Abstract
We studied ubiquinone (Q), Q homologue ratio and steady-state levels of mCOQ transcripts in tissues from mice fed ad libitum or under calorie restriction. Maximum ubiquinone levels on a protein basis were found in kidney and heart, followed by liver, brain and skeletal muscle. Liver and skeletal muscle showed the highest Q9/Q10 ratios with significant inter-individual variability. Heart, kidney, and particularly brain exhibited lower Q9/Q10 ratios and inter-individual variability. In skeletal muscle and heart, the most abundant mCOQ transcript was mCOQ7, followed by mCOQ8, mCOQ2, mPDSS2, mPDSS1 and mCOQ3. In non muscular tissues (liver, kidney and brain) the most abundant mCOQ transcript was mCOQ2, followed by mCOQ7, mCOQ8, mPDSS1, mPDSS2, and mCOQ3. Calorie restriction increased both ubiquinone homologues and mPDSS2 mRNA in skeletal muscle, but mCOQ7 was decreased. In contrast, Q9 and most mCOQ transcripts were decreased in heart. Calorie restriction also modified Q9/Q10 ratio, which was increased in kidney and decreased in heart without alterations of mPDSS1 or mPDSS2 transcripts. We demonstrate for the first time that unique patterns of mCOQ transcripts exist in muscular and non-muscular tissues, and that Q and COQ genes are targets of calorie restriction in a tissue-specific way.
Keywords: Calorie restriction, COQ transcripts, Ubiquinone, Ubiquinone biosynthesis
1. Introduction
Ubiquinone (coenzyme Q, Q) is a prenylated benzoquinone which plays a fundamental role shuttling electrons from Complex I and Complex II to Complex III in the mitochondrial electron transport chain [1,2]. Other functions of Q include the mitochondrial oxidation of dihydroorotate in the pyrimidine biosynthesis pathway [3,4], protection against oxidative stress as the only lipid-soluble antioxidant which is synthesized in all organisms [1,2], and electron transport as a carrier in the transplasma membrane redox system [5]. A number of proposed functions for Q are less defined from a molecular point of view. These include modulating the sensitivity of mitochondrial uncoupling proteins (UCPs) to purine nucleotides by Q redox state, regulating the mitochondrial permeability transition pore activity [7], and inhibiting plasma membrane neutral sphingomyelinase [8]. Q is positioned in a privileged position because it links basic aspects of cell physiology such as energy metabolism, antioxidant protection, and the regulation of cell growth and death.
Although dietary Q is incorporated to some extent in particular tissues, under normal conditions its de novo synthesis accounts for the majority of Q present in peripheral tissues [1,2,9,10]. The precursor of the benzoquinone ring is 4-hydroxybenzoate, whereas the synthesis of the isoprenoid side chain starts from acetyl-coenzyme A to yield farnesyl pyrophosphate (FPP) through the mevalonate pathway [1,2,11]. The trans-prenyltransferase activity, a tetramer composed of PDSS1 and PDSS2 gene products [12,13], catalyzes the condensation of FPP with several units of isopentenyl pyrophosphate (IPP), all in trans configuration, to form a long polyisoprenoid chain whose length, 9 or 10 isoprene units in mammals, determines the actual isoform which is synthesized (Q9 or Q10, respectively) [1,2]. Then, 4-hydroxybenzoate and polyprenyl pyrophosphate are condensed by polyprenyl 4-hydroxybenzoate transferase, encoded by COQ2 [14]. After condensation, at least six enzymes (encoded by COQ3-8) catalyze C-hydroxylations, decarboxylation, O-methylations, and C-methylation to synthesize Q [1,2].
Q distribution is not uniform among different tissues and organs, indicating that Q levels are adapted to the particular physiology of the tissue. Since Q is rapidly broken down, as reflected in its short half-life, actual levels of Q in tissues are likely determined by a coordinated balance between synthesis and degradation, both of which occur in all tissues [1,2]. In mice, rats and humans, maximal Q concentrations are present in kidney and heart, while lower amounts can be detected in liver, brain and skeletal muscle [1,15-19]. Since Q redistribution between organs appears insignificant [2], tissue-specific mechanisms must exist to determine actual levels of Q in any given tissue. However, the control of Q distribution in tissues is not fully understood.
Since Q plays a central role in energy metabolism, alterations in Q biosynthesis may participate in adaptive responses to physiological, experimental or pathological conditions shown to modify Q levels in tissues. Q is increased under cold adaptation and with exercise, while it may be decreased by aging, at least in some tissues [2]. Calorie restriction (CR) is the most robust intervention that improves healthspan and extends lifespan in many animal models [20-22]. Thus, previous investigations have also studied putative changes of Q levels with CR. In this regard, it has been reported that long-term CR increases total Q in mitochondria from skeletal muscle, liver, heart and kidney [23,24].
Because of the complex pathway leading to Q synthesis, simultaneously examining Q levels and several COQ mRNAs or proteins in different tissues can give us novel insights to understand genetic factors which determine tissue-specific distribution of Q and its modulation during metabolic adaptation. However, previous studies have focused on single COQ genes under normal conditions, and data are only available for COQ3 [10], COQ2 [14] and COQ7 [25-28]. The possibility that CR influences Q biosynthesis has not yet been investigated, since none of the previous studies examining Q changes with CR were focused on COQ mRNAs or proteins. We hypothesized that specific patterns of COQ mRNA levels might be identified in tissues, and that these mRNA patterns could be modulated by CR. To test this hypothesis we systematically evaluated the profile of Q9 and Q10 and steady-state COQ mRNAs levels in different tissues of animals fed ad libitum (AL) or under CR. We demonstrate for the first time that muscular and non-muscular tissues display unique patterns of mCOQ transcripts, and that Q and COQ genes are targets of CR in a tissue-specific way, supporting the idea that alterations of Q biosynthesis are part of the metabolic adaptation to CR in mice.
2. Materials and Methods
Unless otherwise indicated, all chemicals were purchased from Sigma (Spain).
2.1. Animals, tissue samples and ethics statement
A cohort of 32 ten week old C57BL/6 were used (Charles River Laboratories, Spain). Male C57BL/6 mice were bred and raised in a vivarium at the Centro Andaluz de Biología del Desarrollo (CABD) under a 12h light/dark cycle (8:00 AM-8:00 PM) under controlled conditions of temperature (22 ± 3°C) and humidity. The animals were fed an AIN93G diet. Food intake was measured during 1 month to determine the average amount of food consumed per day. At 3 months of age, animals were randomly divided into AL (95% of average diary intake) or CR (60% average diary intake) groups. Filtered and acidified water was available ad libitum for all groups and food was replaced with every day between 8:00-9:00 am. Animals were fed for 1 month under these conditions, and then sacrificed after an 18 hour fast by cervical dislocation. Samples from liver, kidney, brain, skeletal muscle (gastrocnemius) and heart were taken, frozen in liquid nitrogen, and stored at −80°C for later analysis. Animals were handled in accordance with the Pablo de Olavide University Ethical Committee rules, and the 86/609/EEC Directive on the protection of animals used for experimental and other scientific purposes.
