Abstract
The eIF4E-binding proteins (4E-BPs) interact with translation initiation factor 4E to inhibit translation. Their binding to eIF4E is reversed by phosphorylation of several key Ser/Thr residues. In Drosophila, S6 kinase (dS6K) and a single 4E-BP (d4E-BP) are phosphorylated via the insulin and target of rapamycin (TOR) signaling pathways. Although S6K phosphorylation is independent of phosphoinositide 3-OH kinase (PI3K) and serine/threonine protein kinase Akt, that of 4E-BP is dependent on PI3K and Akt. This difference prompted us to examine the regulation of d4E-BP in greater detail. Analysis of d4E-BP phosphorylation using site-directed mutagenesis and isoelectric focusing-sodium dodecyl sulfate-polyacrylamide gel electrophoresis indicated that the regulatory interplay between Thr37 and Thr46 of d4E-BP is conserved in flies and that phosphorylation of Thr46 is the major phosphorylation event that regulates d4E-BP activity. We used RNA interference (RNAi) to target components of the PI3K, Akt, and TOR pathways. RNAi experiments directed at components of the insulin and TOR signaling cascades show that d4E-BP is phosphorylated in a PI3K- and Akt-dependent manner. Surprisingly, RNAi of dAkt also affected insulin-stimulated phosphorylation of dS6K, indicating that dAkt may also play a role in dS6K phosphorylation.
Binding of the 40S ribosomal subunit to mRNA is mediated by the eukaryotic initiation factor 4F (eIF4F) complex. eIF4F is a heterotrimer comprising eIF4A, eIF4E, and eIF4G, and this complex is recruited to the mRNA 5′ end via the direct interaction of eIF4E with the m7G cap structure. eIF4G bridges the mRNA with the 40S subunit through its interaction with the ribosome-bound eIF3, and the eIF4A subunit is believed to disrupt the secondary structure present in the 5′ region of the mRNA (43). The eIF4E-binding proteins (4E-BPs) are inhibitors of cap-dependent protein synthesis (37) and are conserved in all metazoans. There are three 4E-BPs in mammals (37, 40) but only one in Drosophila melanogaster (33). The 4E-BPs are low-molecular-weight proteins that interact with eIF4E and prevent its interaction with eIF4G to form the eIF4F complex. All 4E-BPs and eIF4G family members share the consensus eIF4E-binding motif YXXXXLΦ (where X is any residue and Φ is a hydrophobic residue) (29). Upon binding to eIF4E, the 4E-BPs adopt an energetically favorable α-helical structure that mimics the eIF4E-binding site of eIF4G (30). The block to translation caused by the 4E-BPs is reversible by their phosphorylation at certain key residues (4, 15, 17). Mammalian 4E-BP1 (hereafter referred to as 4E-BP1) may be phosphorylated on at least seven sites: Thr37, Thr46, Ser65, Thr70, Ser83, Ser101, and Ser112 (10, 20, 55). Thr37 and Thr46 are coordinately phosphorylated, priming the hierarchical phosphorylation of Thr70, followed by that of Ser65, which ultimately results in 4E-BP1 release from eIF4E (15, 17). Although Ser83 is phosphorylated, it is not required for 4E-BP1 release from eIF4E (6). A recent report suggests that a Ser or Glu residue at position 101 is necessary for phosphorylation of Ser65 and that Ser112 phosphorylation affects 4E-BP1 binding to eIF4E (55). In d4E-BP, Thr37, Thr46, Ser65, and Thr70 are identical to 4E-BP1, but Ser83 is a Thr residue, Ser101 is Gln, and Ser112 is absent (33).
In mammals, the phosphoinositide 3-OH kinase (PI3K) and mammalian target of rapamycin (mTOR) signaling pathways impinge on two known downstream effectors, namely, 4E-BP1 and ribosomal protein S6 kinase 1 (S6K1) (for a review, see reference 18). In the fruit fly, D. melanogaster, genetic studies have underscored the importance of the insulin-PI3K and TOR signaling pathways in controlling the sizes of cells and organs, as well as that of the entire animal (for a review, see reference 45). These evolutionarily conserved pathways are also critical for the control of cell growth in mammals (1, 11, 24, 49, 54). The signaling pathways have been assumed to function similarly in mammalian and invertebrate systems (23), but recent studies of Drosophila S6K (dS6K) argue that this may not be true. Indeed, dS6K-regulated cell growth, which was believed to be triggered following activation of PI3K, has been shown to be independent of dPI3K and its downstream effector, dPKB (41, 42).
The insulin-induced phosphorylation of the single Drosophila 4E-BP (d4E-BP), which is reduced by inhibition of PI3K and TOR signaling with the inhibitors LY294002 and rapamycin, respectively, was previously described(33). However, the recent finding that S6K signaling in Drosophila is independent of PI3K and Akt led us to undertake a detailed study of the regulation of d4E-BP phosphorylation by the insulin, PI3K, and TOR signaling pathways using double-stranded RNA interference (dsRNAi) (7).
MATERIALS AND METHODS
Cell culture and extract preparation.
Drosophila Schneider 2 (S2) cells were grown in Schneider's medium (Invitrogen Canada Inc.) containing 10% heat-inactivated fetal bovine serum. The cells (3 × 106) were grown in 100-mm-diameter plates to 80% confluence. The cells were washed once with phosphate-buffered saline (PBS) and placed in Schneider's medium lacking serum for 36 h. Pretreatment with rapamycin (Calbiochem) (dissolved in ethanol) was performed by preincubating cells for 30 min with Schneider's medium containing the appropriate drug concentration. For insulin treatment, cells were incubated with Schneider's medium containing 1 μg of bovine pancreatic insulin (Invitrogen Canada Inc.)/ml for the appropriate periods. Extracts were prepared by scraping cells into cold cap binding buffer (100 mM KCl, 20 mM HEPES, pH 7.6, 7 mM β-mercaptoethanol, 0.2 mM EDTA, 10% [vol/vol] glycerol, 50 mM β-glycerol phosphate, 50 mM NaF, 100 μM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and complete EDTA-free protease inhibitor [Boehringer] used according to the manufacturer's instructions). The suspensions were then subjected to four freeze-thaw cycles. Cell debris was pelleted by centrifugation, and the protein concentration of the supernatant was determined using the Bio-Rad assay.
Plasmids, transfections, and induction of protein expression.
