Abstract
We investigated the association of polymorphisms in CCR5, the major human immunodeficiency virus (HIV)–1 coreceptor, and copy number of its potent ligand CCL3L1 with tuberculosis in 298 individuals from Colombia. The CCR5-HHD haplotype, a known genetic determinant of increased susceptibility to HIV-AIDS, and a high copy number of CCL3L1, a known genetic determinant of enhanced CCL3/CCL3L1 chemokine expression, each associated with presence of tuberculosis. Furthermore, CCR5-HHD was associated with higher CCR5 gene and surface expression. These results substantiate the strong link between the pro-inflammatory effects of CCR5 and its ligands with active tuberculosis and suggest that chemokine-chemokine receptor genetic determinants may influence tuberculosis in addition to HIV/AIDS.
Tuberculosis (TB) is still widespread, and according to the World Health Organization, 2 billion people are infected with the causative bacillus Mycobacterium tuberculosis [1]. Although ∼10% of infected persons develop active TB, the precise basis by which the majority of infected persons never experience progression to active TB remains unknown [1]. In addition to the environment and bacterium, host genetic differences that influence the inflammatory response are thought to contribute significantly to the final outcome of infection [1].
Genes encoding chemokines and their cognate receptors play a key role in the inflammatory response during TB. Prior studies have shown that (1) CC chemokine receptor 5 (CCR5), the major human immunodeficiency virus (HIV)–1 coreceptor, is highly expressed on T helper (Th) 1 cells (discussed in [2]); (2) Th1 responses play a critical role in immunity against TB [3]; (3) compared with healthy individuals, the levels of CCR5 are elevated in Th cells in patients with pulmonary TB [3, 4]; (4) CCR5 expression is increased in humans and rhesus monkeys with active TB [5, 6]; (5) the ligands of CCR5, such as CCL3 and CCL5, are also increased during TB [3, 4, 6, 7]; and (6) cell-mediated immunity (CMI) is a central determinant of TB resistance [3], and both CCR5 and CCL3L1, the most potent ligand of CCR5 [8], influence CMI (discussed in [9]). However, the association of variations in the chemokine-chemokine receptor pair CCL3L1 and CCR5 with TB in HIV-negative subjects is unknown. Together, these observations prompted us to test the hypothesis that variations in CCR5 and CCL3L1 gene copy number are associated with TB susceptibility. We tested this hypothesis in individuals with and without TB who were recruited from a well-defined northwestern Colombian population.
METHODS
Study Subjects
We evaluated 114 patients with TB and 184 healthy control subjects from the Paisa community in northwestern Colombia (South America), where TB has moderately-high endemicity. Additional characteristics of the study participants have been described, (see [10] and Supplementary Methods). Historical and genetic evidence suggest that this community is genetically isolated, with a documented low degree of admixture and a calculated ancestral ethnic composition of 85% white, 15% Amerindian, and a non-significant African contribution (see Supplementary Methods).
Genotyping
The methods for genotyping the CCR5 polymorphisms that allow for generation of the 9 major CCR5 haplotypes are as described elsewhere [8, 11, 12]. CCL3L1 gene copy number was determined as described elsewhere [8]. We genotyped a validated set of 96 ancestry informative markers (AIMs) using TaqMan genotyping assays (see Supplementary Methods).
Reporter Assays and CCR5 Expression
We introduced promoter sequences specific to HHA, HHB, and HHD CCR5 haplotypes into a firefly luciferase reporter vector and then evaluated the transcriptional strength of these haplotype-specific reporter constructs in in vitro transcription assays using methods described elsewhere [12] and in the Supplementary Methods section. Transcriptional activity was assessed in a Jurkat T-cell line and in anti-CD3/CD28 activated peripheral blood mononuclear cells (PBMCs) from healthy persons (see Supplementary Methods). CCR5 levels on CD4+ T cells were assessed by fluorescence activated cell sorting (see Supplementary Methods).
