Abstract
Single-copy gene and promoter regions have been excised from yeast chromosomes and have been purified as chromatin by conventional and affinity methods. Promoter regions isolated in transcriptionally repressed and activated states maintain their characteristic chromatin structures. Gel filtration analysis establishes the uniformity of the transcriptionally activated state. Activator proteins interact in the manner anticipated from previous studies in vivo. This work opens the way to the direct study of specific gene regions of eukaryotic chromosomes in diverse functional and structural states.
Studies of chromatin in vitro have been performed in the past almost entirely on unfractionated chromosomal material and artificial histone-DNA complexes. While specific chromosomal regions can be detected within unfractionated material by hybridization and PCR, and while histone-DNA complexes share important features with natural chromatin, these approaches suffer from important limitations. The composition, structure, and dynamics of a specific region cannot be directly determined. The modification state of the histones and locations of nucleosomes in artificial complexes often differ from those found in nature. These limitations can be overcome by the excision of specific regions from chromosomal loci and their purification to homogeneity. Indeed, only when chromosomal transactions such as gene activation and transcription have been reconstituted with purified proteins and purified, natural chromatin segments will these transactions be fully understood.
Previous efforts to isolate chromosomal material have been directed towards episomal DNA elements. The maintenance of such elements in cell nuclei requires an origin of DNA replication and a selectable marker. It also relies, in most cases, on stabilization sequences, of viral origin in mammalian cells, and from the 2μ plasmid or centromeres in yeast. These stabilization sequences, which retain episomal DNA in nuclei in vivo, also cause retention of DNA elements in nuclei in vitro, preventing their extraction in soluble form. TRP1/ARS1 plasmids of yeast, which contain only a replication origin and a selectable marker, have been successfully extracted and purified to near homogeneity (11). For lack of a stabilization sequence, however, TRP1/ARS1 plasmids are segregated preferentially to the mother of a cell division and vary widely in copy number in a population of cells, ranging up to about 1,000 copies per cell. As a result, most functions of DNA elements incorporated in TRP1/ARS1 plasmids, which depend on an appropriate level of interacting proteins in the cell, are grossly perturbed.
For a solution of the chromatin isolation problem we have exploited an approach devised by Ansari and coworkers (2), who demonstrated the efficient excision of a chromatin segment flanked by recognition sites for the R recombinase, following induction of recombinase expression. The chromatin segment is converted by recombination to a circular form, which can be separated from bulk chromatin by differential centrifugation. We have applied this approach to genes in repressed and transcriptionally activated states, with the use of affinity chromatography for further purification of chromatin circles. We encountered a number of technical difficulties and describe here the development of general methods by which they may be overcome.
We have applied our methods to the PHO5 gene of the yeast Saccharomyces cerevisiae, whose promoter region undergoes a transition in chromatin structure upon transcriptional activation, as shown by various researchers (1, 15). We previously reported evidence, gained in part by the use of PHO5 gene circles, indicating the complete unfolding of promoter nucleosomes upon transcriptional activation. While nucleosomes are removed from all promoter sequences, the loss is not complete in any of them, suggesting an equilibrium between nucleosome removal and reformation (3). We now report the extension of this work to promoter circles, giving further insight into the nature of the activated state.
MATERIALS AND METHODS
Genetic elements.
All manipulations described here were done by using standard molecular cloning techniques (12).
To generate plasmid pJSS3.1, full-length LexA, plectin spacer, and tandem affinity purification (TAP) tag were amplified by three separate PCRs with Escherichia coli genomic DNA, rat genomic DNA, and plasmid pBS1479 (8) as templates, respectively. Primers were designed to introduce novel restriction sites in frame for subsequent ligation into the complete construct. The final three-part ligation product was subcloned into the E. coli-yeast centromeric shuttle vector p416-GPD (ATCC no. 87360). Yeast cells transformed with plasmid pJSS3.1 express the adaptor molecule constitutively and at moderate levels under the control of a glyceraldehyde phosphate dehydrogenase promoter.
Plasmid pB3 was derived by digesting plasmid pB2 (3) with BaeI and religating, thereby deleting a potential LexA-binding site. Plasmid pB3 was used for inducible expression of the site-specific R recombinase from Zygosaccharomyces rouxii in S. cerevisiae.
To generate plasmid pH1.6, the PHO4 coding sequence was amplified by PCR, using yeast genomic DNA as template, with primers designed to introduce novel restriction sites in frame for subsequent cloning into pET-21d(+) (Novagen), cut with EagI and NcoI. Plasmid pH1.6 was used for expression of recombinant yeast Pho4p protein bearing a C-terminal hexahistidine tag in E. coli.
Yeast strains and media.
Yeast strains used in this study, yM2.1 and yM8.14 for purification of PHO5 gene circles and yM3.2 and yM9.7 for purification of PHO5 promoter circles, have been described previously (3).
Strains were grown in Hartwell's synthetic medium containing 2% (wt/vol) raffinose (SCR). Strains used for circle formation were transformed with plasmid pB3 for inducible expression of R recombinase. For affinity purification, cells were also transformed with plasmid pJSS3.1 for constitutive expression of the recombinant LexA adaptor protein. pB3-transformed strains were grown in SCR medium lacking leucine. Cells transformed with pB3 and pJSS3.1 were grown in SCR medium lacking leucine and uracil. Recombination was induced at a cell density of 5 × 107 cells/ml by adding galactose to a final concentration of 2% (wt/vol). Cells were grown for an additional 1.5 h at 30°C before harvesting.
Purification of chromatin circles.
