Abstract
RIN proteins serve as guanine nucleotide exchange factors for Rab5a. They are characterized by the presence of a RIN homology domain and a C-terminal Vps9 domain. Currently three family members have been described and analyzed. Here we report the identification of a novel RIN family member, Rin-like (Rinl), that represents a new interaction partner of the receptor tyrosine kinase MuSK, which is an essential key regulator of neuromuscular synapse development. Rinl is localized to neuromuscular synapses but shows the highest expression in thymus and spleen. Rinl preferentially binds to nucleotide-free Rab5a and catalyzes the exchange of GDP for GTP. Moreover, Rinl also binds GDP-bound Rab22 and increases the GDP/GTP exchange implicating Rinl in endocytotic processes regulated by Rab5a and Rab22. Interestingly, Rinl shows a higher catalytic rate for Rab22 compared to Rab5a. Rinl is closely associated with the cytoskeleton and thus contributes to the spatial control of Rab5a and Rab22 signaling at actin-positive compartments. Most importantly, overexpression of Rinl affects fluid-phase as well as EGFR endocytosis.
Abbreviations: AChR, acetylcholine receptor; BGT, bungarotoxin; EGFR, epidermal growth factor receptor; GEF, guanine nucleotide exchange factor; GFP, green fluorescent protein; GST, glutathione S-transferase; HRP, horseradish peroxidase; MuSK, muscle specific kinase; RT-PCR, reverse transcriptase polymerase chain reaction; RTK, receptor tyrosine kinase; SH2, Src homology 2
Keywords: Endocytosis, Rab5, GEF, MuSK, Receptor tyrosine kinase
Research highlights
► Rinl is a novel member of the RIN protein family and specifically binds MuSK. ► Rinl catalyzes the nucleotide exchange on Rab5a and Rab22. ► Rinl shows a higher catalytic activity for Rab22 than Rab5a. ► Rinl regulates the localization of Rab5a and Rab22 to actin-positive membrane ruffles. ► Overexpression of Rinl increases both fluid-phase and receptor-mediated endocytosis.
1. Introduction
Cycling of Rab GTPases between the inactive (GDP bound) and active (GTP bound) state depends on nucleotide exchange of GDP to GTP catalyzed by guanine nucleotide exchange factors (GEFs) and the stimulation of the intrinsically slow GTP-hydrolysis activity, which converts Rab bound GTP to GDP. The GDP to GTP exchange activates Rab proteins by inducing a conformation that enables them to interact with effector proteins. Ras-interaction/interference (RIN) proteins, also known as Ras and Rab interactors, belong to the family of Vps9 domain-containing GEFs [1]. The founding member of the RIN protein family, RIN1, was identified as an inhibitor of signaling mediated by the GTPase H-Ras. Via its Ras association (RA) domain RIN1 competes with binding of the Ras effector Raf1 [2]. Further studies revealed that RIN1 contains a functional Vps9 domain indicative for Rab GEFs. This domain was shown to be necessary for binding to Rab5a and for GDP/GTP exchange activity, and indeed RIN1 was characterized as the Ras effector protein that connects Ras signaling to the control of endocytotic processes [3]. The Rab5a GEF activity is shared by all three known RIN proteins as well as the conserved domain structure consisting of an RA domain, a Vps9 domain, a RIN homology (RH) domain and an N-terminal SH2 domain [1]. The Vps9 domain was originally identified in the yeast protein Vps9p, which serves as GEF for Vps21p and its mammalian homologue Rab5a [4]. Nucleotide exchange by GEFs on Rab5a results in increased endocytosis [5].
The consensus Vps9p catalytic domain is present in a variety of multi-domain proteins including the RIN protein family [1]. The crystal structure of the Rabex-5 Vps9 domain showed that the core Vps9 domain is stabilized by an essential helical bundle N-terminal to the Vps9 region, in the case of the RIN proteins corresponding to the RH domain [6]. In addition, a high specificity for the Rab5 subfamily, which consists of Rab5a, Rab5b, Rab5c, Rab21, Rab22 and Rab31 (also termed Rab22B), was demonstrated.
The proteins of the Rab5 subfamily are important regulators of early endocytosis. Rab5a is among the best-studied Rab proteins and is known as the key regulator of endocytotic processes like clathrin-coated vesicle formation, fusion between early endosomes, endosomal cargo recruitment and endosomal motility [7]. Rab21 and Rab22 are less well characterized. Both have been localized to early endosomes and implicated in early endocytosis [8,9]. However, Rab21 is also thought to be involved in transport from the trans Golgi network to endosomes whereas the predominant role of Rab22 has been attributed to recycling processes [10,11].
RIN proteins have been proposed as regulators of early Rab5a-dependent endocytosis due to their binding properties for Rab5a and their Rab5a-specific GEF activity. RIN1 stimulates Rab5a-dependent endosome fusion and ligand-mediated endocytosis of the EGF-receptor (EGFR), the insulin receptor and the EphA4 receptor [12–14]. In addition, RIN3 has been shown to interact with amphiphysin II, which is involved in clathrin- and dynamin-dependent endocytosis [15]. These observations are of special interest since increasing evidence supports a close interplay between signaling and endocytosis of receptor tyrosine kinases (RTK). In particular, it was shown that receptors remain active within endosomes [16]. This led to the hypothesis that membrane trafficking could control signal transduction. Studies on different RTKs have now demonstrated that there are two main types of functions that receptor trafficking has in regulating receptor signaling: it controls the magnitude of the response or it controls the specificity of the response [17].
Here we report the identification of a novel RIN family member, Rin-like (Rinl), which was isolated as interaction partner of the muscle-specific RTK MuSK. Signal transduction events induced by MuSK are crucially linked to the formation of the neuromuscular synapse (NMS) [18]. MuSK is activated by the motor neuron-derived heparansulfate proteoglycan agrin [19]. Agrin does not bind MuSK directly but interacts with Lrp4, a member of the LDL receptor family [20,21]. Upon binding, the Lrp4/MuSK complex presumably undergoes a structural rearrangement that results in a dimerization and subsequent autophosphorylation of MuSK. The subsequent activation of the MuSK kinase induces a signaling cascade leading to the formation of the NMS including postsynaptic differentiation characterized by the accumulation of acetylcholine receptors (AChRs) at synaptic sites and presynaptic differentiation as depicted by the development of active zones. Consistent with this model, agrin, MuSK and lrp4 mutant mice fail to form NMSs, lacking all features of pre- and postsynaptic specializations [22–24].
Rinl interacts with MuSK independent of its phosphorylation and localizes to NMSs. Further, we show that Rinl acts as GEF for Rab5a. Moreover, Rinl interacts with inactive Rab22 and catalyzes the GDP/GTP exchange on Rab22. Co-localization of Rinl with Rab5a or Rab22 in actin-rich membrane ruffles implicates a novel mechanism for the recruitment of Rab5a and Rab22 to the cytoskeleton via binding to Rinl. In addition, increased fluid-phase uptake and EGFR endocytosis upon Rinl overexpression implicates Rinl as regulator of early endocytotic processes.
2. Materials and methods
2.1. Antibodies and reagents
The following antibodies were purchased from commercial sources: anti-phosphotyrosine PY99 (Santa Cruz) and PY-100 (Cell Signaling), anti-myc 9E10 (Sigma-Aldrich), anti-GFP (Santa Cruz). Antibodies against the C-terminal sequence of MuSK were described previously [25]. Alexa 594-conjugated α-bungarotoxin (BGT), Alexa 555-conjugated EGF and Alexa 488-conjugated secondary antibodies were obtained from Invitrogen. Horseradish-peroxidase-coupled and fluorophore-conjugated secondary antibodies were purchased from Jackson ImmunoResearch. TRITC-conjugated phalloidin and cytochalasin D were purchased from Sigma-Aldrich.
2.2. Generation of antibodies against Rinl
Rinl (aa 243–307, Mus musculus) was amplified by PCR (5′-TATGAATTCCACTTTTCCTGTCCT-3′ and 5′-TATAAGCTTTGGGCTGTAGGGATTT-3′) and cloned into modified pMALc and pGEX-4T-3 vectors, which encode for seven C-terminal histidines in addition to MBP or GST respectively [26]. Recombinant proteins were expressed in the Escherichia coli strain XL1 blue in LB medium supplemented with 0.4% glucose by induction with 0.2 mM IPTG and purified via HIS-affinity chromatography. The MBP fusion protein was used to raise polyclonal antisera in rabbits as described previously [26]. Reactive serum was affinity purified on an Affi-Gel 10 (Bio-Rad) column coupled with the GST fusion protein mentioned above. The column was sequentially washed with phosphate buffered saline (PBS), 0.1 M NaHCO3 and 0.1 M NaHCO3/0.5 M NaCl. The bound antibodies were eluted with 7 ml acidic elution buffer (0.1 M glycine, pH 2.5) and fractions of 1 ml were collected. Purified antibodies were tested for immunoblotting and immunoprecipitation as shown in Fig. S4.