Drosophila 4E-BP phosphorylation mutants were generated by PCR mutagenesis using Pwo polymerase (Boehringer). Wild-type and mutant d4E-BP coding regions were inserted into pcDNA-3HA, and the coding regions, including the three-hemagglutinin (3HA) tags, were once more amplified and inserted into the pUAST vector (2). For transfections, S2 cells (3 × 106) were propagated in a 100-mm-diameter plate and left to attach overnight. Next, the cells were cotransfected with equal quantities of pUAST-3HA-d4E-BP and pMK-GAL4 vectors (700 ng or 1 μg) using Effectene (Qiagen) and left to grow for 48 h. The cells were then placed in Schneider's medium lacking serum for 24 h as described above. For induction of protein expression, cells were treated with 0.7 mM copper sulfate (CuSO4) for 30 min. For insulin treatment, the cells were treated for 5 min as described above after the 25-min copper sulfate treatment, for a total of 30 min.
Phosphatase treatment.
Cells were lysed in phosphatase reaction buffer, and the resulting extract (50 μg) was incubated with or without the addition of 10 U of calf intestine alkaline phosphatase (CIP; New England Biolabs) at the required reaction temperature (4 or 30°C). The composition of CIP reaction buffer was 50 mM Tris · HCl, pH 7.5, 1 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride.
IEF-SDS-PAGE.
Cell lysate prepared in cap binding buffer (∼20 μl) was mixed with 27 mg of urea and 7 μl of isoelectric focusing (IEF) sample buffer (17% [vol/vol] Pharmalytes at pH 3 to 10, 4% [vol/vol] Pharmalytes at pH 8 to 10.5, 14% [vol/vol] β-mercaptoethanol, 35% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 1.8% [wt/vol] sodium dodecyl sulfate [SDS]), and incubated at room temperature until the urea was dissolved. Two-dimensional IEF-SDS-polyacrylamide gel electrophoresis (PAGE) was performed as previously described (17), with the exception that the first-dimension gel contained 1% (vol/vol) Pharmalytes at pH 8 to 10.5 instead of 1% (vol/vol) Pharmalytes at pH 2.5 to 5.
Antibodies and immunoblotting analysis.
Antibody 1868 to d4E-BP (33), anti-deIF4A (25), and antibody 1739 to deIF4E (50) were described previously. Anti-phospho-S6K(Thr389) (which recognizes Thr389 of dS6K), anti-Akt, and anti-phospho-4E-BP1(Thr37/46), -4E-BP1(S65), and -4E-BP1(T70) antibodies were obtained from Cell Signaling Technology. Anti-dS6K and anti-dTSC1 were gifts from G. Thomas, Basel, Switzerland, and D. Pan, Dallas, Tex., respectively. Anti-HA.11 was obtained from BAbCO. Polypeptides were resolved on SDS-polyacrylamide gels (15% for d4E-BP, deIF4E, dS6K, and dAkt immunoblotting and 6% for dTsc1 immunoblotting) and transferred to 0.22-μm-pore-size nitrocellulose membranes. The membranes were blocked with 2% (wt/vol) nonfat dry milk in PBS containing 0.5% (vol/vol) Tween-20 (all subsequent incubations were performed in the same solution) for 2 h and incubated for 2 h to overnight with the individual primary antibody 1868 (1:2,000) or 1739 (1:3,000) or antibody directed to deIF4A (1:3,000), phospho-S6K (Thr389; 1:1,000), dS6K (1:200), Akt (1:1,000), dTSC1 (1:5,000), or HA.11 (1:2,500). Incubation with secondary antibody was performed with peroxidase-coupled donkey anti-rabbit immunoglobulin or with peroxidase-coupled sheep anti-mouse immunoglobulin, followed by enhanced chemiluminescence (Amersham).
dsRNA interference.
Fragments from Dp110 (positions 375 to 1075), dAkt1/dPKB (positions 784 to 1510), dPTEN (positions 205 to 912), dPDK1 (positions 2521 to 3008), dTSC1 (positions 435 to 1098), and dTOR (positions 5401 to 6130) were amplified by PCR using gene-specific primers flanked by the T7 RNA polymerase-binding site (5′-GAATTAATACGACTCACTATAGGGAGA-3′). The PCR products were purified using the Ultraclean GelSpin DNA purification kit (MoBIO Laboratories). The fragments (4 μl) were used in 50-μl in vitro transcription reaction mixtures, which also contained 2 mM nucleotide mixture, 50 U of RNasin RNase inhibitor (Promega), and 30 U of T7 RNA polymerase (MBI Fermentas). The synthesized RNA was phenol-chloroform extracted, ethanol precipitated, resuspended in water, and annealed by heating it at 65°C for 30 min, followed by slow cooling to room temperature. Each dsRNA was visualized as a single band on a 1% agarose gel. For RNAi, S2 cells (5 × 106) were propagated in 60-mm-diameter plates and left to attach overnight. Next, the medium was removed and replaced with 1.5 ml of serum-free medium containing the dsRNA. The same effect was produced with either 10 or 20 μg of dsRNA for dPDK1; 10, 15, or 20 μg of dsRNA produced the same effect for dAkt. For Dp110, dPTEN, and dTSC1, 20 μg of dsRNA was used. After 30 min of incubation, 2.5 ml of complete medium was added, and the cells were left to grow for 3 days at 25°C. The cells were then serum starved for 36 h as described above, with the exception that a fresh dose of dsRNA was added to the serum-free medium.
Northern analysis to measure RNAi effects.
No antibodies were available to confirm the knockdown of the expression of Dp110, dPTEN, dPDK1, or dTOR. Therefore, knockdown was analyzed by Northern hybridization of a blot containing 5 μg of total RNA extracted from a plate of S2 cells treated with the dsRNA of interest concomitantly with the other experimental plates. The probes were derived as follows: Dp110 was an 824-nucleotide (nt) fragment from the BglII/PvuII digest of the pUAS-Dp110 vector (a kind gift from Sally Leevers), dPTEN was a 395-nt fragment from the HincII/HindIII digest of pUAST-dPTEN (a kind gift of D. Pan), and dPDK1 was a 656-nt fragment from the XhoI/HindIII digest of the dPDK1 cDNA excised a priori from the pUAST-dPDK1 vector by digestion with EcoRI and XbaI (44). Hybridization was performed with ExpressHyb (BD Biosciences Clontech) according to the manufacturer's instructions.
Quantitation.