Cells from a 12-liter culture were harvested by centrifugation, washed twice with water, pelleted in a sealed 20-ml syringe by centrifugation, and extruded into liquid nitrogen after unsealing the syringe (13). The frozen cells were ground in a liquid nitrogen-dry ice mixture by using a commercial blender (Waring) for 10 min, alternating every 30 s between high and low blending speeds. After evaporation of liquid nitrogen and most of the dry ice, 5 volumes of 200 mM potassium acetate, 2 mM EDTA, 10% (vol/vol) glycerol, 125 μM spermidine, 50 μM spermine, 5 mM 2-mercaptoethanol, and 25 mM HEPES-KOH (pH 7.4) containing protease inhibitors (complete protease inhibitor cocktail; Boehringer) were added. The crude extract was stirred for 15 min at 4°C and was cleared by centrifugation at 72,700 × g for 1 h at 4°C. Chromatin circles in the supernatant were sedimented by centrifugation at 371,000 × g for 2 h (gene circle) or 3 h (promoter circle) at 4°C. The pellet was suspended in 1/10 of the original volume of buffer A (100 mM potassium acetate, 1 mM EDTA, 10% [vol/vol] glycerol, 0.01% NP-40, 125 μM spermidine, 50 μM spermine, 5 mM 2-mercaptoethanol, 25 mM HEPES-KOH [pH 7.4]) containing protease inhibitors.
For gel filtration the suspended pellet was applied to a TSK-G4000SW column (TosoHaas) in a solution containing 100 mM potassium acetate, 2 mM EDTA, 5% (vol/vol) glycerol, 125 μM spermidine, 50 μM spermine, 5 mM 2-mercaptoethanol, 25 mM HEPES-KOH (pH 7.1).
For affinity chromatography the suspended pellet was diluted to a total protein concentration of 2.5 to 5 mg/ml with buffer A and applied to 1/20 of the load volume of immunoglobulin G (IgG)-agarose beads (Sigma) that had been equilibrated with buffer A overnight. Beads were incubated for 2 h at 4°C, washed with 80 column volumes of buffer A without glycerol but containing 400 mM potassium acetate and 100 μg of insulin/ml, and subsequently were washed with 80 column volumes of buffer A without glycerol but containing a solution of 50 mM potassium acetate, 0.1% NP-40, and 100 μg of insulin/ml. Chromatin circles were released by proteolytic cleavage for 1 h at 4°C with 80 μg of recombinant tobacco etch virus (TEV) protease (Gibco BRL)/ml of beads in a solution of 200 mM potassium acetate, 1 mM EDTA, 25 mM HEPES-KOH (pH 7.4). Residual chromatin circles were washed from the beads with 3 column volumes of buffer A. The eluate was adjusted to the composition of buffer A without glycerol but containing 1 mM imidazole, 2 mM calcium chloride, 2 mM magnesium acetate, 0.1% NP-40, 10 mM 2-mercaptoethanol, and 100 μg of insulin/ml and was passed through 1/36 of the applied volume of Talon metal affinity resin (Clontech). This step removed virtually all of the recombinant TEV protease. The flowthrough was applied to a 1/20 volume of calmodulin affinity resin (Stratagene) and was incubated for 1 h at 4°C. The column was washed with 80 column volumes of buffer A without glycerol and EDTA but containing 300 mM potassium acetate, 0.1% NP-40, 2 mM calcium chloride, 1 mM magnesium acetate, 1 mM imidazole, 10 mM 2-mercaptoethanol, and 100 μg of insulin/ml. Chromatin circles were eluted within 3 column volumes of 200 mM potassium acetate, 2 mM EGTA, 1 mM EDTA, 10 mM 2-mercaptoethanol, and 25 mM HEPES (pH 7.4) and were adjusted to a final concentration of 10% glycerol and 100 μg of insulin/ml before freezing.
Purification of recombinant Pho2p and Pho4p proteins.
E. coli BL21 cells were transformed with plasmids pH1.6 or pPHO2-His (4). Cells were cultured in 1.5 liters of Luria-Bertani medium containing 100 μg of ampicillin/ml at 37°C to mid-log phase. Expression of recombinant proteins was induced by addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. Cells were cultured for an additional 3 h and were sedimented by centrifugation. Cells were resuspended in 35 ml of 500 mM sodium chloride, 5 mM 2-mercaptoethanol, 20 mM Tris-HCl (pH 8.0). Bacterial lysates were prepared by using a FRENCH pressure cell press. The lysate was cleared by centrifugation and was passed through a column of 3 ml of Talon metal affinity resin. The column was washed with 30 column volumes of 1 M sodium chloride, 5 mM 2-mercaptoethanol, 20 mM Tris-HCl (pH 8.0). After equilibration with a solution of 150 mM sodium chloride, 5 mM 2-mercaptoethanol, 20 mM Tris-HCl (pH 8.0), bound proteins were eluted with 4.5 ml of the same buffer containing 100 mM imidazole. Eluates were dialyzed against a solution containing 100 mM sodium chloride, 0.5 mM EDTA, 0.5 mM dithiothreitol, 10% glycerol, 20 mM Tris-HCl (pH 8.0) and were stored at −80°C. The purity of the proteins was verified by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and staining with Coomassie brilliant blue (data not shown).
Endonuclease digestion analysis of the chromosomal PHO5 promoter and affinity purified PHO5 chromatin circles.
Preparation of nuclei and endonuclease analysis of the chromosomal PHO5 promoter was performed as described previously (3).
For restriction enzyme endonuclease digestion, 50 amol of affinity-purified chromatin circles was adjusted to a final volume of 50 μl with buffer A containing 100 μg of insulin/ml. The solution was further adjusted to 10 mM magnesium-acetate and 0.58 μM double-stranded oligonucleotide (5′-TACTGTATGTACATACAGTA-3′) in a total volume of 100 μl. Digestion was performed for 30 min at 37°C with 5 and 20 U of the restriction endonucleases indicated in the figure legends. The reaction was terminated by adding 100 μl of stop buffer (500 mM sodium chloride, 20 mM EDTA, 1% SDS, 50 mM Tris-HCl [pH 8.0]) and 1.4 U of Proteinase K (Sigma). After incubation for 30 min at 37°C and phenol-chloroform extraction, DNA was precipitated with ethanol by using 100 μg of glycogen/ml as a carrier.
