Abstract
Purpose
To determine the recurring DNA copy number alterations (CNAs) in classical Hodgkin lymphoma (HL) by microarray-based comparative genomic hybridization (aCGH) using laser capture micro-dissected CD30+ Hodgkin/Reed-Sternberg (HRS) cells.
Experimental Design
Archived tissues from 27 CD30+ HL plus control samples were analyzed by DNA microarrays. The HL molecular karyotypes were compared to the genomic profiles of germinal center B cells and treatment outcome (chemotherapy responsive vs. primary refractory disease).
Results
Gains and losses observed in >35% of HL samples were localized respectively to 22 and 12 chromosomal regions. Frequent gains (>65%) were associated with growth and proliferation, NF-κB activation, cell cycle control, apoptosis, and immune and lymphoid development. Frequent losses (>40%) observed encompassed tumor suppressor genes (SPRY1, NELL1, ID4), transcriptional repressors (TXNIP), SKP2 (ubiquitin ligase component) and an antagonist of NF-κB activation (PPARGC1A). In comparison to the germinal center profiles, the most frequent imbalances in HL were losses in 5p13 (AMACR, GDNF, SKP2), and gains in 7q36 (SHH) and 9q34 (ABL1, CDK9, LCN2, PTGES). Gains (>35%) in the HL chemoresponsive patients housed genes known to regulate T-cell trafficking or NF-κB activation (CCL22, CX3CL1, CCL17, DOK4 and IL10), whereas the refractory samples showed frequent loss of 4q27 (IL2/IL21), 17p12 and 19q13.3 gain (BCL3/RELB).
Conclusion
We identified non-random CNAs in the molecular karyotypes of classical HL. Several recurring genetic lesions correlated with disease outcome. These findings may be useful prognostic markers in the counseling and management of patients and for the development of novel therapeutic approaches in primary refractory HL.
Introduction
The annual incidence of Hodgkin lymphoma (HL) is estimated at three cases per 100,000 persons, making this malignancy one of the most common lymphomas in the Western world (1). The characteristic pathological feature of classical HL is the presence of Hodgkin and Reed-Sternberg (HRS) cells, which usually comprise <3% of the affected mixed cellular lesion. There is compelling evidence suggesting that the pathognomonic HRS cells are an outgrowth of a malignant clone derived from a “reprogrammed” germinal center (GC) B cell that no longer expresses B-lineage specific genes such as POU2F2, POU2AF1 and PU.1 and may express genetic markers characteristic of other hematopoietic lineages like IL21, CCL17, CSF1, ID2, CD3 and CD4 (2–4). Recurrent genetic lesions in critical hematopoietic transcription factors have led to the discovery that constitutive activation of the NF-κB signaling pathway is essential for HRS cell survival and proliferation (4). In particular, gains of REL and deletions or inactivating mutations of TNFAIP3, genes involved in the NF-κB signaling pathway, have been detected in ~40% of classical HL (5–7). To gain further insight into the pathogenesis of HL and its lineage infidelity, comprehensive genomic characterization of HRS cells from primary HL tumor samples is necessary; however, the scarcity of HRS cells in the HL lesions remains a key limiting factor in unraveling the molecular consequences of this malignancy.
Treatment with doxorubicin-based combination chemotherapy regimens leads to complete remission in the majority (70–90%) of HL patients (1). Unfortunately, HL patients who fail front-line or second-line therapies, including stem cell transplantation, are quite often young and have a poor median survival (<3 years) (8). In primary refractory HL, the poor response to initial therapy may be attributed to non-random genetic alterations that underlie disease aggressiveness at onset. Comparison of the genetic aberrations of HRS cells from patients who respond favorably to standard HL treatment protocols to those who fail treatment has the potential to provide critical insight into the clinical behavior of HL and guide future treatment (9). Moreover, to develop novel HL therapies directed against rational targets, we need focused investigations that lead to more thorough understanding of the genetic alterations of HRS, its lineage infidelity consequences and drug resistance.
Until recently, the majority of genetic and molecular biology HL data was gathered from studies using HL continuous cell lines established from patients with relapsed HL (10–13). Distinguishing primary genetic alterations from secondary events in HL pathogenesis remains a challenge. Recently, high-resolution cytogenetic techniques that combine laser capture micro-dissection with microarray-based comparative genomic hybridization (aCGH) technology have provided new opportunities to investigate genome-wide DNA alterations in limited-sized lesions (13–15). Genomic DNA-based aCGH experiments show less variability than mRNA expression arrays, and suspected “cryptic” DNA aberrations can be confirmed easily by fluorescence in situ hybridization (FISH). Importantly, robust prognostic features for many hematological malignancies have been revealed using a combination of clinical parameters and genetic data gathered from conventional cytogenetics and molecular techniques. Given the paucity of genomic analyses of HRS cells, we sought to characterize their DNA copy-number aberrations (CNAs) with bacterial artificial chromosome (BAC)-based aCGH using DNA extracted from laser capture microdissected (LCM) CD30+ HRS cells. The objectives of this study were to determine the recurring CNAs in HL, compare the findings of the malignant HL lesions to those detected in “benign” GC B-cells – the normal counterpart of HRS cells – and define the most common CNA differences between the chemotherapy responsive (CR) and primary refractory (PR) HL samples.
Materials and Methods
Patient and Control Samples
Upon approval from the Institutional Review Board of the City of Hope, primary formalin-fixed paraffin-embedded (FFPE) diagnostic samples were obtained from 27 patients, including 15 patients with CR and 12 patients with PR HL. Clinico-pathological characteristics of the patients are summarized in Table 1. The International Prognostic Score (IPS) scores of our study population were calculated as described (16). In this study, patients with an IPS score of ≤ 2 were given a favorable designation, and patients with a score of ≥ 3 were assigned to the unfavorable group. Control samples included nine FFPE benign lymph node samples (four males and five females) and GC cells from 10 FFPE reactive follicular hyperplasia (RFH) samples (two males and eight females). Each sample was submitted for conventional histopathological processing to confirm HL involvement in the test samples or no evidence of malignancy in the control samples.
Table 1.
Clinical and Pathological Characteristics of the Hodgkin Lymphoma Patients
| # of Patients | Percentage | ||
|---|---|---|---|
| Age1 (range) | 32 (5–64) | 27 | 100 |
| Gender (M/F) | 13/14 | 27 | 48/52 |
| REAL/WHO classification | Nodular Sclerosis | 23 | 85 |
| Mixed Cellularity | 3 | 11 | |
| Classical HD, NOS1 | 1 | 4 | |
| Stage | Early (IA–IIB) | 16 | 59 |
| Advanced (IIIA–IVB) | 11 | 41 | |
| A vs. B symptoms | A | 13 | 48 |
| B | 14 | 52 | |
| Bulky Disease | 7 | 26 | |
| BM involvement | 1 | 4 | |
| IPS score2 | Favorable | 8 | 30 |
| Unfavorable | 17 | 63 | |
| Indeterminate | 2 | 7 | |
| Initial Treatment | ABVD | 20 | 74 |
| Stanford V | 3 | 11 | |
| BEACOPP3 | 3 | 11 | |
| COPP/ABV | 1 | 4 | |
| Treatment Response | Sensitive | 15 | 56 |
| Refractory | 12 | 44 | |
| Transplant | Autologous | 11 | 41 |
| Allogeneic | 1 | 4 |
Age, median (yr); NOS, not otherwise specified
IPS is based upon seven potential unfavorable features at diagnosis: serum albumin ≤ 4 g/dL (40 g/L), hemoglobin ≤ 10.5 g/dL (105 g/L), male gender, ≥45 years, stage IV, WBC ≥ 15,000/µL, and lymphocyte count ≤ 600/µL and/or ≤ 8 % of the WBC count (16).
