Abstract
Epidemiological results tend to suggest that adults with Down syndrome (DS) have a reduced incidence of cancer, but some studies have reached the opposite conclusion. In this study, we offer direct biological evidence in support of the notion that DS reduces incidence of multiple types of cancer. Previous studies showed that introduction of the ApcMin mutation into the Ts65Dn mouse model of DS by interbreeding caused formation of intestinal adenomas at a significantly reduced incidence compared to control (euploid) animals that did not have trisomy. To a large degree, this reduction was determined to reflect an increased dosage of the Ets2 tumor repressor gene due to trisomy. Studies of tumor grafts using Ts65Dn suggested angiogenesis as a mechanism that mediated reduced tumor growth, metastasis and mortality in DS individuals. To confirm and extend these findings, we employed the complex cancer mouse model, NPcis, which is heterozygous for the Trp53 and Nf1 genes and through loss of heterozygosity develops lymphomas, sarcomas or carcinomas with 100% penetrance. In this aggressive model, trisomy did not prevent cancer but it nevertheless extended host survival relative to euploid littermates. However, protection in this case was not attributable to either Ets2 dosage or to reduced angiogenesis. Together, our findings indicate that the genetic complexity underlying DS supports multiple mechanisms that contribute to reduced mortality from cancer.
Keywords: Down syndrome, cancer, angiogenesis, Ets2
Introduction
Down syndrome (DS), a genetic disorder resulting from trisomy for human chromosome 21 (Hsa21), affects all cells in the body. Many effects are deleterious, including a significantly increased risk of acute megakaryoblastic leukemia in children with DS (1). In contrast, some (though by no means all) epidemiological studies have observed a lower incidence of cancer in this population after childhood (2, 3). The observation that the incidence of many different types of cancer may be reduced in DS suggests that there might be a general mechanism by which trisomy affects tumorigenesis. Increased tumor immune surveillance, sensitivity to apoptosis in response to early tumorigenic events or inhibition of angiogenesis could represent such mechanisms. Epidemiological data alone cannot identify gene candidates nor provide biological proof of mechanism.
Mice that are trisomic for genes whose orthologs are on Hsa21 have proven to be very useful in understanding the etiology of DS phenotypes (4). In particular, the widely studied Ts65Dn mouse is trisomic for orthologs of about half of the genes (104 of 231) that are conserved between mouse and Hsa21 (5, 6). Several Hsa21 genes have been identified as candidates for tumor repression (7–9).
Endostatin, a cleavage product of the Collagen 18A1 gene, blocks tumor growth by inhibiting angiogenesis (8). Similarly, over-expression of Rcan1 (formerly known as Dscr1) and Dyrk1a, both of which are trisomic in Ts65Dn mice, has been reported to inhibit growth of B16 melanoma as well as Lewis Lung Carcinoma xenografts in Ts65Dn mice by blocking angiogenesis (7). Recent studies using Tc1 mice, which are trisomic for 81% of Hsa21 genes, attribute reduced angiogenesis in B16 xenografts and epithelial explants to four Hsa21 genes, ADAMTS1, ERG1, JAM-B and PTTG1IP (10). Although the role of Rcan1 in regulating VEGF to control angiogenesis has been known for some time (11), its contributions to this process in trisomic xenografts are unclear because Tc1 mice have normal dosage (2 copies) of Rcan1, but show the same reduction in xenograft angiogenesis as do Ts65Dn mice with three copies (10). Dosage of the Ets2 gene plays a critical role in repression of intestinal adenomas in the ApcMin model independent of either Rcan1 or Col18a1 dosage (9). This effect may result from increased sensitivity of cells to p53-mediated apoptosis when Ets2 is over-expressed (12). Thus, multiple genes and mechanisms appear to be involved in the repression of tumors in DS.
To address the generality of protection against cancer by trisomy, we used a complex cancer model, the NPcis mouse (NP). These mice carry null alleles for the adjacent tumor suppressor genes, Nf1 and Trp53, and develop several kinds of cancer due to LOH of the normal alleles (13, 14). We crossed NP mice to Ts65Dn and compared survival, tumor growth properties, angiogenesis and the role of Ets2 function on euploid and trisomic backgrounds to determine whether trisomy was protective in this complex tumor model.
Methods
Mice
C57BL/6J-NPcis mice (here referred to as NP) were kindly provided by Dr. Karlyne M. Reilly (15). B6EiC3Sn a/A-Ts (1716)65Dn (Ts65Dn) mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and maintained by breeding females to male (C57BL/6J × C3H//HeJ) F1 mice. C57BL/6J-Ets2+/− mice were generously provided by Dr. Michael Ostrowski (16). Mouse tail DNA was used for genotyping. NPcis and Ets2+/− mice were genotyped by PCR as described (15, 16). Ts65Dn mice were screened by PCR and confirmed using fluorescent in situ hybridization (FISH) (17, 18). All procedures were approved by the Johns Hopkins Animal Care and Use Committee.
Tumor Analysis
Mice carrying the NP mutation were maintained until tumors developed. Mice were sacrificed if they had any of the following signs: 1, visible tumor size greater than 2 cm in diameter; 2, no obvious tumor, but with little movement and rough hair; 3, no obvious tumor, but with abnormal behavior indicating a possible brain tumor. All major organs were examined grossly. Tumor and brain tissues were fixed in formalin then embedded in paraffin. Hematoxylin and Eosin stained slides were used to categorize tumor type, and partial tumors were further characterized by immunostaining.