Statistical Analysis
The association of CCR5 haplotypes or CCL3L1 gene copy number with the risk of TB was evaluated using multiple logistic regression analyses. On the basis of the mean CCL3L1 copy number found in the control group (which was 2), we stratified the study subjects into 3 groups: those with <2, 2, or >2 CCL3L1 copies. We examined the potential contribution of population admixture to genotype-phenotype associations by using 96 AIMs with the EIGENSTRAT method and STRUCTURE software (see Supplementary Methods).
RESULTS
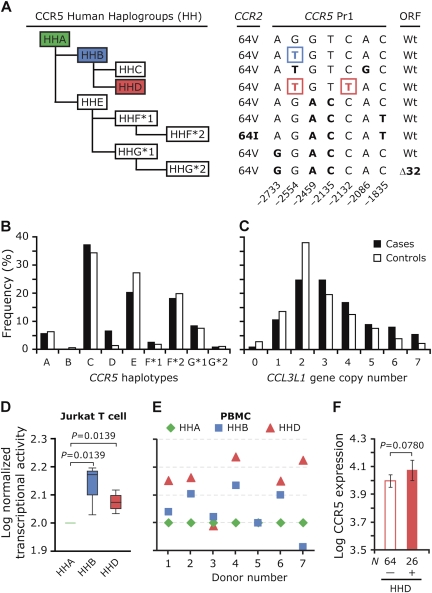
On the basis of linkage disequilibrium patterns, the major polymorphisms in CCR2-CCR5 were categorized into 9 CCR5 haplotypes, as described elsewhere ([12]; Figure 1A). We found that all of the polymorphisms typed were in Hardy-Weinberg equilibrium in control subjects. The CCR5-HHC haplotype was the most common haplotype in both case patients and control subjects, followed by HHE and HHF*2 (Figure 1B). In control subjects, 2 copies of CCL3L1 was the median gene copy number, whereas in case patients 2 and 3 CCL3L1 copies were the most frequent values (Figure 1C).
Figure 1.
Distribution of CCR5/CCL3L1 genotypes and influence of CCR5 promoter polymorphisms on gene and surface expression. A, Schematic represents the evolutionary relationship among CCR5 human haplogroups (HH) that contain the major CCR5 haplotypes designated as HH-A, -B, -C, -D, -E, F*1, F*2, G*1, and G*2 [12]. Haplotype classification is based on the linkage disequilibrium of the indicated 9 polymorphisms in CCR2 (V64I), CCR5 promoter 1 (Pr1), and the CCR5 open reading frame (ORF; Δ32) [12]. CCR5-HHA is the ancestral haplotype based on the evolutionary conservation at the indicated nucleotide positions with the chimpanzee CCR5 [12]. Nucleotide numbering is according to Mummidi et al [12]. The signature single-nucleotide polymorphisms (SNPs) that distinguish CCR5 haplotypes from the ancestral haplotype are in boldface font. The haplotypes studied in this report are highlighted in colored boxes and are color-matched. B and C, Distribution of CCR5 haplotypes (B) and CCL3L1 gene copy number (C) in case patients (n = 114) and control subjects (n = 184). D, Box-whisker plot showing haplotype-specific differences in CCR5 gene transcriptional activity in Jurkat T cells. Cells were cotransfected with HHA-, HHB-, and HHD-specific firefly luciferase reporter constructs and Renilla luciferase construct, and dual luciferase assays were performed. Data shown are from 4 independent experiments, and the transcriptional activity was log-transformed after normalization to HHA (see Supplementary Methods). Significance values shown at the top for the indicated comparisons were obtained using the Mann-Whitney U test. E, Haplotype-specific differences in CCR5 transcriptional activity in peripheral blood mononuclear cells (PBMCs). Haplotype-specific HHA (green), HHB (blue), and HHC (red) promoter constructs were nucleofected into PBMCs stimulated with anti-CD3/CD28 antibodies and transcriptional activity was assessed. All data were normalized to the transcriptional activity of the HHA-containing reporter vector. F, CCR5 surface expression on CD4+HLA-DR+ cells in a cohort of 90 human immunodeficiency virus–uninfected sex workers from South Africa. CCR5 expression was log-transformed. The height of the bar indicates the mean CCR5 surface expression in subjects who possess (+) or lack (-) the CCR5-HHD haplotype, and the error bars indicate the standard error of the mean. The P values were calculated using the Student t-test.