Pediatric protocol CCG 59704.
FFPE Tissue Processing and Laser Capture Micro-dissection (LCM)
Five-micrometer serial sections from the FFPE tissue blocks were fixed onto PALM membrane slides (PEN-membrane; Zeiss, Jena, Germany) and processed as previously described (13, 14). A series of experiments designed to assess the impact of DNA source (e.g., archival material, including frozen and FFPE), quantity, and amplification on array comparative genomic hybridization were preformed to establish the FFPE aCGH protocol (13). Briefly, the slides were pretreated as follows for immunostaining: 1 hour at 65°C in a dry oven, 1 minute in xylene at room temperature, 5 minutes in 100% ethanol (x2), 5 minutes in 3% H2O2, and rinsed in dH2O. Antigen retrieval was performed at 98°C for 30 minutes using the decloaking chamber and decloaker universal heat retrieval buffer (both from Biocare Medical, Concord, CA). Test sample slides were stained with monoclonal mouse anti-human CD30 (Dako Inc., Carpinteria, CA) using the DakoCytomation Autostainer (Dako) per manufacturer’s instructions. Control samples were stained with CD20. A total of 150 CD30+ HRS cells per test sample and 150 CD20+ GC cells per control sample were LCM into microfuge caps containing water.
Whole Genome Amplification (WGA)
Isolated LCM cells were randomly fragmented and amplified in the LCM collection tubes using the GenomePlex single-cell whole-genome amplification kit (WGA4, Sigma-Aldrich, St Louis, MO), according to product instructions. The quality and quantity of the isolated amplified DNA were assessed electrophoretically and spectrophotometrically. Samples were processed prior to aCGH using the GenElute PCR clean-up kit (Sigma-Aldrich). A single sample was run in triplicate to evaluate assay reproducibility.
BAC-Based aCGH Labeling and Hybridization
One µg of genomic DNA was labeled using Bioarray Kit (Enzo Life Sciences, Farmingdale, NY), prior to subsequent hybridization to a RPCI 19K BAC array (Roswell Park, Buffalo, NY) using a Gentac Hybridization Station (Genomic Solutions, Ann Arbor, MI). Sex-mismatched pooled DNA from 20 healthy donors was used as reference DNA. The minimal tiling RPCI BAC array contained ~19,000 BAC clones was used (backbone consisted of ~4600 BAC clones with a BAC center-to-center distance of 165 kb) (human genome build 36.1) (13). Slides were scanned on the Genepix 4200A scanner (Molecular Devices, Sunnyvale, CA).
Control Assays, Selection Criteria and Outcome Predictor
Two control studies were performed to establish experimental reproducibility and quality control associated with multiple features of our study. These control studies had the objective of establishing standards for processing FFPE tissue, WGA, or eliminate problematic microarray calls associated with cross-hybridizing BACs or poor labeling/hybridization efficiency. First, nine FFPE benign lymph node control samples were processed in an identical fashion to the test samples. Second, 1.05 ng of peripheral blood DNA (DNA equivalent of 150 cells) obtained from six normal, healthy individuals (three males and three females) was amplified and analyzed by aCGH. Deviant calls from either of these control studies found in two or more microarrays were masked before the HL test samples were processed. Selection criteria for the reportable CNA calls in the HL samples was >5 BACs (~700 kb in size) observed in ≥ 35% in either the sensitive or resistant samples with a p-value < 0.15 and not present in <20% (no more than one patient) of the benign control samples.
To determine which genes are presumably critical in the HL malignant process, GC B cells from 10 lymph node samples showing reactive follicular hyperplasia (RFH) were processed identically to the test samples and compared with the HL CNAs. Genes associated with RFH had to meet the following selection criteria: large (>700 kb) CNAs observed ≥ 15% more common in RFH group (two or more samples) with a p-value <0.15, and observed in <20% of the benign control group. The CNAs associated with RFH were filtered prior to the comparative analysis of the chemosensitive and primary refractory disease CNAs. This final analysis generated a list of genes that could be used as a potential outcome predictor to discriminate between CR and PR HL at presentation. To test the outcome predictor in a preliminary manner, four additional primary HL lesions were analyzed in a blinded yet similar manner to the original 27 HL test samples and categorized using the proposed predictor model.
Data Analysis
Data images were quantified by ImaGene (BioDiscovery, Inc., El Segundo, CA), and the chromosome regions of abnormal copy number across the genome were detected by Nexus (BioDiscovery, Inc). To explore gene function and interactions, the CNAs were uploaded into Ingenuity Pathway Analysis tools 3.0 (IPA) (http://www.ingenuity.com) for alternative and canonical pathway mapping. IPA calculates a per-network significance score by applying the hyper-geometric distribution calculated via the Fisher's Exact Test for 2×2 contingency tables. The p-value is calculated by comparing the number of user-specified genes of interest that participate in a given pathway, relative to the total number of occurrences of these genes in all pathway annotations stored in the Ingenuity Pathways Knowledge Base (IPKB).
FISH Confirmation Studies
Locus-specific FISH analyses were performed to confirm the recurring CNAs detected by aCGH. The probes used for FISH analysis were carefully chosen and mapped within the genomic coordinates of the specific chromosome region showing gain or loss. In each case, a control DNA FISH probe from the opposite chromosome arm was included to confirm the CNAs in relation to the ploidy level of the HRS cell. To validate CNAs by FISH, CD30-stained IHC slides with AEC detection were scanned, localized and recorded on the slide using the Bioview Duet image analyzer (BioView, Ltd., Rehovot, Israel) prior to FISH analysis as described previously (17).
Statistical Evaluation
We background-corrected and normalized the quantified images with print-tip loess after removing bad spots. We evaluated all pairwise correlations between normalized replicates by scatterplots and Pearson's correlations on the log2 ratios.
Results
Defining the threshold and reproducibility of CNAs
From each patient and control FFPE sample, 150 cells (CD30+ HRS cells from test samples and CD20+ cells from control samples) were isolated from 5-µm thick tissue sections by LCM, amplified using WGA, and hybridized to a 19K whole-genome BAC array. Two control studies were performed to reduce deviant calls associated with poor hybridization efficiency or amplification bias associated with WGA or FFPE tissues. In the first experiment, 1.05 ng of peripheral blood DNA (DNA equivalent of 150 cells) obtained from six normal individuals (three males and three females), were WGA amplified and analyzed by aCGH. In the second study, nine formalin-fixed benign lymph node control samples (four males and five females) were processed similar to the test samples. A total of 235 of the 16,952 (1.4%) autosomal BACs showed a false positive (gain or loss) result in the normal control samples; these BACs were masked from the test data analyses. In addition, no chromosomal site was called positive if its respective BACs showed gain or loss in more than two arrays from normal control samples. Based on these control data, and to minimize the risk of reporting false-positive results, thresholds for calling gains and losses were set at >35% of test samples. Reproducibility of the FFPE was performed by analyzing 150 HRS cells from a single HL patient that was isolated independently from the block and processed by the LCM-WGA-aCGH method in triplicate. Supplemental figure 1 shows the high correlation between the normalized log2 ratio values of each possible pair of replicates (Pearson r = 0.90, P-values uniformly <0.0001), confirming reproducibility of the assay and the array.