Immunohistology
Brain samples were stained with polyclonal rabbit anti-GFAP antibody (1:500, Dako, Denmark). Sarcomas were stained with polyclonal rabbit anti-S100 (1:400, Dako, Denmark). Lymphomas were stained with polyclonal rabbit anti-CD3 (1:100, Dako) and polyclonal mouse anti-Pax5 (1:500, BD Pharmingen, San Diego). Rabbit IgG was detected by biotin-SP-conjugated goat anti-rabbit IgG (H+L) (1:500, Jackson Immunoresearch, West Grove), biotin was detected using the vectastain ABC peroxidase kit (Vector Lab, Burlingame) and mouse IgG was detected with the M.O.M. peroxidase kit (Vector Lab, Burlingame) and DAB substrate development (Vector lab, Burlingame). Endogenous peroxidases were blocked by 3% hydrogen peroxide in methanol. Slides were counterstained with hematoxylin, dehydrated through series of ethanol and xylene, and preserved with clarion mounting medium. TUNEL (Roche, Indianapolis) was done following the manufacturer’s instruction.
Microvessel density (MVD) count
Paraffin-embedded, 6 μm tumor sections were immunostained for CD31 (1:50, Abcam, Cambridge). The number of discrete microvessels was counted using image analysis software (NIS-Elements BR3.0). Discrete microvessels were identified as any highlighted endothelial cell separated from adjacent microvessels.
DNA extraction from paraffin embedded samples (19)
Four 30-μm sections were cut from formalin-fixed and paraffin-embedded tumor samples and incubated in xylene at 45°C for 15 minutes. Pellets were collected by centrifugation at 14,000 rpm for 10 minutes and washed successively with 100%, 90% and 70% ethanol, resuspended in 1 M NaSCN and incubated at 37°C overnight. The extract was digested with proteinase K at 55 °C, extracted with phenol followed by ethanol precipitation and resuspended for analysis.
Taqman probes for gene dosage determination
Apob probes and primers are from Liu, Schmidt et al from the Jackson Laboratory (20). Taqman assays for Rcan1, Mx1 and Jam2 genomic sequences were designed using ABI online software (21) (sequence in Supplementary information).
Xenografts
TCL.Np1 and TCL.Np2 lines were established from resected mouse sarcomas as previously described (15). The pigmented, B16-F10 (CRL-6475) melanoma cells were obtained directly from ATCC and made into multiple low passage freezer stocks and was shown to contain three mouse genes by species-specific PCR. For xenografts, 50,000 cells were injected into flanks of H2-compatible mice. Four weeks later, xenografts were resected, weighed, fixed in formalin, embedded in paraffin and stained for TUNEL, CD31 and Ki67 for analysis. Five images were taken with each slide and analyzed using NIS-Elements BR3.0 software. The number of stained cells divided by the sum of all cells was reported.
Statistical analysis of NPcis tumours on different strain backgrounds
Data were reported as mean ± s.d. P values were calculated using Student’s t-test (Excel). Log-rank test, chi-square test and Fisher’s exact test were computed using R with survival package (22). Kruskal-Wallace was done manually in Excel according to definition (23). Power analysis was done using University of Iowa statistics package (24).
Results
Trisomy extends survival of NPcis mice
Ts65Dn mice cannot be inbred more than 3 generations and are maintained as an advanced intercross, (C57BL/6J × C3H/HeJ)Fn (genetic background is 50% B6, 50% C3H on average). Because both the NPcis and Ets2 null alleles are maintained on a B6 background, we generated mice with a range of B6 representation in their genomes. In accordance with a previous report (25), we found similar survival times on either the B6 or a mixed 75% B6, 25% C3H background (log-rank test, P = 0.232, n = 37 and 36, respectively) (Supplementary Fig. 1A, B). Based on this result we included data from both stages of the cross in the following analyses.
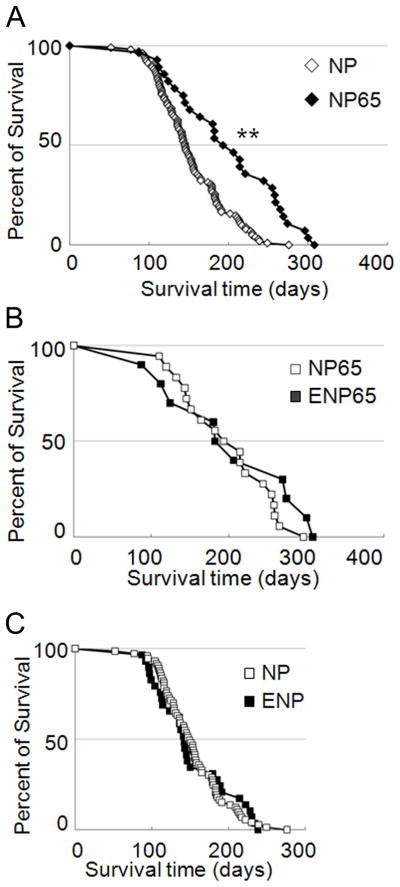
The NPcis mutation was transmitted at Mendelian frequencies. Survival curves for NP and NP65 mice showed that life span was extended significantly in trisomic mice (median survival was 145 and 188 days, log-rank test, P = 2.4e-5, n = 102 and 28, respectively) (Fig. 1A). Thus, trisomy has a protective effect against mortality from cancer in this complex model. Ets2 dosage had no effect on survival in the NPcis model (Fig. 1B, C). Survival time was not significantly different between NPcis mice carrying a null allele of Ets2 (ENP, one copy of Ets2) and NP mice with two copies (Log-rank test, P = 0.90). NP65 mice (3 copies of Ets2) had a similar life span to ENP65 (2 copies of Ets2) (Log-rank test, P = 0.22).
Figure 1. Survival of NPcis mice is extended by trisomy and not affected by Ets2 dosage.