For micrococcal nuclease digestion, 100 amol of affinity-purified chromatin circles was adjusted to a final volume of 100 μl with buffer A containing 100 μg of insulin/ml. The solution was further adjusted to 1 mM magnesium acetate, 0.5 mM calcium chloride, and 100 μg of denatured salmon sperm DNA (Sigma)/ml in a total volume of 200 μl. Digestion was performed for 5 min at 37°C with 0.2, 0.6, and 1.8 U of micrococcal nuclease (Sigma). The digestion was terminated by the addition of 200 μl of stop buffer and 2.8 U of Proteinase K (Sigma), and DNA was extracted as described above.
Electrophoretic mobility shift assay.
Affinity-purified chromatin circles (500 amols) in a volume of 120 μl were adjusted to 10 mM magnesium-acetate and 0.58 μM double-stranded oligonucleotide (5′-TACTGTATGTACATACAGTA-3′) in a total volume of 250 μl. Digestion was performed for 30 min at 37°C with 50 U of the restriction enzymes indicated in the figure legends. Poly(dI-dC) • poly(dI-dC) (Amersham Biosciences) was added to a concentration of 50 ng/μl. Aliquots of each reaction mixture were incubated with and without 30 and 3 nM, 150 and 15 nM, and 750 and 75 nM recombinant Pho4p and Pho2p proteins, respectively. After incubation for 20 min at room temperature protein complexes were separated in a 2% agarose gel in 0.5× Tris-borate-EDTA with 6.5 V/cm for 220 min at 4°C.
Blot analysis and quantitation.
Nucleic acids from agarose gels were blotted onto nylon membranes (Schleicher & Schuell) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) as described previously (12). DNA probes to hybridize nucleic acids blotted onto nylon membranes were generated by random prime labeling by using [α-32P]dATP (Amersham Biosciences). Quantitative analysis was performed by using a PhosphorImager and ImageQuant software (Amersham Biosciences).
RESULTS
When yeast cells are grown in the presence of phosphate, the PHO5 promoter is transcriptionally repressed. The promoter chromatin is characterized by four translationally positioned nucleosomes (15), two of which obscure important functional elements, a binding site for the Pho4 transcriptional activator protein (UASP2, U2 in Fig. 1A), and the TATA box (T in Fig. 1A). Upon phosphate starvation a signal transduction pathway is activated, leading to an unknown mechanism for promoter chromatin rearrangement and subsequent transcriptional activation (7). In the course of promoter activation, nucleosomes are removed from UASP2 and the TATA box (3, 9).
FIG. 1.
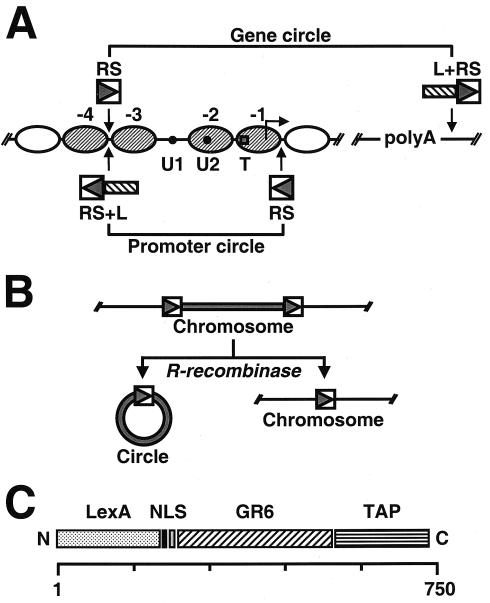
(A) Schematic representation of the genetically modified PHO5 locus. Ovals represent positioned nucleosomes (−1 to −4) on the promoter under repressing conditions. Black dots represent transcription factor binding sites UASP1 (U1) and UASP2 (U2). A square represents the TATA box (T). Arrowheads point to the sites where recognition elements for R recombinase from Z. rouxii (RS) and the LexA operator cluster (L) were inserted into the genomic locus. PHO5 domains released by homologous recombination are indicated. (B) Schematic representation of site-specific homologous recombination. (C) Schematic representation of the recombinant adaptor molecule. LexA, full-length LexA protein from E. coli; NLS, simian virus 40 nuclear localization signal; GR6, Garnier Robson helix 6 from rat plectin; TAP, tandem affinity purification tag. The scale on the bottom reflects the number of amino acids.
For the isolation of PHO5 chromatin, we framed the chromosomal locus with recognition sites (RS) for the R recombinase from Zygosaccharomyces rouxii (Fig. 1A). RS elements flanked either the PHO5 promoter region (encompassing nucleosomes N-1 to N-3), bearing the elements essential for transcriptional regulation, or the entire PHO5 gene (Fig. 1A). R recombinase was under control of the GAL1 promoter. Following induction with galactose, the chromatin regions between the RS elements were released as circles of 750 and 2,200 bp for promoter and gene, respectively (Fig. 1B). The efficiency of release was typically 75 to 80% (Fig. 2B). We included a cluster of LexA-binding sites adjacent to an RS site in an orientation that would assure its incorporation in the chromatin circles (L in Fig. 1A) to enable the use of a LexA adaptor protein for binding the circles to a solid support (3). Initial experiments with a glutathione S-transferase-LexA fusion protein as adaptor for binding the circles to a glutathione-agarose support were unsuccessful, with the retention of almost no circles on the support. There proved to be two main problems limiting the purification of circles by this approach. First, leaky expression of R recombinase in the absence of galactose resulted in the premature excision and, in some cases, total loss of the chromosomal locus during cell culture. Second, chromatin circles were not retained on glutathione-agarose in the presence of the GST-LexA adaptor. We solved the problem of leaky recombinase expression by flanking a LEU2 selection marker on the recombinase expression vector with RS elements. Premature expression of the recombinase results in excision of the selection marker, reversion to leucine auxotrophy, and elimination from the population of cells grown in the absence of leucine. The problem of retention on the glutathione-agarose support proved to depend on the size of the DNA. Oligonucleotides and DNA fragments up to about 800 bp in length and bearing LexA-binding sites were efficiently retained on the support. Larger DNAs and naked circles, however, were not bound at all. This problem could be overcome by the use of anti-GST antibody for binding to a Protein A-agarose support (3). Evidently, the conformation of DNA greater than about 800 bp creates a requirement for a longer adaptor for interaction with a support.
