of drug metabolism, PXR, CAR, and their dimerization partner RXRα (31–33). Such repression can lead to profound changes in the expression of important drug metabolizing enzymes, such as CYP3As, also known to be altered under diverse pathological conditions (34, 35). However, studies that examine the mechanistic link between disease and decreased expression of drug metabolizing enzymes have commonly employed LPS, turpentine or direct administration of cytokines, to elicit or mimic acute inflammatory states. Little information is available regarding alterations in transcription factors, nuclear receptors and important drug metabolizing enzymes in complex disease settings involving a chronic inflammatory response, such as is often observed in cancer patients. Thus, these studies provide the first mechanistic information concerning CYP3A repression using an in vivo cancer model. We show only significantly decreased CAR mRNA levels in tumor-bearing mice as opposed to the broad transcriptional repression of many transcription factors under acute inflammatory conditions. However, the function of both CAR and PXR was impaired, as determined by CYP3A4 regulatory transgene induction in the presence of the EHS tumor. In tumor-bearing mice both PXR and CAR ligands failed to induce CYP3A4 transgene expression to the same extent as in control animals. Functional CAR impairment was further confirmed by the reduced degree of induction of mouse Cyp3a11 and Cyp2b10 expression in response to TCPOBOP. The observation that the decrease in PXR-mediated Cyp3a11 induction in tumor mice was not as great as that seen with the CYP3A4 transgene could be due to species-specific differences in PXR DNA binding elements between mouse Cyp3a11 and human CYP3A4. To date, no transcriptional enhancer equivalent to the human CYP3A4 xenobiotic-responsive element (XREM) (36) has been identified in the mouse Cyp3a gene cluster. Nonetheless, evidence of impaired CAR and PXR function may provide a partial explanation for repression of CYP3A-mediated metabolism. Furthermore, their functional impairment could also potentially impact on a number of important drug metabolising and disposition enzymes as well as contributing to perturbed energy balance (37).
Decreased nuclear and increased cytoplasmic RXRα seen with Western blot analysis and immunofluorescence suggests that in the presence of cancer the activity of RXRα is decreased. RXRα is the obligate heterodimerization partner of class II nuclear receptors, such as PXR, CAR, VDR, PPARs, FXR, RAR, TR and LXR (18). Therefore, cytoplasmic retention of RXRα may contribute to the functional impairment of PXR and CAR seen in tumour-bearing animals. Furthermore, as the obligate heterodimerization partner of class II NRs, decreased nuclear availability of RXRα widens the scope of tumor-mediated perturbations in the liver, beyond drug metabolism. Reduced nuclear availability of RXRα, which has been previously demonstrated only in acute inflammation (29, 38) suggests that similar pathways could also be operative in the presence of cancer exhibiting a chronic inflammatory phenotype. Thus, tumor-mediated inflammatory signaling in the liver is likely to influence nuclear receptor function, resulting in dysregulated metabolic processes. The role of specific cytokines in this process could be explored with blocking antibodies or other interventions to disrupt downstream signaling pathways. Such an approach would distinguish between direct effects of cytokines from compensatory changes in overall metabolic balance associated with tumor growth.
The potential for the hepatic expression of nuclear receptors to be altered by extra-hepatic cancer has not been previously considered. In the present study, all 49 mouse nuclear receptors were profiled in an attempt to gain a better understanding of affected metabolic pathways. Tumor effects were observed among 16 endocrine, adopted orphan and orphan receptors, with the majority of affected nuclear receptors showing decreased expression. Changes in endocrine nuclear receptors can have complex and profound effects on physiology and energy metabolism. Altered MR, TRβ, RARα, AR and VDR seen in tumor-bearing animals implies alterations in electrolyte and fluid balance, metabolic rate and oxidative metabolism, cell physiology, reproductive function and general homeostasis (39–42). Repression of orphan nuclear receptors such as, Reverb α and β which have a diverse function in regulating cell physiology and circadian rhythm (43) indicates broad tumor-related disturbances in hepatic physiology. Decreased expression of PPARα, LXRβ and CAR in tumor-bearing mice translates into disturbed regulation of fatty acid oxidation, cholesterol homeostasis and, as discussed above, xenobiotic metabolism. Impaired CAR action may also contribute to perturbed energy balance as it has been shown to play a role in adaptation to metabolic stress (37). Functional assessment of hepatic PPARα and PPARγ showed some impairment of their action for selected but not all target genes and provided further evidence of tumor effects on nuclear receptor activity. It would be interesting to carry out expression profiling by microarray analysis of livers from tumour-bearing mice to characterise the impact of alterednuclear receptors on hepatic metabolism in cancer.