2.2. Quantification of steady-state levels of COQ transcripts
Quantification of mRNA steady-state copy number was carried out by quantitative real time RT-PCR (qrtRT-PCR), where samples were run against synthetic standards. Total RNA was obtained from tissues using the Absolutely RNA RT-PCR Miniprep Kit (Stratagene, La Jolla, CA) and then reverse transcribed using the cDNA First Strand Synthesis kit (MRI Fermentas, Hanover, MD). Primers were selected using genomic sequences obtained from Genbank (National Center for Biotechnology Information (NCBI)) and Primer 3 software [29]. Specific primer sequences for COQ and housekeeping genes (glyceraldehyde-3-phosphate dehydrogenase – mGAPDH, hypoxanthine ribosyltransferase – mHPRT, and 18S ribosomal RNA – m18S) measured in this study, with indication of GenBank accession numbers and product sizes, are given in Table I.
Table I.
Primers for qrtRT-PCR used in the study
| PRIMERS | SENSE (5′-3′) | ANTI-SENSE (5′-3′) | PRODUCT SIZE (BP) |
GENBANK ACCESSION NO. |
|---|---|---|---|---|
| mHPRT | CAGTCAACGGGGGACATAAA | AGAGGTCCTTTTCACCAGCAA | 183 | NM_013556 |
| mGAPDH | ATGGCCTTCCGTGTTCCTAC | GCCTGCTTCACCACCTTCTT | 104 | XM_194302 |
| m18S | TGACTCAACACGGGAAACCT | AACCAGACAAATCGCTCCAC | 125 | X00686 |
| mSPS1 | CATCAAAGGACACCAGCAATGT | GCACCACAATAATCGGTCTAAAGG | 208 | NM_019501 |
| mDLP1 | ATGCTGACCTCCAGCCTTTT | GTCACACCTTTGCCAGCTTT | 196 | NM_027772 |
| mCOQ2 | GCCCACCAGGACAAGAAAGAC | AGCCACAGCAGCGTAGTAGG | 171 | NM_027978 |
| mCOQ3 | GTGAGCCACCTGGAAATGTT | CCCACGTATGAGTGCCTTTT | 163 | NM_172687 |
| mCOQ7 | TGATGGAAGAGGACCCTGAGAAG | GCCTGTATCGTGGTGTTCAAGC | 89 | NM_009940 |
| mCOQ8 | AGCAAGCCACACAAGCAGATG | CCAGACCTACAGCCAGACCTC | 106 | NM_023341 |
Primers were used in a standard PCR with cDNA from the tissue of interest as template using the 2× Master Mix PCR reagent (MRI Fermentas). Products were run on agarose gels to confirm that only one band was amplified and no primer dimers were formed. PCR products were then column-purified (QIAGEN, Valencia, CA) and sequenced to confirm target specificity. The concentration of purified products generated by standard PCR was then determined using Molecular Probe’s Picogreen DNA quantification kit, and serial dilutions of PCR products were carried out to obtain standards containing 101, 102, 103, 104, 105, 106 and 108 copies of synthetic template. To determine the starting copy number of cDNA, samples were amplified by qrtRT-PCR using SYBR PCR Master Mix (Bio-Rad Laboratories, Hercules, CA). Thermocycling and fluorescence detection was performed according to standard protocols using a Bio-Rad real-time PCR machine. The signal obtained from a given tissue cDNA was compared with that of a standard curve run on the same plate. In addition, total RNA samples that were not reverse transcribed and a no-DNA control were run on each plate to control for genomic DNA contamination and to monitor potential exogenous contamination, respectively.
To determine if the selected housekeeping genes were appropriate as internal controls, the stability of expression was calculated using the GeNorm 3.3 Visual basic application for Microsoft Excel (http://medgen.ugent.be/~jvdesomp/genorm/) to calculate the average pairwise variation of a particular gene with all other control genes (M value) as previously developed and validated [30]. In our study, calculation of the M values for the three control genes assessed revealed that mGAPDH and m18S levels were the least stable under all conditions. Therefore, M values were recalculated using mHPRT where all M values were <1.5. Therefore, the copy numbers for this gene within each sample were used as normalization factors (NF). Results were then reported as [median (minimum–maximum) of COQ genes copy number minus background] / NF.
2.3. Lipid extraction and Q quantification
A total lipid fraction was obtained from the tissues (about 30 mg fresh weight) by homogenization in 90 μl of Hank’s solution to which 10 μl of 10% SDS had been added to facilitate disruption of the membranes. Afterwards, two volumes of 95% ethanol–5% isopropanol were added and the mixture was vigorously vortexed. Q was recovered from SDS-alcoholic solution by extraction with five volumes of hexane. After hexane evaporation under vacuum, the lipid extract was dissolved in a 1:1 mixture of methanol and n-propanol and then used for quantification of Q9 and Q10 by reversed-phase HPLC separation with a C18 column (25 × 0.45 cm, 5 μm particle size).
HPLC separation was accomplished at 1 ml/min with a mobile phase composed of a mixture of methanol and n-propanol (1:1) containing lithium perchlorate (2.12 g/L). Monitoring was carried out with a Coulochem II electrochemical detector (ESA, Chelmsford, MA) fitted with a Model 5010 analytical cell with the electrodes set at potentials of −500 mV and +300 mV. Quinones were detected from the signal obtained at the second electrode. Concentrations were calculated by integration of peak areas and comparison with external standards (Sigma, Madrid, Spain).
2.4. SDS-PAGE and immunodetection of Coq2p and Coq3p
For immunodetection of Coq2p and Coq3p polypeptides, protein (about 100 μg) was denatured by heating at 45°C during 15 min in SDS-dithiothreitol loading buffer (10% sucrose, 2 mM EDTA, 1.5% (w/v) SDS, 20 mM dithiothreitol, 0.01% (w/v) bromophenol blue and 60 mM Tris–HCl (pH 6.8)) containing 1 mM PMSF and 20 μg/μl each of chymostatin, leupeptin, antipain and pepstatin A. Proteins were separated by SDS-PAGE (10% acrylamide) and then transferred to nitrocellulose sheets. Blots were stained with Ponceau S for visualization of protein lanes. Density of staining with Ponceau’s was used as a control for protein loading as described [31]. Immunostaining of western blots was carried out with chicken polyclonal anti-sera raised against the protein of interest and diluted at 1:2000. A horseradish peroxidase-conjugated anti-chicken IgG secondary antibody (Sigma) diluted at 1:5000 was used to reveal binding sites by enhanced chemiluminiscence. Photographic films and Ponceau S-stained blots were scanned in a GS-800 calibrated densitometer (Bio-Rad) to obtain digital images. Quantification of intensity reaction was carried out using Quantity One software (Bio-Rad). Data obtained from quantification of stained bands (in arbitrary units) were normalized to that of the corresponding lane stained with Ponceau S in order to correct for any difference in protein loading between samples.
2.5. Statistical analysis
For gene expression and Q quantification studies, samples from all conditions within an experiment were processed at the same time, and therefore the in vivo effects of diet were assessed by Student’s t test (vs. corresponding controls). The effects of diet on different tissues in C57BL/6 mice were assessed by one-way ANOVA followed by analysis of significant differences with Tukey’s test for multiple comparisons. In case this test was found inappropriate for our purposes we used Duncan’s non-parametric test. P < 0.05 was considered significant. All values are expressed as mean ± SEM. All statistical analyses were performed using the SPSS 11.0 software package (Chicago, USA) and STATISTICA 8.0. P values less than 0.05 were considered significant.