The proportion of men was higher among case patients ( = 50 patients [56.82%]) than among control subjects ( = 38 patients [43.18%]). The mean patient age (± standard deviation) was 39.83 ± 16.17 years, whereas the mean age (± standard deviation) of the control subjects was 46.41 ± 16.04 years. Younger age (odds ratio [OR], = 0.98; 95% confidence interval [CI,] = , 0.97–0.99; P = .003) and male sex (OR, = 3.08; 95% CI, = 1.82–5.22; P < .001) was significantly associated with presence of TB. We therefore included these 2 variables along with the CCR5 haplotypes and a copy number of CCL3L1 that was less than or greater than 2 copies in a single multiple logistic regression model, to determine whether variations in CCR5 and CCL3L1 were independently associated with TB. In the full model, CCR5-HHD (OR, = 4.32; 95% CI, = 1.27–14.7; P = .019) and possession of >2 CCL3L1 copies (OR, = 1.84; 95% CI, = 1.02–3.31; P = .043) were significantly and independently associated with the presence of TB (Table 1, full model).
Table 1.
Association of CCR5 Haplotypes and CCL3L1 Gene Copy Number With Tuberculosis
| Full model, OR (95% CI) |
||||||
| CCR5/CCL3L1 | Unadjusted |
Adjusteda |
||||
| OR | (95% CI) | P | OR | 95% CI | P | |
| CCR5 HHA | 1.01 | (0.42–2.40) | .985 | 1.28 | (0.52–3.15) | .591 |
| CCR5 HHC | 1.17 | (0.57–2.41) | .666 | 1.26 | (0.59–2.70) | .555 |
| CCR5 HHD | 4.13 | (1.27 - 13.5) | .019 | 4.32 | (1.27 - 14.7) | .019 |
| CCR5 HHE | 0.70 | (0.36–1.37) | .299 | 0.78 | (0.38–1.59) | .493 |
| CCR5 HHF*1 | 1.24 | (0.36–4.22) | .732 | 1.13 | (0.30–4.20) | .860 |
| CCR5 HHF*2 | 0.90 | (0.44–1.86) | .778 | 1.04 | (0.48–2.25) | .919 |
| CCR5 HHG*1 | 1.21 | (0.55–2.67) | .632 | 1.44 | (0.62–3.33) | .399 |
| CCR5 HHG*2 | 0.81 | (0.13–4.89) | .815 | 1.04 | (0.14–7.63) | .965 |
| CCL3L1 <2 copies | 1.04 | (0.46–2.32) | .931 | 1.18 | (0.51–2.76) | .695 |
| CCL3L1 >2 copies | 1.94 | (1.11 – 3.38) | .019 | 1.84 | (1.02 – 3.31) | .043 |
| Final Modelb | ||||||
| CCR5/CCL3L1 | ||||||
| CCR5 HHD | 4.15 | (1.42 -12.13) | .009 | 3.95 | (1.28 -12.18) | .017 |
| CCL3L1 >2 copies | 1.95 | (1.19 – 3.17) | .007 | 1.75 | (1.04 – 2.92) | .034 |
NOTE. Boldface type indicates statistical significance. CI, confidence interval; OR, odds ratio.
Adjusted for age and sex.
The results for the final model are from step-wise multiple logistic regression analysis with a retention criterion of .1.