Band intensities were quantified using the image analysis software Image-J, a Java program inspired by NIH-Image that runs on any computer platform (http://rsb.info.nih.gov/nih-image/about.html).
RESULTS
Phosphorylation of d4E-BP.
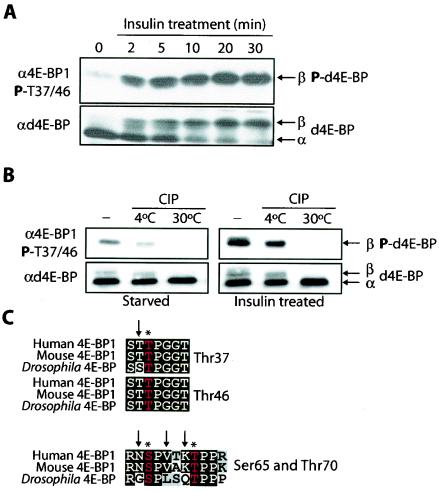
To characterize d4E-BP phosphorylation after insulin stimulation, Drosophila S2 cells were deprived of serum. In SDS-PAGE, d4E-BP was detected primarily as a fast-migrating α form and a small amount of β form (Fig. 1A, bottom) (33). After insulin treatment, a rapid shift toward the β form was observed, which was completed by 30 min (Fig. 1A, bottom). We tested the reactivities of phosphospecific antiserum directed against 4E-BP1 phosphopeptides corresponding to human phosphorylated Thr37/46, Ser65, and Thr70 (15, 17). Anti-phospho-4E-BP1(Ser65) and -4E-BP1(Thr70) failed to detect d4E-BP before and after insulin treatment (data not shown). In serum-starved cells, however, anti-phospho-4E-BP1(Thr37/46) detected a faint signal corresponding to the β form, which intensified immediately after insulin treatment (Fig. 1A, top). This effect was blocked by treating the cells with rapamycin, indicating that this phosphorylation is susceptible to the inhibition of dTOR (data not shown) (33).
FIG. 1.
Insulin treatment of S2 cells and potential phosphorylation sites of d4E-BP. (A) Time course of d4E-BP phosphorylation in response to insulin. S2 cells were starved for 36 h and stimulated with bovine insulin (1 μg/ml) for the indicated times. (B) Phosphatase treatment. Extracts from starved or insulin-treated S2 cells were treated with CIP for 30 min at the indicated temperatures. The effect of insulin treatment on d4E-BP in panels A and B was assessed by immunoblotting with anti-phospho-4E-BP1(Thr37/46) (α4E-BP1) (top) and by gel shift of the α phosphoisoform to β as visualized by anti-d4E-BP (αd4E-BP) (bottom) using extracts from S2 cells. P, phosphorylated. (C) Sequence alignment of the putative phosphorylation sites of d4E-BP and human and murine 4E-BP1. Identical (solid boxes) and conserved (shaded boxes) amino acids are highlighted. Conserved phosphorylation sites are colored red and marked by asterisks. The arrows indicate differences in (for Thr37, Ser65, and Thr70) or identity of (for Thr46) the amino acids surrounding the putative phosphorylation sites of d4E-BP.
To determine whether the β-form is phosphorylated, we incubated extracts from starved or insulin-treated S2 cells with CIP. Both the α and β forms were detected in untreated extracts, as well as in treated extracts kept on ice. Phosphatase-treated extracts incubated at 30°C contained only the α form (Fig. 1B, bottom) and lacked the phosphoreactive β form (Fig. 1B, top). Although Thr37, Thr46, Ser65, and Thr70 are conserved between humans and flies, only the amino acids flanking Thr46 are perfectly conserved between 4E-BP1 and d4E-BP (Fig. 1C). The residues flanking the serine/threonine proline sites of Ser65 and Thr70 in d4E-BP are different from their 4E-BP1 counterparts (Fig. 1C), which could explain the lack of reactivity of these antibodies. It is also possible, however, that these residues are not phosphorylated in response to insulin treatment. For 4E-BP1, the residues surrounding Thr37 and Thr46 are identical (Fig. 1C), and therefore the antibodies directed against the individual Thr37 and Thr46 phosphopeptides cross-react (15). The region surrounding Thr46 in d4E-BP is perfectly conserved, whereas the Thr37 site has a mismatch in the −1 position (Fig. 1C). Thus, the phosphospecific phospho-4E-BP1(Thr37/46) antibodies most likely recognize phospho-Thr46 in d4E-BP.
Analysis of d4E-BP mutant phosphorylation sites.
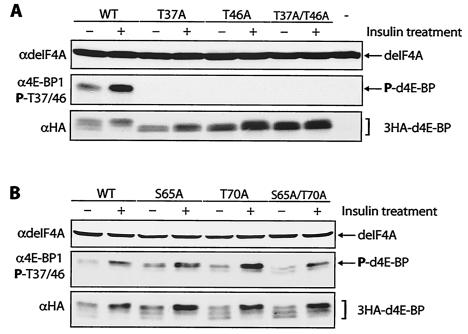
To study the phosphorylation states of different residues of d4E-BP, HA-tagged d4E-BPs (3HA-d4E-BPs) were generated and transfected into S2 cells. First, we mutated Thr37 and Thr46 to Ala, either individually or in combination. Transfected wild-type d4E-BP migrated as three distinct bands (Fig. 2A, bottom). This protein is phosphorylated to some extent in serum-starved cells, but after insulin treatment, the amount of phosphorylated protein is increased 1.7-fold (Fig. 2A, middle). This effect is paralleled by a characteristic upward shift of d4E-BP after phosphorylation (Fig. 2A, bottom). Mutation of either Thr37 or Thr46 to Ala abolished both the insulin-stimulated upward shift and the phosphorylation of d4E-BP, as detected with the antibody to phospho-4E-BP1(Thr37/46) (Fig. 2A, middle). These results suggest that the phosphorylations of these residues in Drosophila 4E-BP are interdependent, as in human 4E-BP1 (15).
FIG. 2.