2.3. Plasmids
The MuSK cytoplasmic domain carrying the mutations Y553F or Y750, 754, 755F (KD) was subcloned into the yeast bait vector pBTM116. It was previously demonstrated that MuSK Y750, Y754, 755F, which affects the activation loop of the kinase domain, behaves as kinase-deficient mutant similar to a K608A mutant, which blocks ATP binding within the autocatalytic loop [27,28]. The MMT bait construct contains the TrkA cytoplasmic domain including the MuSK juxtamembrane region in pBTM116 [27]. To generate BTM/tpr-met-JM, the juxtamembrane region of MuSK (Rattus norvegicus, aa 528–570) was subcloned into BTM/tpr-met using the primers 5′-AATGAATTCAGAGAGTCGGCAGC-3′ and 5′-TAGTCGACTACGGATACTCCAGGCTGAG-3′. Kinase-dead (K608A) and kinase-active MuSK (LS745,746MT) carrying a myc-tag were provided by Dr. Burden (NYU School of Medicine, New York, [20]).
The Rinl 5′-end was amplified from mouse muscle cDNA using the following primers: 5′-CTGAGCAGCCTTGAGTCTTGTCTT-3′ (located upstream of the predicted start codon) and 5′-CCTGGCCAGAGCACGGATGT-3′. Full length Rinl was then cloned into pcDNA3 (Invitrogen) with or without an N-terminal myc-tag and into pEGFP-C3 (Clontech). Truncations ΔSH2 (aa 200–563) and ΔVps9 (aa 1–394) with an N-terminal myc-tag were subcloned into pcDNA3. For yeast two-hybrid and GST-pulldown experiments Rinl and truncation mutants were cloned into pACT2 (Clontech) and pGEX (GE Healthcare) vectors respectively: GST-Rinl (aa 1–563 in pGEX-5X-2), GST-Y2H (aa 200–563 in pGEX-3), GST-RH/ΔVps9 (aa 200–479 in pGEX-2), GST-RH/Vps9 (aa 248–563 in pGEX-2).
pLexA-Rab5a (wt, S34N, Q79L) plasmids were a gift from Dr. Zerial (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden), for GFP-tagged constructs Rab5a wt and mutants were cloned into pEGFP-C2 (Clontech) using EcoRI. pEGFP-Rab22 (wt, S19N, Q64L) was provided by Dr. Donaldson (NIH, Bethesda). Rab22 inserts were amplified by PCR (Rab22 MunI fw 5′-AAACAATTGATGGCGCTGAGGGAGCTCAAA-3′ and Rab22 SalI rev 5′-AAAGTCGACGCAGCAGCTTCGCTGCGGCTC-3′) and cloned into pLexA linearized with EcoRI and SalI. GFP-Rab7a (wt, T22N, Q67L) constructs were a gift from Dr. Bucci (University of Salento, Lecce).
All constructs used in this study were checked by sequencing.
2.4. Cell culture
HEK 293T cells, COS7 and HeLa cells were maintained in DMEM supplemented with glutamine, 4.5 mg/ml glucose, 10% fetal bovine serum (FBS) and 100 μg/ml penicillin/streptomycin. HEK 293T cells were transfected using the protocol described by Chen and Okayama [29]. COS7 and HeLa cells were transfected using TurboFect (Fermentas). Briefly, COS7 and HeLa cells were plated on glass coverslips and transfected the next day with 1 μg DNA and 1.5 μl TurboFect in 2 ml DMEM.
2.5. Yeast two-hybrid screen
The MuSK cytoplasmic domain (Rattus norvegicus, aa 517–868) was subcloned into the modified pBTM116 vector, BTM/tpr-met (a gift from Dr. Birchmeier, Max-Delbrueck Center for Molecular Medicine, Berlin), which contains an active c-Met kinase lacking the multiple docking site [30]. Insertion of MuSK produces a fusion between c-Met and MuSK. The MuSK bait was screened against a mouse muscle cDNA library (a gift from Dr. Chamberlain, University of Michigan Medical School, Ann Arbor) cloned into the pACT2 prey vector [31]. The BTM/tpr-met-MuSK bait plasmid was introduced into the L40 yeast strain. Large scale transformations with the prey library were performed according to Vojtek and Hollenberg [32]. A total of 7.2 × 106 transformants were screened for growth on –HIS. Forty-six positive clones were recovered. After retransformation of the recovered cDNA plasmids six clones grew on –HIS and produced a positive signal in a β-gal assay with MuSK but were negative for control plasmids (BTM/tpr-met, BTM/cMet and BTM/lamin). To analyze the interaction between MuSK and Rinl or Rinl and Rab5a/Rab22 respectively, the bait and prey plasmids were transformed into the yeast strain L40. Single colonies were restreaked on –LEU/TRP or –LEU/TRP/HIS. A β-gal assay screening for lacZ expression was performed as described previously [32].
2.6. GST-pulldown experiments
All GST fusion proteins were expressed in the bacterial strain Rosetta by induction with 0.5 mM IPTG. The harvested bacteria were resuspended in ice-cold buffer R (PBS, 1% Triton X-100, 1 mg/ml lysozyme, 0.2 mM phenylmethylsulfonyl fluoride (PMSF)). Cells were lysed on ice for 1 hour, 0.25% sarkosyl was added and samples were sonicated. Cleared lysates were incubated with glutathione agarose beads (Sigma) for 1 hour under constant rotation at 4 °C, followed by three washing steps with buffer R (without lysozyme). Protein concentration and quality were assayed by SDS-PAGE and subsequent Coomassie staining. Recombinant proteins were stored at − 80 °C until use. Cell lysates were incubated with immobilized GST fusion proteins for 4–18 hours at 4 °C. Beads were washed three times with NP-40 lysis buffer (1% Nonidet P-40, 5 mM EGTA, 50 mM NaCl, 30 mM triethanolamine [pH 7.5], 50 mM NaF). Proteins bound to the beads were subjected to SDS-PAGE and immunoblotting.
2.7. Lysate preparation, immunoprecipitation and immunoblotting
Lysates were prepared from cultured cells or adult mouse tissues using NP-40 lysis buffer supplemented with fresh proteinase inhibitors (1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 0.2 mM PMSF, 1 mM sodium orthovanadate). Protein concentrations were determined using Roti-Nanoquant (Roth) protein assay reagent.
For co-immunoprecipitation experiments, HEK 293T cells were co-transfected with indicated constructs. Cleared lysates were precipitated with polyclonal affinity-purified antibodies directed to either MuSK or Rinl overnight. The next day protein A agarose (Roche) was added for 1–3 hours, the beads were washed three times with NP-40 lysis buffer with increased (150 mM) NaCl concentration and precipitated protein complexes were analyzed by SDS-PAGE and immunoblotting.
2.8. RT-PCR and quantitative PCR
RNA was isolated from C57BL/6 mice using TRI reagent (Sigma) according to the manufacturer's protocol and reverse transcribed to cDNA. The following primers were used for subsequent PCR reactions: Rinl-1154fw (5′-GAAGATCTTGGCCCCGCTGT-3′), Rinl-1645rev (5′-CTGGTAGTGAGCAATGTGGT-3′), actin1 (5′-TTCTACAATGAGCTGCGTGTGG-3′), actin2 (5′-CTCGGTCAGGATCTTCATGAGG-3′), Gapdh/fw (5′-TGCATCCTGCACCACCAACT-3′), Gapdh/rev (5′-ATGCCTGCTTCACCACCTTC-3′). Quantitative real-time PCR (qPCR) was performed with IQTM SYBR Green Supermix (Bio-Rad) using a Bio-Rad iCycler. Rinl levels were normalized to Gapdh using the formula: Rinl/Gapdh = 2(CTx − CTrel) / 2(CTx − CTrel). Spleen was used as reference tissue (rel) with a value set to 1. PCR reactions were performed in duplicates or triplicates.
2.9. Immunofluorescence
Eighteen hours after transfection, COS7 cells were fixed with 4% paraformaldehyde (PFA)/PBS for 10 minutes at room temperature (RT), permeabilized with 0.1% Triton/PBS for 5 minutes at RT and incubated with blocking solution (PBS containing 10% FBS) followed by incubation with appropriate primary antibodies for 1 hour at RT. Cells were washed with PBS and incubated with fluorophore-conjugated secondary antibodies with or without TRITC-labeled phalloidin. Cells were mounted in Mowiol and visualized using a Leica TCS SP5 spectral confocal microscope with a HCX PL APO CS 63×/1.4 oil objective. For the disruption of the actin cytoskeleton, transfected cells were treated with 1 μM cytochalasin D or the solvent DMSO in DMEM for 30 minutes before the cells were fixed and stained with antibodies. The distribution of Rab5a or Rab22 in Rinl positive cells was categorized into four different classes and plotted as the average ± s.e.m. of three independent experiments (sum of all subcategories is 100%).
Mouse muscle cryosections were stained as previously described [33]. Stained muscle sections were viewed on a confocal microscope as described above.