A, Median survival was significantly longer for NP65 (trisomic) mice than for NP (euploid) (Log-rank test, **P=2.4e-05, n = 102 and 28 for euploid and Ts65Dn, respectively). B, Euploid mice with one (ENP, n=29) or two (NP, n=73) copies of Ets2 had the same median survival (Log-rank test, P=0.90). C, Ets2 dosage had no effect on median survival of trisomic mice with two (ENP65, n=10) or three (NP65, n=18) copies (Log-rank test, P=0.22).
Extended survival by NP females compared to males has been observed in previous studies (26) and was evident here, as well, for both trisomic and euploid mice (Supplementary Fig. 1C–F). The reasons for this sex difference remain unclear. Trisomic female NP65 mice survived significantly longer than did euploid NP females (p<0.0002) (Supplementary Fig. 1E). The number of male Ts65Dn mice recovered was substantially less than the number of trisomic females (Supplementary Tables 1 and 2, Supplementary Fig. 2) and the relatively small number of males did not power meaningful statistical comparison after parsing by genotype, tumor type and sex. Median survival of male NP65 mice was similar to that of male NP mice (144 and 136 days, respectively) (Supplementary Fig. 1F).
To assure that the survival data was not skewed by the lower proportion of males in the trisomic set (37% vs. 50% males in the euploid set), we randomly selected a subset of euploid males to combine with euploid females so as to match the sex ratios in the trisomic set (i.e., 37% males) and repeated the analysis in Fig. 1A. A similarly robust, significant extension of survival was found in each of three iterations of this exercise. As there is no evidence for a sex bias in tumor protection in people with DS (3), nor statistical support for skewing here, we proceeded with combined euploid vs. trisomic datasets. Our analysis does not exclude the possibility of a small, trisomy-specific, sex-influenced differential response to the NPcis mutation.
Trisomy alters the tumor spectrum in NPcis mice
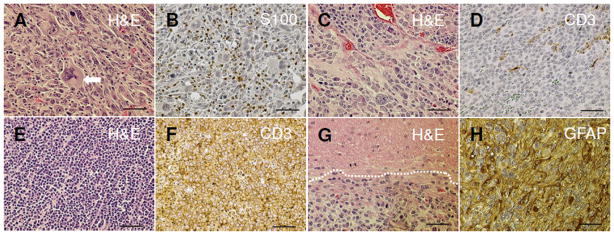
We assessed gross and histological characteristics of all tumors and determined their frequencies (Fig. 2). As expected, we observed multiple soft tissue sarcomas, adrenal tumors, lymphomas and brain lesions (14). Most of the sarcomas that we observed were malignant peripheral nerve sheath tumors (MPNSTs). Histologically, MPNSTs are composed of spindle-shaped cells and high-grade MPNSTs exhibit high cellularity with marked nuclear pleomorphism (Fig. 2A) (27). Twenty-nine of the 34 soft tissue sarcomas (85.3%) stained positively for S100B (Fig. 2B).
Figure 2. Four tumor types were characterized in trisomic and euploid NPcis mice.
A, MPNST composed of atypical spindle-shaped cells arranged in fascicles, nuclear pleomorphism indicated by arrow (H&E). B, An MPNST showing S-100 protein-positive cells (Schwann cells, brown cells are stained with the DAB substrate). C, H&E stain of an adrenal tumor. D, A drenal tumors were negative for CD3. E, Lymphoma, H&E. F, T-cell lymphomas were positive for CD3. G, A strocytoma showing an irregular mass of tumor (H&E). H, A strocytoma stained positively with GFAP. Scale bar, 50 μm.
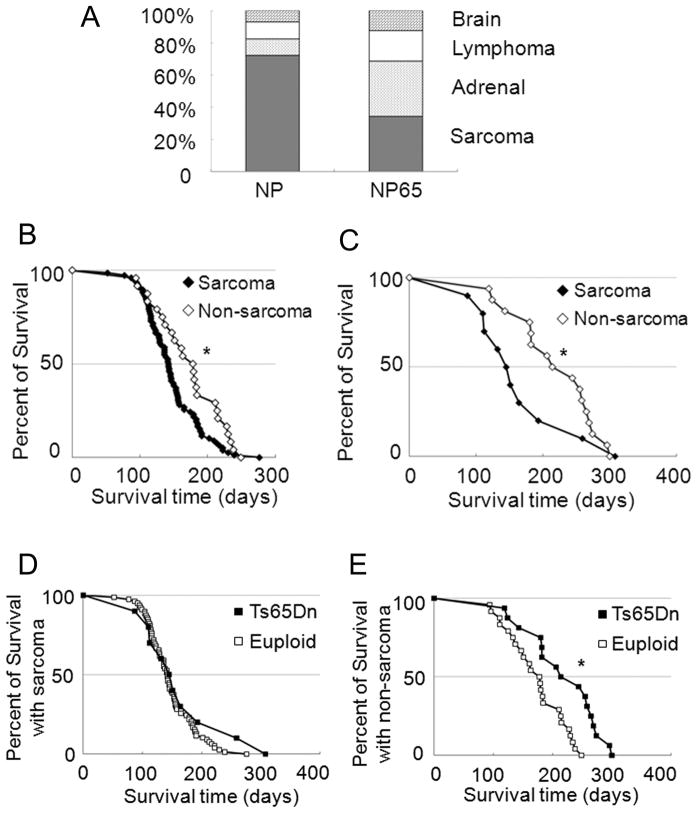
The incidence of sarcoma in trisomic mice was significantly reduced (Fisher’s exact test, P = 0.0001). Eleven out of 32 (34.4%) tumors were sarcomas in the trisomy group, while in the euploid group, 92 out of 126 (73%) were sarcoma (Fig. 3A, Supplementary Table 3, Supplementary Fig. 2). Both euploid and trisomic mice without sarcoma survived longer than mice that developed sarcoma (Fig. 3B, C). Ts65Dn didn’t extend survival of mice with sarcoma (median survival of 142 and 145 days in NP and NP65 respectively, Fig. 3D, Supplementary Fig. 3), but did extend survival significantly in mice with other cancers (median survival of 178 days and 214 days in NP and NP65, respectively) (Fig. 3E, Supplementary Fig. 3). The median survival time was longer in NP65 than in NP for each non-sarcoma tumor type (Supplementary Table 3).