CNAs detected in 27 primary FFPE diagnostic HL samples
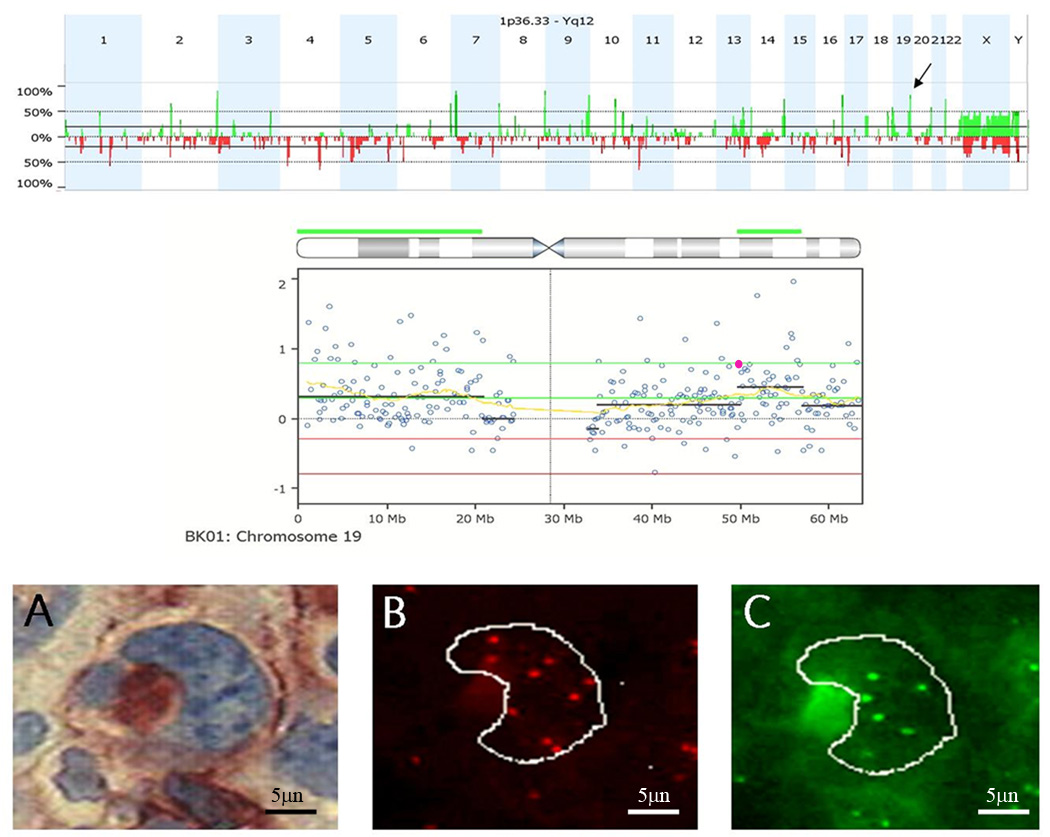
Primary tissue blocks were obtained from 15 CR and 12 PR patients. The clinical and pathological characteristics of the patients are listed in Table 1. Recurring chromosome imbalances were observed among all 27 HL samples. The most frequent gains (>65%) mapped to 2q37.3, 7p21.1, 8q24.3, 16q23.3–q24.3, 9q34.13-q34.3,14q32.33, 19q13.33, 20q13.33 (Supplemental data, Table 1) and included genes associated with growth and proliferation (AHR, BAI1, BOP1, COMMD5, LY6E, PTP4A3, SLURP1, CBFA2T3, SLC7A5, NOTCH1, FOXF1, TRAF2, IRF8, S100B, MYH14), cell cycle (AKT1, CDK10, SUMO3), drug metabolism (CYP11B1, CYP11B2, SLC19A1), angiogenesis and cell adhesion (COL18A1, CDH4, ITGB2, FOXC2), apoptosis regulation (FOXF1, GPR132), immune and lymphatic development (CBFA2T3, IL17C, IRF8, CLEC11A, RXRA, SPIB, ICOSLG) and invasion, metastasis or cancer-relatedness (VAV2, PSCA, PTP4A3, GINS2, FUT7, TUBB2C, KLK, POLD1, TFF2). Losses observed in >40% of HL samples encompassed SPRY1, NELL1, SLC1A3, GDNF, IL7R, SKP2, ID4, PPARGC1A, and TXNIP. Because ploidy differences are inherent to HRS cells, to accurately validate the CNAs observed by aCGH, HRS-specific FISH confirmation studies using locus-specific probes that mapped within the genetic coordinates of the recurring CNAs were paired with a DNA control probe that localized to the opposite chromosome arm. These chromosome region-specific FISH analyses confirmed the presence or absence of aCGH-defined CNAs in CD30+ HRS for 15 regions in three to five patient samples each for each paired FISH probe set (Figure 1).
Figure 1.
FISH confirmation study to validate CNAs detected by aCGH. Top) Genome aggregate for 12 primary refractory HL samples. Arrow indicates gain of 19q13.31 in 83% of refractory samples. Middle) Chromosome 19 drill down for an Individual sample. Each circle represents an individual BAC on the array. Green line, threshold for gain. Red line, threshold for CNA loss. The yellow line represents the moving average. The black lines represent cluster values of gain or loss defined by a circular binary segmentation algorithm. The highlighted BAC (pink) was selected for FISH validation studies. RP11–876a24 BAC contains the genes for PVR and CEACAM19. Bottom) Representative FISH images for sample BK01. (a) Bright field image of CD30+ cell. (b) The corresponding rhodamine filter image. RP11–876a24 was labeled with digoxigenin and detected with Rhodamine anti-dig. Nine copies of this clone were detected. (c) FITC image of internal control probe, only 6 copies were detected (net gain of three additional copies of PVR in cell).
Comparison of aCGH CNAs in 15 chemosensitive & 12 primary refractory HL samples
The genomic differences observed at >35% in the HL chemo-responsive patients clustered within two chromosomal bands on chromosome 16 (16q13 and 16p11.2) and showed gains of genes known to regulate T-cell trafficking or NF-κB activation (CCL22, CX3CL1, CCL17, DOK4 and IL10) (Table 2). In comparison to the CR samples, the 12 PR patients showed frequent loss of genes localized to 1q21.2, 4q27, 5p15, 11p14.3, and 17p12 with recurrent gains of chromosome bands 2q37.2, 13q31.2, 19q13.3 and 22q13.33. These data were also compared with the IPS scores of our study population (Table 1). Of interest, the 39 unique genes we identified in regions with CNAs comparing patients with favorable versus unfavorable IPS scores showed remarkable overlap with the 63 unique genes identified in chemosensitive and primary resistant genomic profiles (Table 2), with 36 unique genes appearing in both lists. The probability of this occurring by chance follows a hypergeometric distribution (18, 19), and the associated p-value was p < 0.0001.
Table 2.