FIG. 2.
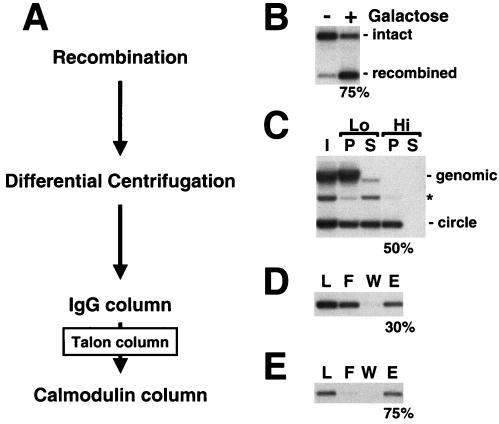
(A) Purification scheme for chromatin circles. (B) Site-specific homologous recombination. The PHO5 locus before (−) and after (+) addition of galactose to induce recombinase expression. Genomic DNA was extracted, digested with ApaI and NciI, subjected to 1% agarose gel electrophoresis, and analyzed by blot hybridization with a probe upstream of the PHO5 locus. Positions of the intact and recombined loci are indicated on the right. (C) Differential centrifugation of yeast whole-cell lysates. I, input; P, pellet; S, supernatant; Lo, low-speed spin; Hi, high-speed spin. DNA was extracted, subjected to 1% agarose gel electrophoresis, and analyzed by blot hybridization with a probe specific for the PHO5 locus. Positions of genomic DNA and circle DNA are indicated on the right. The asterisk marks the position of nicked circle DNA that arose as an artifact during DNA preparation. (D and E) IgG and calmodulin affinity chromatography of chromatin circles. L, load; F, flowthrough; W, wash fraction; E, eluate. DNA was analyzed as described for panel C. Numbers on the bottom of each panel give the percentage of recovery of chromatin circles in each purification step, as determined by quantitative blot hybridization.
Recombinant adaptor and chromatin circle purification.
While the combination of GST-LexA and anti-GST antibody, added after cell lysis to chromatin circle preparations, was effective as an adaptor for circle purification, higher yields were obtained when the adaptor was expressed and bound to the circles in vivo. To this end we constructed a recombinant adaptor (Fig. 1C) comprising the entire coding sequence of LexA from E. coli at its N terminus, followed by a simian virus 40 nuclear localization signal, a long spacer bearing the Garnier Robson helix 6 from rat plectin (14), and a C-terminal TAP tag (10). The spacer was chosen on the basis of its predicted extended helical structure. Other spacers were tried but gave lower yields of circles. The TAP tag enables two steps of affinity purification (10) (Fig. 2A). Constitutive expression of the adaptor did not interfere with PHO5 expression or the overall configuration of chromatin at the PHO5 locus (data not shown).
Cells bearing circles with bound adaptors were lysed in a liquid nitrogen-dry ice mixture in a commercial blender to limit shearing of chromosomal DNA. More than 95% of the chromosomal DNA could then be removed by centrifugation at low speed (2) (Lo in Fig. 2C). Chromatin circles remaining in the supernatant could be sedimented in a second spin at higher speed (Hi in Fig. 2C). The recovery in the pellet was typically about 50% for gene circles (Fig. 2C) and about 65% for the smaller promoter circles (data not shown). The pellet contained about 10% of the total protein of the cell lysate and included nuclease, protease, topoisomerase, and chromatin remodeling activities, precluding its use in biochemical studies without further purification (data not shown).
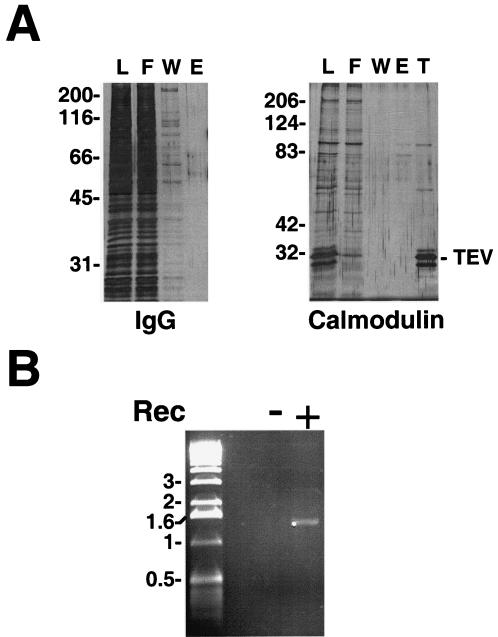
Chromatin circles from the high-speed pellet were bound to an IgG-agarose column through the Protein A moiety of the TAP tag on the recombinant adaptor. After being washed on the column the circles were released by cleavage with His-tagged TEV protease (Fig. 2D). The majority of the contaminating protein and nucleic acid was removed at this step (Fig. 3A and data not shown). The eluate was passed through a Talon metal affinity column to remove the His-tagged protease (Fig. 3A, lane T), and the circles were then adsorbed to calmodulin beads in the presence of calcium through the calmodulin-binding component of the TAP tag. After being washed on the column the circles were released with EGTA (Fig. 2E). The recovery after the two steps of affinity purification was ∼20% for gene circles (Fig. 2D and E and Table 1) and ∼45% for promoter circles (data not shown). Gel electrophoresis revealed circular DNA, free from contamination with other nucleic acids. No DNA was detected when the same procedure was applied to a yeast strain lacking the recombinase expression vector (Fig. 3B).