3. Results and Discussion
3.1. Ubiquinone distribution in mouse tissues
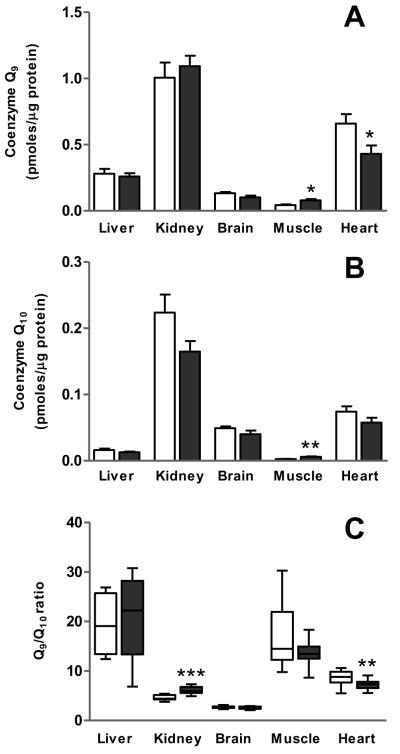
We first evaluated Q distribution in different mouse tissues. For this study we chose both mitotic (liver, kidney) and postmitotic (brain, skeletal muscle, heart) tissues and organs. As depicted in Figure 1A, the highest concentration of Q9 on a protein basis was found in kidney, followed by heart, liver, brain and skeletal muscle. In the case of Q10, the order (from maximal to minimal Q concentration) was kidney, heart, brain, liver, and skeletal muscle (Figure 1B). Differences in Q distribution among tissues were very high, which agrees with previous reports for the same tissues and organs obtained from mice [16-18] or rats [19]. For instance, when we compared kidney versus skeletal muscle, differences could be as high as 20-fold in the case of Q9 and 100-fold in the case of Q10. Lower content of Q in gastrocnemius muscle than in heart was expected since gastrocnemius contains abundant fast-twitch glycolytic fibres and much less mitochondria than heart [32]. In accordance, differences between skeletal muscle and heart are much lower when measuring Q9 and Q10 in isolated mitochondria [18,19,23].
Figure 1.
Tissue distribution of Q homologues and Q9/Q10 ratio. Tissues (liver, kidney, brain, skeletal muscle and heart) were taken from 4 mo old mice that had been fed ad libitum or under 1 mo CR. Quinones were recovered by organic extraction with hexane, and separated and quantified by HPLC. A) Q9 levels. B) Q10 levels. C) Q9/Q10 ratio. Data are means ± SEM (n=16). Significant differences between diets (ad libitum vs CR) are indicated by asterisks (* p<0.05 ; ** p<0.01 ; ***p<0.001). Open bars, ad libitum; Closed bars : CR.
Q9/Q10 ratios also displayed significant variability among tissues and organs in the mouse. Highest Q9/Q10 ratios were observed in liver and skeletal muscle (mean values of about 19 and 15, respectively), although inter-individual variability was also very high. A lower Q9/Q10 ratio (mean value of 8.6) was found in heart, which also showed lower individual variability. Kidney and brain exhibited both the lowest Q9/Q10 ratios (mean values of 4.6 and 2.7, respectively) and the lowest variability among individuals, particularly in the case of brain, where all Q9/Q10 values were contained within an extremely narrow range (Figure 1C). This observation supports the idea that the homeostatic range for Q isoforms varies among tissues and organs, and the maintenance of a given Q9/Q10 ratio may be important for proper function of organs, particularly brain, showing a relatively high Q10 content.
3.2. Steady-state levels of COQ transcripts in tissues
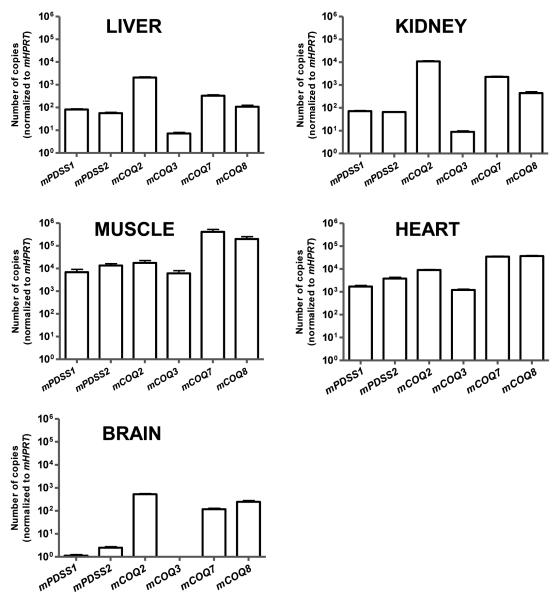
One of our major aims was to systematically evaluate steady-state levels of COQ transcripts in different mouse tissues. No previous study has applied qrtRT-PCR to provide a reliable estimate of steady-state levels of several COQ transcripts. Previous studies have also not investigated the relationship between levels of several COQ transcripts and Q distribution in the same tissues. We focused our study on genes encoding proteins of known or putative function in Q biosynthesis for which inactivating mutations that result in Q deficiency have been observed in humans [2]. These included mPDSS1 and mPDSS2, the two subunits of polyprenyl diphosphate synthase; mCOQ2, the polyprenyl-4-hydroxybenzoate transferase which catalyzes the first enzymatic step that is specific for Q biosynthesis, and mCOQ8 /ADCK3, a putative kinase [1,2]. In addition, we also included in our study two additional COQ genes with well-established function in Q biosynthesis: mCOQ3 and mCOQ7. COQ3 encodes 2,3-dihydroxy-5-polyprenylbenzoate-methyl transferase, which is responsible for the two O-methylations steps in the production of Q, the last of these yielding the final product [10,33,34]. COQ7/Clk-1 encodes an hydroxylase which catalyzes the conversion of 5-demethoxy-Q to hydroxy-Q, the penultimate step of Q biosynthesis [35], and plays a prominent role in determining life span in vertebrate and invertebrate animal models [36].
We demonstrate here for the first time that tissues can be grouped according to steady-state levels of COQ transcripts (Figure 2) into two basic patterns: muscular and non muscular. Among the transcripts studied, mCOQ7 was the most abundant in gastrocnemius and heart, followed by mCOQ8, mCOQ2, mPDSS2, mPDSS1 and mCOQ3. However, despite exhibiting a similar pattern of mCOQ transcripts, the two muscular tissues differed by almost one order of magnitude in Q levels (see Figures 1 and 2). The striking similarity between the transcript patterns of gastrocnemius and heart supports the idea that the difference of Q content is due to post-transcriptional mechanisms which drive Q biosynthesis rate, the amount of mitochondria, and probably the catabolic rates of Q degradation [2]. The two mitotic organs studied, liver and kidneys, displayed an almost identical pattern of steady-state COQ transcripts levels. Maximal abundance was observed for mCOQ2, followed by mCOQ7, mCOQ8, mPDSS1, mPDSS2, and mCOQ3. The pattern of COQ transcripts in brain was similar to those of liver and kidneys, although the absolute number of copies of COQ transcripts was lower (Figure 2). Of note, compared with other mCOQ mRNAs, steady-state levels of mCOQ3 transcripts were very low in non muscular tissues.