Consistent CNAs observed in chemotherapy responsive vs. primary refractory HL, not observed in germinal center cells.
| Band | Region | Event | Length (bp) |
Freq. in Ref |
Freq.in Sen. |
Genes in Sen pts |
|---|---|---|---|---|---|---|
| 16q13 | chr16:55,456,619-56,127,978 | Gain | 671,359 | 25% | 60% | ARL2BP, CCDC102A, CCL17, CCL22, CETP, CIAPIN1, COQ9, CPNE2, CX3CL1, DOK4, HERPUD1, NIP30, NLRC5, PLLP, POLR2C, RSPRY1, SLC12A3 |
| 16p11.2 | chr16:32,592,341-33,172,220 | Loss | 579,879 | 17% | 53% | LOC729355, TP53TG3 |
| Band | Region | Event |
Length (bp) |
Freq. in Ref |
Freq. in Sen. |
Genes in Ref Pts |
| 1q21.2 | chr1:148,603,613-148,818,760 | Loss | 215,147 | 42% | 7% | ADAMTSL4, ECM1, KIAA0460, MCL1, TARS2 |
| 2q37.2 | chr2:236,067,474-236,698,859 | Gain | 631,385 | 50% | 27% | CENTG2 |
| 4q27 | chr4:120,634,997-123,761,662 | Loss | 3,126,665 | 50% | 20% | ADAD1,ANXA5, BBS7, C4orf31,CCNA2, EXOSC9, GPR103, IL2,IL21,KIAA1109, MAD2L1, PDE5A, TMEM155, TNIP3, TRPC3 |
| 5p15.31 | chr5:7,449,342-7,883,194 | Loss | 433,852 | 42% | 0% | ADCY2 |
| 5p15.2 | chr5:9,088,137-9,683,463 | Loss | 595,326 | 42% | 7% | SEMA5A,TAS2R1 |
| 11p14.3 | chr11:22,171,297-22,257,975 | Loss | 86,678 | 58% | 7% | TMEM16E |
| 13q31.2 | chr13:87,122,870-87,129,871 | Gain | 7,001 | 58% | 27% | SLITRK5 |
| 17p12 | chr17:11,864,859-15,526,918 | Loss | 3,662,059 | 58% | 13% | CDRT1, CDRT15, CDRT4, COX10, ELAC2, FAM18B2, FLJ45831,HS3ST3A1, HS3ST3B1, MAP2K4, MYOCD, PMP22, RICH2, TEKT3, TRIM16 |
| 19q13.31 | chr19:49,894,197-49,955,141 | Gain | 60,944 | 50% | 20% | BCL3,RELB, CEACAM19 |
| 22q13.33 | chr22:49,092,963-49,100,010 | Gain | 7,047 | 50% | 20% | FAM116B |
Comparison of HL samples with benign reactive follicular hyperplasia (RFH)
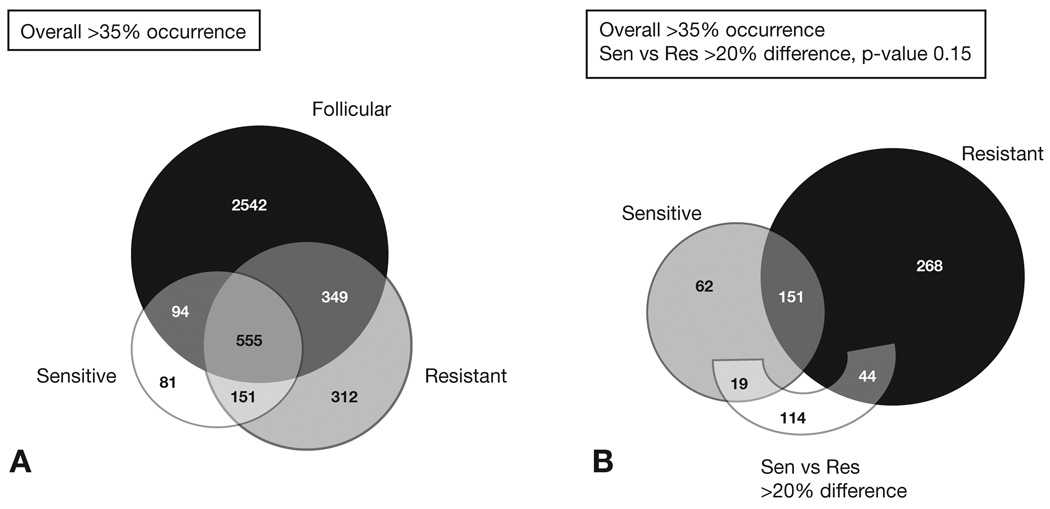
To delineate which genes are more likely to play a critical role in HL oncogenesis, normal GC cells from 10 RFH samples were analyzed and compared to the HL results. As described in the methods, strict criteria were established to call recurring gains and losses associated with CR HL, PR HL and benign RFH. Figure 2A shows the number of genes concordant and discordant among these three groups observed in >35% of the samples tested in each group. The first assessment of these data identified 151 genes frequently altered in both CR and PR HL samples but not detected in the benign RFH samples, 81 genes commonly detected in the CR HL and 312 genes more frequently observed in the PR HL tumors and not in the other two groups. The ten most frequent genetic alterations detected among the 151 HL CNAs included losses of genes in 5p13 (AMACR, GDNF, SKP2), gains of 7q36 (SHH), and gains of 9q34 (ABL1, CDK9, ENG, LCN2, PTGES, TSC1). A second assessment of the CR and PR HL gene subsets was performed requiring a frequency difference of >20% and a p-value of 0.15 between the sensitive and resistant samples (Figure 2B). In this analysis, 19 genes were clearly more common in chemosensitive samples, whereas 44 genes were more prevalent in the primary refractory tumors (Table 2).
Figure 2.
Comparison of HL samples with benign follicular hyperplasia (BFH). A) Concordant and discordant genes among RFH Follicular and the chemo-responsive (CR) Sensitive and the primary refractory (PR) HL Resistant samples. A total of 151 genes frequently detected in both CR and PRHL samples but not detected in the RFH samples, 81 genes were commonly detected in the CR HL and 312 genes more frequently observed in the PR HL tumors and not in the other two groups. B) A second assessment of the CR and PR genes showed the differences and similarity between the sensitive and resistant samples.
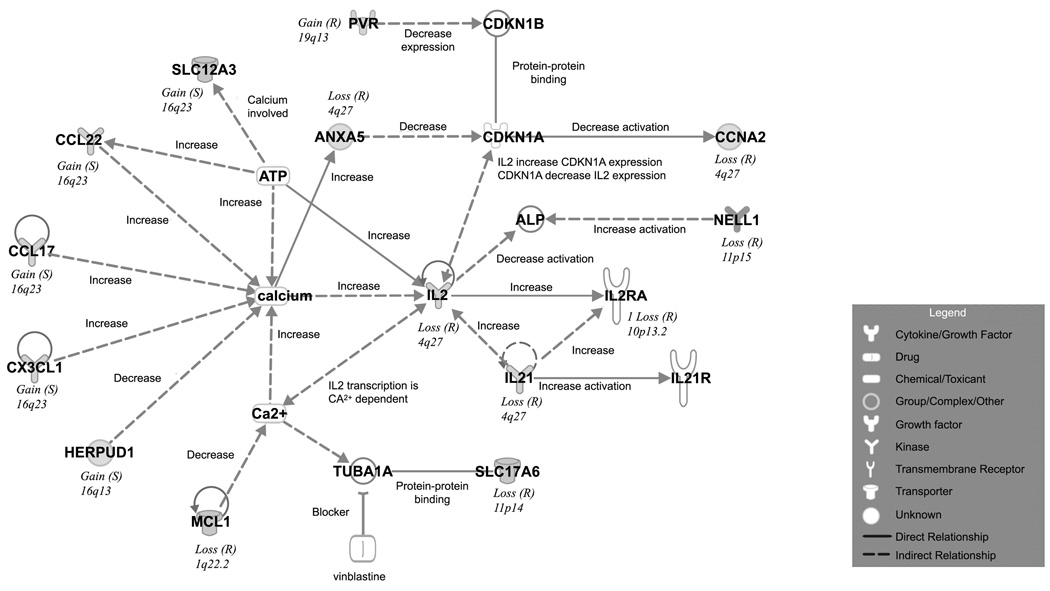
CNAs interaction networks and predictive modeling
Possible genetic interactions for those genes listed in Table 2 were examined in the context of the curated list of published molecular interactions in IPKB. In addition to an association with the NF-κB signaling pathway, IPA revealed connections between genes in the calcium signaling and IL-2/IL-21 signaling pathway (Figure 3). Key genes from these studies were used to construct a putative outcome model for predicting chemo-sensitive or primary refractory disease (Figure 4). Of interest, all seven patients showing loss of the SLC17A6 (solute carrier family 17, sodium-dependent inorganic phosphate co-transporter member 6) gene had PR disease; however, the four patients showing gains of the SLC12A3 (solute carrier family 12 sodium/chloride transporters, member 3) gene were CR. Second, patients showing no 16q13 gain, loss of 4q27 or 17p12 were commonly associated with a refractory phenotype (nine of 11 patients). Third, patients showing 16q13 gain with either few CNAs (10 of 12), or no loss of 4q27 or 17p12 (three of three) were usually CR.