Ligands that modulate nuclear receptor activity have significant potential in therapeutic applications. From our studies we can speculate that enhancing the activation of the nuclear receptors PXR or CAR prior to chemotherapy may ameliorate toxicity in those patients showing poor drug-metabolism. It is even more appealing to speculate that targeted therapies focused on RXR function may provide novel means of restoring not only pathways in drug metabolism but also other vital hepatic functions regulated by its essential binding. RXR is activated by its endogenous ligand 9-cis retinoic acid (44) and several RXR-selective agonists, known as `rexinoids', have been developed (45, 46). It has yet to be determined if rexinoid treatment will result in increased nuclear availability in tumor mouse hepatocytes to allow heterodimerization with other class II nuclear receptors.
In summary, our findings suggest that extra-hepatic tumors can decrease transcriptional expression of hepatic CYP3A genes in part by reductions in C/EBPβ protein and impaired function of PXR and CAR. Furthermore, our results suggest that decreased nuclear availability of RXRα may explain impaired activity of both CAR and PXR and lead to functional impairment of other nuclear receptor regulated pathways that rely on RXRα heterodimerization. Thus, altered hepatic nuclear receptor function may be one mechanism underlying tumor-mediated cancer cachexia, which involves a complex array of perturbed metabolic functions. With a better understanding of the mechanistic links between extra-hepatic tumors and impaired nuclear receptor action in the liver, therapies based on inhibiting or stimulating specific nuclear receptors represents a promising intervention approach to potentially reduce aberrant toxic side effects associated with anti-cancer treatments and possibly aid in the prevention of metabolic abnormalities that lead to cancer cachexia. While this study has focused on such processes in the context of cancer, the findings of altered basal transcription factors and impaired hepatic nuclear receptor action may be relevant to many other clinical settings involving chronic inflammation and cachexia.
Statement of Translational Relevance.
The findings provide insight into the mechanisms underlying reduced drug clearance in the setting of cancer and underscore the challenges in therapeutic drug dosing. This could aid the design of intervention strategies to normalize drug clearance and metabolic pathways in cancer patients at risk of chemotherapy-induced toxicity or cancer cachexia.
Acknowledgments
This work was supported, in whole or in part, by National Health and Medical Research Council of Australia Project Grants 352419 (GRR, SJC, CL) and 402493 (CL, MD). This work was also supported by National Institutes of Health Grants NICHD HD027183 and 5U19 DK062434-08 from NIDDK (to R. M.E) in addition funds from the to the Samuel Waxman Cancer Research Foundation and the Salk Center for Nutritional Genomics and the Leona M. & Harry B. Helmsley Charitable Trust.
Recipient of financial support from the Haematology and Oncology Trust Fund Concord RG Hospital and a Concord Hospital Volunteers PhD Scholarship.