Figure 2.
Patterns of steady-state levels of mCOQ transcripts in mouse tissues. RNA samples were extracted from liver, kidney, brain, skeletal muscle and heart obtained from control animals (fed ad libitum), and then used for reverse transcription and quantification of the absolute number of mRNA copies by qrtRT-PCR. Primers are depicted in Table I. Results obtained for mCOQ mRNAs were normalized to those of mHPRT as internal control. Data are means ± SEM (n=16).
Very few studies have addressed the tissue distribution of COQ transcripts, and none have analyzed simultaneously several COQ genes by means of a quantitative and reliable technique, such as qrtRT-PCR. Previous studies have focused on single COQ genes, and data are only available for COQ2, COQ3, and COQ7. Forsgren et al. [14] cloned the human COQ2 gene and studied tissue distribution of hCOQ2 transcript using dot blot with a radioactive probe in a commercial array for human mRNA. Maximal levels of hCOQ2 mRNA were detected in muscle and heart tissue, which agrees with our results. However, the abundance of hCOQ2 transcript was considerable lower in kidney, as well as in other non muscular tissues. Here we show that mCOQ2 transcripts can be found at comparable levels in kidney and muscular tissues. This discrepancy could be due to species specificity (human vs mouse). The study of Marbois et al. [10] was focused on tissue distribution of COQ3 mRNA in the rat, using Northern blot with a radioactive probe. Steady-state levels of COQ3 mRNA were studied in brain, heart, kidney, liver, skeletal muscle and testis. Rat COQ3 mRNA was particularly abundant in testis, heart and thigh muscle, followed in order of abundance by kidney, brain and liver, which agrees with our study. Finally, a number of studies have addressed the distribution of COQ7 mRNA in several human and mouse tissues. These studies have demonstrated that hCOQ7 mRNA is found predominantly in heart and skeletal muscle, whereas very low levels are present in brain, placenta, liver, kidney, pancreas and lung [25,37]. In mice, mCOQ7 mRNA was particularly abundant in heart, skeletal muscle and kidney [27], which also agrees with our results.
We report here that the mRNAs encoding for the two subunits of the polyprenyl diphosphate synthase (mPDSS1 and mPDSS2) were particularly abundant in muscular tissues. PDSS1 and PDSS2 have been proposed to play an important role in determining Q levels because the polyprenyl diphosphate synthase is the branch-point enzyme that utilizes FPP in the terminal part of Q biosynthesis, a process which is considered limited by the availability of polyisoprenoid chain precursor [2]. However, steady-state levels of mPDSS1 and mPDSS2 transcripts were similar in kidney and liver, two organs whose Q content differed substantially (see Figure 1), suggesting the existence of post-transcriptional levels of regulation. It is also possible that a limited amount of mPDSS1 and mPDSS2 transcripts is sufficient to provide enough polyisoprenoid chain precursor in these two mitotic organs, the higher concentration of Q in kidney being determined by a higher amount of mitochondria [2]. Transgenic mice harboring an overexpression of mutated PDSS2 isoform showed a kidney defect associated with Q deficiency that was partially rescued by Q supplementation [13].
3.3. Tissue patterns of Coq2 and Coq3 polypeptides
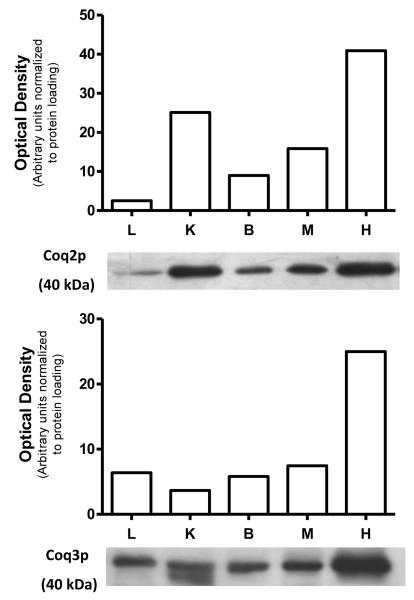
Of note, mCOQ3 transcripts varied substantially among the different tissues irrespective of their Q levels (see Figures 1 and 2). The lack of correlation between mCOQ3 transcripts and Q levels in tissues has been explained on the basis of the existence of post-transcriptional controls [10]. However, no study has been designed to date to test this hypothesis. Thus, we were interested in analyzing tissue levels of mCoq3 polypeptide to investigate if protein levels followed those of mRNA. Our study at the protein level also included mCoq2p. Although increased in kidney, heart and skeletal muscle, steady-state levels of mCOQ2 in tissues were more homogeneous than those of mCOQ3 (see Figure 2).
As depicted in Figure 3, mCoq2p levels were higher in tissues and organs which showed the highest levels of mCOQ2 transcripts. Maximal levels of Coq2p were detected in heart and kidney, the two organs showing the highest levels of Q, followed by skeletal muscle. The lowest levels of mCoq2p were detected in brain and liver, which also showed lower steady-state levels of mCOQ2 transcripts. With respect to mCoq3p, maximal levels were detected in heart, while this polypeptide was found at significantly lower amounts in liver, kidney, skeletal muscle and brain. It is noteworthy that differences between the latter four tissues in terms of steady-state levels of mCOQ3 transcripts were of up to 3 orders of magnitude (if we compare skeletal muscle versus brain) whereas similar levels of mCoq3p were found in these tissues, demonstrating a strong regulation at a post-transcriptional level, as previously suggested [10]. The relatively low level of mCoq3p in kidney is very interesting because this organ showed by far the highest levels of Q9 and Q10. Although Coq3p catalyzes two steps in Q biosynthesis, the last of them yielding the final product [10,33,34], the low levels of mCOQ3 RNA and mCoq3p in kidney is consistent with a non limiting role for Coq3p in Q biosynthesis. In accordance, it has been suggested that Q levels in tissues are regulated at the initial steps in the terminal part of Q biosynthesis. Competitive inhibition of Coq2p by p-hydroxybenzoate analogues readily decreases Q levels in cellular systems [38,39], supporting an important role for Coq2p in the overall regulation of Q synthesis. Furthermore, there was no correlation between levels of Coq7p (which catalyzes the penultimate step of Q biosynthesis) and those of Q in a mouse model of transgene mCOQ7 expression [28]. However, the Coq7 reaction is apparently a point of regulation of Q biosynthesis pathway in yeast by post-transcriptional modification of either Coq7p or any component of the biosynthesis pre-complex [40].
Figure 3.
Levels of Coq2p and Coq3p in mouse tissues. Samples of liver, kidney, brain, skeletal muscle and heart were obtained from control animals (fed ad libitum), and then processed for SDS-PAGE and immunodetection of the polypeptides of interest by Western blot. Staining was repeated in three different preparations. This figure depicts the results of a representative blot.