Figure 3. Ploidy affects the pattern and growth of tumors that develop in NPcis mice.
A, The frequencies of lymphoma and of brain tumors were not significantly different in trisomic mice compared to euploid. A significantly larger fraction of adrenal tumors (P= 0.0014, Fisher’s exact test) and fewer sarcomas (P= 0.0001, Fisher’s exact test) were observed in trisomic, NP65 mice than in their euploid counterparts. B, Euploid mice without sarcoma (open diamond, n=24) lived longer than mice with sarcoma (filled diamond, n=78, *P = 0.026 by Log-rank test). C, Trisomic mice without sarcoma (n=16) lived longer than those with sarcoma (n=10, *P = 0.042 by Kruskal-Wallis test). D, Median survival times were the same in trisomic and euploid mice with sarcoma (P=0.168). E) Trisomic mice with non-sarcoma tumors lived longer than euploid mice with non-sarcoma tumors (*P=0.0017).
Adrenal tumors were the most frequent type in NP65 (Fig. 2C). These tumors were located adjacent to and invading the kidney, and were bloody with hemorrhage and necrosis. They displayed large round cells with eosinophilic cytoplasm, high nuclear grade and diffuse architecture and were negative for lymphoma markers (Fig. 2D). The incidence of adrenal tumors as a percent of total was significantly higher in trisomic NP65 mice compared to euploid (Fisher’s exact test, P = 0.0014). Among the NP65 group, 11 out of 32 (34.4%) were adrenal tumors while the frequency for euploid mice was 12 out of 126 (10%) (Fig. 3A).
All but three of the lymphomas identified here were T-cell lymphomas composed of cells with a round shape and prominent basophilic nucleoli (Fig. 2E). They were strongly positive for the T-cell marker, CD3 (Fig. 2F), and negative for the B-cell marker, Pax5. B-cell lymphomas were identified in spleen, two in NP and one in NP65. Trisomy did not significantly alter the incidence of lymphoma (11% in euploid, 18.8% in trisomy, P = 0.24 by Fisher’s exact test).
Brain lesions including astrocytoma and neuroblastoma have been observed in NP mice (13, 14). We screened brains microscopically to detect abnormal cell growth and anaplastic morphology (Fig. 2G). GFAP staining confirmed the diagnosis of astrocytoma (Fig. 2H) in all but two of the brain tumors, which had characteristics of neuroblastoma. The incidence was not significantly different between trisomic and euploid mice (6.4% euploid, 12.5% trisomy, P = 0.26 by Fisher’s exact test) and was lower than reported previously (25). This is due in part to the fact that we screened for tumors in brain but not in the spinal cord and thus would expect to see fewer than if both tissues were considered. In addition, the NPcis haplotype was always inherited from males in this study, whereas it has been observed that the progeny of males have less astrocytoma due to an unknown epigenetic factor (25).
A substantial fraction of mice had more than one tumor at the time they were sacrificed. This frequency was not affected by ploidy (Fisher’s exact test, P = 0.48) as 31 of 102 in euploid mice and 10 out of 26 trisomic mice developed more than one tumor (Supplementary Table 1).
Tumor growth properties are affected by trisomy
Extended survival of NP65 mice compared to their euploid counterparts can be explained in part by the shift in tumor types away from sarcoma and toward adrenal tumors, since median survival time with sarcoma was significantly shorter. We examined several additional growth parameters of tumors that could contribute to the extended survival of NP65 mice.
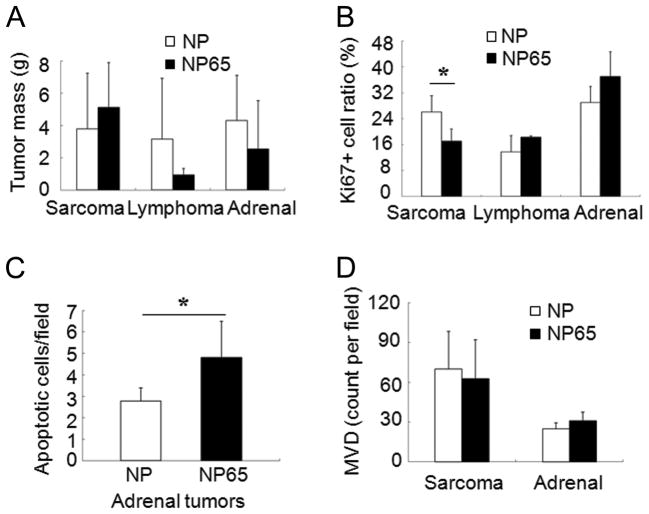
Tumor mass was highly variable even within a given type of tumor, so although some trends were recognized, tumor size was not significantly different between the NP and NP65 groups (Fig. 4A). Proliferation, measured as percent Ki67+ cells, was significantly reduced in sarcomas that occurred in trisomic mice compared to euploid (P =0.023). However, we did not observe a significant proliferation reduction in lymphomas or in adrenal tumors (Fig. 4B, Supplementary Fig. 4A). We found no difference in the frequency of apoptosis by TUNEL staining between NP and NP65 in either sarcoma or lymphoma; however, apoptosis was significantly elevated in adrenal tumors in trisomic mice (P = 0.04, Fig. 4C, Supplementary Fig. 4B). The average mass of adrenal tumors was somewhat lower in NP65 mice, although this trend was not statistically significant.