Analysis of mutations at d4E-BP phosphorylation sites. (A) Mutation of Thr37 and/or Thr46 to Ala (T37A and T46A, respectively) prevents the insulin-induced phosphorylation of d4E-BP. As expected, the double mutant (T37A/T46A) also showed no phosphorylation. The last lane (-) is an untransfected control. WT, wild type; +, treated; −, untreated; P, phosphorylated. (B) Mutation of Ser65 and/or Thr70 to Ala (S65A and T70A, respectively) had no effect on phosphorylation. As expected, the double mutant (S65A/T70A) also had no effect. S2 cells transfected with 3HA-tagged d4E-BP mutant cDNAs were starved for 24 h before insulin treatment. The effect of insulin was assessed with anti-phospho-4E-BP1(Thr37/46) (α4E-BP1 P-T37/46) (middle), and protein expression levels were compared using anti-HA-11 (αHA) (bottom). Total protein loading levels were compared using anti-deIF4A (αdeIF4A).
To delineate the order of d4E-BP phosphorylation, we mutated Ser65 and Thr70 to Ala as described above. In human 4E-BP1, these residues must be phosphorylated following Thr37 and Thr46 phosphorylation, first on Thr70 and then on Ser65, before 4E-BP1 is released from eIF4E (17). Neither mutation, either alone or in combination, affected insulin-mediated phosphorylation of the 4E-BP mutants, as detected with the antibody to phospho-4E-BP1(Thr37/46) (Fig. 2B, middle). Furthermore, all mutants displayed the characteristic upward shift of the wild-type d4E-BP after phosphorylation (Fig. 2B, bottom). These results suggest that neither Ser65 nor Thr70 affects the phosphorylation of d4E-BP at Thr37 and Thr46.
Analysis of d4E-BP phosphorylation by IEF-SDS-PAGE.
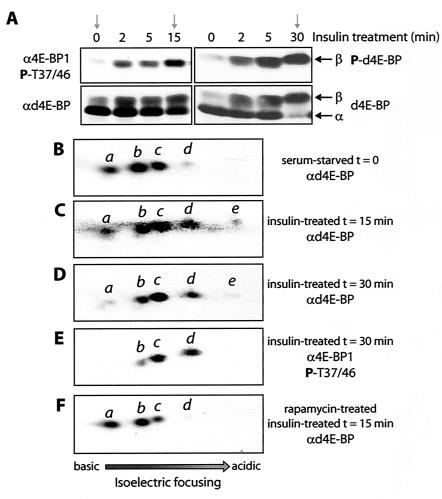
To better understand d4E-BP phosphorylation, we used two-dimensional IEF combined with immunoblotting with phosphospecific antibodies (IEF-SDS-PAGE), a method developed to show the hierarchical phosphorylation of 4E-BP1 (17). This method separates the d4E-BP molecules based on their charges and reflects the number of phosphate residues added. Extracts from serum-starved S2 cells, insulin-treated cells, and rapamycin-treated cells were analyzed using this method. The starved cells expressed little of the β form relative to the α form; the 15-min sample had both the α and β forms, and the 30-min sample contained the β form almost exclusively (Fig. 3A). Rapamycin treatment prevented insulin-induced d4E-BP phosphorylation (data not shown) (33). In extracts from serum-starved or rapamycin-treated cells, d4E-BP exists as three predominant isoforms, as well as a fourth, less-abundant isoform (Fig. 3B and F, isoforms a to d). The α form therefore consists of either a nonphosphorylated and three phosphorylated isoforms of d4E-BP or of four phosphorylated isoforms, depending on the phosphorylation state of isoform a, which is not known. After insulin treatment, a stronger signal for isoform d was detected, and a fifth weak and more acidic isoform, e, was also detected (Fig. 3C and D). The longer 30-min treatment generally caused the signal intensity for isoforms a and b to weaken relative to the 15-min treatment. Duplicate blots were probed with anti-phospho-4E-BP1(Thr37/46). In starved or rapamycin-treated cells, no signal was detected. Thus, there is very little, if any, of the β form in these extracts. After insulin treatment, isoforms b to d were detected, but not isoform e (the 30-min treatment is shown in Fig. 3E).
FIG. 3.
Analysis by IEF-SDS-PAGE of d4E-BP phosphorylation. (A) Selected S2 cell extracts for IEF-SDS-PAGE (arrows). Rapamycin-treated cell extract (not shown) is similar to unstimulated cell extract. (B to F) IEF-SDS-PAGE of extracts from serum-starved (B), insulin-treated (15 min) (C), insulin-treated (30 min) (D and E), and rapamycin- and insulin-treated (15 min) (F) cells were probed with the indicated antibodies. d4E-BP (B to F) was assessed by blotting it with anti-d4E-BP (αd4E-BP) or anti-phospho-4E-BP1(Thr37/46) (α4E-BP1 P-T37/46) as indicated. The individual isoforms are represented by the letters a to e. P, phosphorylated.
Despite different migration patterns on SDS-PAGE, the insulin treatment time points give almost identical patterns on IEF-SDS-PAGE. Thus, in IEF-SDS-PAGE, the shift to the β form is represented by each d4E-BP isoform moving to the spot occupied by the next isoform so that there is isoform overlap, except for isoform d, which becomes e (see Fig. 6B for proposed models). Hence, our results suggest a simple mode of regulation for d4E-BP after insulin stimulation. In starved or rapamycin-treated cells, the bulk of the α form is composed of a mixture of isoforms a to c, indicating that d4E-BP is phosphorylated at a number of sites that do not prevent its interaction with deIF4E. The most likely candidates are Thr37, Ser65, and Thr70, since they are conserved from Drosophila to humans. After insulin stimulation, a regulatory interplay between Thr37 and Thr46 culminates in phosphorylation of Thr46, which causes d4E-BP to dissociate from deIF4E. Thus, these results support the conclusion that phosphorylations at Thr37 and Thr46 represent major events in the regulation of d4E-BP after insulin stimulation.
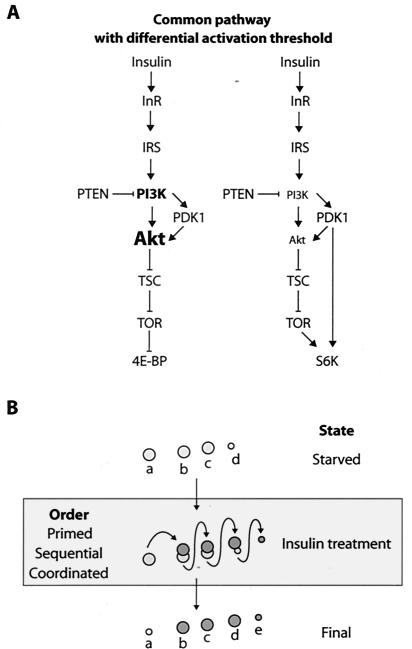
FIG. 6.