In the final step-wise regression model, we found that, independent of age and sex, CCR5-HHD haplotype (OR, = 3.95; 95% CI, = 1.28–12.18; P = .017) and possession of >2 CCL3L1 copies (OR, = 1.75; 95% CI, = 1.04–2.92; P = .034) were significantly associated with the presence of TB (Table 1, final model). Concordant results were obtained when we analyzed only those control subjects (n = 85) who had a tuberculin reaction of ≥10 mm (data not shown).
Because the CCR5-HHD haplotype is commonly found in persons of African ancestry but is also present in individuals of European and Hispanic ancestry [11], we examined whether the associations observed for this haplotype were because of potential population admixture. However, we found that genetic stratification was an unlikely explanation for the observed association of CCR5-HHD with risk of TB (Supplementary Figure 1). Collectively, these observations suggested that both the CCR5-HHD haplotype and a high CCL3L1 gene copy number were strongly associated with the presence of TB in the study population.
A high CCL3L1 gene copy number is a genetic determinant of increased CCL3/CCL3L1 expression [8], and this may provide a possible functional basis by which this chemokine genotype affects TB pathogenesis. It is unknown whether the single nucleotide polymorphism (SNP) at −2132T that distinguishes the CCR5-HHD haplotype from all other haplotypes (Figure 1A) has an impact on gene expression. To evaluate this, we considered the following framework. CCR5-HHA haplotype is the ancestral haplotype, and CCR5 haplotypes can be broadly dichotomized into those containing −2459G/−2135T (HH-A, -B, -C, and -D) versus −2459A/−2135C (HH-E, -F, and -G) (Figure 1A) [12]. We demonstrated previously that, compared with the promoter sequences of −2459A/−2135C-containing haplotypes (HHE, HHF, or HHG), the HHA promoter sequence exhibited the least transcriptional activity [12]. Because −2459G/−2135T is common to HHA, HHB, HHC, and HHD haplotypes (Figure 1A), one possibility was that the promoters of these 4 haplotypes have similar transcriptional strengths. An alternative possibility was that the combinatorial content of promoter SNPs that differentiate HHB, HHC, and HHD from the ancestral HHA (Figure 1A) uniquely affects gene expression.
We found previously that, in the K562 cell line, HHA- and HHC-specific promoter sequences had similarly low transcriptional activity [12]. Here, we found that, in the Jurkat T-cell line, CCR5-HHB and CCR5-HHD promoter sequences had higher transcriptional activity than did HHA (Figure 1D). We conducted similar transcriptional analyses in primary PBMCs obtained from healthy persons. These analyses revealed that although there was some donor-to-donor variability, promoter sequences of HHB—and especially those of HHD—had higher transcriptional activity than did those of HHA in anti-CD3/CD28 activated PBMCs (Figure 1E). To assess whether the higher gene expression associated with CCR5-HHD translates to increased protein expression, we evaluated a cohort of 90 black, HIV-negative female sex workers from South Africa who were part of the CAPRISA 002 acute infection study (see Supplementary Methods). In these women, compared with women who lacked HHD, those possessing CCR5-HHD had higher CCR5 expression on CD4+HLA-DR+ T cells (Figure 1F). Of note, although CCR5-HHB is rare, like CCR5-HHD, it is also more prevalent among persons of African ancestry [11] and consistent with the results of our transcriptional studies, the 2 women who had HHB-containing genotypes also had high CCR5 surface expression (data not shown).
In previous studies, we demonstrated that the G→T transversion at position −2554 (common to HHB, HHC, and HHD) associates with differences in binding of the NF-κB family of transcription factors [12]. Here, we found that nuclear factors from Jurkat T cells bound differentially to −2132T and −2132C alleles (Supplementary Figure 2). Although bioinformatic analyses suggested the binding of FOXO family transcription factors to the cis-regions in the close vicinity of HHD-specific SNP at −2132, competition electrophoretic mobility shift assays with oligonucleotides containing FOXO consensus sequences were not conclusive (Supplementary Figure 2).