2.10. EGF uptake
Twenty to 24 hours post transfection cells were starved for 3 hours in DMEM, labeled with Alexa 555-conjugated EGF for 30 minutes at 4 °C and washed four times with ice-cold DMEM supplemented with 1 mg/ml BSA. Then the cells were put back at 37 °C for endocytosis to occur. At indicated time points cells were fixed with 1% PFA in PBS and embedded in Mowiol. Cells were analyzed by confocal microscopy as described above. Representative images are shown.
2.11. HRP uptake
HEK 293T cells were transfected with the following constructs: pEGFP, pEGFP-Rab5a wt, pEGFP-Rinl as stated. About 20 hours after transfection cells were sorted for GFP expression by FACS. GFP-positive cells were seeded on polyornithine coated 12-well plates overnight. The next day HRP uptake was analyzed as previously described [34]. Briefly, cells were incubated in internalization medium (IM; DMEM, 1% FBS, 24 mM HEPES [pH 7.5]) containing 4 mg/ml HRP for 1 hour at 37 °C. After internalization, cells were washed once with warm IM followed by three washes with ice-cold PBS supplemented with 1 mM CaCl2, 1 mM MgCl2, 2 mg/ml BSA. Finally, cells were washed once with cold PBS and extracted for 15 minutes on ice with lysis buffer (1% w/v Triton X-100, 20 mM HEPES [pH 7.5], 0.2 mM PMSF). Total HRP activity of the lysates was determined in duplicates of triplicate samples using the substrate 3,3′,5,5′-tetramethylbenzidine (Sigma). HRP activity was normalized to the protein concentration of individual lysates.
2.12. GEF activity assay
Rab5a (Cane lupus, aa 15–185), Rab7a (Cane lupus, aa 1–185), Rab22 (Cane lupus, aa 1–173) and Rinl (Mus musculus, aa 260–563) were cloned into pGEX-4T-3 and expressed in the bacterial strain CK600K in Standard I medium (Merck) by induction with 100 μM IPTG at an OD600 of 0.8 at 25 °C overnight. Bacteria were harvested by centrifugation and washed with 0.9% NaCl. Rab proteins were purified, cleaved from GST-tag and loaded with mGDP (2′-/3′-O-(N′-Methylanthraniloyl)guanosine-5′-O-diphosphate) essentially as described for Rap [35]. Rinl was purified the same way except that the buffer was MgCl2-free and contained 5 mM EDTA, and the GST-tag was not cleaved. Fluorescence measurements were performed as described [35]. In brief, 200 nM of the G-protein loaded with mGDP were incubated in buffer G (50 mM Tris–HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 5 mM DTT, 5% glycerol) in the presence of 20 μM GDP and increasing amounts of Rinl. For kinetic analysis the obtained curves were fitted as single exponential decay to obtain the rate constant kobs. kobs were plotted against the concentration of Rinl and analyzed by linear fitting, whereby the slope was defined as the catalytic efficiency of the GEF reaction.
3. Results
3.1. Rinl: a novel interaction partner of MuSK
To identify proteins that bind to MuSK, a mouse muscle cDNA library was screened with the MuSK cytoplasmic domain using the yeast two-hybrid system. Screening of 7.2 × 106 transformants yielded six positive clones that interacted strongly with MuSK. One of these clones, termed CL-6, was identified independently three times. It consisted of a 2204 bp cDNA encoding 363 amino acids, an in-frame stop codon, the 3′ UTR and the poly(A) tail. The isolated cDNA was identical to ENSMUST00000059857 lacking however the first 199 amino acids. We therefore isolated the missing 5′ sequence by RT-PCR. Analysis of the total cDNA revealed an open reading frame that encoded 563 amino acids in which the original CL-6 sequence corresponded to amino acids 200–563 (Fig. S1A).
Homology search revealed a sequence similarity and structural homology between the novel MuSK interacting protein and the family of RIN proteins, which currently consists of RIN1, 2 and 3 (Fig. S1B). Therefore, this protein was designated Rin-like (Rinl) (gene locus: Rinl). Similar to the so-far known family members Rinl contains an SH2 domain, an RH domain and a Vps9 domain. In contrast, the RA domain and proline-rich regions found in RIN1, 2 and 3 are not present in Rinl.
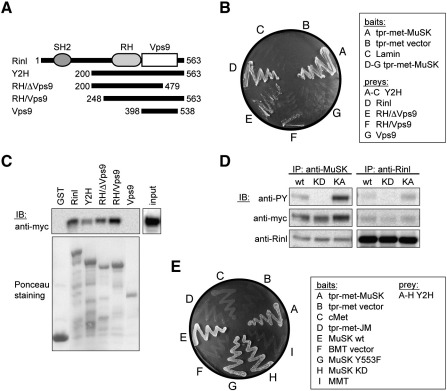
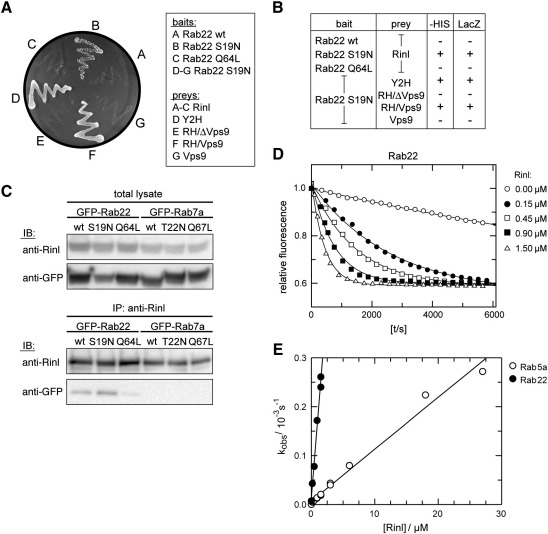
To determine the region(s) within Rinl necessary for interaction with MuSK, we generated several Rinl deletion mutants fused to the Gal4 activation domain (Fig. 1A). Using the reporter genes HIS and lacZ, interactions with the full-length MuSK cytoplasmic domain were assayed in yeast. As shown in Fig. 1B, deletion mutants lacking either the SH2 domain (Y2H and RH/Vps9) or the Vps9 domain (RH/ΔVps9) were able to interact with MuSK. In contrast, no interaction was detected with a deletion construct that encodes the Vps9 domain only (Vps9). We confirmed these findings by GST pulldowns of MuSK expressed in HEK 293T cells using the same Rinl deletion constructs fused to GST (Fig. 1C). These results suggest that the Rinl/MuSK interaction depends on the RH domain of Rinl.
Fig. 1.
Rinl is a specific binding partner of MuSK. (A) A scheme of Rinl and corresponding truncation mutants. (B) Yeast was transformed with bait and prey constructs as indicated. Interaction was assayed by growth of yeast clones on –HIS. (C) Rinl GST-deletion constructs were purified from bacteria and used for a pulldown of MuSK-myc expressed in HEK 293T cells. Proteins were detected by immunoblotting (IB) using anti-myc antibodies. GST protein input was assayed by Ponceau staining. 5% of the total MuSK-myc input is shown in the right panel. (D) MuSK-myc and Rinl were expressed in HEK 293T cells and co-immunoprecipitated either with anti-MuSK antibodies or anti-Rinl antibodies. Immunoprecipitated proteins were assayed by immunoblotting using anti-myc and anti-Rinl antibodies, respectively. Phosphorylated MuSK was detected with anti-phosphotyrosine (PY). wt, wild-type; KD, kinase-dead; KA, kinase-active. IP, immunoprecipitation. (E) Yeast was transformed with the indicated MuSK bait constructs and the Rinl (Y2H) prey construct. Interaction was followed by growth of yeast clones on –HIS.
The MuSK cytoplasmic domain consists of a short juxtamembrane region, a tyrosine kinase domain and an eight amino acid C-terminal tail. Upon agrin stimulation MuSK becomes tyrosine-phosphorylated, which induces recruitment of downstream factors and consequently downstream signaling [25,27,36]. To test whether the interaction between MuSK and Rinl is phosphorylation dependent, we expressed MuSK wild-type, MuSK kinase-active or MuSK kinase-dead together with Rinl in HEK 293T cells. Co-immunoprecipitation of Rinl with MuSK was detected independent of MuSK phosphorylation (Fig. 1D and Fig. S2). To narrow down the site of interaction within MuSK we generated MuSK deletion and point mutants fused to the lexA binding domain. Binding of Rinl to these MuSK mutant proteins was assayed in yeast using the reporter genes HIS and lacZ (Fig. 1E). Mutation of tyrosines in the autoactivation loop of the kinase domain (MuSK KD) do not affect Rinl/MuSK interaction. Similarly, the juxtamembrane tyrosine Y553 is dispensable for Rinl binding to MuSK. We did not include any truncations into the kinase domain since deletions of C-terminal parts will destroy the correct folding of the protein and consequently negatively influence our binding studies. We used however constructs carrying only the MuSK juxtamembrane region (JM) or the MuSK juxtamembrane NPXY motif inserted in the TrkA cytoplasmic domain (MMT). These fusion proteins are unable to interact with Rinl. Taken together, we conclude that Rinl binds to the MuSK kinase domain and that this binding is independent of MuSK phosphorylation.