Models of the insulin-signaling pathway in Drosophila and regulation of d4E-BP by phosphorylation. (A) A common pathway regulates 4E-BP and S6K. The regulation of 4E-BP is dependent on signaling from Inr-PI3K-Akt-TSC and TOR. The regulation of S6K is also effected by PDK1 and the same upstream elements. The different size of the Akt typeface is to illustrate that the intensity of signaling toward TSC affects 4E-BP and S6K differentially. (B) The pattern of phosphorylation of d4E-BP as observed by IEF-SDS-PAGE may result from one of three possible models. (i) In the primed model, d4E-BP is already phosphorylated at Thr37 and is subsequently phosphorylated on one additional site, Thr46, after insulin stimulation. (ii) In the sequential model, d4E-BP is phosphorylated on Thr37 and then Thr46 (or vice versa). (iii) In the coordinated model, d4E-BP is phosphorylated coordinately on Thr37 and Thr46.
dsRNAi targeting of genes in the insulin-signaling pathway.
Drosophila S6K-regulated cell growth is independent of dPI3K- and dPKB-mediated signaling (42) but is affected by the tumor suppressors dTSC1 and -2 (13, 41). In contrast, it was reported that d4E-BP is a downstream target of PI3K (33). This is rather puzzling, because both dS6K and d4E-BP are downstream targets of TOR. Most of these studies were based on the use of pharmacological agents. In an effort to solve this conundrum, we used an alternative approach to inhibition by pharmacological agents, RNAi experiments for protein components in the Drosophila insulin-like signaling pathway.
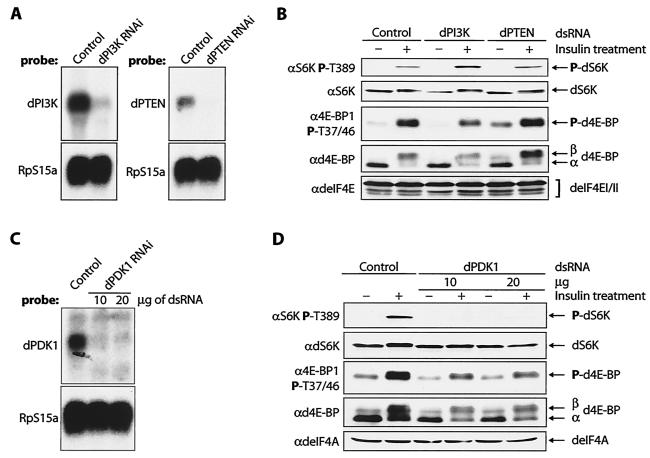
First, we reduced the expression of the upstream components of the signaling pathway in S2 cells: the Dp110 catalytic subunit of dPI3K, as well as the phosphatase dPTEN, which antagonizes dPI3K activity by hydrolyzing the 3′ phosphate of phosphatidylinositol-3,4,5-P3 (PIP3) (Fig. 4A). dPI3K and dPTEN act in combination as a dedicated growth rheostat that controls the production of PIP3 (35). RNAi of Dp110 reduced the insulin-stimulated phosphorylation of d4E-BP ∼1.5-fold, whereas knockdown of dPTEN caused an increase in the basal level of phosphorylated d4E-BP of ∼3.4-fold in serum-starved cells (Fig. 4B, middle). This is consistent with dPI3K and dPTEN being upstream regulators of d4E-BP phosphorylation. The phosphorylation of dS6K at Thr398 after insulin treatment of Dp110-RNAi cells was more robust (∼2.2-fold) than in control cells (Fig. 4B, top). However, the significance and mechanism of this increase are not clear. Phosphorylation of dS6K at Thr398 in dPTEN-RNAi cells was similar to that in control cells before and after insulin treatment (Fig. 4B, top). These results are consistent with the notion that the threshold activity of dPI3K required for dS6K activation is relatively low, while that required for d4E-BP phosphorylation is higher.
FIG. 4.
Effects of RNAi of dPI3K, dPTEN, and dPDK1 on d4E-BP phosphorylation in S2 cells. (A) Northern analysis confirms that the dPI3K (Dp110) and dPTEN transcript amounts are reduced compared with control cells. (B) RNAis of dPI3K and dPTEN have different effects on d4E-BP phosphorylation. Whereas dPI3K RNAi reduces the insulin-stimulated phosphorylation of d4E-BP, dPTEN RNAi increases the phosphorylation of d4E-BP in unstimulated cells. The phosphorylation of dS6K Thr398 is increased by RNAi of dPI3K but not by RNAi of dPTEN. RNAi of Dp110 and dPTEN was performed at least three times. +, treated; −, untreated. (C) Northern analysis confirms that the dPDK1 transcript is reduced compared with control cells. (D) A reduction in dPDK1 amounts reduces the endogenous levels of d4E-BP but does not affect its insulin-induced phosphorylation. dPDK1 RNAi does, however, block phosphorylation of dS6K at Thr398. RNAi of dPDK1 was performed four times. The effect on d4E-BP was assessed by blotting with anti-phospho-4E-BP1(Thr37/46) (α4E-BP1 P-T37/46) and by gel shift of the α phosphoisoform to β with anti-d4E-BP (αd4E-BP). The phosphorylation of dS6K was detected with anti-phospho-S6K Thr389 (αS6K P-T389). Antibodies against translation factors deIF4E (αdeIF4E) and deIF4A (αdeIF4A) were used to confirm equal loading of the gels in panels B and D, respectively. RpS15a is the probe for ribosomal protein S15a, which was used to confirm equal loading in panels A and C. P, phosphorylated.
As a consequence of the PIP3 increase that occurs during insulin signaling, the Ser/Thr protein kinase dAkt is recruited to the plasma membrane and activated by phosphorylation of the activation loop site by dPDK1. We therefore examined the phosphorylation of d4E-BP and dS6K as a consequence of the reduction of the level of dPDK1. Treatment with dsRNA directed against dPDK1 caused a dramatic decrease in the amount of dPDK1 mRNA (>90%) (Fig. 4C) and a 60% overall decrease in the levels of d4E-BP detected in S2 cells (Fig. 4D, fourth gel from top). After normalization of d4E-BP protein levels, we saw that the insulin-induced phosphorylation of d4E-BP was not, however, reduced by RNAi of dPDK1 (Fig. 4D, third gel from top). The decrease in protein levels and the absence of a reduction in d4E-BP phosphorylation differ from the findings with PDK1-null embryonic stem cells, in which basal 4E-BP1 phosphorylation was increased, although insulin-like growth factor treatment did not further stimulate phosphorylation relative to controls (56). Drosophila PDK1 is required for activation of dS6K (42), and consistent with this observation, phosphorylation of Thr398 after insulin treatment was abolished in dPDK1-RNAi cells (Fig. 4D, top). This result is consistent with the direct phosphorylation at Thr389 of S6K1 by PDK1. The differences in the effects of the knockdown of dPDK1 on d4E-BP and dS6K will be further addressed below.