3.4. The effect of calorie restriction on Q tissue levels
One month of 40% CR produced changes in the absolute levels of Q in muscular tissues (Figure 1). An increase of both Q9 and Q10 was detected in skeletal muscle, whereas a decrease of Q9 with no change in Q10 was detected in heart. In addition, the Q9/Q10 ratio was increased in kidney but decreased in heart. Previous reports have shown that long-term CR increased Q9 content of mitochondria isolated from skeletal muscle [23], liver, heart and kidney [24], although another study reported that CR induced a decrease of Q9 and Q10 in liver mitochondria [41]. The increase of Q9 by CR in skeletal muscle mitochondria [23] agrees with our results, although we also observed an increase of Q10, and these changes were detected very early, after one month of CR. Our results are in agreement with Bevilacqua et al. [42], who demonstrated that skeletal muscle (but not liver) mitochondria are rapidly adapted to short-term (2-wk and 2-mo) CR in the rat with a significant decrease of ROS generation [43]. It is possible that CR effects on Q in liver are a late event since life-long CR attenuated the increase of Q9/Q10 ratio in rat liver plasma membrane from aged rats [44].
Very few pharmacological treatments or physiological conditions are known to produce significant increases of Q in tissues, and most of them are related with oxidative stress conditions. An early work described increases in the contents of Q in liver and, to some extent, in kidney and muscle of rats treated with peroxisome proliferators, without modifying cholesterol levels [45]. More recently, it was reported that camptothecin and other chemotherapeutic drugs, such as etoposide, doxorubicin, and methotrexate, increased Q10 levels in tumour cells as part of an antioxidant response [46]. Finally, the mono and diepoxy derivatives of α- γ-tocotrienols are strong stimulators of Q biosynthesis [2]. The CR-induced increase of Q9 and Q10 in skeletal muscle is unique because it is associated with decreased oxidative stress [42]. In addition to alterations in the abundance of mCOQ transcripts (see below), a more reduced status might lead to higher Q levels in skeletal muscle due to reduced oxidative degradation.
The effects of CR on Q levels and Q9/Q10 ratio in heart are noteworthy because these modifications might be associated with alterations in mitochondrial function. Interestingly, ubisemiquinone-derived ROS are believed to be involved in Ca2+-stimulated permeabilization of mitochondria due to the opening of the permeability transition pore (PTP) [47], and quinones are well known modulators of the PTP [48,49]. Interestingly, short-term (2 mo) CR produced a small (14%) decrease in ROS production by heart ventricular subsarcolemmal mitochondria isolated from 6 mo male F344 rats [50], and counteracted the age-related loss in their Ca2+ retention capacity [51]. It is tempting to speculate that the CR-induced changes of the Q system we show here could be related with some of the beneficial effects of CR on the mitochondrial physiology in heart. However, it has to be acknowledged that CR produced opposite effects on Q levels in kidney, which points to the existence of different mechanisms of regulation in this mitotic organ compared with heart.
The Q9/Q10 ratio is determined by the polyisoprenoid substrate available for the reaction catalyzed by the polyprenyl 4-hydroxybenzoate transferase. The actual products of the trans-prenyltransferase activity (nona- or decaprenylpyrophosphate) are determined by the ratio between FPP and IPP, giving more decaprenylpyrophosphate in the presence of high IPP-Mg2+ [52]. Thus, our observations that CR increased Q9/Q10 ratio in kidney but decreased it in heart might indicate the existence of alterations in IPP/FPP ratio under CR conditions.
3.5. Effect of calorie restriction on steady-state COQ transcripts and Coq2 and Coq3 polypeptides
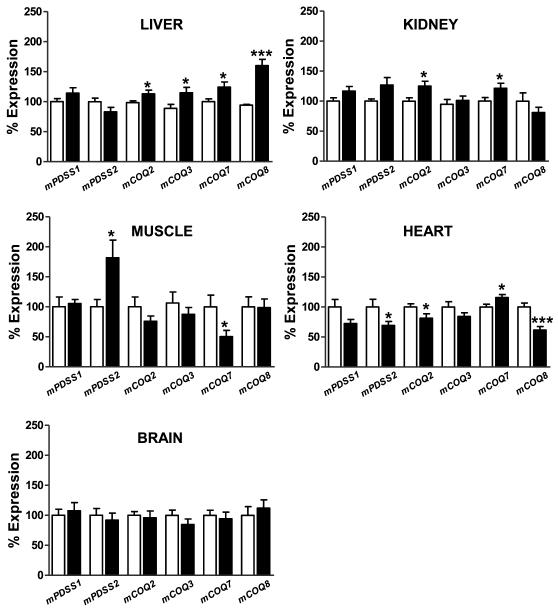
We next studied how calorie restriction affected steady-state levels of COQ transcripts in tissues. As depicted in Figure 4, the abundance of mCOQ transcripts was affected by CR in a tissue-specific manner. The response of the two mitotic tissues to CR included a modest increase of some mCOQ transcripts: mCOQ2, mCOQ3, mCOQ7 and mCOQ8 in liver, and mCOQ2 and mCOQ7 in kidney. If we compare these results with those obtained for Q levels, we can conclude that the limited changes of mRNAs detected in liver and kidney were not sufficient to produce significant alterations of Q levels. No changes in mPDSS1 or mPDSS2 transcripts were observed in kidney from CR mice. A mutation in PDSS1 has been demonstrated to decrease significantly Q10 without affecting Q9 levels in humans [53].
Figure 4.
The effect of short-term (1 mo) CR on steady-state abundance of mCOQ mRNAs. RNA samples were extracted from liver, kidney, brain, skeletal muscle and heart obtained from animals fed ad libitum or under CR (1 mo). Samples were processed as described in Figure 2. Depicted results show the absolute number of copies of mCOQ mRNAs present in tissues obtained from calorie-restricted animals, normalized to mHPRT and then referred to the values obtained for the same tissue obtained from control animals (set to 100% in each case). Data are means ± SEM (n=16). Open bars, ad libitum; Closed bars : CR.
Muscular tissues were affected by CR in a different way. In skeletal muscle mPDSS2 was increased, whereas COQ7 was decreased. The increase of mPDSS2 in skeletal muscle coincided with a significant increase of Q9 and Q10 levels (see above). Camptothecin-induced increase of Q10 levels in cellular systems was associated with up-regulation of Coq7p expression, as part of an antioxidant response [46]. Increase of Q10 biosynthesis by chemotherapeutic drugs is dependent on NF-κB that binds specifically to two κB binding sites present in the 5′-flanking region of the COQ7 gene, inducing both the COQ7 expression and Q10 biosynthesis [54]. Here, we show that the CR-induced increase of Q levels in skeletal muscle is not dependent on increased abundance of mCOQ7 mRNA but, on the contrary, decreased steady-state levels of mCOQ7 with CR were found in this tissue. The fact that CR can increase both steady-state levels of mPDSS2 and total Q in skeletal muscle may be of importance because skeletal muscle is one of the specific sites of Q deficiency and tissue damage caused by a mutation in the PDSS2 gene in humans [55]. In heart, the general response was a decrease in many of the COQ transcripts analyzed, except for COQ7, which showed a slight increase. The decrease of Q9 with CR in heart is consistent with significant decreases of mPDSS2, mCOQ2, and mCOQ8 transcripts. A trend towards lower levels was also observed for mPDSS1 and mCOQ3 in heart from CR mice, although differences were not statistically significant. Remarkably, no change in steady-state levels of any of the COQ transcripts was observed in brain, which coincided with the lack of changes elicited by CR in Q levels.