3.2. Rinl is ubiquitously expressed with highest expression in lymphoid organs and specifically accumulated at NMSs
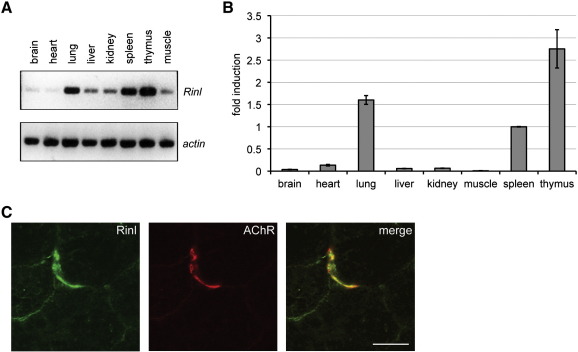
We performed RT-PCR using RNA isolated from adult mouse tissue. Rinl is expressed in all tested tissues with the highest expression in lung, spleen and thymus (Fig. 2A). These data were confirmed by qPCR showing that Rinl is up to 40 times more expressed in thymus than muscle (Fig. 2B). We also studied the expression of Rinl during development and found a slight increase in expression in spleen and thymus within the first weeks after birth but no profound developmental regulation of Rinl gene expression in brain and muscle (Fig. S3A and B). To further extend these expression studies we investigated Rinl protein expression in thymus, spleen and muscle. Analysis by immunoblotting revealed a similar expression profile as detected by RT-PCR. High expression was found in thymus and spleen compared to a weak expression in muscle (Fig. S3C).
Fig. 2.
Rinl expression in different tissues and localization at the NMS. (A) RT-PCR was performed from different mouse tissue samples. Actin was used as quality control. (B) qPCR from cDNAs generated from different tissue RNAs was performed. Spleen was set to 1. s.e.m. is shown. (C) Mouse muscle sections were stained with anti-Rinl antibodies and Alexa 594-conjugated α-BGT. Images were taken on a confocal microscope. Scale bar, 20 μm.
Proteins important for NMS development are usually enriched at synaptic sites. We therefore tested whether Rinl is localized at NMSs. We stained muscle sections with antibodies against Rinl and Alexa-594 conjugated α-BGT (Fig. 2C and S4). As shown in Fig. 2C, a weak but specific enrichment of Rinl at NMSs is detectable.
3.3. Rinl acts as GEF for Rab5a
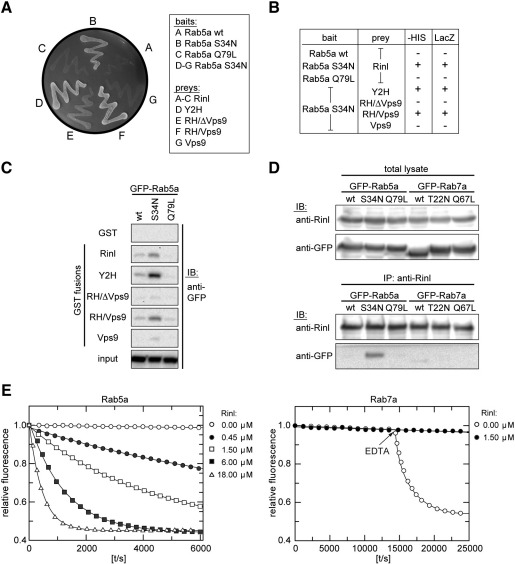
The family of RIN proteins is characterized by its Vps9 domain and its binding affinity for Rab5a. All so far known RIN proteins are able to catalyze the GDP to GTP exchange on Rab5a [3,15,37]. First experiments were performed using the yeast two-hybrid system to determine whether Rinl binds to Rab5a. The dominant-negative form Rab5a S34N showed a strong interaction with Rinl. In contrast, Rinl did not interact with Rab5a wild-type and the constitutively-active Rab5a Q79L variant (Fig. 3A). Dominant-negative mutations are known to reduce the nucleotide affinity of the G-protein, resulting in increased concentrations of nucleotide free G-protein which binds with high affinity to its GEFs. The observed interaction profile thus suggests that Rinl might be a Rab5a GEF. To map the site of interaction we used the Rinl mutant constructs described in Fig. 1A and found that the N-terminal region is dispensable for Rinl/Rab5a interaction. However, deletion of the Vps9 domain or the RH domain abolished the binding of Rinl to Rab5a S34N suggesting that both the RH and Vps9 domain are required for interaction (Fig. 3A and B). These findings were confirmed by GST pulldowns of Rab5a expressed in COS7 cells using the same Rinl deletion constructs fused to GST (Fig. 3C). We further tested the interaction between Rinl and Rab5a by co-immunoprecipitation from HEK 293T cells transfected with Rinl and Rab5a wild-type, Rab5a S34N or Rab5a Q79L, respectively. Rab5a S34N efficiently co-immunoprecipitates with Rinl, whereas Rab5a wild-type and Rab5a Q79L show a weak or no interaction (Fig. 3D). In contrast, Rinl does not bind to the late endosomal marker Rab7a, independent of its activation status.
Fig. 3.
Rinl preferentially interacts with nucleotide free Rab5a and acts as GEF for Rab5a. (A) Yeast was transformed with the indicated Rab5a bait constructs and Rinl full-length and deletion prey constructs. Interaction was screened by growth on –HIS. (B) Table showing the activation of the reporter genes HIS and lacZ. (C) Rinl GST-deletion constructs were purified from bacteria and used for a pulldown of GFP-Rab5a (wt, S34N, Q79L) expressed in COS7 cells. Proteins were detected by immunoblotting (IB) using anti-GFP antibodies. 5% of the total Rab5a lysates are shown as input. (D) Extracts of transiently transfected HEK 293T cells were used to immunoprecipitate Rinl with anti-Rinl antibodies. Co-immunoprecipitated Rab5a was detected by immunoblotting with anti-GFP. No Rab7a co-immunoprecipitation is detected. 10% of the input is shown (total lysate). IP, immunoprecipitation. (E) 200 nM Rab5a or Rab7a loaded with mGDP were incubated with increasing amounts of Rinl in the presence of 20 μM unlabeled GDP. The exchange of mGDP for GDP was measured as decay in fluorescence signal. In case of Rab7a proper nucleotide loading was demonstrated by the addition of EDTA at the indicated time point, which induces the release of nucleotides and a rapid decay in the fluorescence signal.
As Rinl contains a classical Vps9 domain and binds preferentially to nucleotide free Rab5a, we next asked whether Rinl indeed acts as GEF for Rab5a. Recombinant proteins were expressed in bacteria and used in an in vitro assay to measure nucleotide exchange. In brief, the Rab GTPase was loaded with the fluorescent nucleotide analog mGDP. The fluorescence intensity of mGDP is approximately twice as high if bound to the hydrophobic environment of a protein as if exposed freely to the buffer solution. The exchange of mGDP in the presence of excess unlabeled GDP can thus be measured in real time as fluorescence signal decay. As shown in Fig. 3E, the addition of Rinl accelerates nucleotide exchange in a concentration dependent manner by several orders of magnitude. In contrast, no exchange activity of Rinl for Rab7a could be detected. To validate proper nucleotide loading of Rab7a, the Mg2+ chelator EDTA was added to the control reaction at the indicated time point. This induces the release of nucleotides from G-proteins as their binding is Mg2+ dependent. These data demonstrate that Rinl similar to the other members of the RIN protein family acts as GEF for Rab5a.