Figure 3.
Calcium Signaling and IL2/IL21 Pathway Interactions in HL. Top Scoring network of interactions among the differential CNAs in CR versus PR Hodgkin lymphoma. The interactions indicate differences in the Ca2+ and IL2/LI21 Signaling Pathways. Inset: legend functional classes of the genes noted. Solid line, direct interaction. Dotted line, indirect interaction.
Figure 4.
Gene Profile and Prediction Outcome Model. R: primary refractory patient, S: chemo-responsive patient, s: initially chemo-responsive but relapsed 10 years after disease onset, X: the prediction didn’t match with the medical record.
Based on the relatively high frequency of IL21/IL2 loss at 4q27 and BCL3 overrepresentation in our PR HL cohort, we hypothesized samples without gains in 16q13 and losses of IL21/IL2 at 4q27,11p14.3/SLC17A6 or 17p12 with or without concurrent gains of BCL3 portend a highly likely drug-resistant HL phenotype. To test this CR vs PR outcome model in a preliminary manner, we first re-categorized all 27 patients according to the proposed model and predicted the outcome correctly in 23 (85%) of 27 patients (Figure 4). Four newly acquired HL samples were then tested in a blind fashion. One sample showed loss of 4q27 without BCL3 gains and was correctly assigned primary refractory; the remaining three cases were correctly designated “chemosensitive,” however, one “chemosensitive” patient (BK21) who initially responded to treatment suffered relapse of disease 10 years later.
Discussion
The identification of recurring cytogenetic and molecular alterations in HL has been thwarted by the infrequent occurrence of HRS cells in affected lesions (4). To gain further insight into the molecular genetic alterations in HL, we used a combined LCM-WGA-aCGH approach to analyze the genomic composition of HRS cells from 27 diagnostic samples from patients with classical HL. As expected, the molecular karyotypes of the HRS cells revealed numerous CNAs with recurring gains more common than losses due to the ploidy variation associated with mononucleated Hodgkin and multinucleated Reed-Sternberg cells. To refine our analysis, we limited this study to include the most common CNAs, defined as gains and losses in >35% of the HL samples, and confirmed by chromosome region-specific FISH analyses in CD30+ HRS.
Previous studies have shown that multiple signaling pathways and transcription factors are de-regulated in HRS cells, including NF-κB, JAK-STAT, and PI3K-AKT (4, 7, 20). Interestingly, regions with the most frequent gains in this study housed NOTCH1 and TRAF2, transcription factors, which mediate NF-κB activation (21, 22). Gains at the AKT1 locus support a role for constitutive activation of phosphatidyl-inositide 3 kinase in HL (20, 23) and further support data linking activated AKT1 to NF-κB signaling, p53 inhibition and enhanced resistance to apoptosis (24). Overrepresentation of the FOXC2 gene, a known transcriptional regulator of CXCR4 and Delta-like 4 (DLL4) (25) implies the down regulation of the Notch inhibitor Deltex1 to promote constitutive activation of NOTCH1 activity in HRS cells (21). In this study, gains of the forkhead (FOX) transcription factor FOXF1 gene were consistently observed with gains of SHH (sonic hedgehog homolog) supporting the supposition that NF-κB regulates Shh expression to further advance NF-κB -mediated proliferation and apoptosis resistance (26, 27). Other critical gains detected included the CDK10 cyclin-dependent kinase gene, which plays a role in cellular proliferation; GPR132, a member of the G-protein couple receptor (GPCR) family that slows cell proliferation and repairs damaged DNA; and SUMO3, which encodes a small ubiquitin-related modifier involved in nuclear transport, DNA replication, breakage and repair, mitosis, signal transduction and the cellular SUMOylation system (28).
Prominent losses observed in HRS cells targeted genes with prominent roles in cell cycle progression and tumor suppression. Both SKP2 (5p13.2) and ID4 (6p22.3) have been linked to the pathogenesis of lymphoma (29, 30). Inactivation by hypermethylation of ID4 (inhibitor of DNA binding 4) in follicular and diffuse large cell lymphoma suggests a tumor suppressor (TSG) role in malignant lymphoma (30). Similar studies in other cancers also indicate that SPRY1 and NELL1, two receptor tyrosine kinase signaling genes (31, 32) also have tumor suppressor roles. SKP2 (F-box protein S-phase kinase-associated protein 2 gene) encodes a critical component of the ubiquitin ligase complex that regulates the G1/S transition of the cell cycle by mediating target proteins such as CDKN1B cyclin-dependent kinase inhibitor and RELB (33). Consistent with the phenotype of HRS cells, SKP2-knockout experiments in mice show a hypo-proliferative phenotype with serious cellular defects, including nuclear enlargement, polyploidy, an increased number of chromosomes and chromosomal instability (34). Two other notable losses observed in HL were the thioredoxin interacting protein (TXNIP) gene, a gene required for natural killer cell maturation and PPARGC1A, a gene reported to antagonize NF-κB activation and the inflammatory gene expression of PTGS2 (also known as cyclooxygenase-2 or COX2) (35).
A comparison between the HRS and normal GC molecular karyotypes showed a shared relationship with gains in four transcription factors associated with hematopoietic stem cells and early lymphocyte development. IRF8 is a key transcription factor that modulates the myeloid versus lymphoid lineage choice by hematopoietic stem cells via interactions with the ETS family proteins encoded by SPI1 or SPIB. SPI1 encodes an ETS-domain transcription factor (PU.1) that activates gene expression during myeloid and B-lymphoid cell development, whereas SPIB acts as a lymphoid-specific enhancer to promote development of plasmacytoid dendritic cells or natural interferon (IFN)-producing cells (36, 37). Specifically within the GC, IRF8 is part of the transcriptional network governing B-cell lineage differentiation including the transcriptional regulation of activation-induced cytidine deaminase and BCL6 (37). Finally, ICOSLG is a ligand for the T-cell-specific cell surface receptor ICOS, and the product of this gene acts as a costimulatory signal for lymphoid proliferation and cytokine secretion to mediate local tissue responses to inflammatory conditions. Of interest, ICOS is highly expressed on tonsillar T-cells, which are closely associated with B cells in the apical light zone of GC (38). Taken together, the commonality of CNAs observed between GC and HRS cells substantiate a hematopoietic stem-cell-derived progenitor cell relationship (4, 39).