Figure 4. Growth characteristics of endogenous tumors.
A. The average mass of tumors of each type suggested trends in euploid versus trisomic mice, but differences were not statistically significant. Mice that developed multiple types of tumors are not included in this graph. For sarcoma, avg. mass for euploid is 3.80 g. (n=72) vs. trisomy, 5.11 g (n=10); lymphoma, euploid 3.16 g (n=11) vs. trisomy 0.94 g. (n=3); adrenal tumor, euploid 4.31 g (n=10) vs. trisomy 2.55 g (n=11). B. The percentage of proliferating, Ki67+ cells was significantly reduced in sarcomas in trisomic mice, but not different between euploid and trisomic mice in other tumor types (n = 17, 2 and 7 in euploid and 8, 2 and 5 in Ts65Dn for sarcoma, lymphoma and adrenal tumor, respectively). C. The mean number of apoptotic cells per field was 4.8 (n = 6) in trisomic adrenal tumors and 2.8 (n = 5) in euploid (T-test, *P = 0.037). D. MVD (avg. number of blood microvessels per field) was the same in sarcomas from euploid and trisomic mice (n = 28 euploid and 10 trisomic tumors, P= 0.52, T-test) and in adrenal tumors (n = 8 euploid and 8 trisomic tumors, P= 0.18 by T-test).
The extra chromosome was retained in trisomic tumors
Extended survival time of trisomic NPcis mice, the observation that trisomic mice carrying the ApcMin mutation form fewer intestinal tumors (9) and the reduction in mortality from cancer in individuals with DS (3) raises the question of whether loss of the extra chromosome is required as an early step in order for cells to become transformed. Trisomy in the Ts65Dn mouse model occurs due to an extra freely segregating chromosome as it does in most people with DS.
To isolate tumor DNA with minimal contamination by neighboring non-transformed cells, we took thick sections of paraffin-embedded tumors and removed central portions of the tumor. DNA was extracted and assessed for the presence of Trp53 and Nf1 genes. Since tumors are initiated by loss of heterozygosity, deleting the single copies of these genes, their presence is indicative of contamination of the tumor sample by non-transformed cells. As expected, we saw no or a significantly reduced signal for the wild type forms of both genes in all tumor samples (Supplementary Fig. 5).
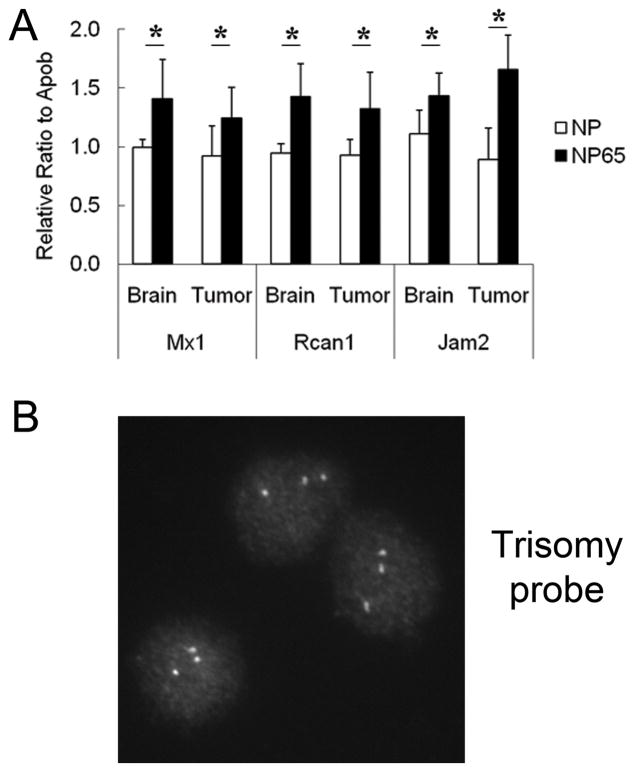
We then measured gene copy number as a surrogate for trisomy in NP65 tumors. DNA was extracted from six tumors (five with matched brain samples) from euploid mice, and from twelve tumor samples (ten with matched brain samples) from Ts65Dn mice. Three genes that span the trisomic chromosome in Ts65Dn, Jam2, Rcan1 and Mx1, were quantified using a Taqman assay and dosage levels for these genes were compared to a disomic control gene, Apob. We reasoned that the ratios of the Mmu16 genes to control should be 1:1 or 1.5:1 in non-transformed brain tissue of euploid or trisomic mice, respectively, and that the ratio in trisomic tumors would be 1:1 if the extra chromosome were lost systematically or very early in the transformation process and 1.5:1 if it were not.
The mean ratio of Mx1 to Apob in brain from one individual was arbitrarily set to 1.0 and all euploid mice showed a value close to 1.0, while trisomic mice showed an average ratio of 1.4 in brain and 1.2 in tumors (Fig. 5A). Rcan1 and Jam2 results followed a similar pattern. Rcan1:Apob ratios were near 1.0 in euploid and 1.4 in trisomic brain, with a ratio of 1.3 in trisomic tumors. Jam2:Apob ratios were 1.1 in euploid and 1.4 in trisomic brain, with a ratio of 1.6 in trisomic tumors.
Figure 5. Trisomic tumors retain higher dosage for chromosome 16.
A, Trisomic and euploid gene copy number was determined using Taqman assay. Normalized ratios of Mmu16 genes (Jam2, Rcan1 and Mx1) to control genes (Apob) were elevated in samples from both trisomic normal brain and tumor samples, indicating that extra Mmu16 information is retained in the tumors (*P < 0.05). B, Cells recovered from a disaggregated sarcoma that arose in Ts65Dn retain three copies of distal Mmu16 by FISH (more than 50 cells counted).