3.4. Rinl colocalizes with dominant-negative Rab5a to cytoskeletal-rich membrane ruffles
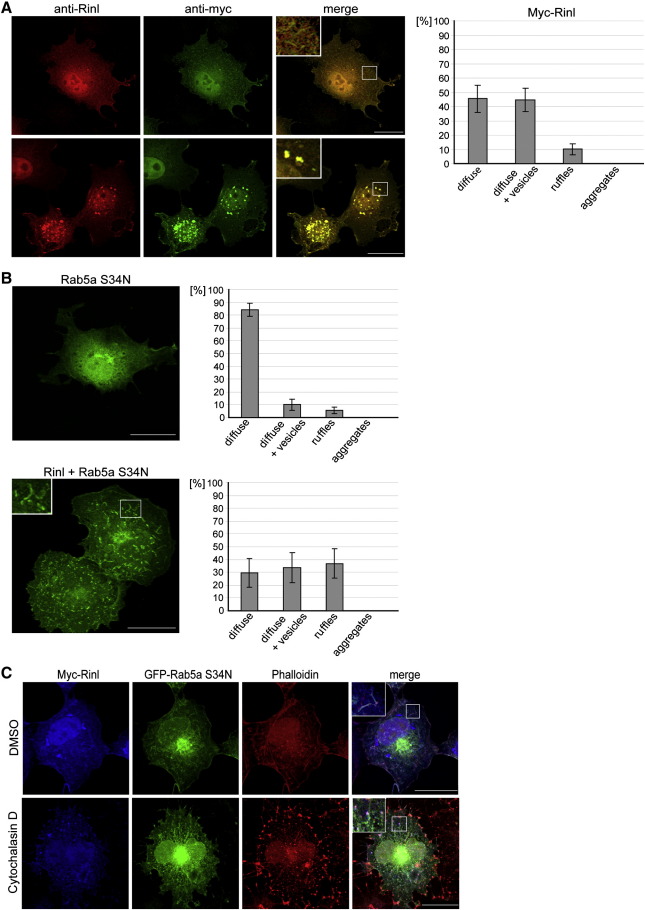
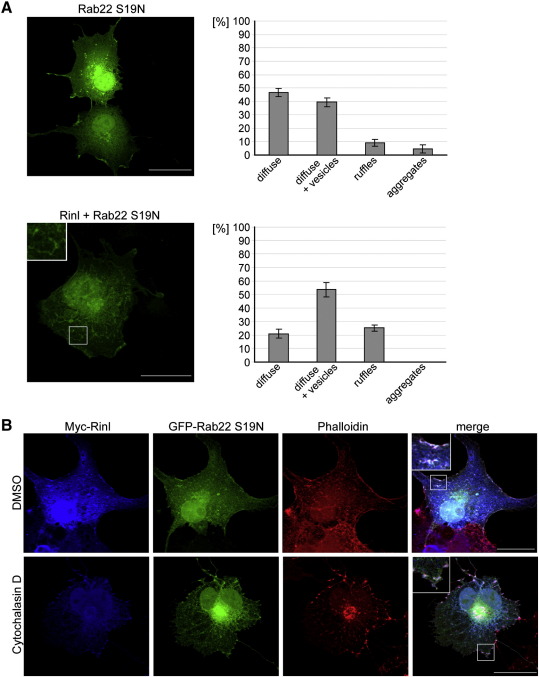
To examine the subcellular distribution of Rinl COS7 cells were transfected with myc-tagged Rinl (Fig. 4A). We found Rinl localized to a variety of different compartments. To analyze the distribution in more detail we divided Rinl localization into four categories and quantified the distribution within these categories. As shown in Fig. 4A, around 90% of Rinl is localized either diffusely and/or in vesicles. The remaining 10% of Rinl show a ruffle-like distribution. This localization pattern is the same for GFP-tagged Rinl or untagged Rinl (Fig. S5). Rinl shows no co-localization with early endosomes, recycling and late endosomes or the Golgi (Fig. S6). Interestingly, upon co-expression of Rinl and Rab5a S34N, Rinl and Rab5a are co-localized predominantly to membrane ruffles (Fig. 4B and S7A). Similarly, a weak co-localization between Rinl and Rab5a wt is detectable at membrane-ruffles. This co-localization is increased when Rinl lacking the SH2 domain is co-expressed with Rab5a wt (Fig. S7B). Most importantly, Rab5a S34N becomes redistributed from a diffuse cytoplasmic localization to either ruffle-like or vesicular-like structures. Membrane ruffles have been implicated in endocytotic processes associated with cytoskeletal rearrangements. To test whether actin is concentrated within the Rinl/Rab5a-positive ruffles we stained cells with phalloidin. Fig. 4C shows a co-localization of actin with Rinl and Rab5a. Moreover, disruption of the cytoskeleton using cytochalasin D blocked the formation of membrane ruffles and caused an accumulation of Rinl and Rab5a in actin-positive aggregates.
Fig. 4.
Subcellular localization of Rinl and co-localization with Rab5a and actin. (A) myc-tagged Rinl was transiently expressed in COS7 cells and stained with antibodies against Rinl and myc. Representative confocal images are shown. Scale bar, 25 μm. Transfected cells were assayed for their Rinl expression patterns. Expression patterns were divided into four categories. A quantification is shown (n > 100). Error bars, s.e.m. (B) GFP-Rab5a S34N alone or together with myc-tagged Rinl were co-expressed in COS7 cells. Scale bar, 25 μm. A quantification of the Rab5a S34N distribution was performed as described in A (n = 50). Error bars, s.e.m. (C) Transiently transfected COS7 cells were treated with cytochalasin D or DMSO for 30 minutes. Cells were stained with anti-myc antibodies to detect Rinl and with TRITC-conjugated phalloidin to label the actin cytoskeleton. Representative confocal images are shown. Scale bar, 25 μm.
3.5. Rinl acts as GEF for Rab22 and co-localizes with Rab22 to actin-rich domains
Rab22 is the closest homologue of Rab5 with 52% sequence identity [38]. Like Rab5a, Rab22 has been localized to early endosomes but its role during endocytosis is so far not well understood. Since Vps9 domain containing proteins have been reported to activate Rab5a and its homologues Rab21 and Rab22, we tested the interaction of Rinl with Rab22 [6]. Using the yeast two-hybrid system we detect a strong interaction between Rinl and the dominant-negative form of Rab22. Moreover, the binding of Rinl to Rab22 is dependent on the RH and Vps9 domain but independent of the N-terminal region (Fig. 5A and B). The Rinl/Rab22 interaction was confirmed by co-immunoprecipitation experiments in transfected HEK 293T cells. Dominant-negative Rab22 S19N efficiently co-immunoprecipitates with Rinl upon co-expression, whereas Rab22 wild-type and constitutively active Rab22 Q64L show a weak or no interaction (Fig. 5C). To determine whether Rinl acts indeed as GEF for Rab22, the GEF activity of Rinl toward Rab22 was measured in vitro (Fig. 5D). Rinl strongly accelerates nucleotide exchange of Rab22 and can therefore be classified as a GEF of Rab22. For a more quantitative comparison of the Rinl activity toward Rab5a and Rab22, the rates of the nucleotide exchange reaction, kobs, were determined from measurements as presented in Figs. 3E and 5D. The dependency of kobs on the Rinl concentration is presented in Fig. 5E. As can be seen, nucleotide exchange toward Rab22 is more sensitive to the concentration of Rinl, which thus displays a higher catalytic rate for Rab22 than for Rab5a. Moreover, Rinl and Rab22 S19N co-localize to similar compartments upon co-expression in COS7 cells (Fig. S7). Like for Rab5a, Rinl only weakly co-localizes with Rab22 wt whereas Rinl lacking the SH2 domain robustly co-localizes with Rab22 wt (Fig. S7B). Rinl induces a redistribution of diffusely localized Rab22 to ruffle-like structures (Fig. 6A). Similar to Rab5a and Rinl, also Rab22 and Rinl co-localize to actin-positive structures (Fig. 6B).
Fig. 5.
Rinl preferentially interacts with GDP-bound Rab22 and acts as GEF for Rab22. (A) Yeast was transformed with Rab22 bait constructs (wt, S19N, Q64L) and Rinl full-length or deletion prey plasmids. Growth on –HIS is shown. (B) The interaction between Rab22 and Rinl was assayed by growth on –HIS and LacZ expression (β-gal assay). (C) Extracts of transiently transfected HEK 293T cells were used to immunoprecipitate Rinl with anti-Rinl antibodies. Co-immunoprecipitated Rab22 was detected by immunoblotting (IB) with anti-GFP. Rab7a was used as negative control. 10% of the input is shown in the top panel (total lysate). (D) 200 nM of mGDP loaded Rab22 were incubated with increasing amounts of Rinl in the presence of 20 μM unlabeled GDP. The exchange of mGDP for GDP was measured in real time as decay in fluorescence signal. (E) The velocity of the nucleotide exchange reaction (kobs) toward Rab5a and Rab22 were determined and plotted against the concentration of Rinl.
Fig. 6.
Rab22 and Rinl co-localize to actin-positive membrane ruffles. (A) GFP-Rab22 S19N alone or together with myc-tagged Rinl were expressed in COS7 cells. Representative confocal images are shown. Scale bar, 25 μm. Rab22 expression, when transfected alone or together with Rinl, was assayed by confocal microscopy. Subcellular expression patterns were quantified as described in Fig. 5. n > 50; Error bars, s.e.m. (B) Transiently transfected COS7 cells were treated with cytochalasin D or DMSO for 30 minutes. Cells were stained with anti-myc antibodies to detect Rinl and with TRITC-conjugated phalloidin to label the actin cytoskeleton. Scale bar, 25 μm.