A further comparison of the GC and HL aCGH results defined a subset of 151 CNAs consistently associated with HL. The ten most frequent gene alterations in HL localized to three chromosomal regions and involved genes associated with inflammation, cell cycle progression and NF-κB /PI3K signaling: losses of AMACR, GDNF, SKP2 at 5p13, gain of SHH/7q36, and gains of ABL1, CDK9, ENG, LCN2, PTGES, and TSC1 at 9q34. As noted earlier, loss of SKP2 appears to be critical for the HRS hypoproliferative phenotype. The short-lived protein Cdk9 (the product of the CDK9 or cyclin-dependent kinase 9 gene) is also regulated by the SCF (SKP2) ubiquitin ligase complex, suggesting loss of SKP2 and gains of CDK9 allows for a stronger association with TRAF2 and continued NF-κB activation (40). LCN2 (lipocalin 2, previously known as NGAL) mediates inflammatory responses, and has binding sites for NF-κB, the hematopoietic transcription factors GATA-1 and SPI1 (PU.1), as well as the matrix metalloproteinases (MMP), which protect them from degradation and promote tumor cell invasion and metastasis (41). A link between MMP and LCN2 in this study is intriguing since Steidl and colleagues recently reported that over-expression of MMP11 is consistently associated with a tumor-associated macrophage expression profile and shortened survival in HL (9). Finally, gains of PTGS2 (prostaglandin-endoperoxide synthase-2, formerly known as COX2) are thought to enhance tumor angiogenesis, suppress anti-tumor immunity, and possibly play an important role for cytokine-driven PTGS2 NF-κB activation (42). Our genome-wide discrimination analyses on CNAs revealed distinct clusters that correlated closely with the favorable and unfavorable IPS scores in our HL patients suggesting the possibility of different chemoresponsive genomic profiles at disease presentation. To test this hypothesis, we compared the two distinct HL subgroups, selected 20 genes from 10 variable CNAs, and uploaded the data into Ingenuity Pathway Analysis tools 3.0 to explore gene function and interactions. The data suggested that a key difference between the CR and PR molecular karyotypes may be opposing calcium signaling and IL2/IL21 pathway interactions within the tumor microenvironment. The CR patients showed frequent gains of 16q13, a chromosome region known to house genes that regulate T-cell trafficking or NF-κB activation (CCL22, CX3CL1, CCL17, DOK4 and IL10). HRS cells are known to secrete CCL17 and CCL22 which attracts TH2 and regulatory T cells; report higher numbers of CD4+CD25 (high) FOXP3+ regulatory T cells are reported to predict for improved survival and prognosis in non-Hodgkin lymphoma (43). Further characterization of the tumor microenvironment may provide insight on the differential regulation of regulatory T cell function and survival in HL.
In contrast, PR HL showed frequent loss of genes at 4q27, 5p15, 11p14.3, 17p12 and gains at 13q31.2 and 19q13.31. Losses of two cytokine genes, IL2 and IL21 within 4q27, were evident in ~50% of the PR HRS cells. IL21 exhibits potent antitumor responses, is homologous to IL2, and shares the common cytokine receptor gamma chain of the IL2 receptor family (44). Although IL21 expression is usually restricted to subsets of CD4+ T cells and natural killer cells (44), two recent studies describe aberrant expression of IL21 activating STAT3 and STAT5 (signal transducer of activation of transcription 3 and 5) in HL continuous cell lines and a subset of primary HRS cells (3, 45). Moreover, Scheeren et al. (45) have shown that constitutive activation of STAT5 by IL21 results in activated NF-κB activity in immortalized B cells that have lost their B-cell phenotype and therefore mimic HRS cells, supporting a link between STAT5 and NF-κB in a subset of HL.
Conversely, the loss of IL21 in PR HL, as seen in our study, may prompt additional mutations in HRS cells to maintain cell survival in a cytokine-independent manner. One such complementary mutation may be gain of BCL3-RELB at 19q13.3. Previous to this report, only rare primary cases of HL (~15%) showed BCL3 gains or translocations in HRS cells (15, 46); however, gains of BCL3 appear to be more common in HL cell lines (12). In this study, we show that BCL3 gains are common in PR HL but infrequent findings in CR HL (~50% vs. 15%, respectively). Of interest, BCL3 not only regulates the NF-κB signaling pathway by modulating the DNA-binding activity of NF-κB transcription factors involved in apoptosis and cell growth, it also inhibits p53-induced apoptosis (47). Because IL21 is a potent inducer of BCL3 (48), the absence of IL21 may trigger BCL3 gene amplification as a mechanism for HRS cell survival. Interestingly, gains or over-expression of BCL3 have been associated with poor prognosis in other lymphoid malignancies including HTLV-1-induced ATLL and multiple myeloma (48, 49). Finally, losses within chromosome band 17p12 could possibly result in genomic rearrangements leading to over-expression of CD68, a gene housed within the neighboring chromosome band, 17p13.1. Increased numbers of CD68+ macrophages in HL have been correlated with short disease-free survival and a high likelihood of relapse post autologous stem cell transplantation (9). Based on our aCGH HL findings, we propose HL samples without gains in 16q13 and losses of IL21/IL2 at 4q27, 11p14.3/SLC17A6 and/or 17p12 and possibly concurrent gains of BCL3 portend a chemorefractory HL phenotype. A preliminary evaluation of the model appeared to support our hypothesis and the finding of 16p13 gains in the CR samples suggest that genes housed in this region may predict for a chemosensitive HL phenotype. More detailed explorations of these loci are needed to identify precise and reproducible prognostic genetic markers in HL.
Even though BAC aCGH technology is robust and provides high signal-to-noise ratios when working with challenging samples such as HRS cells captured by LCM from FFPE tissue, there are some limitations to its use for development of new clinical prognostic assays in HL. Since aCGH does not detect inactivating mutations and our selection criteria precluded the detection of small deletions, loss of the small ~16 kb TNFAIP3 (tumor necrosis factor, alpha-induced protein 3 gene or A20) at 6q23.3 was not detected in this study. We also did not observe amplification of the 42 kb REL gene within 2p16.1 as reported by others in ~30 to 40% of classical HL (5, 46); however gains within chromosome band 19q13.3 which houses both BCL3 and RELB were observed. Most likely, our strict inclusion criteria of investigating only gains and losses observed in >35% of samples tested and within a five-BAC region (> 700 kb) precluded detection of the small gene gains and losses, suggesting the necessity for higher-resolution microarray platforms in future studies.
Clarification of the genetic alterations and molecular mechanisms that underlie the oncogenic HRS cell are likely to improve our current understanding of HL. In this study, we identified DNA CNAs in HL by aCGH using DNA extracted from micro-dissected CD30+ HRS cells, demonstrating the potential utility of this approach for defining prognostic parameters. Despite the limited sample size, this HL investigation is one of the largest to date analyzing the molecular karyotypes of well-characterized, primary diagnostic HL samples with long-term follow-up. The molecular data showed a clear relationship between CNAs of key genes associated with the NF-κB signaling pathway, the tumor microenvironment, cell cycle regulation and apoptosis; however, the functional consequences of many of these genetic lesions and their complex interactions in HL tumorigenesis remain to be elucidated. Finally, the genetic differences observed between the chemosensitive and primary refractory HL molecular karyotypes, in this study and others (5–7, 9), suggest important pathogenetic and prognostic differences are present at disease onset. Given the paucity of primary refractory HL tumor samples available for study and the rarity of HRS cells within these lesions, it is imperative that we unite our efforts for the collection of retrospective HL samples with long term follow-up data for ongoing studies to confirm, refine, and define relevant biomarkers in primary refractory HL. Further studies exploiting these genetic lesions and defining their role in the pathogenesis of HL will be essential in developing new targeted therapeutic approaches.