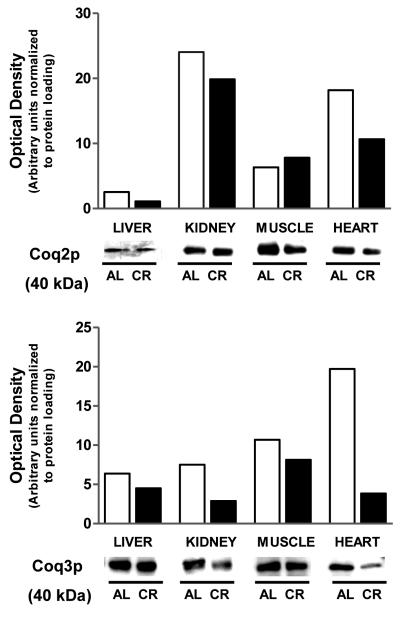
We also studied putative variations in the amount of Coq2 and Coq3 polypeptides induced by CR. For this analysis we only included the organs or tissues that exhibited some change associated with CR, either in Q or COQ transcripts levels. As depicted in Figure 5, the general tendency was either no change, or decreases of Coq2 and Coq3 polypeptides with CR. Although Coq3p was decreased by CR in kidney, Coq2p was unchanged. As stated above, steady-state abundance of mCOQ3 or Coq3p do not correlate with Q levels, and the possibility exists that relatively low levels of Coq3p are sufficient to provide enough Q biosynthesis. The lack of CR-induced changes for mCOQ2 and mCOQ3 transcripts in skeletal muscle is consistent with the absence of changes at the level of protein, which agrees with the idea that the increase of Q levels by short-term CR in skeletal muscle is regulated at an early point, most likely by the availability of polyisoprenoid chain precursor [2] and/or by a less oxidant status [42] which leads to decreased oxidative degradation. In heart, the decrease in most mCOQ transcripts coincided with decreases of Coq2p and Coq3p, suggesting that lower levels of Q9 in heart after 1 mo of CR may be the result of decreased biosynthesis.
Figure 5.
The effect of short-term (1 mo) CR on steady-state abundance of Coq2p and Coq3p. Samples of liver, kidney, brain, skeletal muscle and heart were obtained from control (fed ad libitum) and from calorie-restricted animals, and then processed for SDS-PAGE and immunodetection of the polypeptides of interest by Western blot. Staining was repeated in three different preparations. This figure depicts the results of a representative blot. Open bars, ad libitum; Closed bars : CR.
In summary we demonstrate for the first time that muscular and non-muscular tissues display unique patterns of mCOQ transcripts, and that Q and COQ genes are targets of CR in a tissue-specific way, supporting the idea that alterations of Q biosynthesis are part of the metabolic adaptation to CR in mice.
5. Acknowledgements
This work was supported by NIH grant 1R01AG028125-01A1 (to JJR, PN and JMV), Junta de Andalucía Proyectos de Excelencia grant CVI-00648 (to PN), Junta de Andalucía Proyectos de Excelencia grant P09-CVI-4887 (to JMV), Junta de Andalucía Proyectos Internacionales grant (to JMV), and BIO-276 (Junta de Andalucía and the University of Córdoba, to JMV). CP was funded by CVI-00648 grant. We thank Raúl Luque for providing housekeeping genes sequences and for transmitting us his experience with qrtRT-PCR. We thank Almudena Velázquez for handling of laboratory animals.
6. List of non standard abbreviations
- AL
Ad libitum
- CR
calorie restriction
- FPP
farnesyl pyrophosphate
- IPP
isopentenyl pyrophosphate
- NF
normalization factor
- PTP
permeability transition pore
- Q
coenzyme Q, ubiquinone
- UCPs
mitochondrial uncoupling proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References
- [1].Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta. 2004;1660:171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- [2].Bentinger M, Tekle M, Dallner G. Coenzyme Q – Biosynthesis and functions. Biochem. Biophys. Res. Commun. 2010;396:74–79. doi: 10.1016/j.bbrc.2010.02.147. [DOI] [PubMed] [Google Scholar]
- [3].González-Aragón D, Ariza J, Villalba JM. Dicoumarol impairs mitochondrial electron transport and pyrimidine biosynthesis in human myeloid leukemia HL-60 cells. Biochem. Pharmacol. 2007;73:427–439. doi: 10.1016/j.bcp.2006.10.016. [DOI] [PubMed] [Google Scholar]
- [4].López-Martín JM, Salviati L, Trevisson E, Montini G, DiMauro S, Quinzii C, Hirano M, Rodríguez-Hernández A, Cordero MD, Sánchez-Alcázar JA, Santos-Ocaña C, Navas P. Missense mutation of the COQ2 gene causes defects of bioenergetics and de novo pyrimidine synthesis. Hum. Mol. Genet. 2007;16:1091–1097. doi: 10.1093/hmg/ddm058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Villalba JM, Navarro F, Córdoba F, Serrano A, Arroyo A, Crane FL, Navas P. Coenzyme Q reductase from liver plasma membrane: purification and role in transplasma membrane electron transport. Proc. Natl. Acad. Sci. USA. 1995;92:4887–4892. doi: 10.1073/pnas.92.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jarmuszkiewicz W, Navet R, Alberici LC, Douette P, Sluse-Goffart CM, Sluse FE, Vercesi AE. Redox state of endogenous coenzyme Q modulates the inhibition of linoleic acid-induced uncoupling by guanosine triphosphate in isolated skeletal muscle mitochondria. J. Bioenerg. Biomembr. 2004;36:493–502. doi: 10.1023/B:JOBB.0000047331.25248.7a. [DOI] [PubMed] [Google Scholar]
- [7].Papucci L, Schiavone N, Witort E, Donnini M, Lapucci A, Tempestini A, Formigli L, Zecchi-Orlandini S, Orlandini G, Carella G, Brancato R, Capaccioli S. Coenzyme Q10 prevents apoptosis by inhibiting mitochondrial depolarization independently of its free radical scavenging property. J. Biol. Chem. 2003;278:28220–28228. doi: 10.1074/jbc.M302297200. [DOI] [PubMed] [Google Scholar]
- [8].Villalba JM, Navas P. Plasma membrane redox system in the control of stress-induced apoptosis. Antiox. Redox Signal. 2000;2:213–230. doi: 10.1089/ars.2000.2.2-213. [DOI] [PubMed] [Google Scholar]
- [9].Elmberger PG, Kalèn A, Appelkvist E–L, Dallner G. In vitro and in vivo synthesis of dolichol and other main mevalonate products in various organs of the rat. Eur. J. Biochem. 1987;168:1–11. doi: 10.1111/j.1432-1033.1987.tb13379.x. [DOI] [PubMed] [Google Scholar]
- [10].Marbois BN, Xia Y–R, Lusis AJ, Clarke CF. Ubiquinone biosynthesis in eukaryotic cells: Tissue distribution of mRNA encoding 3,4-dihydroxy-5-polyprenylbenzoate methyltransferase in the rat and mapping of the COQ3 gene to mouse chromosome 4. Arch. Biochem. Biophys. 1994;313:83–88. doi: 10.1006/abbi.1994.1362. [DOI] [PubMed] [Google Scholar]