We then disaggregated a sarcoma from an NP65 mouse. FISH was done with a BAC that represents a Mmu16 segment contained in the Ts65Dn chromosome. Three signals were identified in 100% of the spreads examined (Fig. 5B).
The slightly lower than theoretical 1.5 level in trisomic tumors could suggest that some cells have lost the extra chromosome or that the chromosomes containing the control gene were differentially amplified relative to Mmu16 in aneuploid tumor cells. Even if some cells lose the marker chromosome, the observation that the overall ratio in trisomic tumors is significantly higher than that in euploid in all three comparisons and the results from FISH of primary tumor cells indicate that loss of the extra chromosome is not a necessary or frequent early event in tumors arising from trisomic cells.
Neither endogenous tumors nor xenografts showed angiogenesis deficits
Reduced growth of xenografts in Ts65Dn as compared to euploid hosts has been reported multiple times and ascribed to reduced angiogensis in tumors that arise in trisomic hosts. The reduction in angiogenesis has in turn been attributed to dosage effects of at least six different trisomic genes (7, 10, 28). NP65 mice provided the opportunity to examine angiogenesis in endogenous tumors arising in NP and NP65 mice, where not only the microenvironment, but also the transformed cells themselves were trisomic.
We identified capillaries in euploid and trisomic tumors with a CD31 antibody as described (Supplementary Fig. 6). MVD was determined using two different procedures, a widely employed “hot spot” method (29) and a general survey of vessel formation throughout the tumors. Similar results were obtained with either method. Contrary to expectations from previous xenograft experiments, we observed no difference in MVD between either sarcomas or adrenal tumors that arose in trisomic hosts compared to euploid (Fig. 4D).
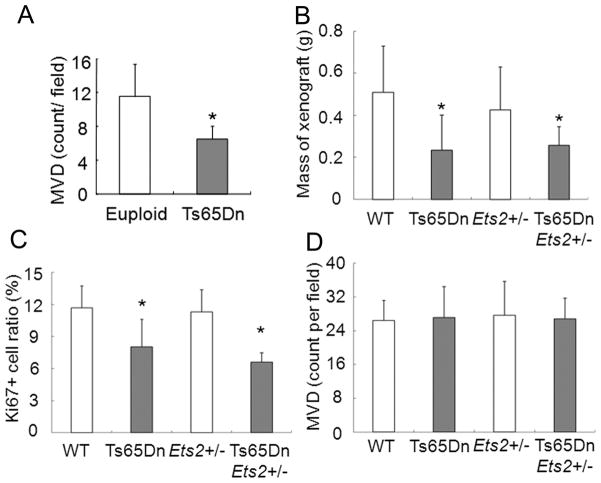
Given previous observations that Ts65Dn mice showed reduced MVD in xenografts produced from B16 and Lewis Lung Carcinoma cell lines, it was somewhat surprising that endogenous tumors in trisomic mice did not show any reduction in angiogenesis. We performed the xenograft assay using B16F10 cells (ATCC) to produce 14 xenografts in euploid and 16 in Ts65Dn mice. As reported, angiogenesis was reduced in B16 xenografts made in Ts65Dn mice (7) (Fig. 6A).
Figure 6. Characterization of xenografts in trisomic or euploid hosts.
For A, a total of 14 xenografts were made with B16F10 in euploid and 16 Ts65Dn mice. A, MVD in B16 xenografts is significantly lower in Ts65Dn than in euploid (P = 0.0001, T-test). For B, C and D, the number of tumors per host genotype are: euploid (WT), n = 16; Ets2+/−, n = 11; Ts65Dn, Ets2+/−, n = 4; and Ts65Dn, n = 9. B, TCL.NP xenografts in trisomic mice are significantly smaller than those in euploid hosts and the size is independent of Ets2 copy number in either host (Anova, F=4.555, *P = 0.0083). C, The fraction of dividing cells is significantly reduced in TCL.NP xenografts into Ts65Dn hosts. (Anova, F=9.552, *P = 0.0001). D, MVD is not different among TCL.NP xenografts in the four host genotypes.
To extend this observation, we carried out a xenograft experiment with two independent sarcoma cell lines, TCL.Np1 and TCL.Np2, that we established from sarcomas originating in euploid NP mice. Sixteen xenografts of TCL.Np1 or 2 were produced in euploid mice and nine in Ts65Dn. We found the tumor mass of TCL.Np xenografts in Ts65Dn was significantly less than in euploid mice (average mass of 256.3 mg vs. 508.5 mg, P = 0.0006) (Fig. 6B). The mass reduction in Ts65Dn was correlated with a reduced Ki67+ cell ratio, i.e., Ts65Dn hosts had a significantly lower percentage of dividing cells than did the larger tumors in euploid hosts (Fig. 6C). We found no difference in MVD between tumors arising in trisomic or euploid hosts (Fig. 6D). TUNEL staining also did not reveal a significant difference in the level of apoptosis between the different hosts. Thus it appears that proliferation rates and not inhibition of angiogenesis contribute to reduced xenograft growth in Ts65Dn for these newly derived, relatively low passage sarcoma cell lines.
In further experiments, we also compared growth of xenografts of the TCL.Np1 and 2 tumor cell lines in NP, ENP, NP65 and ENP65 mice. We observed no effect of Ets2 dosage on tumor mass or MVD (Fig. 6B–D).
Discussion
Epidemiological studies of DS suggest that trisomic individuals are protected against multiple types of cancer (3, 9). Here, we provide biological evidence for this protective effect. The NPcis mouse develops sarcomas, carcinomas and lymphomas (13) and mortality is 100% in this extremely aggressive cancer model. Trisomy did not reduce the penetrance of tumorigenesis, however, lifespan was extended significantly.