Added note in support of our findings
During the manuscript review process, an interesting and similar aCGH in classical HL study using laser microdissected HRS cells was published by Steidl et al (Blood 116(3):418–427, 2010). Both studies agree that gains and losses of genes involved in the NF-kB signaling pathway are recurrent findings in classical HL. Unlike our study, Steidl et al (2010) report on the significance of gains and overexpression of the multidrug resistance gene ABCC1 in their treatment failure cohort, which consisted of lymph node samples collected from both treatment-naïve and treatment-exposed HL patients. We did not find a correlation between gains of ABCC1 and drug resistance; however, our population consisted only of newly-diagnosed pre-treatment samples and we used a slightly different method (FFPE not frozen samples). Although our study size is smaller, we only utilized samples collected at HL disease presentation (no relapsed samples) to eliminate bias associated with treatment for our comparison of chemosensitive versus primary refractory HL. This more homogenous patient population in combination with stringent gain and loss threshold requirements makes our data equally compelling.
Supplementary Material
Acknowledgments
We thank Aaron Theisen for his critical reading of the manuscript and helpful suggestions.
Grant Support: This work was supported in part by a Lymphoma SPORE Developmental Research Award, Cancer Center grants at Roswell Park Cancer Institute (NCI CA016056) and the City of Hope (NCI CA33572 and CA-30206), The Tim Nesvig Lymphoma Research Fund, and a private donation in memory of Pearl Ruttenberg.
Footnotes
Disclosure of Potential Conflicts of Interest: ML Slovak is currently an employee of Signature Genomic Laboratories.
REFERENCES
- 1.Diehl V, Thomas RK, Re D. Part II: Hodgkin's lymphoma--diagnosis and treatment. Lancet Oncol. 2004;5:19–26. doi: 10.1016/s1470-2045(03)01320-2. [DOI] [PubMed] [Google Scholar]
- 2.Schwering I, Brauninger A, Klein U, et al. Loss of the B-lineage-specific gene expression program in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2003;101:1505–1512. doi: 10.1182/blood-2002-03-0839. [DOI] [PubMed] [Google Scholar]
- 3.Lamprecht B, Kreher S, Anagnostopoulos I, et al. Aberrant expression of the Th2 cytokine IL-21 in Hodgkin lymphoma cells regulates STAT3 signaling and attracts Treg cells via regulation of MIP-3alpha. Blood. 2008;112:3339–3347. doi: 10.1182/blood-2008-01-134783. [DOI] [PubMed] [Google Scholar]
- 4.Kuppers R. The biology of Hodgkin's lymphoma. Nat Rev Cancer. 2009;9:15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 5.Joos S, Menz CK, Wrobel G, et al. Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood. 2002;99:1381–1387. doi: 10.1182/blood.v99.4.1381. [DOI] [PubMed] [Google Scholar]
- 6.Martin-Subero JI, Gesk S, Harder L, et al. Recurrent involvement of the REL and BCL11A loci in classical Hodgkin lymphoma. Blood. 2002;99:1474–1477. doi: 10.1182/blood.v99.4.1474. [DOI] [PubMed] [Google Scholar]
- 7.Schmitz R, Hansmann ML, Bohle V, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horning S, Fanale M, Sd Defining a population of Hodgkin lymphoma patients for novel therapeutics: an international effort [abstract] Ann Oncol. 2008;20:118. [Google Scholar]
- 9.Steidl C, Lee T, Shah SP, et al. Tumor-Associated Macrophages and Survival in Classic Hodgkin's Lymphoma. N Engl J Med. 2010;362:875–885. doi: 10.1056/NEJMoa0905680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giefing M, Arnemann J, Martin-Subero JI. Identification of candidate tumour suppressor gene loci for Hodgkin and Reed-Sternberg cells by characterisation of homozygous deletions in classical Hodgkin lymphoma cell lines. Br J Haematol. 2008;142:916–924. doi: 10.1111/j.1365-2141.2008.07262.x. [DOI] [PubMed] [Google Scholar]
- 11.Feys T, Poppe B, De Preter K, et al. A detailed inventory of DNA copy number alterations in four commonly used Hodgkin's lymphoma cell lines. Haematologica. 2007;92:913–920. doi: 10.3324/haematol.11073. [DOI] [PubMed] [Google Scholar]
- 12.Mathas S, Johrens K, Joos S, et al. Elevated NF-kappaB p50 complex formation and Bcl-3 expression in classical Hodgkin, anaplastic large-cell, and other peripheral T-cell lymphomas. Blood. 2005;106:4287–4293. doi: 10.1182/blood-2004-09-3620. [DOI] [PubMed] [Google Scholar]
- 13.Nowak NJ, Miecznikowski J, Moore SR, et al. Challenges in array comparative genomic hybridization for the analysis of cancer samples. Genet Med. 2007;9:585–595. doi: 10.1097/gim.0b013e3181461c4a. [DOI] [PubMed] [Google Scholar]
- 14.Moore SR, Persons DL, Sosman JA, et al. Detection of copy number alterations in metastatic melanoma by a DNA fluorescence in situ hybridization probe panel and array comparative genomic hybridization: a southwest oncology group study (S9431) Clin Cancer Res. 2008;14:2927–2935. doi: 10.1158/1078-0432.CCR-07-4068. [DOI] [PubMed] [Google Scholar]
- 15.Hartmann S, Martin-Subero JI, Gesk S, et al. Detection of genomic imbalances in microdissected Hodgkin and Reed-Sternberg cells of classical Hodgkin's lymphoma by array-based comparative genomic hybridization. Haematologica. 2008;93:1318–1326. doi: 10.3324/haematol.12875. [DOI] [PubMed] [Google Scholar]
- 16.Hasenclever D, Diehl V. A prognostic score for advanced Hodgkin's disease. International Prognostic Factors Project on Advanced Hodgkin's Disease. N Engl J Med. 1998;339:1506–1514. doi: 10.1056/NEJM199811193392104. [DOI] [PubMed] [Google Scholar]
- 17.Bedell V, Forman SJ, Gaal K, Pullarkat V, Weiss LM, Slovak ML. Successful application of a direct detection slide-based sequential phenotype/genotype assay using archived bone marrow smears and paraffin embedded tissue sections. J Mol Diagn. 2007;9:589–597. doi: 10.2353/jmoldx.2007.070050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fury W, Batliwalla F, Gregersen PK, Li W. Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf Proc IEEE Eng Med Biol Soc. 2006;1:5531–5534. doi: 10.1109/IEMBS.2006.260828. [DOI] [PubMed] [Google Scholar]
- 19.Bankhead A, 3rd, Sach I, Ni C, et al. Knowledge based identification of essential signaling from genome-scale siRNA experiments. BMC Syst Biol. 2009;3:80. doi: 10.1186/1752-0509-3-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutton A, Reynolds GM, Dawson CW, Young LS, Murray PG. Constitutive activation of phosphatidyl-inositide 3 kinase contributes to the survival of Hodgkin's lymphoma cells through a mechanism involving Akt kinase and mTOR. J Pathol. 2005;205:498–506. doi: 10.1002/path.1725. [DOI] [PubMed] [Google Scholar]