FIG. 3.
(A) Protein analyses for IgG and calmodulin affinity purification. Samples at different stages of purification were analyzed by SDS-polyacrylamide gel electrophoresis and silver staining. L, F, W, and E are as described in the legend to Fig. 2D and E; T, Talon beads. Numbers on the left of each panel give the positions of molecular size standards (Mr × 10−3). The position of recombinant TEV protease is indicated on the right. (B) DNA analysis of concentrated calmodulin affinity eluates. Calmodulin affinity eluates obtained from 12 liters of yeast culture of strains capable of (+) or deficient in (−) galactose-induced recombinase expression (Rec) were concentrated by centrifugation. DNA was extracted from two thirds of the sample, subjected to 1% agarose gel electrophoresis, and detected by ethidium bromide staining. Numbers on the left give the positions of DNA size standards (in kilobase pairs).
TABLE 1.
Affinity purification of a 2.2-kb chromosomal domain from the yeast PHO5 locus
| Step | Recoverya | PHO5 circle: unrelated genomic locusb | PHO5 locus: total proteinc |
|---|---|---|---|
| Input | 1 | 7.7 × 10−7 | |
| Recombination | 75 | ||
| Differential centrifugation | 50 | 40 | 2.9 × 10−6 |
| Affinity purification | 22.5 | 180,000 | 0.07 |
| Total recovery | 8.5 | ||
| Total enrichment | 180,000 | 84,000 |
Numbers are the recovery of PHO5 DNA for individual stages of the purification in the percentage of the respective input. Total recovery is calculated as the percentage of the initial PHO5 DNA present at the end of the purification. DNA amounts were determined by quantitative blot hybridization.
Numbers are the ratios of absolute DNA amounts from PHO5 to genomic DNA at individual stages of the purification. DNA amounts were determined by quantitative PCR with primers to amplify PHO5 sequences and four unrelated genomic loci.
Numbers are the ratios of grams of PHO5 DNA to grams of total protein at individual stages of the purification. Total enrichment is calculated by dividing the number obtained for the affinity eluate by the number obtained for the input of the purification. DNA amounts were determined by quantitative blot hybridization. Protein amounts were determined in a Bradford assay.
From 13 liters of cell culture (1012 cells) we recovered 130 fmols of PHO5 gene circles, corresponding to an overall yield of 8.5% (22% for promoter circles; data not shown) (Table 1). Quantitative PCR, with primers for several chromosomal loci unrelated to PHO5, showed a 180,000-fold enrichment of the PHO5 region, corresponding to a 20- to 30-fold excess of PHO5 DNA over any other genomic DNA in the purified material. The chromatin circles were enriched 84,000-fold with respect to total cellular protein (Table 1) and contained no detectable nuclease, protease, topoisomerase, or chromatin remodeling activity (data not shown). The total amount of contaminating proteins was, however, still 10- to 20-fold above the amount of histones associated with the isolated DNA circles (data not shown).
Nucleosomes on purified chromatin circles.
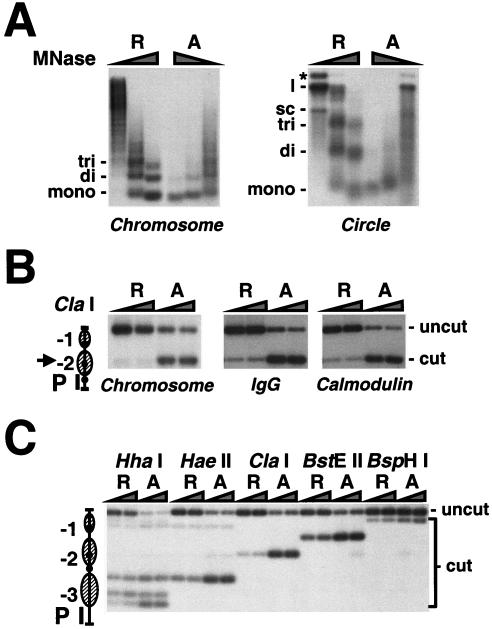
The distribution of nucleosomes on purified chromatin circles was determined by digestion with micrococcal nuclease and restriction endonucleases. The products of micrococcal nuclease digestion of promoter circles were revealed by blot hybridization with a probe spanning the entire PHO5 promoter region (Fig. 4A, circle). Circles derived from transcriptionally repressed promoters gave products corresponding to nucleosome monomer, dimer, and trimer, as expected from the locations of nucleosomes mapped previously at the PHO5 chromosomal locus (Fig. 1A). Digestion at the same enzyme concentrations of circles derived from transcriptionally active promoters gave a different picture (Fig. 4A). The activated circles were more sensitive to nuclease attack, released no nucleosome dimers or trimers, and were almost immediately converted to monomers, as expected from our previous evidence for a single nucleosome remaining, on average, on the activated promoter (3). The patterns of digestion for both repressed and activated promoter circles were similar to those obtained for the PHO5 promoter or gene flanked by RS elements at its chromosomal locus or for purified gene circles (see Fig. 4A, Chromosome, and reference 3).
FIG. 4.