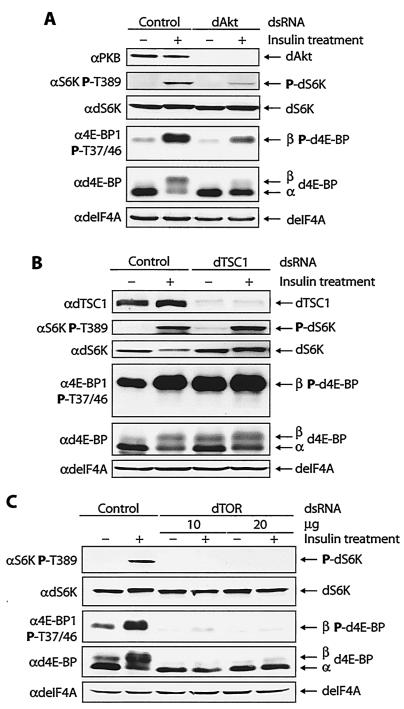
Treatment of S2 cells with dsRNA targeting dAkt abrogated dAkt expression in S2 cells (Fig. 5A, top). Knockdown of dAkt resulted in a 2.1-fold decrease in d4E-BP phosphorylation after insulin treatment (Fig. 5A, fourth gel from top), consistent with dAkt being an upstream regulator of d4E-BP phosphorylation. Surprisingly, dS6K phosphorylation at Thr398 after insulin treatment was also reduced relative to control cells (∼3.3-fold less) (Fig. 5A, second gel from top), which is inconsistent with previous reports (41, 42). This result demonstrates that in our system, dS6K is a downstream target of dPI3K/dAkt signaling (see below).
FIG. 5.
Effects of RNAi of dAkt, dTSC1, and dTOR on d4E-BP phosphorylation in S2 cells. (A) A reduction in the amount of dAkt blocks signaling to d4E-BP and reduces insulin-induced phosphorylation. The phosphorylation of dS6K at Thr398 is also reduced. +, treated; −, untreated; P, phosphorylated. (B) A reduction in the amount of dTsc1 augments signaling to d4E-BP, causing an increase in the basal levels of phosphorylated d4E-BP. The phosphorylation of dS6K at Thr398 is also increased. (C) A reduction in the amount of dTOR blocks signaling to d4E-BP and dS6K and abolishes the insulin-induced phosphorylation of both proteins. RNAis of dAkt, dTSC1, and dTOR were performed at least three times. The effects on d4E-BP and dS6K were assessed as for Fig. 4.
The tumor suppressors dTsc1 and -2 form the tuberous sclerosis complex (TSC). dAkt directly phosphorylates the dTsc2 subunit, which prevents TSC from functioning as a growth inhibitor (39). RNAi of either TSC subunit produces identical effects and increases signaling to dS6K (13). To determine whether TSC is involved in regulating d4E-BP, we targeted dTsc1 with dsRNA. Consistent with TSC being an upstream regulator of d4E-BP phosphorylation, knockdown of dTsc1 in S2 cells resulted in elevated levels (2.1-fold) of phosphorylated d4E-BP in serum-starved cells relative to control cells (Fig. 5B, fourth gel from top). In starved dTsc1-RNAi cells, the level of dS6K phosphorylation at Thr398 was elevated compared with control cells (2.6-fold) (Fig. 5B, second gel from top). This observation is consistent with previous reports (13, 41), indicating that dS6K is negatively regulated by TSC.
Finally, we targeted dTOR for RNAi. Treatment with dsRNA mimicked the effects of rapamycin on d4E-BP and dS6K phosphorylation. The phosphorylation of d4E-BP and dS6K after insulin treatment was abolished (Fig. 5C). Moreover, the basal level of phosphorylated d4E-BP in serum-starved cells was reduced in dTOR-RNAi cells (Fig. 5C, third and fourth gels from top). Taken together, our RNAi experiments confirm that d4E-BP is a downstream target of the PI3K-Akt-TSC-TOR signaling cascade.
DISCUSSION
Conserved regulatory dependency between Thr37 and Thr46.
Insulin treatment caused a strong increase in the immunoreactivity of d4E-BP to antibodies directed against human phospho-4E-BP1(Thr37/46). Since the residues in the region of Thr46, but not Thr37, are perfectly conserved between 4E-BP1 and d4E-BP, it is probably Thr46 that is hyperphosphorylated in d4E-BP after insulin treatment. Regulation of d4E-BP phosphorylation thus appears to differ from that of mammalian 4E-BP1. Phosphorylation of Thr37 and Thr46 of 4E-BP1 is only modestly induced after serum stimulation in serum-starved HEK293 cells (15). In serum-starved HEK293 cells treated with rapamycin, the phosphorylation of 4E-BP1 at Thr37 and Thr46 is reduced, but upon serum addition, it is restored to its original state, whereas phosphorylation of Ser65 and Thr70 remains blocked (15, 17). Thus, Ser65 and Thr70 are the rapamycin-sensitive sites of 4E-BP1. In contrast, in S2 cells, phosphorylation of d4E-BP at Thr46 is robustly induced after insulin treatment, and rapamycin completely blocks its phosphorylation. Therefore, unlike 4E-BP1, phosphorylation at Thr46 is a major insulin-stimulated and rapamycin-sensitive event involved in d4E-BP regulation. A dependency between Thr37 and Thr46 that is analogous to that of 4E-BP1 is important for d4E-BP phosphorylation. For 4E-BP1, phosphorylations at Thr37 and Thr46 are intimately linked, as these two phosphorylation events are regulated coordinately by mTOR (15). The link between Thr37 and Thr46 in d4E-BP is conserved, but it is not known if Thr37 acts as a priming event for the subsequent phosphorylation of Thr46 or if Thr37 and Thr46 are regulated coordinately, similar to 4E-BP1. Hence, phosphorylation of d4E-BP may be explained by three possible models, which are depicted in Fig. 6B.