DISCUSSION
We found that the CCR5-HHD haplotype and a copy number of CCL3L1 that is greater than the average found in a northwestern Colombian population was associated with a greater likelihood of active TB. These results support our hypothesis that CCR5 and its potent ligand CCL3L1 may contribute to TB pathogenesis.
The primary host immune response, which includes chemokine-dependent leukocyte recruitment and consequent granuloma formation, favors the host by providing local control of mycobacterial spread. The precise factors that tilt the balance from contained infection to active TB are unknown. However, there is increasing recognition that TB is an immunopathological disease, in which active TB develops due to uncontrolled host inflammatory responses to the pathogen (discussed in [7]). Thus, the concept was advanced that among immunocompetent hosts, the outcome of TB is a result of quantitatively and genetically controlled differences in the magnitude of the inflammatory responses, rather than a direct consequence of mycobacterial colonization [7]. This is in accord with burgeoning data suggesting a critical role of the proinflammatory effects of chemokines and their receptors in TB pathogenesis, including CCR5 and its ligands. Notably, the CCR5 ligand–CCR5 receptor axis plays a key role in amplifying immune responses (discussed in [2]).
The gene encoding CCL3L1 is subject to gene copy number variation (range, 0–12), with higher copies in persons of African ancestry [8]. Because CCL3L1 copy number is associated with increased CCL3/CCL3L chemokine production [8], it is conceivable that a high copy number may associate with proinflammatory state that leads to enhanced immunopathology that favors development of active TB. This possibility is underscored by the observation that increased CCL3 levels favor TB progression [7] and that the pro-inflammatory effects of CCL3 may contribute to tissue inflammation independently of M. tuberculosis load [6, 7]. Conceivably, in the context of persons concurrent HIV infection and TB, the protective HIV-suppressive effects of high CCL3L1 chemokine expression may be offset by its pro-inflammatory effects on TB.
Our reporter gene studies indicate that the combinatorial SNP makeup of the CCR5-HHD haplotype is associated with enhanced CCR5 gene expression in activated PBMCs. As a corollary, CCR5-HHD is also associated with higher surface expression on activated (HLA-DR+) CD4+ T cells. Together, these findings provide a possible functional basis for the observed association of CCR5-HHD with active TB and underscore the importance of accounting for the combinatorial content of SNPs in the CCR5 promoter haplotypes when evaluating their impact on gene/surface expression.
Interestingly, CCR5 HHD-containing genotypes have also been associated with adverse outcomes in HIV/AIDS, a clinical setting in which higher levels of CCR5 are a strong correlate of increased susceptibility to HIV infections and AIDS. Of note, we and others have shown that compared with those genotypes that do not contain HHD, CCR5 HHD-containing genotypes are associated with an increased risk of acquiring HIV infection [13, 14], having higher plasma viral loads [15], experiencing a significant increase in vaginal shedding of HIV-1–infected cells [16], and having faster rates of progression to AIDS and death [11, 16]. Thus, it is possible that CCR5-HHD haplotype may serve as a common genetic factor for both HIV infection and TB. Intriguingly, the CCR5-HHD haplotype is found more commonly in persons of African ancestry, and whether its dual influence on HIV infection and TB susceptibility contributes to the growing burden of concurrent HIV infection and TB in Africa requires further evaluation. Taken together, the results of our association studies substantiate the accumulating data suggesting a strong role for CCR5 and its ligands in TB pathogenesis.
Supplementary Data
Supplementary methods and figures are available at http://jid.oxfordjournals.org/ online.
Funding
This work was supported by the Veterans Administration Center on AIDS and HIV infection of the South Texas Veterans Health Care System, and the NIH (R37046326), a VA MERIT award, the Elizabeth Glaser Scientist Award, the Burroughs Wellcome Clinical Scientist Award in Translational Research, and the Doris Duke Distinguished Clinical Scientist Award to S. K. A. The work was also supported by a VA MERIT award to S. M.