(A) Micrococcal nuclease accessibility of the PHO5 promoter in isolated nuclei and in purified chromatin circles. Nuclei isolated from yeast strains repressed (R) and activated (A) in PHO5 expression were digested with 1, 5, 10, or 20 U of micrococcal nuclease (MNase)/ml for 20 min (Chromosome). PHO5 promoter circles (100 amols of DNA) from the same strains purified by IgG affinity purification were digested with 0.5, 1.5, and 4.5 U of micrococcal nuclease for 5 min (Circle). DNA was extracted, subjected to 2% agarose gel electrophoresis, and analyzed by blot hybridization with a probe spanning nucleosomes N-1 to N-3. The positions of DNA fragments that have been protected by mono-, di-, and trinucleosomes and the positions of supercoiled (sc), linear (l), and nicked (*) DNA are given on the left of each panel. (B and C) Restriction endonuclease accessibility of the PHO5 promoter in isolated nuclei and in purified chromatin circles. Nuclei isolated from yeast strains repressed (R) and activated (A) in PHO5 expression were digested with 50 or 200 U of ClaI restriction endonuclease for 1 h (Chromosome). DNA was extracted, digested with HaeIII, subjected to 1.5% agarose gel electrophoresis, and analyzed by blot hybridization with the probe indicated (P). PHO5 gene circles from the same yeast strains purified by IgG, IgG and calmodulin affinity chromatography (B), or PHO5 promoter circles purified by IgG affinity chromatography (C) were digested with 5 or 20 U of the restriction endonuclease indicated for 30 min. The DNA was extracted, digested with EcoRI (B) or HpaI (C), subjected to 1.5% agarose gel electrophoresis, and analyzed by blot hybridization with the probe indicated. An arrowhead points to the position of the ClaI cutting site in the PHO5 promoter, depicted on the left. Positions of uncut and cut DNA are indicated on the right.
A hallmark of the chromatin transition at the PHO5 promoter is an increase in accessibility of a ClaI restriction endonuclease site near the dyad axis of nucleosome −2 upon transcriptional activation (1). The chromatin circles showed such an increase, with ClaI accessibilities through the course of purification similar to those observed for chromatin in isolated nuclei (Fig. 4B). Analysis of many additional restriction sites showed only one significant difference between purified gene circles and chromatin in isolated nuclei, a change in accessibility of an HaeII site within the linker region between nucleosomes −3 and −2 (3). This site was more protected in purified repressed circles than at the chromosomal locus, which can be explained by a slight shift of nucleosome −3 towards nucleosome −2 in the circles. Such a shift was also seen for isolated promoter circles (Fig. 4C). Similar observations have been made for PHO5 promoter DNA complexed with histones in vitro (16) and for plasmids carrying the PHO5 gene (6). We conclude that the differences in chromatin structure between repressed and activated PHO5 promoters are unaffected by excision from the chromosomal locus and are stable under the conditions of isolation and manipulation in vitro.
Probability of the repressed state under activating conditions.
As already mentioned, the results of micrococcal nuclease digestion are consistent with our conclusion from previous work regarding the retention of a single nucleosome, on average, on the activated PHO5 promoter. The question arises whether any promoters remain repressed under activating conditions. Might some promoters retain a full complement of three nucleosomes? Or have essentially all promoters lost one or more nucleosomes?
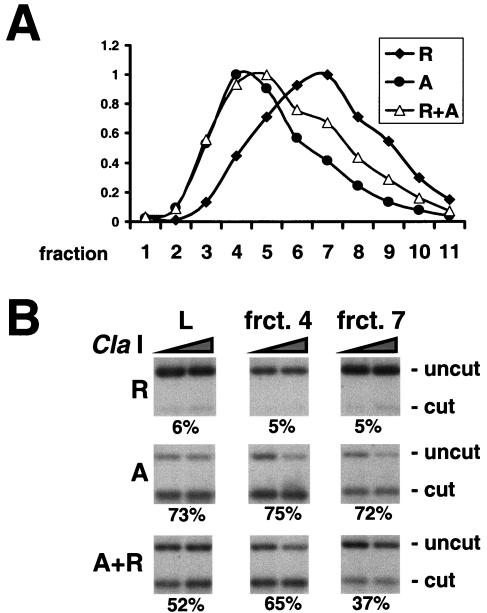
We could distinguish between these possibilities by gel filtration analysis of purified promoter circles. Activated and repressed promoter circle preparations formed distinct but overlapping gel filtration profiles, with peaks in fractions 4 and 7, respectively (Fig. 5A). If the activated circle preparation also contained repressed circles, these should be enriched in fraction 7, corresponding to the peak for repressed circles. The accessibility of the ClaI site was, however, the same for fractions 4 and 7, indicating the presence of only activated circles (Fig. 5B). In a control experiment, repressed and activated circle preparations were mixed in a 1:2 ratio. Following gel filtration of the mixture the accessibility of the ClaI site differed from the manner expected, demonstrating that activated and repressed circles were indeed enriched in fractions 4 and 7 (Fig. 5B). We conclude that under activating conditions the fraction of promoters bearing three nucleosomes (corresponding to the repressed state) is much less than one in three.
FIG. 5.
(A) Gel filtration of PHO5 promoter circles. Chromatin circles from yeast strains repressed (R) and activated (A) in PHO5 expression and grown in the absence of the recombinant LexA adaptor molecule were partially purified by differential centrifugation. A TSK-G4000SW column was used to separate promoter circles from the individual preparations and from a 2:1 mixture of activated and repressed promoter circles (A+R). DNA eluting in different fractions (1 to 11) from the column was extracted, subjected to 1% agarose gel electrophoresis, and analyzed by blot hybridization with a probe spanning nucleosomes N-1 to N-3. The DNA concentration relative to that in the respective peak fraction, determined from the radioactivity in the blot, is plotted for each gel filtration experiment on the ordinate. The numbers of the fractions are indicated on the abscissa. (B) Endonuclease accessibility in promoter circles subjected to gel filtration. PHO5 promoter circles present in the load (L) and fractions (frct.) 4 and 7 of the respective gel filtration were digested with ClaI as described in the legend to Fig. 4B. DNA was extracted, digested with HpaI, subjected to 2% agarose gel electrophoresis, and analyzed by blot hybridization with a probe spanning nucleosome N-3. Numbers on the bottom of each panel give the percentage of accessibility as the mean of the plateau values. Positions of uncut and cut DNA are indicated on the right.