- 21.Jundt F, Acikgoz O, Kwon SH, et al. Aberrant expression of Notch1 interferes with the B-lymphoid phenotype of neoplastic B cells in classical Hodgkin lymphoma. Leukemia. 2008;22:1587–1594. doi: 10.1038/leu.2008.101. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Blackwell K, Shi Z, Habelhah H. The RING domain of TRAF2 plays an essential role in the inhibition of TNFalpha-induced cell death but not in the activation of NF-kappaB. J Mol Biol. 2010;396:528–539. doi: 10.1016/j.jmb.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgakis GV, Li Y, Rassidakis GZ, Medeiros LJ, Mills GB, Younes A. Inhibition of the phosphatidylinositol-3 kinase/Akt promotes G1 cell cycle arrest and apoptosis in Hodgkin lymphoma. Br J Haematol. 2006;132:503–511. doi: 10.1111/j.1365-2141.2005.05881.x. [DOI] [PubMed] [Google Scholar]
- 24.Jeong SJ, Pise-Masison CA, Radonovich MF, Park HU, Brady JN. Activated AKT regulates NF-kappaB activation, p53 inhibition and cell survival in HTLV-1-transformed cells. Oncogene. 2005;24:6719–6728. doi: 10.1038/sj.onc.1208825. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi H, Kume T. Foxc transcription factors directly regulate Dll4 and Hey2 expression by interacting with the VEGF-Notch signaling pathways in endothelial cells. PLoS One. 2008;3:e2401. doi: 10.1371/journal.pone.0002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakashima H, Nakamura M, Yamaguchi H, et al. Nuclear factor-kappaB contributes to hedgehog signaling pathway activation through sonic hedgehog induction in pancreatic cancer. Cancer Res. 2006;66:7041–7049. doi: 10.1158/0008-5472.CAN-05-4588. [DOI] [PubMed] [Google Scholar]
- 27.Kasperczyk H, Baumann B, Debatin KM, Fulda S. Characterization of sonic hedgehog as a novel NF-kappaB target gene that promotes NF-kappaB-mediated apoptosis resistance and tumor growth in vivo. FASEB J. 2009;23:21–33. doi: 10.1096/fj.08-111096. [DOI] [PubMed] [Google Scholar]
- 28.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seki R, Ohshima K, Fujisaki T, et al. Prognostic significance of S-phase kinase-associated protein 2 and p27kip1 in patients with diffuse large B-cell lymphoma: effects of rituximab. Ann Oncol. 2009 doi: 10.1093/annonc/mdp481. [DOI] [PubMed] [Google Scholar]
- 30.Hagiwara K, Nagai H, Li Y, Ohashi H, Hotta T, Saito H. Frequent DNA methylation but not mutation of the ID4 gene in malignant lymphoma. J Clin Exp Hematop. 2007;47:15–18. doi: 10.3960/jslrt.47.15. [DOI] [PubMed] [Google Scholar]
- 31.Jin Z, Mori Y, Yang J, et al. Hypermethylation of the nel-like 1 gene is a common and early event and is associated with poor prognosis in early-stage esophageal adenocarcinoma. Oncogene. 2007;26:6332–6340. doi: 10.1038/sj.onc.1210461. [DOI] [PubMed] [Google Scholar]
- 32.Kwabi-Addo B, Wang J, Erdem H, et al. The expression of Sprouty1, an inhibitor of fibroblast growth factor signal transduction, is decreased in human prostate cancer. Cancer Res. 2004;64:4728–4735. doi: 10.1158/0008-5472.CAN-03-3759. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16:323–333. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama K, Nagahama H, Minamishima YA, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackerman WEt, Zhang XL, Rovin BH, Kniss DA. Modulation of cytokine-induced cyclooxygenase 2 expression by PPARG ligands through NFkappaB signal disruption in human WISH and amnion cells. Biol Reprod. 2005;73:527–535. doi: 10.1095/biolreprod.104.039032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidlin H, Diehl SA, Nagasawa M, et al. Spi-B inhibits human plasma cell differentiation by repressing BLIMP1 and XBP-1 expression. Blood. 2008;112:1804–1812. doi: 10.1182/blood-2008-01-136440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Lee CH, Qi C, et al. IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112:4028–4038. doi: 10.1182/blood-2008-01-129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hutloff A, Dittrich AM, Beier KC, et al. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature. 1999;397:263–266. doi: 10.1038/16717. [DOI] [PubMed] [Google Scholar]
- 39.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 40.MacLachlan TK, Sang N, De Luca A, Puri PL, Levrero M, Giordano A. Binding of CDK9 to TRAF2. J Cell Biochem. 1998;71:467–478. [PubMed] [Google Scholar]
- 41.Yan L, Borregaard N, Kjeldsen L, Moses MA. The high molecular weight urinary matrix metalloproteinase (MMP) activity is a complex of gelatinase B/MMP-9 and neutrophil gelatinase-associated lipocalin (NGAL). Modulation of MMP-9 activity by NGAL. J Biol Chem. 2001;276:37258–37265. doi: 10.1074/jbc.M106089200. [DOI] [PubMed] [Google Scholar]
- 42.Ackerman WEt, Summerfield TL, Vandre DD, Robinson JM, Kniss DA. Nuclear factor-kappa B regulates inducible prostaglandin E synthase expression in human amnion mesenchymal cells. Biol Reprod. 2008;78:68–76. doi: 10.1095/biolreprod.107.061663. [DOI] [PubMed] [Google Scholar]
- 43.Ke X, Wang J, Li L, Chen IH, Wang H, Yang XF. Roles of CD4+CD25(high) FOXP3+ Tregs in lymphomas and tumors are complex. Front Biosci. 2008;13:3986–4001. doi: 10.2741/2986. [DOI] [PubMed] [Google Scholar]
- 44.Spolski R, Leonard WJ. Interleukin-21: basic biology and implications for cancer and autoimmunity. Annu Rev Immunol. 2008;26:57–79. doi: 10.1146/annurev.immunol.26.021607.090316. [DOI] [PubMed] [Google Scholar]
- 45.Scheeren FA, Diehl SA, Smit LA, et al. IL-21 is expressed in Hodgkin lymphoma and activates STAT5: evidence that activated STAT5 is required for Hodgkin lymphomagenesis. Blood. 2008;111:4706–4715. doi: 10.1182/blood-2007-08-105643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin-Subero JI, Wlodarska I, Bastard C, et al. Chromosomal rearrangements involving the BCL3 locus are recurrent in classical Hodgkin and peripheral T-cell lymphoma. Blood. 2006;108:401–402. doi: 10.1182/blood-2005-09-3843. author reply 2–3. [DOI] [PubMed] [Google Scholar]
- 47.Kashatus D, Cogswell P, Baldwin AS. Expression of the Bcl-3 proto-oncogene suppresses p53 activation. Genes Dev. 2006;20:225–235. doi: 10.1101/gad.1352206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brenne AT, Fagerli UM, Shaughnessy JD, Jr, et al. High expression of BCL3 in human myeloma cells is associated with increased proliferation and inferior prognosis. Eur J Haematol. 2009;82:354–363. doi: 10.1111/j.1600-0609.2009.01225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim YM, Sharma N, Nyborg JK. The proto-oncogene Bcl3, induced by Tax, represses Tax-mediated transcription via p300 displacement from the human T-cell leukemia virus type 1 promoter. J Virol. 2008;82:11939–11947. doi: 10.1128/JVI.01356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.