The reasons for extended survival were complex. One reason for this pattern was that Ts65Dn mice were resistant to sarcoma. The resulting shift in the pattern of tumors away from a rapidly lethal sarcoma that represented the majority of tumors in euploid mice meant that trisomic mice were likely to survive longer. It is worth noting that the peripheral nerve sheath is a neural crest derived tissue and in Ts65Dn mice, reduced delamination, migration and proliferation of cranial crest underlies the mid-face skeletal hypoplasia characteristic of these mice and of DS (30). Regardless, this shift away from sarcoma was not the whole explanation for the cancer phenotype, since trisomic mice with tumor types other than sarcoma (30) survived longer than their euploid littermates without sarcoma (Fig. 3, Supplementary Fig. 3).
Repression of tumorigenesis by trisomy could involve any of a number of mechanisms, and two possible sources of protection have been described previously. Increased expression for a number of genes on Hsa21 has been correlated with reduced angiogenesis of xenografts implanted in trisomic as compared to euploid hosts (7, 10, 28, 31). As a protective mechanism, reduced angiogenesis would likely affect later stages of tumor progression and metastasis and thus might be expected to be associated more with reduced mortality than with reduced incidence of cancer. We found that the effect of trisomy on angiogenesis in xenografts was cell line specific. Newly established tumor cell lines that grow more slowly than the B16 and LLC lines traditionally used in these studies did not show a reduction in MVD when grafted into trisomic hosts compared to euploid. Instead, we observed that reduced xenograft mass in trisomic hosts was correlated with reduced cell proliferation relative to the same tumors in euploid mice. Of more relevance, endogenous tumors showed no differences in angiogenesis between trisomic and euploid mice.
A second protective mechanism more tightly linked to incidence can be seen in the occurrence of intestinal tumors in ApcMin mice. Tumor load is significantly reduced in Ts65Dn mice carrying this mutation and this outcome is closely associated with dosage of the Ets2 gene (9). We posited that increasing Ets2 dosage in trisomic cells might sensitize cells to Trp53-mediated apoptosis (32), thereby enhancing elimination of tumorigenic cells very early in the process of transformation. This mechanism would not be operative in tumors formed in the NPcis model, since loss of Trp53 is an initiating step in tumor formation. Indeed, Ets2 dosage made no contribution to survival in NPcis mice. Further, we found no effect of Ets2 dosage on tumor mass or MVD in xenografts of NP tumor cell lines on a euploid or trisomic background.
We also examined the possibility that loss of the extra chromosome is a necessary step in neoplastic transformation of trisomic cells. Any additional step in the transformation process would be assumed to reduce or delay the onset of tumorigenesis; indeed, removing one such barrier to transformation is the basis of the high tumor frequency in the NPcis model. However, trisomic sarcomas retained a relative gene dosage of trisomic to disomic genes of ~1.5 and also retained the supernumerary chromosome by FISH analysis.
The NPcis cancer model is highly complex, and the responses in trisomic mice are equally so, suggesting that protection against cancer mortality in DS is influenced by multiple trisomic genes affecting a number of cellular properties. Lifespan was significantly extended in trisomic mice with a variety of cancers and this extension was independent of known mechanisms of trisomic tumor repression. The occurrence of tumor repression in DS provides an important approach to cancer prophylaxis that has been identified thanks to the genetic legacy of persons with Down syndrome.
Supplementary Material
Acknowledgments
This work was supported by PHS award 1R01 HD038384 from the National Institute of Child Health and Human Development and the National Cancer Institute (RHR).
The authors thank Karlyne Reilly for providing NPcis mice and for many thoughtful suggestions during the course of this work. Cory Brayton provided important insights into tumor pathology.
References
- 1.Zipursky A, Poon A, Doyle J. Leukemia in Down syndrome: a review. Pediatr Hematol Oncol. 1992;9:139–49. doi: 10.3109/08880019209018329. [DOI] [PubMed] [Google Scholar]
- 2.Satge D, Sommelet D, Geneix A, Nishi M, Malet P, Vekemans M. A tumor profile in Down syndrome. Am J Med Genet. 1998;78:207–16. [PubMed] [Google Scholar]
- 3.Yang Q, Rasmussen SA, Friedman JM. Mortality associated with Down’s syndrome in the USA from 1983 to 1997: a population-based study. Lancet. 2002;359:1019–25. doi: 10.1016/s0140-6736(02)08092-3. [DOI] [PubMed] [Google Scholar]
- 4.Moore CS, Roper RJ. The power of comparative and developmental studies for mouse models of Down syndrome. Mamm Genome. 2007;18:431–43. doi: 10.1007/s00335-007-9030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardiner K, Fortna A, Bechtel L, Davisson MT. Mouse models of Down syndrome: how useful can they be? Comparison of the gene content of human chromosome 21 with orthologous mouse genomic regions. Gene. 2003;318:137–47. doi: 10.1016/s0378-1119(03)00769-8. [DOI] [PubMed] [Google Scholar]
- 6.Reeves R, Irving N, Moran T, Wohn A, Kitt C, Sisodia S, et al. A mouse model for Down Syndrome exhibits learning and behaviour deficits. Nature Genetics. 1995;11:177–83. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 7.Baek KH, Zaslavsky A, Lynch RC, Britt C, Okada Y, Siarey RJ, et al. Down’s syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature. 2009;459:1126–30. doi: 10.1038/nature08062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ergun S, Kilic N, Wurmbach JH, Ebrahimnejad A, Fernando M, Sevinc S, et al. Endostatin inhibits angiogenesis by stabilization of newly formed endothelial tubes. Angiogenesis. 2001;4:193–206. doi: 10.1023/a:1014027218980. [DOI] [PubMed] [Google Scholar]