Our results suggest a simpler mode of regulation of d4E-BP by phosphorylation compared with the hierarchical phosphorylation of 4E-BP1. The phosphorylation of 4E-BP1 is the best understood, but not all mammalian 4E-BPs are regulated in a similar manner. 4E-BP2 is phosphorylated on fewer residues (27) and is dephosphorylated more slowly than 4E-BP1 (B. Raught and N. Sonenberg, unpublished data). Also, 4E-BP3 is weakly stimulated by insulin treatment, causing poor release from eIF4E. This has been attributed to the lack of the four-residue RAIP motif found in the N terminus of 4E-BP1 and -2 but not in 4E-BP3 (53). The RAIP motif seems to be required for the efficient overall phosphorylation of 4E-BP1, and intriguingly, this motif is also lacking in d4E-BP. Another important motif for 4E-BP1 regulation is the TOR signaling (TOS) motif (47), which is conserved (FQLDL, at the C terminus) in d4E-BP. Hence, a consequence of the lack of the RAIP motif may be the simpler regulation of d4E-BP to improve its release from eIF4E when d4E-BP is recruited to the dTOR/dRaptor complex through the TOS motif (34, 48). Moreover, because of a divergent eIF4E-binding site, d4E-BP does not interact as strongly with deIF4E as 4E-BP1 (33). It is conceivable that because of the poorer interaction of d4E-BP with deIF4E, phosphorylation at Thr37 and Thr46 is sufficient to bring about its release from eIF4E. Our IEF-SDS-PAGE results indicate that in serum-starved S2 cells, some d4E-BP is already phosphorylated. Although these additional sites do not prevent the interaction between deIF4E and d4E-BP, it is possible that they contribute to the release from deIF4E once Thr46 becomes phosphorylated.
Drosophila 4E-BP is a downstream target of PI3K-PTEN-Akt-TSC-TOR signaling.
A large body of work has established the paramount importance of the InR-IRS-PI3K-PTEN signaling module in the control of cell growth (8, 23, 45, 57). This module coordinates cellular metabolism with the nutritional state (3). The primary outcome of its activation is the modulation of the PI3K/PTEN cycle and consequent PIP3 production (35). The increase in PIP3 facilitates the recruitment of pleckstrin homology domain-containing proteins, such as dAkt and dPDK1, to the plasma membrane. dPTEN mutant flies die because of increased membrane translocation and activation of dAkt. Phosphorylation of the Tsc2 subunit of TSC by Akt results in inhibition of the complex by causing its dissociation or by blocking its interaction with other proteins (for reviews, see references 31 and 32). TSC inhibits S6K and 4E-BP1 by repressing the GTPase Rheb, preventing it from activating mTOR through an unknown mechanism (14). Thus, mTOR, is clearly a critical regulator of S6K and 4E-BP1 in mammals and Drosophila (14, 21, 36, 46, 52, 58).
Is d4E-BP regulated by a PI3K/Akt-independent pathway similar to that described for dS6K (42)? Our analysis of signaling to d4E-BP using RNAi indicates that it is not. It is more likely that d4E-BP is a direct downstream target of the dInR-dPI3K-dPTEN-dAkt-dTSC-dTOR signaling cascade. Thus, a linear pathway from InR to Akt that is important for 4E-BP regulation is conserved between Drosophila and mammals (9, 16).
PDK1 signaling to 4E-BP versus S6K in Drosophila.
dPDK1 is critical for regulating growth by phosphorylating dAkt and dS6K (5, 44). RNAi of dPDK1 did not significantly affect insulin-induced phosphorylation of d4E-BP. However, consistent with the direct phosphorylation of dS6K by dPDK1 (42, 44), the phosphorylation of dS6K at Thr398 was completely blocked by RNAi of PDK1. Thus, our results favor a model in which d4E-BP regulation is effected through dAkt, even when dPDK1 levels are dramatically reduced, whereas dS6K requires both dAkt and dPDK1. The differential effects of dPDK1 RNAi on d4E-BP and dS6K phosphorylation can be explained as follows: dPDK1 levels may be reduced below a threshold that is required to phosphorylate dS6K but is still adequate to activate dAkt, allowing d4E-BP phosphorylation. Since dS6K requires direct phosphorylation by dPDK1, it may be more susceptible to variations in its levels. On the other hand, d4E-BP, which relies on a signal relayed by dAkt, may be less affected by variations in dPDK1. In mammalian PDK1-hypomorphic mutants, a kinase activity that is 10-fold lower than normal still results in normal Akt and S6K1 activation, yet these animals are greatly reduced in size (26). This observation supports the notion that reduced PDK1 activity may differentially activate downstream targets.
Regulation of dS6K signaling by dAkt but not dPI3K/dPTEN?
In Drosophila, coexpression of dS6K with dPI3K does not cause additive cellular overgrowth, unlike coexpression of dAkt and dPI3K (42). RNAi of dPTEN in Kc 167 cells and overexpression of dPTEN in Drosophila larvae had little effect on dS6K activity (41). Moreover, removal of both dS6K and dPTEN in cell clones did not prevent the dPTEN-dependent overgrowth phenotype (41). Together, these results and the results of our dPI3K and dPTEN RNAi experiments would seemingly support the notion that dS6K-dependent cell growth is not influenced by dPI3K and dPTEN. However, a different effect of dPTEN RNAi on dS6K has been reported by another group. Gao et al. observed an increase in dS6K phosphorylation following RNAi of dPTEN (13). Consistent with this observation, Lizcano et al. (28) (published while this paper was under review) demonstrated that RNAi directed against dPI3K and dPTEN could modulate dS6K phosphorylation. A reasonable explanation for these discrepancies is that the knockdown of dPI3K and dPTEN achieved in our experiments was not sufficient to completely deplete these proteins and affect dS6K phosphorylation.