- [11].Villalba JM, Parrado C, Santos-González M, Alcaín FJ. Therapeutic use of coenzyme Q10 and coenzyme Q10-related compounds and formulations. Expert. Opin. Invest. Drugs. 2010;19:535–554. doi: 10.1517/13543781003727495. [DOI] [PubMed] [Google Scholar]
- [12].Saiki R, Nagata A, Kainou T, Matsuda H, Kawamukai M. Characterization of solanesyl and decaprenyl diphosphate synthases in mice and humans. FEBS J. 2005;272:5606–5622. doi: 10.1111/j.1742-4658.2005.04956.x. [DOI] [PubMed] [Google Scholar]
- [13].Peng M, Falk MJ, Haase VH, King R, Polyak E, Selak M, Yudkoff M, Hancock WW, Meade R, Saiki R, Lunceford AL, Clarke CF, Gasser DL. Primary coenzyme Q deficiency in Pdss2 mutant mice causes isolated renal disease. PLoS Genet. 2008;4:e1000061. doi: 10.1371/journal.pgen.1000061. doi:10.1371/journal.pgen.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Forsgren M, Attersand A, Lake S, Grünler J, Swiezewska E, Dallner G, Climent I. Isolation and functional expression of human COQ2, a gene encoding a polyprenyl transferase involved in the synthesis of CoQ. Biochem. J. 2004;382:519–526. doi: 10.1042/BJ20040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Aberg F, Appelkvist EL, Dallner G, Ernster L. Distribution and redox state of ubiquinones in rat and human tissues. Arch. Biochem. Biophys. 1992;295:230–234. doi: 10.1016/0003-9861(92)90511-t. [DOI] [PubMed] [Google Scholar]
- [16].Lass A, Forster MJ, Sohal RS. Effects of coenzyme Q10 and α-tocopherol administration on their tissue levels in the mouse: Elevation of mitochondrial α-tocopherol by coenzyme Q10. Free Radic. Biol. Med. 1999;26:1375–1383. doi: 10.1016/s0891-5849(98)00330-x. [DOI] [PubMed] [Google Scholar]
- [17].Tang PH, Miles MV, Miles L, Quinlan J, Wong B, Wenisch A, Bove K. Measurement of reduced and oxidized coenzyme Q9 and coenzyme Q10 levels in mouse tissues by HPLC with coulometric detection. Clin. Chim. Acta. 2004;341:173–184. doi: 10.1016/j.cccn.2003.12.002. [DOI] [PubMed] [Google Scholar]
- [18].Sohal RS, Kamzalov S, Sumien N, Ferguson M, Rebrin I, Heinrich KR, Forster MJ. Effect of coenzyme Q10 intake on endogenous coenzyme Q content, mitochondrial electron transport chain, antioxidative defenses, and life span of mice. Free Radic. Biol. Med. 2006;40:480–487. doi: 10.1016/j.freeradbiomed.2005.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kwong LK, Kamzalov S, Rebrin I, Bayne A–CV, Jana CK, Morris P, Forster MJ, Sohal RS. Effects of coenzyme Q10 administration on its tissue concentrations, mitochondrial oxidant generation, and oxidative stress in the rat. Free Radic. Biol. Med. 2002;33:627–638. doi: 10.1016/s0891-5849(02)00916-4. [DOI] [PubMed] [Google Scholar]
- [20].Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu. Rev. Med. 2003;54:131–152. doi: 10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- [21].Pollak M. Do cancer cells care if their host is hungry? Cell Metab. 2009;9:401–403. doi: 10.1016/j.cmet.2009.04.006. [DOI] [PubMed] [Google Scholar]
- [22].Witte AV, Fobker M, Gellner R, Knecht S, Floel A. Caloric restriction improves memory in elderly humans. Proc. Natl. Acad. Sci. USA. 2009;106:1255–1260. doi: 10.1073/pnas.0808587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lass A, Kwong L, Sohal RS. Mitochondrial coenzyme Q content and aging. BioFactors. 1999;9:199–205. doi: 10.1002/biof.5520090215. [DOI] [PubMed] [Google Scholar]
- [24].Kamzalov S, Sohal R. Effect of age and caloric restriction on coenzyme Q and α-tocopherol levels in the rat. Exp. Gerontol. 2004;39:1199–1205. doi: 10.1016/j.exger.2004.04.007. [DOI] [PubMed] [Google Scholar]
- [25].Asaumi S, Kuroyanagi H, Seki N, Shirasawa T. Orthologues of the Caenorhabditis elegans longevity gene clk-1 in mouse and human. Genomics. 1999;58:293–301. doi: 10.1006/geno.1999.5838. [DOI] [PubMed] [Google Scholar]
- [26].Takahashi M, Asaumi S, Honda S, Suzuki Y–I, Nakai D, Kuroyanagi H, Shimizu T, Honda Y, Shirasawa T. Mouse coq7/clk-1 orthologue rescued slowed rhythmic behavior and extended life span of clk-1 longevity mutant in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2001;286:534–540. doi: 10.1006/bbrc.2001.5439. [DOI] [PubMed] [Google Scholar]
- [27].Jiang N, Levavasseur F, McCright B, Shoubridge EA, Hekimi S. Mouse CLK-1 is imported into mitochondria by an unusual process that requires a leader sequence but no membrane potential. J. Biol. Chem. 2001;276:29218–29225. doi: 10.1074/jbc.M103686200. [DOI] [PubMed] [Google Scholar]
- [28].Nakai D, Shimizu T, Nojiri H, Uchiyama S, Koike H, Takahashi M, Hirokawa K, Shirasawa T. Coq7/clk1 regulates mitochondrial respiration and the generation of reactive oxygen species via coenzyme Q. Aging Cell. 2004;3:273–281. doi: 10.1111/j.1474-9728.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- [29].Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- [30].Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bello RI, Alcaín FJ, Gómez-Díaz C, López-Lluch G, Navas P, Villalba JM. Hydrogen peroxide and cell density-regulated expression of cytochrome b5 reductase in HeLa cells. J. Bioenerg. Biomembr. 2003;35:169–179. doi: 10.1023/a:1023702321148. [DOI] [PubMed] [Google Scholar]
- [32].Hamalainen N, Pette D. The histochemical profiles of fast fiber types IIB, IID and IIA in skeletal muscles of mouse, rat and rabbit. J. Histochem. Cytochem. 1993;41:733–743. doi: 10.1177/41.5.8468455. [DOI] [PubMed] [Google Scholar]
- [33].Poon WW, Barkovich RJ, Hsu AY, Frankel A, Lee PT, Shepherd JN, Myles DC, Clarke CF. Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J. Biol. Chem. 1999;274:21665–21672. doi: 10.1074/jbc.274.31.21665. [DOI] [PubMed] [Google Scholar]