The abbreviations used are
- CYP3A
cytochrome P450 3A
- CCS
cancer cachexia syndrome
- EHS
Engelbreth–Holm–Swarm sarcoma
- PXR
pregnane X receptor
- CAR
constitutive androstane receptor
- RXR
retinoid X receptor
- C/EBP (LIP/LAP)
CCAAT-enhancer-binding protein (Liver Activating Protein/Liver Inhibitory Protein)
- HNF4
hepatocyte nuclear factor
- PPAR
peroxisome proliferator-activated receptor
- PCN
pregnenolone-16α-carbonitrile
- TCPOBOP
1,4-Bis[2-(3,5-dichloropyridyloxy)]benzene
- LXR
liver X receptor
- FXR
farnesoid X receptor
REFERENCES
- 1.Rivory LP, Slaviero KA, Clarke SJ. Hepatic cytochrome P450 3A drug metabolism is reduced in cancer patients who have an acute-phase response. British Journal of Cancer. 2002;87:277–80. doi: 10.1038/sj.bjc.6600448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slaviero KA, Clarke SJ, McLachlan AJ, Blair EY, Rivory LP. Population pharmacokinetics of weekly docetaxel in patients with advanced cancer. Br J Clin Pharmacol. 2004;57:44–53. doi: 10.1046/j.1365-2125.2003.01956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma R, Cunningham D, Smith P, Robertson G, Dent O, Clarke SJ. Inflammatory (B) symptoms are independent predictors of myelosuppression from chemotherapy in Non-Hodgkin Lymphoma (NHL) patients--analysis of data from a British National Lymphoma Investigation phase III trial comparing CHOP to PMitCEBO. BMC Cancer. 2009;9:153. doi: 10.1186/1471-2407-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slaviero KA, Clarke SJ, Rivory LP. Inflammatory response: an unrecognised source of variability in the pharmacokinetics and pharmacodynamics of cancer chemotherapy. Lancet Oncology. 2003;4:224–32. doi: 10.1016/s1470-2045(03)01034-9. [DOI] [PubMed] [Google Scholar]
- 5.Kacevska M, Robertson GR, Clarke SJ, Liddle C. Inflammation and CYP3A4-mediated drug metabolism in advanced cancer: impact and implications for chemotherapeutic drug dosing. Expert Opin Drug Metab Toxicol. 2008;4:137–49. doi: 10.1517/17425255.4.2.137. [DOI] [PubMed] [Google Scholar]
- 6.Brian WR, Sari MA, Iwasaki M, Shimada T, Kaminsky LS, Guengerich FP. Catalytic activities of human liver cytochrome P-450 IIIA4 expressed in Saccharomyces cerevisiae. Biochemistry. 1990;29:11280–92. doi: 10.1021/bi00503a018. [DOI] [PubMed] [Google Scholar]
- 7.Stedman C, Robertson G, Coulter S, Liddle C. Feed-forward regulation of bile acid detoxification by CYP3A4: studies in humanized transgenic mice. J Biol Chem. 2004;279:11336–43. doi: 10.1074/jbc.M310258200. [DOI] [PubMed] [Google Scholar]
- 8.Charles KA, Rivory LP, Brown SL, Liddle C, Clarke SJ, Robertson GR. Transcriptional repression of hepatic cytochrome P450 3A4 gene in the presence of cancer. Clin Cancer Res. 2006;12:7492–7. doi: 10.1158/1078-0432.CCR-06-0023. [DOI] [PubMed] [Google Scholar]
- 9.Sharma R, Kacevska M, London R, Clarke SJ, Liddle C, Robertson G. Downregulation of drug transport and metabolism in mice bearing extra-hepatic malignancies. British journal of cancer. 2008;98:91–7. doi: 10.1038/sj.bjc.6604101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skipworth RJ, Stewart GD, Dejong CH, Preston T, Fearon KC. Pathophysiology of cancer cachexia: much more than host-tumour interaction? Clin Nutr. 2007;26:667–76. doi: 10.1016/j.clnu.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Dewys WD, Begg C, Lavin PT, Band PR, Bennett JM, Bertino JR, et al. Prognostic effect of weight loss prior to chemotherapy in cancer patients. Eastern Cooperative Oncology Group. Am J Med. 1980;69:491–7. doi: 10.1016/s0149-2918(05)80001-3. [DOI] [PubMed] [Google Scholar]