Supplementary Material
Acknowledgments
We thank Alvaro Gaitan for advice, Florian Hladik for assistance with flow cytometry analyses, Salim S. Abdool Karim and the CAPRISA 002 study, and study participants of the CAPRISA cohort. We regret that, because of space constraints we were unable to cite other excellent work.
References
- 1.Moller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb) 2010;90:71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Camargo JF, Quinones MP, Mummidi S, et al. CCR5 expression levels influence NFAT translocation, IL-2 production, and subsequent signaling events during T lymphocyte activation. J Immunol. 2009;182:171–82. doi: 10.4049/jimmunol.182.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Algood HM, Chan J, Flynn JL. Chemokines and tuberculosis. Cytokine Growth Factor Rev. 2003;14:467–77. doi: 10.1016/s1359-6101(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 4.Pokkali S, Das SD. Augmented chemokine levels and chemokine receptor expression on immune cells during pulmonary tuberculosis. Hum Immunol. 2009;70:110–5. doi: 10.1016/j.humimm.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Pokkali S, Das SD, Expression RL. of CXC and CC type of chemokines and its receptors in tuberculous and non-tuberculous effusions. Cytokine. 2008;41:307–14. doi: 10.1016/j.cyto.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Qiu L, Huang D, Chen CY, et al. Severe tuberculosis induces unbalanced up-regulation of gene networks and overexpression of IL-22, MIP-1alpha, CCL27, IP-10, CCR4, CCR5, CXCR3, PD1, PDL2, IL-3, IFN-beta, TIM1, and TLR2 but low antigen-specific cellular responses. J Infect Dis. 2008;198:1514–9. doi: 10.1086/592448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyadova IV, Tsiganov EN, Kapina MA, et al. In mice, tuberculosis progression is associated with intensive inflammatory response and the accumulation of gr-1 cells in the lungs. PLoS One. 2010;5:e10469. doi: 10.1371/journal.pone.0010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–0. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 9.Catano G, Chykarenko ZA, Mangano A, et al. Concordance of CCR5 genotypes that influence cell-mediated immunity and HIV disease progression rates. J Infect Dis. 2011;203:263–72. doi: 10.1093/infdis/jiq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez LM, Anaya JM, Vilchez JR, et al. A polymorphism in the inducible nitric oxide synthase gene is associated with tuberculosis. Tuberculosis (Edinb) 2007;87:288–94. doi: 10.1016/j.tube.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez E, Bamshad M, Sato N, et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci U S A. 1999;96:12004–9. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mummidi S, Bamshad M, Ahuja SS, et al. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA: potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J Biol Chem. 2000;275:18946–61. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- 13.Kostrikis LG, Neumann AU, Thomson B, et al. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J Virol. 1999;73:10264–71. doi: 10.1128/jvi.73.12.10264-10271.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malhotra R, Hu L, Song W, et al. Retrovirology. 2011. Association of chemokine receptor gene (CCR2-CCR5) haplotypes with acquisition and control of HIV-1 infection in Zambians. 8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang J, Wilson CM, Schaen M, Myracle A, Douglas SD, Kaslow RA. CCR2 and CCR5 genotypes in HIV type 1-infected adolescents: limited contributions to variability in plasma HIV type 1 RNA concentration in the absence of antiretroviral therapy. AIDS Res Hum Retroviruses. 2002;18:403–12. doi: 10.1089/088922202753614164. [DOI] [PubMed] [Google Scholar]
- 16.John GC, Bird T, Overbaugh J, et al. CCR5 promoter polymorphisms in a Kenyan perinatal human immunodeficiency virus type 1 cohort: association with increased 2-year maternal mortality. J Infect Dis. 2001;184:89–92. doi: 10.1086/321006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.