- 9.Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down’s syndrome. Nature. 2008;451:73–5. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds LE, Watson AR, Baker M, Jones TA, D’Amico G, Robinson SD, et al. Tumour angiogenesis is reduced in the Tc1 mouse model of Down’s syndrome. Nature. 2010;465:813–7. doi: 10.1038/nature09106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minami T, Horiuchi K, Miura M, Abid MR, Takabe W, Noguchi N, et al. Vascular endothelial growth factor- and thrombin-induced termination factor, Down syndrome critical region-1, attenuates endothelial cell proliferation and angiogenesis. J Biol Chem. 2004;279:50537–54. doi: 10.1074/jbc.M406454200. [DOI] [PubMed] [Google Scholar]
- 12.Wolvetang EJ, Bradfield OM, Hatzistavrou T, Crack PJ, Busciglio J, Kola I, et al. Overexpression of the chromosome 21 transcription factor Ets2 induces neuronal apoptosis. Neurobiol Dis. 2003;14:349–56. doi: 10.1016/s0969-9961(03)00107-4. [DOI] [PubMed] [Google Scholar]
- 13.Cichowski K, Shih TS, Schmitt E, Santiago S, Reilly K, McLaughlin ME, et al. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–6. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 14.Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, Parada LF. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–9. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reilly KM, Loisel DA, Bronson RT, McLaughlin ME, Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26:109–13. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- 16.Wei G, Srinivasan R, Cantemir-Stone CZ, Sharma SM, Santhanam R, Weinstein M, et al. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009;114:1123–30. doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore CS, Lee JS, Birren B, Stetten G, Baxter LL, Reeves RH. Integration of cytogenetic with recombinational and physical maps of mouse chromosome 16. Genomics. 1999;59:1–5. doi: 10.1006/geno.1999.5812. [DOI] [PubMed] [Google Scholar]
- 18.Lorenzi H, Duvall N, Cherry SM, Reeves RH, Roper RJ. PCR prescreen for genotyping the Ts65Dn mouse model of Down syndrome. Biotechniques. 2010;48:35–8. doi: 10.2144/000113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speicher MR, du Manoir S, Schrock E, Holtgreve-Grez H, Schoell B, Lengauer C, et al. Molecular cytogenetic analysis of formalin-fixed, paraffin-embedded solid tumors by comparative genomic hybridization after universal DNA-amplification. Hum Mol Genet. 1993;2:1907–14. doi: 10.1093/hmg/2.11.1907. [DOI] [PubMed] [Google Scholar]
- 20.Liu D, Schmidt C, Billings T, Davisson MT. Quantitative PCR Protocol [homepage on the Internet] 2011 [cited 2010 Jan 10]; Available from: http://www.jax.org/cyto/quanpcr.html.
- 21.ABI. Custom TaqMan® Assay Design Tool [homepage on the Internet] 2009 [cited 2010 Jan 10]; Available from: https://www5.appliedbiosystems.com/tools/cadt/
- 22.R-Development-Core-Team. R: A language and environment for statistical computing [computer software] 2010 [cited 2010 Jun 10]; Available from: http://www.r-project.org/
- 23.Lowry R. Concepts & Applications of Inferential Statistics [homepage on the Internet] c1998–2011 [cited 2010 Nov 6]; Available from: http://faculty.vassar.edu/lowry/webtext.html.
- 24.Lenth RV. Java Applets for Power and Sample Size [Computer software] c2006–09 [cited 2011 Feb 23]; Available from: http://www.stat.uiowa.edu/~rlenth/Power.
- 25.Reilly KM, Tuskan RG, Christy E, Loisel DA, Ledger J, Bronson RT, et al. Susceptibility to astrocytoma in mice mutant for Nf1 and Trp53 is linked to chromosome 11 and subject to epigenetic effects. Proc Natl Acad Sci U S A. 2004;101:13008–13. doi: 10.1073/pnas.0401236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walrath JC, Fox K, Truffer E, Gregory Alvord W, Quinones OA, Reilly KM. Chr 19(A/J) modifies tumor resistance in a sex- and parent-of-origin-specific manner. Mamm Genome. 2009;20:214–23. doi: 10.1007/s00335-009-9179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stemmer-Rachamimov AO, Louis DN, Nielsen GP, Antonescu CR, Borowsky AD, Bronson RT, et al. Comparative pathology of nerve sheath tumors in mouse models and humans. Cancer Res. 2004;64:3718–24. doi: 10.1158/0008-5472.CAN-03-4079. [DOI] [PubMed] [Google Scholar]
- 28.Folkman J. Angiogenesis-dependent diseases. Semin Oncol. 2001;28:536–42. doi: 10.1016/s0093-7754(01)90021-1. [DOI] [PubMed] [Google Scholar]
- 29.Ryeom S, Baek KH, Rioth MJ, Lynch RC, Zaslavsky A, Birsner A, et al. Targeted deletion of the calcineurin inhibitor DSCR1 suppresses tumor growth. Cancer Cell. 2008;13:420–31. doi: 10.1016/j.ccr.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 30.Roper RJ, VanHorn JF, Cain CC, Reeves RH. A neural crest deficit in Down syndrome mice is associated with deficient mitotic response to Sonic hedgehog. Mech Dev. 2009;126:212–9. doi: 10.1016/j.mod.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zorick TS, Mustacchi Z, Bando SY, Zatz M, Moreira-Filho CA, Olsen B, et al. High serum endostatin levels in Down syndrome: implications for improved treatment and prevention of solid tumours. Eur J Hum Genet. 2001;9:811–4. doi: 10.1038/sj.ejhg.5200721. [DOI] [PubMed] [Google Scholar]
- 32.Wolvetang EJ, Wilson TJ, Sanij E, Busciglio J, Hatzistavrou T, Seth A, et al. ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum Mol Genet. 2003;12:247–55. doi: 10.1093/hmg/ddg015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.