3.6. Rinl recruits Rab5a and Rab22 to the cytoskeleton via the Vps9 domain
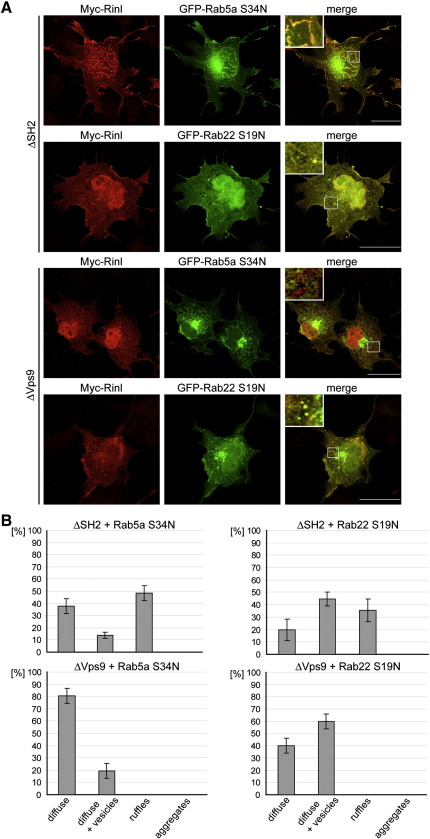
Since we detected a significant redistribution of dominant-negative Rab5a and Rab22 upon co-expression with Rinl, we next asked how this localization is influenced by Rinl deletion mutants. For that we used constructs that either lack the N-terminal SH2 domain or the C-terminal Vps9 domain. These truncations were co-expressed with Rab5a S34N or Rab22 S19N and the localization assayed by immunostaining. Rinl lacking the SH2 domain (ΔSH2) localizes to vesicles and ruffles together with Rab5a and Rab22 (Fig. 7). In particular, Rinl ΔSH2 has a more pronounced localization to ruffles compared to full-length Rinl (data not shown). Moreover, Rinl ΔSH2 shows a higher degree of co-localization with Rab5a or Rab22 to ruffles than full-length Rinl and Rab5a/Rab22 (compare Figs. 4B and 6A to 7B). This suggests that the SH2 domain might have an inhibitory effect on the interaction between Rinl and Rab proteins. In contrast, Rinl lacking the Vps9 domain (ΔVps9) does not co-localize with Rab5a S34N and Rab22 S19N (Fig. 7). Rab5a and Rab22 are unchanged upon co-expression with Rinl ΔVps9 and remain diffusely distributed and/or in vesicles. Rinl ΔVps9 itself is localized mainly throughout the cytoplasm or in vesicles but does not attach to actin-rich membrane ruffles. Therefore, the Vps9 domain is not only responsible for Rab5a/Rab22 activation but also mediates the localization of Rinl and Rab proteins to the cytoskeleton.
Fig. 7.
Rinl-dependent recruitment of Rab5a and Rab22 to the cytoskeleton. (A) myc-tagged Rinl (ΔSH2 and ΔVps9) and GDP-bound Rab5a or Rab22 were co-transfected into COS7 cells. Cells were stained with anti-myc antibodies and visualized by confocal microscopy. Representative images are shown. Scale bar, 25 μm. (B) The distribution of Rab5a and Rab22 was quantified as described in Fig. 5. n > 35; Error bars, s.e.m.
3.7. Rinl regulates fluid-phase and receptor-mediated endocytosis
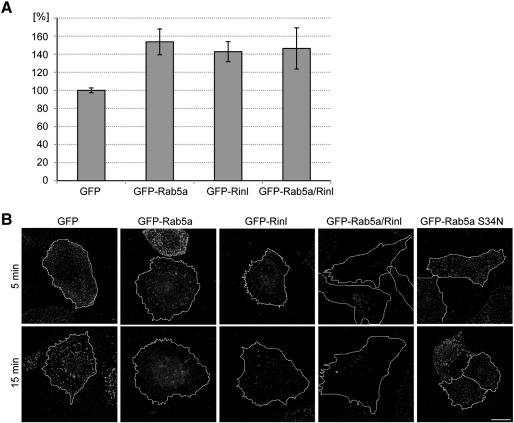
Since RIN family members have previously been implicated in endocytotic processes we set out to study the function of Rinl during fluid-phase endocytosis. For that we examined the HRP uptake in HEK 293T cells expressing Rinl, Rab5a wt, Rab5a wt together with Rinl, or GFP as a control. Expression of Rinl and Rab5a induced the internalization of HRP (Fig. 8A). The co-expression of Rab5a wt and Rinl induced a similar degree of HRP uptake than expression of Rinl or Rab5a wt alone.
Fig. 8.
Rinl increases the internalization of HRP and EGFR. (A) HEK 293T cells expressing GFP-Rinl, GFP-Rab5a wt, GFP-Rinl and GFP-Rab5a wt or GFP alone were loaded with HRP. HRP uptake was quantified from two independent experiments done in triplicates. Error bars, s.e.m. (B) HeLa cells were transfected with GFP, GFP-Rinl, GFP-Rab5a wt, GFP-Rinl and GFP-Rab5a wt or GFP-Rab5a S34N. Cells were incubated with Alexa 555-conjugated EGF at 4 °C and EGF uptake at 37 °C was imaged after 5 and 15 minutes. Representative confocal images are shown. Scale bar, 25 μm. The GFP signals of the equivalent cells are shown in Fig. S8.
It has previously been demonstrated that RIN1 acts as regulator of EGFR endocytosis [12,39]. We therefore asked whether Rinl also affects EGFR internalization. We used Alexa 555-conjugated EGF to label endogenous EGFR in HeLa cells expressing Rinl, Rab5a wt, Rab5a S34N, Rinl together with Rab5a wt, or GFP as a control. EGF uptake at 37 °C was imaged at various times. As shown in Figs. 8B and S8, in control cells EGFR endocytosis is rapidly induced within 5 minutes and perinuclear accumulations of EGF-positive vesicles is prominent by 15 minutes. Expression of Rab5a wt or Rinl leads to an acceleration of EGFR endocytosis and a decrease in EGF-positive vesicles. This increase in EGFR endocytosis is especially pronounced in cells co-expressing Rab5a wt and Rinl. In contrast, cells expressing Rab5a S34N show a reduced EGFR endocytosis. These results support a functional role of Rinl during early endocytotic processes.
4. Discussion
The GTPase Rab5a is the key regulator during early endocytotic processes. Therefore, it is of particular interest to identify mechanisms and molecules that modulate Rab5a action. In this study we report the identification and characterization of Rinl, a novel Rab5a GEF, which shows a high homology to the family of RIN proteins. Rinl was isolated via its interaction with the RTK MuSK. It specifically interacts with MuSK through the central portion of the protein containing the RH domain. Rinl interacts and co-localizes with Rab5a and catalyzes GDP/GTP exchange on Rab5a. Similar biochemical and enzymatic properties were demonstrated toward Rab22. Furthermore, we identified the Vps9 domain of Rinl as a critical molecular determinant that controls the recruitment of Rab5a and Rab22 to cytoskeletal membrane compartments. Most importantly, Rinl stimulates fluid-phase uptake and EGFR endocytosis.
The formation of the NMS is crucially linked to signal transduction events induced by the muscle-specific RTK MuSK [18]. MuSK activation via agrin/Lrp4 induces a signaling cascade that leads to post- as well as presynaptic differentiation. Several MuSK binding proteins have been identified which include adaptor proteins like Dok7, kinases like Abl and scaffolding proteins including Magi-1c and ColQ [18]. More recently the trafficking protein NSF and the E3 ubiquitin ligases PDZRN3 and PAUL were isolated as MuSK interaction partner, which are thought to regulate MuSK endocytosis and degradation, respectively [40–42]. Here we identified Rinl as novel interaction partner of MuSK. Binding of Rinl to MuSK requires the internal RH domain of Rinl but is independent of MuSK phosphorylation. This suggests that Rinl binding is not associated with MuSK activation. However, a regulation of Rinl/MuSK interaction dependent on the subcellular localization appears likely since we show a specific localization of Rinl to vesicles and membrane ruffles. In support of this hypothesis we detect a specific co-localization of MuSK and Rinl at actin-positive membrane ruffles (data not shown). The identification of Rinl as binding partner of MuSK appeared of distinct interest since it was recently demonstrated that MuSK endocytosis regulates MuSK signaling [42]. However so far, we have not been able to correlate Rinl action and MuSK function. Rinl is very weakly expressed in muscle cells questioning the importance of Rinl in cultured muscle cells. Nevertheless, this does not exclude the possibility that Rinl plays a role during MuSK-dependent signal transduction in vivo, either at the NMS or in the brain, where MuSK function during memory formation has been implicated recently [43]. A gene targeting approach in mice will be necessary to answer these questions. Furthermore, a functional compensation by one of the other RIN family members cannot be ruled out at the moment.
The RIN family members have been implicated in early endocytotic events [1]. In particular, the role of RIN1 during receptor endocytosis has been demonstrated many-fold [12–14,39]. Here we are able to show that Rinl overexpression accelerates fluid-phase endocytosis as well as EGFR endocytosis. These data support our in vitro data and provide first hints on the physiological role of Rinl. RIN1 has been postulated as crucial regulator of EGFR endocytosis and signaling [12,39,44]. Our findings raise the questions whether Rinl represents a similarly important regulator of EGFR endocytosis and whether Rinl and RIN1 play complementary roles during EGFR endocytosis due to their differential expression patterns.