- [34].Jonassen T, Clarke CF. Isolation and functional expression of human COQ3, a gene encoding a methyltransferase required for ubiquinone biosynthesis. J. Biol. Chem. 2000;275:12381–12387. doi: 10.1074/jbc.275.17.12381. [DOI] [PubMed] [Google Scholar]
- [35].Stenmark P, Grünler J, Mattsson J, Sindelar PJ, Nordlund P, Berthold DA. A new member of the family of di-iron carboxylate proteins. Coq7 (clk-1), a membrane bound hydroxylase involved in ubiquinone biosynthesis. J. Biol. Chem. 2001;276:33297–33300. doi: 10.1074/jbc.C100346200. [DOI] [PubMed] [Google Scholar]
- [36].Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: Loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Vajo Z, King LM, Jonassen T, Wilkin DJ, Ho N, Munnich A, Clarke CF, Francomano CA. Conservation of the Caenorhabditis elegans timing gene clk-1 from yeast to human: a gene required for ubiquinone biosynthesis with potential implications for aging. Mammalian Genome. 1999;10:1000–1004. doi: 10.1007/s003359901147. [DOI] [PubMed] [Google Scholar]
- [38].González-Aragón D, Burón MI, López-Lluch G, Hermán MD, Gómez-Díaz C, Navas P, Villalba JM. Coenzyme Q and the regulation of intracellular steady-state levels of superoxide in HL-60 cells. BioFactors. 2005;25:31–41. doi: 10.1002/biof.5520250105. [DOI] [PubMed] [Google Scholar]
- [39].Forsman U, Sjöberg M, Turunen M, Sindelar PJ. 4-Nitrobenzoate inhibits coenzyme Q biosynthesis in mammalian cell cultures. Nat. Chem. Biol. 2010;6:515–517. doi: 10.1038/nchembio.372. [DOI] [PubMed] [Google Scholar]
- [40].Padilla S, Tran UC, Jiménez-Hidalgo M, López-Martín JM, Martín-Montalvo A, Clarke CF, Navas P, Santos-Ocaña C. Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis. Cell. Mol. Life Sci. 2009;66:173–186. doi: 10.1007/s00018-008-8547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Armeni T, Principato G, Quiles JL, Pieri C, Bompadre S, Battino M. Mitochondrial dysfunctions during aging: Vitamin E deficiency or calorie restriction – Two different ways of modulating stress. J. Bioenerg. Biomembr. 2003;35:181–191. doi: 10.1023/a:1023754305218. [DOI] [PubMed] [Google Scholar]
- [42].Bevilacqua L, Ramsey JJ, Hagopian K, Weindruch R, Harper ME. Effects of short- and medium-term calorie restriction on muscle mitochondrial proton leak and reactive oxygen species production. Am. J. Physiol. Endocrinol. Metab. 2004;286:E852–E861. doi: 10.1152/ajpendo.00367.2003. [DOI] [PubMed] [Google Scholar]
- [43].Ramsey JJ, Hagopian K, Kenny TM, Koomson EK, Bevilacqua L, Weindruch R, Harper ME. Proton leak and hydrogen peroxide production in liver mitochondria from energy-restricted rats. Am. J. Physiol. Endocrinol. Metab. 2004;286:E31–E40. doi: 10.1152/ajpendo.00283.2003. [DOI] [PubMed] [Google Scholar]
- [44].De Cabo R, Cabello R, Rios M, López-Lluch G, Ingram DK, Lane MA, Navas P. Calorie restriction attenuates age-related alterations in the plasma membrane antioxidant system in rat liver. Exp. Gerontol. 2004;39:297–304. doi: 10.1016/j.exger.2003.12.003. [DOI] [PubMed] [Google Scholar]
- [45].Aberg F, Zhang Y, Teclebrhan H, Appelkvist EL, Dallner G. Increases in tissue levels of ubiquinone in association with peroxisome proliferation. Chem. Biol. Interact. 1996;99:205–218. doi: 10.1016/0009-2797(95)03670-9. [DOI] [PubMed] [Google Scholar]
- [46].Brea-Calvo G, Rodríguez-Hernández A, Fernández-Ayala DJ, Navas P, Sánchez-Alcázar JA. Chemotherapy induces an increase in coenzyme Q10 levels in cancer cell lines. Free Radic. Biol. Med. 2006;40:1293–1302. doi: 10.1016/j.freeradbiomed.2005.11.014. [DOI] [PubMed] [Google Scholar]
- [47].Kowaltowski AJ, Castilho RF, Vercesi AE. Ca2+-induced mitochondrial membrane permeabilization: role of coenzyme Q redox state. Am. J. Physiol. 1995;269:C141–C147. doi: 10.1152/ajpcell.1995.269.1.C141. [DOI] [PubMed] [Google Scholar]
- [48].Fontaine E, Ichas F, Bernardi P. A ubiquinone-binding site regulates the mitochondrial permeability transition pore. J. Biol. Chem. 1998;273:25734–25740. doi: 10.1074/jbc.273.40.25734. [DOI] [PubMed] [Google Scholar]
- [49].Walter L, Nogueira V, Leverve X, Heitz MP, Bernardi P, Fontaine E. Three classes of ubiquinone analogs regulate the mitochondrial permeability transition pore through a common site. J. Biol. Chem. 2000;275:29521–29527. doi: 10.1074/jbc.M004128200. [DOI] [PubMed] [Google Scholar]
- [50].Judge S, Judge A, Grune T, Leeuwenburgh C. Short-term CR decreases cardiac mitochondrial oxidant production but increases carbonyl content. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;286:R254–R259. doi: 10.1152/ajpregu.00502.2003. [DOI] [PubMed] [Google Scholar]
- [51].Hofer T, Servais S, Seo AY, Marzetti E, Hiona A, Upadhyay SJ, Wohlgemuth SE, Leeuwenburgh C. Bioenergetics and permeability transition pore opening in heart subsarcolemmal and interfibrillar mitochondria: Effects of aging and lifelong calorie restriction. Mech. Ageing Dev. 2009;130:297–307. doi: 10.1016/j.mad.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Runquist M, Ericsson J, Thelin A, Chojnacki T, Dallner G. Isoprenoid biosynthesis in rat liver mitochondria. J. Biol. Chem. 1994;269:5804–5809. [PubMed] [Google Scholar]
- [53].Mollet J, Giurgea I, Schlemmer D, Dallner G, Chretien D, Delahodde A, Bacq D, de Lonlay P, Munnich A, Rötig A. Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J. Clin. Invest. 2007;117:765–772. doi: 10.1172/JCI29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Brea-Calvo G, Siendones E, Sánchez-Alcázar JA, de Cabo R, Navas P. Cell survival from chemotherapy depends on NF-kappaB transcriptional up-regulation of coenzyme Q biosynthesis. PLoS One. 2009;4:e5301. doi: 10.1371/journal.pone.0005301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lopez LC, Schuelke M, Quinzii CM, Kanki T, Rodenburg RJ, Naini A, Dimauro S, Hirano M. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am. J. Hum. Genet. 2006;79:1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]