- 12.Bruera E. ABC of palliative care. Anorexia, cachexia, and nutrition. BMJ. 1997;315:1219–22. doi: 10.1136/bmj.315.7117.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fearon KC. Cancer cachexia: developing multimodal therapy for a multidimensional problem. Eur J Cancer. 2008;44:1124–32. doi: 10.1016/j.ejca.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 14.Stephens NA, Skipworth RJ, Fearon KC. Cachexia, survival and the acute phase response. Curr Opin Support Palliat Care. 2008;2:267–74. doi: 10.1097/SPC.0b013e3283186be2. [DOI] [PubMed] [Google Scholar]
- 15.Maglich JM, Sluder A, Guan X, Shi Y, McKee DD, Carrick K, et al. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0029. RESEARCH0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans RM. The nuclear receptor superfamily: a rosetta stone for physiology. Mol Endocrinol. 2005;19:1429–38. doi: 10.1210/me.2005-0046. [DOI] [PubMed] [Google Scholar]
- 18.Karpen SJ. Nuclear receptor regulation of hepatic function. Journal of hepatology. 2002;36:832–50. doi: 10.1016/s0168-8278(02)00129-0. [DOI] [PubMed] [Google Scholar]
- 19.Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol. 2003;65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- 20.Ip E, Farrell GC, Robertson G, Hall P, Kirsch R, Leclercq I. Central role of PPARalpha-dependent hepatic lipid turnover in dietary steatohepatitis in mice. Hepatology. 2003;38:123–32. doi: 10.1053/jhep.2003.50307. [DOI] [PubMed] [Google Scholar]
- 21.Robertson GR, Field J, Goodwin B, Bierach S, Tran M, Lehnert A, et al. Transgenic mouse models of human CYP3A4 gene regulation. Molecular pharmacology. 2003;64:42–50. doi: 10.1124/mol.64.1.42. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Hansen PA, Xi L, Chandraratna RA, Burant CF. Distinct mechanisms of glucose lowering by specific agonists for peroxisomal proliferator activated receptor gamma and retinoic acid X receptors. J Biol Chem. 2005;280:38317–27. doi: 10.1074/jbc.M505853200. [DOI] [PubMed] [Google Scholar]
- 23.Sigrist S, Bedoucha M, Boelsterli UA. Down-regulation by troglitazone of hepatic tumor necrosis factor-alpha and interleukin-6 mRNA expression in a murine model of non-insulin-dependent diabetes. Biochemical pharmacology. 2000;60:67–75. doi: 10.1016/s0006-2952(00)00299-9. [DOI] [PubMed] [Google Scholar]
- 24.Compe E, Drane P, Laurent C, Diderich K, Braun C, Hoeijmakers JH, et al. Dysregulation of the peroxisome proliferator-activated receptor target genes by XPD mutations. Mol Cell Biol. 2005;25:6065–76. doi: 10.1128/MCB.25.14.6065-6076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swarm RL. Transplantation of a Murine Chondrosarcoma in Mice of Different Inbred Strains. Journal of the National Cancer Institute. 1963;31:953–75. [PubMed] [Google Scholar]
- 26.Futaki S, Hayashi Y, Yamashita M, Yagi K, Bono H, Hayashizaki Y, et al. Molecular basis of constitutive production of basement membrane components. Gene expression profiles of Engelbreth-Holm-Swarm tumor and F9 embryonal carcinoma cells. J Biol Chem. 2003;278:50691–701. doi: 10.1074/jbc.M304985200. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Jimenez CP, Jover R, Donato MT, Castell JV, Gomez-Lechon MJ. Transcriptional regulation and expression of CYP3A4 in hepatocytes. Curr Drug Metab. 2007;8:185–94. doi: 10.2174/138920007779815986. [DOI] [PubMed] [Google Scholar]
- 28.Jover R, Bort R, Gomez-Lechon MJ, Castell JV. Down-regulation of human CYP3A4 by the inflammatory signal interleukin-6: molecular mechanism and transcription factors involved. Faseb J. 2002;16:1799–801. doi: 10.1096/fj.02-0195fje. [DOI] [PubMed] [Google Scholar]
- 29.Ghose R, Zimmerman TL, Thevananther S, Karpen SJ. Endotoxin leads to rapid subcellular re-localization of hepatic RXRalpha: A novel mechanism for reduced hepatic gene expression in inflammation. Nucl Recept. 2004;2:4. doi: 10.1186/1478-1336-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmerman TL, Thevananther S, Ghose R, Burns AR, Karpen SJ. Nuclear export of retinoid X receptor alpha in response to interleukin-1beta-mediated cell signaling: roles for JNK and SER260. J Biol Chem. 2006;281:15434–40. doi: 10.1074/jbc.M508277200. [DOI] [PubMed] [Google Scholar]