Transcription factor binding to purified chromatin circles.
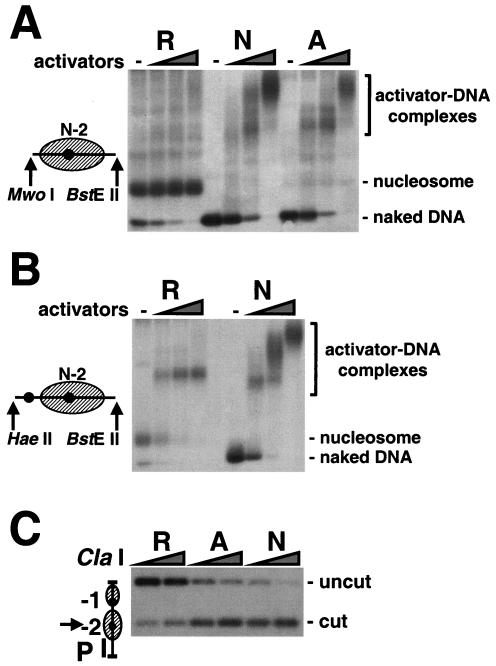
Transcriptional activation of the PHO5 promoter involves the binding of Pho2p and Pho4p to the enhancer sequences UASP1 and UASP2 (denoted U1 and U2 in Fig. 1). UASP1 is exposed in a linker region between nucleosomes −2 and −3 in the repressed state, while UASP2 lies within nucleosome −2 in the repressed state and becomes exposed upon activation (Fig. 1). Pho2p/Pho4p binding to UASP1 might directly lead to the exposure of UASP2 and subsequent binding to UASP2. We could investigate this possibility with the use of purified promoter chromatin. Cleavage with restriction endonucleases MwoI and BstEII released a 173-bp fragment from the nucleosome −2 region of both repressed and activated promoter circles (Fig. 6A). As expected, native gel electrophoresis revealed that this region contains a nucleosome under repressing conditions, with only a trace of naked DNA detected. In contrast, almost all of the fragments released from activated circles were apparently naked, in keeping with previous results from sedimentation analysis (3). Upon addition of purified recombinant Pho2p and Pho4p proteins, the gel electrophoretic mobility of repressed nucleosome −2 was unaffected, while the naked DNA from activated circles was shifted to slower migrating complexes, as was the trace of naked DNA in the repressed preparation. A 173-bp fragment cleaved from naked PHO5 DNA showed a similar behavior. Nucleosome −2 evidently prevents Pho2p/Pho4p binding to UASP2 in the repressed state.
FIG. 6.
(A and B) Transcription factor binding to natural PHO5 chromatin in vitro. Gene circles purified by IgG affinity chromatography from yeast strains repressed (R) and activated (A) in PHO5 expression and a bacterial plasmid bearing the naked PHO5 promoter sequence (N) were digested with MwoI and BstEII (A) or HaeII and BstEII (B). Restriction digests were incubated for 20 min with or without 30 and 3 nM, 150 and 15 nM, and 750 and 75 nM recombinant Pho4p and Pho2p proteins, respectively. DNA-protein complexes were subjected to native agarose gel electrophoresis and were analyzed by blot hybridization with a probe spanning nucleosome −2. Fragments released upon endonuclease digestion are depicted on the left. Positions of activator-DNA complexes, the nucleosome, and naked DNA are indicated on the right. (C) ClaI accessibility of PHO5 promoter DNA in the presence of transcription factors. Gene circles purified by IgG affinity chromatography from yeast strains repressed and activated in PHO5 expression and a bacterial plasmid bearing the PHO5 promoter sequence were incubated with 750 and 75 nM of recombinant Pho4p and Pho2p proteins, respectively. ClaI accessibility was assessed as described in the legend to Fig. 4B. Positions of uncut and cut DNA are indicated on the right.
Does Pho2p/Pho4p binding to UASP1 expose the nucleosome −2 region for binding to UASP2? We repeated the electrophoretic mobility shift analysis except with the use of HaeII and BstEII to release a 228-bp fragment containing not only the nucleosome −2 region but also UASP1 (Fig. 6B). Again, most of the DNA released from repressed circles migrated with the mobility of a nucleosome (Fig. 6B, first lane). In this experiment, however, the addition of Pho2p and Pho4p resulted in a complex of electrophoretic mobility lower than that of a nucleosome but different from those of the complexes formed with naked DNA (Fig. 6B, compare lanes R and N). Despite the apparent binding of Pho2p/Pho4p to UASP1, the ClaI site in nucleosome −2 remained highly protected from digestion (Fig. 6C, lanes R). Control experiments showed that Pho2p/Pho4p binding did not interfere with restriction enzyme digestion (Fig. 6C, lanes A and N). Evidently, Pho2p/Pho4p binding to UASP1 alone is not sufficient to dislodge nucleosome −2.
DISCUSSION
The isolation of natural chromatin offers many advantages over the assembly of artificial histone-DNA complexes: the specific positioning of nucleosomes seen in the vicinity of promoters in vivo may not be readily reconstituted in vitro; the state of posttranslational modification of the histones, delicately modulated in vivo, may be difficult to reproduce in vitro; and non-histone proteins may also be involved in the structure and function of promoters in vivo. The methods we have developed for chromatin isolation, starting from the findings of Ansari and coworkers (2), should be general. The difficulties we encountered and the procedures we devised to overcome them should be applicable in most cases. The chief limitation of our approach is the small quantity of purified chromatin obtainable from single-copy genes and chromosomal loci. While precluding some methods of analysis, this limitation should be no impediment to the use of the purified material as substrate for enzymatic and conformational transactions or as an object of structural analysis by such methods as electron microscopy and mass spectrometry.