The current members of the RIN protein family share an SH2 domain, an RH region, a Vps9 domain and an RA domain. RIN1 was isolated by its ability to bind to H-Ras via the RA domain thereby competing with Raf1 and inhibiting Ras action [2,45]. In addition, the RA domain has been implicated in the interaction with the EGFR via Ras, which leads to a recruitment of RIN1 to the activated receptor linking internalized EGFR to Rab5a-positive endosomes [3]. Rinl lacks an RA domain. Therefore it appears unlikely that Rinl acts as an effector for Ras proteins. However, a recruitment to activated RTKs can still occur through the N-terminal SH2 domain, which would then bring the intracellular receptors to early endosomes. Such an SH2-dependent interaction has been shown for RIN1 and EGFR as well as RIN1 and EphA4 [12,13].
The RIN proteins belong to the family of Vps9 domain containing proteins. These proteins are characterized by their ability to bind Rab5a and to catalyze the GDP/GTP exchange on Rab5a [1]. Crystallization studies on Rabex-5 have shown that an N-terminal helical bundle in addition to the Vps9 domain is required for GEF activity, the so-called HB-Vps9 tandem [6]. The helical bundle conforms to the RH domain in the RIN proteins. Consistent with the Rabex-5 data, it was shown that a splice-variant of RIN1 lacking part of the helical bundle is unable to interact with dominant-negative Rab5a. Likewise, Rinl truncations deleting the RH domain/helical bundle do not bind dominant-negative Rab5a implicating similar biochemical properties for all Vps9 domain containing proteins. It was also reported that full-length Vps9 domain containing proteins including the RIN proteins have a reduced binding activity and/or GEF activity for Rab5a [6,46]. This suggests that autoinhibition by regulatory elements in the N- and/or C-terminus play a role. Similarly, we find a reduced binding activity for Rab5a and Rab22 in full-length Rinl constructs compared to Rinl constructs carrying only the RH and Vps9 domain. Therefore, the N-terminal SH2 domain might represent such an inhibitory element. Protein interactions via the SH2 domain would then relief the autoinhibition and induce exchange activity. So far however, it remains unclear which proteins bind to the SH2 domain of Rinl.
Analysis of the Rabex-5 HB-Vps9 tandem revealed a high specificity toward the Rab5 subfamily but also showed a selective exchange activity for the different Rab5 subfamily members: strong GEF activity for Rab5 and Rab21, a weak activity for Rab22 [6]. This specificity and selectivity is achieved on one hand by conserved exchange determinants on a common surface of the Vps9 domain and on the other hand by invariant aromatic residues in the switch regions of the Rab GTPases. Delprato and colleagues also reported a similar catalytic activity and Rab specificity for RIN1 [6]. In this study we show that Rinl specifically binds Rab5a and Rab22, and there preferentially the nucleotide free forms. Rinl acts as GEF for Rab22 and Rab5a, surprisingly however, presents a higher efficiency for Rab22. This distinguishes Rinl from RIN1 and suggests that different Vps9 domains have different specificity profiles for the Rab5 subfamily. This of course also raises the question whether this specificity is also represented at a physiological level whereby different RIN proteins regulate different Rab5 subfamily dependent processes. Four amino acids (D313, P317, Y354 and T357) in the Vps9 domain in Rabex-5 have been shown to be essential for exchange activity and these residues are highly conserved among different Vps9 domains [6]. Likewise, the Rinl Vps9 domain contains these conserved amino acids (D456, P460, Y497 and T500). Other residues, which have been shown to lie within the binding site for the GTPase differ between Rabex-5 and Rinl. These residues are expected to be responsible for the differences in the specificity profile.
When studying the subcellular distribution of Rinl protein we noticed a characteristic localization in vesicles and membrane ruffles. Moreover, co-expression of Rinl and Rab5a or Rab22 mutants with lowered nucleotide affinity leads to a redistribution of diffusely expressed Rab5a and Rab22 to actin-positive membrane compartments. The localization of Rinl to membrane ruffles as well as the recruitment of Rab5a and Rab22 to the cytoskeleton are dependent on the Vps9 domain. It is unclear at the moment whether the Vps9 domain interacts directly or indirectly (via a so-far unknown actin binding protein) with the cytoskeleton. Rab mutants with lowered nucleotide affinity are known to bind GEFs with high affinity, thereby trapping the active GEF in an unproductive complex [47]. The ability of Rinl to recruit Rab5a S34N and Rab22 S19N indicates that Rinl interacts with the actin cytoskeleton in an active state. Thus it is expected that Rinl activates Rab proteins locally at the cytoskeleton. Actin cytoskeleton remodeling has been implicated in early endocytosis as well as recycling [48]. Furthermore, membrane ruffling is a characteristic of actin remodeling and is closely associated with regions of active endocytosis [49]. Our findings therefore support a model whereby Rinl-dependent activation of Rab5a and Rab22 at the cytoskeleton regulates early endocytotic and/or recycling processes. Future experiments will have to show how actin remodeling, exchange activity by Rinl and Rab5a/Rab22-dependent endocytosis are connected.
The following are the supplementary materials related to this article.
Supplementary material
Acknowledgments
We are grateful to Jeffrey Chamberlain for providing us with the mouse muscle prey library. We would like to thank Walter Birchmeier, Cecilia Bucci, Steve Burden, Judy Donaldson, Said Hashemolhosseini and Marino Zerial for providing plasmids. Many thanks also go to Marije Rensen-De Leeuw for technical assistance. Wilfried Ellmeier provided useful comments on the manuscript. This work was supported by the Austrian Science Fund (P19223-B09 to R.H.) and by the Chemical Sciences of the Netherlands Organization for Scientific Research (NOW-CW to H.R.). B.W. was supported by the DOC-fFORTE Programme from the Austrian Academy of Sciences. M.P. was supported by the TI Pharma Project T3-106.
References
- 1.Carney D.S., Davies B.A., Horazdovsky B.F. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Han L., Colicelli J. A human protein selected for interference with Ras function interacts directly with Ras and competes with Raf1. Mol. Cell. Biol. 1995;15:1318–1323. doi: 10.1128/mcb.15.3.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tall G.G., Barbieri M.A., Stahl P.D., Horazdovsky B.F. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev. Cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 4.Hama H., Tall G.G., Horazdovsky B.F. Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J. Biol. Chem. 1999;274:15284–15291. doi: 10.1074/jbc.274.21.15284. [DOI] [PubMed] [Google Scholar]
- 5.Vitale G., Alexandrov K., Ullrich O., Horiuchi H., Giner A., Dobson C., Baykova O., Gournier H., Stenmark H., Zerial M. The GDP/GTP cycle of Rab5 in the regulation of endocytotic membrane traffic. Cold Spring Harb. Symp. Quant. Biol. 1995;60:211–220. doi: 10.1101/sqb.1995.060.01.024. [DOI] [PubMed] [Google Scholar]
- 6.Delprato A., Merithew E., Lambright D.G. Structure, exchange determinants, and family-wide rab specificity of the tandem helical bundle and Vps9 domains of Rabex-5. Cell. 2004;118:607–617. doi: 10.1016/j.cell.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Zerial M., McBride H. Rab proteins as membrane organizers. Nat. reviews. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 8.Kauppi M., Simonsen A., Bremnes B., Vieira A., Callaghan J., Stenmark H., Olkkonen V.M. The small GTPase Rab22 interacts with EEA1 and controls endosomal membrane trafficking. J. Cell Sci. 2002;115:899–911. doi: 10.1242/jcs.115.5.899. [DOI] [PubMed] [Google Scholar]
- 9.Simpson J.C., Griffiths G., Wessling-Resnick M., Fransen J.A., Bennett H., Jones A.T. A role for the small GTPase Rab21 in the early endocytic pathway. J. Cell Sci. 2004;117:6297–6311. doi: 10.1242/jcs.01560. [DOI] [PubMed] [Google Scholar]
- 10.Weigert R., Yeung A.C., Li J., Donaldson J.G. Rab22a regulates the recycling of membrane proteins internalized independently of clathrin. Mol. Biol. Cell. 2004;15:3758–3770. doi: 10.1091/mbc.E04-04-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opdam F.J., Echard A., Croes H.J., van den Hurk J.A., van de Vorstenbosch R.A., Ginsel L.A., Goud B., Fransen J.A. The small GTPase Rab6B, a novel Rab6 subfamily member, is cell-type specifically expressed and localised to the Golgi apparatus. J. Cell Sci. 2000;113(Pt 15):2725–2735. doi: 10.1242/jcs.113.15.2725. [DOI] [PubMed] [Google Scholar]