- 31.Ruminy P, Gangneux C, Claeyssens S, Scotte M, Daveau M, Salier JP. Gene transcription in hepatocytes during the acute phase of a systemic inflammation: from transcription factors to target genes. Inflamm Res. 2001;50:383–90. doi: 10.1007/PL00000260. [DOI] [PubMed] [Google Scholar]
- 32.Beigneux AP, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. Reduction in cytochrome P-450 enzyme expression is associated with repression of CAR (constitutive androstane receptor) and PXR (pregnane X receptor) in mouse liver during the acute phase response. Biochemical and biophysical research communications. 2002;293:145–9. doi: 10.1016/S0006-291X(02)00196-1. [DOI] [PubMed] [Google Scholar]
- 33.Pascussi JM, Gerbal-Chaloin S, Pichard-Garcia L, Daujat M, Fabre JM, Maurel P, et al. Interleukin-6 negatively regulates the expression of pregnane X receptor and constitutively activated receptor in primary human hepatocytes. Biochemical & Biophysical Research Communications. 2000;274:707–13. doi: 10.1006/bbrc.2000.3219. [DOI] [PubMed] [Google Scholar]
- 34.Morgan ET. Impact of infectious and inflammatory disease on cytochrome P450-mediated drug metabolism and pharmacokinetics. Clin Pharmacol Ther. 2009;85:434–8. doi: 10.1038/clpt.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renton KW. Regulation of drug metabolism and disposition during inflammation and infection. Expert Opin Drug Metab Toxicol. 2005;1:629–40. doi: 10.1517/17425255.1.4.629. [DOI] [PubMed] [Google Scholar]
- 36.Goodwin B, Hodgson E, Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Molecular Pharmacology. 1999;56:1329–39. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- 37.Maglich JM, Lobe DC, Moore JT. The nuclear receptor CAR (NR1I3) regulates serum triglyceride levels under conditions of metabolic stress. J Lipid Res. 2009;50:439–45. doi: 10.1194/jlr.M800226-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Ghose R, Mulder J, von Furstenberg RJ, Thevananther S, Kuipers F, Karpen SJ. Rosiglitazone attenuates suppression of RXRalpha-dependent gene expression in inflamed liver. Journal of hepatology. 2007;46:115–23. doi: 10.1016/j.jhep.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gombart AF, Luong QT, Koeffler HP. Vitamin D compounds: activity against microbes and cancer. Anticancer research. 2006;26:2531–42. [PubMed] [Google Scholar]
- 40.Poon MM, Chen L. Retinoic acid-gated sequence-specific translational control by RARalpha. Proc Natl Acad Sci U S A. 2008;105:20303–8. doi: 10.1073/pnas.0807740105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szafran AT, Szwarc M, Marcelli M, Mancini MA. Androgen receptor functional analyses by high throughput imaging: determination of ligand, cell cycle, and mutation-specific effects. PLoS ONE. 2008;3:e3605. doi: 10.1371/journal.pone.0003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vlaeminck-Guillem V, Wemeau JL. Physiology and pathophysiology of thyroid hormone receptors: the contributions of murine models. Ann Endocrinol (Paris) 2000;61:440–51. [PubMed] [Google Scholar]
- 43.Burris TP. Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22:1509–20. doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, et al. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 45.Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- 46.Takamatsu K, Takano A, Yakushiji N, Morohashi K, Morishita K, Matsuura N, et al. The first potent subtype-selective retinoid X receptor (RXR) agonist possessing a 3-isopropoxy-4-isopropylphenylamino moiety, NEt-3IP (RXRalpha/beta-dual agonist) ChemMedChem. 2008;3:780–7. doi: 10.1002/cmdc.200700313. [DOI] [PubMed] [Google Scholar]