The use of purified chromatin circles for studies of transcriptional activation in vitro is made possible by our finding that the chromatin structures of both repressed and activated states are stable to isolation and manipulation in vitro.
We previously reported on the properties of PHO5 gene circles containing both the promoter and open reading frame (3). Here we extend the analysis to circles containing the promoter alone, comprising only 750 bp of DNA and bearing three nucleosomes in the repressed state or, on average, one nucleosome in the activated state. The results are significant in two regards. First, they show that the promoter defines an autonomous domain that can be uncoupled from adjacent chromosomal regions without loss of characteristic structural properties (present results and unpublished data). Second, an activated promoter circle preparation contains no detectable repressed circles. Thus, the presence of a single nucleosome on average on activated circles does not reflect the contamination of essentially naked, activated circles by repressed circles bearing a full complement of three nucleosomes. This conclusion accords well with our previous finding that the single nucleosome on the activated circle can occupy all three positions but with very different probabilities (3). Contamination by repressed circles would have led to the same probability of occurrence at all positions.
Our native gel electrophoresis of individual promoter nucleosomes supports the conclusion from previous work that the nucleosome −2 region of the PHO5 promoter is essentially nucleosome free in the activated state (3). This analysis further confirms the supposition that a nucleosome in this region prevents the binding of Pho2p and Pho4p to UASp2 in the repressed state, while the absence of the nucleosome enables binding in the activated state (17). Our results are in conflict with those of others showing that Pho2p and Pho4p can bind to UASp2 assembled in a nucleosome in vitro (16). There are a number of possible reasons for this discrepancy. First, nucleosomes formed in vitro may differ from those assembled in vivo as a result of histone modification or the presence of additional interacting proteins. Second, assembly in vitro may be imperfect, leaving a fraction of UASp2 nucleosome free and available for binding Pho2p and Pho4p. In the case of purified PHO5 chromatin we can detect some naked nucleosome −2 DNA released from a repressed promoter (Fig. 6A, lanes R), pointing to some heterogeneity even of chromatin assembled in vivo. Finally, at the very high concentrations of Pho2p and Pho4p attainable in vitro, these proteins may directly displace histones from their binding sites, as has been shown for TATA-binding protein and TFIIA binding to a reconstituted nucleosome (5). Indeed, we have observed that in the presence of micromolar concentrations of Pho4p and Pho2p the nucleosome −2 region released from repressed PHO5 chromatin migrates with the same mobility in a native gel as naked DNA treated with the same concentrations of these proteins (data not shown). This might explain why overexpression of Pho4p can compensate for the deletion of UASp1, which is required for disruption of nucleosome −2 under inducing conditions in vivo (17). Our results recapitulate this requirement for UASp1, suggesting that an additional factor(s) is involved in the chromatin transition at PHO5.
Acknowledgments
Plasmids pRS415-RecR and pABX22 were a kind gift from M. Gartenberg. Plasmid pPHO2-His was a kind gift from D. Stillman.
This research was supported by NIH grant GM36659 to R.D.K.
J.G. and H.B. contributed equally to this work.
REFERENCES
- 1.Almer, A., H. Rudolph, A. Hinnen, and W. Horz. 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari, A., T. H. Cheng, and M. R. Gartenberg. 1999. Isolation of selected chromatin fragments from yeast by site-specific recombination in vivo. Methods 17:104-111. [DOI] [PubMed] [Google Scholar]
- 3.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell. 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 4.Brazas, R. M., and D. J. Stillman. 1993. The Swi5 zinc-finger and Grf10 homeodomain proteins bind DNA cooperatively at the yeast HO promoter. Proc. Natl. Acad. Sci. USA 90:11237-11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godde, J. S., and A. P. Wolffe. 1995. Disruption of reconstituted nucleosomes. The effect of particle concentration, MgCl2 and KCl concentration, the histone tails, and temperature. J. Biol. Chem. 270:27399-27402. [DOI] [PubMed] [Google Scholar]
- 6.Haswell, E. S., and E. K. O'Shea. 1999. An in vitro system recapitulates chromatin remodeling at the PHO5 promoter. Mol. Cell. Biol. 19:2817-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenburg, M. E., and E. K. O'Shea. 1996. Signaling phosphate starvation. Trends Biochem. Sci. 21:383-387. [PubMed] [Google Scholar]
- 8.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 9.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell. 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 10.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 11.Roth, S. Y., and R. T. Simpson. 1991. Yeast minichromosomes. Methods Cell Biol. 35:289-314. [PubMed] [Google Scholar]
- 12.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Schultz, M. C., S. Y. Choe, and R. H. Reeder. 1991. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc. Natl. Acad. Sci. USA 88:1004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinbock, F. A., and G. Wiche. 1999. Plectin: a cytolinker by design. Biol. Chem. 380:151-158. [DOI] [PubMed] [Google Scholar]
- 15.Svaren, J., and W. Horz. 1997. Transcription factors versus nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22:93-97. [DOI] [PubMed] [Google Scholar]
- 16.Terrell, A. R., S. Wongwisansri, J. L. Pilon, and P. J. Laybourn. 2002. Reconstitution of nucleosome positioning, remodeling, histone acetylation, and transcriptional activation on the PHO5 promoter. J. Biol. Chem. 277:31038-31047. [DOI] [PubMed] [Google Scholar]
- 17.Venter, U., J. Svaren, J. Schmitz, A. Schmid, and W. Horz. 1994. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 13:4848-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]