- 12.Barbieri M.A., Kong C., Chen P.I., Horazdovsky B.F., Stahl P.D. The SRC homology 2 domain of Rin1 mediates its binding to the epidermal growth factor receptor and regulates receptor endocytosis. J. Biol. Chem. 2003;278:32027–32036. doi: 10.1074/jbc.M304324200. [DOI] [PubMed] [Google Scholar]
- 13.Deininger K., Eder M., Kramer E.R., Zieglgansberger W., Dodt H.U., Dornmair K., Colicelli J., Klein R. The Rab5 guanylate exchange factor Rin1 regulates endocytosis of the EphA4 receptor in mature excitatory neurons. Proc. Natl Acad. Sci. U.S.A. 2008;105:12539–12544. doi: 10.1073/pnas.0801174105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunker C.M., Giambini H., Galvis A., Hall J., Kruk I., Veisaga M.L., Barbieri M.A. Rin1 regulates insulin receptor signal transduction pathways. Exp. Cell Res. 2006;312:1106–1118. doi: 10.1016/j.yexcr.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Kajiho H., Saito K., Tsujita K., Kontani K., Araki Y., Kurosu H., Katada T. RIN3: a novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J. Cell Sci. 2003;116:4159–4168. doi: 10.1242/jcs.00718. [DOI] [PubMed] [Google Scholar]
- 16.Lai W.H., Cameron P.H., Wada I., Doherty J.J., 2nd, Kay D.G., Posner B.I., Bergeron J.J. Ligand-mediated internalization, recycling, and downregulation of the epidermal growth factor receptor in vivo. J. Cell Biol. 1989;109:2741–2749. doi: 10.1083/jcb.109.6.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorkin A., von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat. reviews. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H., Xiong W., Lin M. To build a synapse: signaling pathways in neuromuscular junction assembly. Dev. Camb. Engl. 2010;137:1017–1033. doi: 10.1242/dev.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass D.J., Bowen D.C., Stitt T.N., Radziejewski C., Bruno J., Ryan T.E., Gies D.R., Shah S., Mattsson K., Burden S.J., DiStefano P.S., Valenzuela D.M., DeChiara T.M., Yancopoulos G.D. Agrin acts via a MuSK receptor complex. Cell. 1996;85:513–523. doi: 10.1016/s0092-8674(00)81252-0. [DOI] [PubMed] [Google Scholar]
- 20.Kim N., Stiegler A.L., Cameron T.O., Hallock P.T., Gomez A.M., Huang J.H., Hubbard S.R., Dustin M.L., Burden S.J. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell. 2008;135:334–342. doi: 10.1016/j.cell.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B., Luo S., Wang Q., Suzuki T., Xiong W.C., Mei L. LRP4 serves as a coreceptor of agrin. Neuron. 2008;60:285–297. doi: 10.1016/j.neuron.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeChiara T.M., Bowen D.C., Valenzuela D.M., Simmons M.V., Poueymirou W.T., Thomas S., Kinetz E., Compton D.L., Rojas E., Park J.S., Smith C., DiStefano P.S., Glass D.J., Burden S.J., Yancopoulos G.D. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–512. doi: 10.1016/s0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- 23.Gautam M., Noakes P.G., Moscoso L., Rupp F., Scheller R.H., Merlie J.P., Sanes J.R. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85:525–535. doi: 10.1016/s0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- 24.Weatherbee S.D., Anderson K.V., Niswander L.A. LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Dev. Camb. Engl. 2006;133:4993–5000. doi: 10.1242/dev.02696. [DOI] [PubMed] [Google Scholar]
- 25.Watty A., Neubauer G., Dreger M., Zimmer M., Wilm M., Burden S.J. The in vitro and in vivo phosphotyrosine map of activated MuSK. Proc. Natl Acad. Sci. U.S.A. 2000;97:4585–4590. doi: 10.1073/pnas.080061997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mossier B., Togel M., Fuchs K., Sieghart W. Immunoaffinity purification of gamma-aminobutyric acidA (GABAA) receptors containing gamma 1-subunits. Evidence for the presence of a single type of gamma-subunit in GABAA receptors. J. Biol. Chem. 1994;269:25777–25782. [PubMed] [Google Scholar]
- 27.Herbst R., Burden S.J. The juxtamembrane region of MuSK has a critical role in agrin-mediated signaling. EMBO J. 2000;19:67–77. doi: 10.1093/emboj/19.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glass D.J., Apel E.D., Shah S., Bowen D.C., DeChiara T.M., Stitt T.N., Sanes J.R., Yancopoulos G.D. Kinase domain of the muscle-specific receptor tyrosine kinase (MuSK) is sufficient for phosphorylation but not clustering of acetylcholine receptors: required role for the MuSK ectodomain? Proc. Natl Acad. Sci. U.S.A. 1997;94:8848–8853. doi: 10.1073/pnas.94.16.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaeper U., Gehring N.H., Fuchs K.P., Sachs M., Kempkes B., Birchmeier W. Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses. J. Cell Biol. 2000;149:1419–1432. doi: 10.1083/jcb.149.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lumeng C., Phelps S., Crawford G.E., Walden P.D., Barald K., Chamberlain J.S. Interactions between beta 2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nat. Neurosci. 1999;2:611–617. doi: 10.1038/10165. [DOI] [PubMed] [Google Scholar]
- 32.Vojtek A.B., Hollenberg S.M. Ras–Raf interaction: two-hybrid analysis. Meth. Enzymol. 1995;255:331–342. doi: 10.1016/s0076-6879(95)55036-4. [DOI] [PubMed] [Google Scholar]
- 33.Nizhynska V., Neumueller R., Herbst R. Phosphoinositide 3-kinase acts through RAC and Cdc42 during agrin-induced acetylcholine receptor clustering. Dev. Neurobiol. 2007;67:1047–1058. doi: 10.1002/dneu.20371. [DOI] [PubMed] [Google Scholar]
- 34.Schnatwinkel C., Christoforidis S., Lindsay M.R., Uttenweiler-Joseph S., Wilm M., Parton R.G., Zerial M. The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2004;2:E261. doi: 10.1371/journal.pbio.0020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rehmann H. Characterization of the activation of the Rap-specific exchange factor Epac by cyclic nucleotides. Meth. Enzymol. 2006;407:159–173. doi: 10.1016/S0076-6879(05)07014-X. [DOI] [PubMed] [Google Scholar]
- 36.Zhou H., Glass D.J., Yancopoulos G.D., Sanes J.R. Distinct domains of MuSK mediate its abilities to induce and to associate with postsynaptic specializations. J. Cell Biol. 1999;146:1133–1146. doi: 10.1083/jcb.146.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito K., Murai J., Kajiho H., Kontani K., Kurosu H., Katada T. A novel binding protein composed of homophilic tetramer exhibits unique properties for the small GTPase Rab5. J. Biol. Chem. 2002;277:3412–3418. doi: 10.1074/jbc.M106276200. [DOI] [PubMed] [Google Scholar]
- 38.Pereira-Leal J.B., Seabra M.C. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 39.Kong C., Su X., Chen P.I., Stahl P.D. Rin1 interacts with signal-transducing adaptor molecule (STAM) and mediates epidermal growth factor receptor trafficking and degradation. J. Biol. Chem. 2007;282:15294–15301. doi: 10.1074/jbc.M611538200. [DOI] [PubMed] [Google Scholar]
- 40.Bromann P.A., Weiner J.A., Apel E.D., Lewis R.M., Sanes J.R. A putative ariadne-like E3 ubiquitin ligase (PAUL) that interacts with the muscle-specific kinase (MuSK) Gene Expr. Patterns. 2004;4:77–84. doi: 10.1016/s1567-133x(03)00146-7. [DOI] [PubMed] [Google Scholar]
- 41.Lu Z., Je H.S., Young P., Gross J., Lu B., Feng G. Regulation of synaptic growth and maturation by a synapse-associated E3 ubiquitin ligase at the neuromuscular junction. J. Cell Biol. 2007;177:1077–1089. doi: 10.1083/jcb.200610060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu D., Yang Z., Luo Z., Luo S., Xiong W.C., Mei L. Muscle-specific receptor tyrosine kinase endocytosis in acetylcholine receptor clustering in response to agrin. J. Neurosci. 2008;28:1688–1696. doi: 10.1523/JNEUROSCI.4130-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Osta A., Tsokas P., Pollonini G., Landau E.M., Blitzer R., Alberini C.M. MuSK expressed in the brain mediates cholinergic responses, synaptic plasticity, and memory formation. J. Neurosci. 2006;26:7919–7932. doi: 10.1523/JNEUROSCI.1674-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbieri M.A., Fernandez-Pol S., Hunker C., Horazdovsky B.H., Stahl P.D. Role of rab5 in EGF receptor-mediated signal transduction. Eur. J. Cell Biol. 2004;83:305–314. doi: 10.1078/0171-9335-00381. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Waldron R.T., Dhaka A., Patel A., Riley M.M., Rozengurt E., Colicelli J. The RAS effector RIN1 directly competes with RAF and is regulated by 14-3-3 proteins. Mol. Cell. Biol. 2002;22:916–926. doi: 10.1128/MCB.22.3.916-926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delprato A., Lambright D.G. Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat. Struct. Mol. Biol. 2007;14:406–412. doi: 10.1038/nsmb1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bos J.L., Rehmann H., Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 48.Doherty G.J., McMahon H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 49.Orth J.D., McNiven M.A. Get off my back! Rapid receptor internalization through circular dorsal ruffles. Cancer Res. 2006;66:11094–11096. doi: 10.1158/0008-5472.CAN-06-3397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material