The role of dAkt in regulating dS6K is subject to debate. In Drosophila, Akt plays a predominant role in mediating the effects of increased PIP3 levels (51), and all Akt-mediated growth signals are thought to be transduced via Tsc1/2 (39). Tsc2 is directly phosphorylated by Akt (for a review, see reference 31), implying that S6K is downstream of Akt in the PI3K signaling pathway. Our observation that RNAi of dAkt reduces dS6K phosphorylation at Thr398 supports a direct link among dAkt, dTSC, and dS6K but contradicts the finding that TSC modulates dS6K activity in a dAkt-independent manner (41, 42). The recent data of Lizcano et al. (28) also support our conclusion of a link between dAkt and dS6K. Clones of cells doubly mutant for dPTEN and dTsc1 display an additive overgrowth phenotype, suggesting that the tumor suppressors act on two independent pathways, from dPTEN to dAkt and from dTSC to dS6K (12, 41). Our findings demonstrate clear effects of dPTEN, dAkt, and dTSC on d4E-BP, which does not preclude the possibility that two pathways regulate d4E-BP; however, a simpler interpretation is that a single pathway is important for its regulation. A possibility is that d4E-BP requires higher dAkt activity than dS6K in order to be phosphorylated. In circumstances of low PI3K activation, low levels of PIP3 are produced, resulting in weaker dAkt activity that is sufficient for dS6K activation but not for d4E-BP phosphorylation. A differential threshold of activation could be the source of the discrepancies between our results and those of Lizcano et al. (28). This model is strongly supported by recent data showing that in cells lacking both Akt1 and Akt2 isoforms, the low level of Akt activity remaining is sufficient for robust S6K1 phosphorylation, but phosphorylation of 4E-BP1 is dramatically reduced (38).
Alternatively, our results could also be explained by the existence of a negative feedback loop between dPI3K and dS6K that dampens insulin signaling by suppressing dAkt activity. This negative feedback loop has been described (19, 41, 42). Similar observations were made in mammals, as insulin-induced activation of Akt is inhibited in Tsc2-deficient mouse embryonic fibroblasts (22). Thus, depletion of dAkt may trigger this negative feedback loop, which diminishes dS6K phosphorylation and activation. Interestingly, engagement of this feedback mechanism can also provide an explanation for the reduction in total d4E-BP levels observed in dPDK1 RNAi-treated cells. Under these conditions, the reduction of dS6K signaling is accompanied by a concomitant reduction in growth signaling on the dPI3K-dAkt branch of the pathway. Thus, a reduced level of d4E-BP is required to accommodate the reduced need for deIF4E inhibition.
Acknowledgments
We thank C. Lister for excellent technical assistance and T. Radimerski, G. Thomas, D. Pan, and S. J. Leevers for gifts of reagents. We thank T. Radimerski and N. Hay for critically reading the manuscript.
This work was supported by grants from the National Cancer Institute of Canada and a Howard Hughes Medical Institute grant. M.M. was supported by a graduate fellowship from the Cancer Research Society.
REFERENCES
- 1.Backman, S. A., V. Stambolic, A. Suzuki, J. Haight, A. Elia, J. Pretorius, M. S. Tsao, P. Shannon, B. Bolon, G. O. Ivy, and T. W. Mak. 2001. Deletion of PTEN in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat. Genet. 29:396-403. [DOI] [PubMed] [Google Scholar]
- 2.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 3.Britton, J. S., W. K. Lockwood, L. Li, S. M. Cohen, and B. A. Edgar. 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2:239-249. [DOI] [PubMed] [Google Scholar]
- 4.Brunn, G. J., P. Fadden, T. A. J. Haystead, and J. C. Lawrence, Jr. 1997. The mammalian target of rapamycin phosphorylates sites having a (Ser/Thr)-Pro motif and is activated by antibodies to a region near its COOH terminus. J. Biol. Chem. 272:32547-32550. [DOI] [PubMed] [Google Scholar]
- 5.Cho, K. S., J. H. Lee, S. Kim, D. Kim, H. Koh, J. Lee, C. Kim, J. Kim, and J. Chung. 2001. Drosophila phosphoinositide-dependent kinase-1 regulates apoptosis and growth via the phosphoinositide 3-kinase-dependent signaling pathway. Proc. Natl. Acad. Sci. USA 98:6144-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, K. M., L. P. McMahon, and J. C. Lawrence, Jr. 2003. Two motifs in the translational repressor PHAS-I required for efficient phosphorylation by mammalian target of rapamycin and for recognition by raptor. J. Biol. Chem. 278:19667-19673. [DOI] [PubMed] [Google Scholar]
- 7.Clemens, J. C., C. A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B. A. Hemmings, and J. E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97:6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon, I., and M. Raff. 1999. Size control in animal development. Cell 96:235-244. [DOI] [PubMed] [Google Scholar]
- 9.Dufner, A., M. Andjelkovic, B. M. Burgering, B. A. Hemmings, and G. Thomas. 1999. Protein kinase B localization and activation differentially affect S6 kinase 1 activity and eukaryotic translation initiation factor 4E-binding protein 1 phosphorylation. Mol. Cell. Biol. 19:4525-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fadden, P., T. A. Haystead, and J. C. Lawrence, Jr. 1997. Identification of phosphorylation sites in the translational regulator, PHAS-I, that are controlled by insulin and rapamycin in rat adipocytes. J. Biol. Chem. 272:10240-10247. [DOI] [PubMed] [Google Scholar]
- 11.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, X., and D. Pan. 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15:1383-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, X., Y. Zhang, P. Arrazola, O. Hino, T. Kobayashi, R. S. Yeung, B. Ru, and D. Pan. 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 4:699-704. [DOI] [PubMed] [Google Scholar]
- 14.Garami, A., F. J. Zwartkruis, T. Nobukuni, M. Joaquin, M. Roccio, H. Stocker, S. C. Kozma, E. Hafen, J. L. Bos, and G. Thomas. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11:1457-1466. [DOI] [PubMed] [Google Scholar]
- 15.Gingras, A. C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gingras, A. C., S. G. Kennedy, M. A. O'Leary, N. Sonenberg, and N. Hay. 1998. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 12:502-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gingras, A. C., B. Raught, S. P. Gygi, A. Niedzwiecka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieszynska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gingras, A. C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807-826. [DOI] [PubMed] [Google Scholar]
- 19.Haruta, T., T. Uno, J. Kawahara, A. Takano, K. Egawa, P. M. Sharma, J. M. Olefsky, and M. Kobayashi. 2000. A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol. Endocrinol. 14:783-794. [DOI] [PubMed] [Google Scholar]
- 20.Heesom, K. J., M. B. Avison, T. A. Diggle, and R. M. Denton. 1998. Insulin-stimulated kinase from rat fat cells that phosphorylates initiation factor-4E binding protein 1 on the rapamycin-insensitive site (serine-111). Biochem. J. 336:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoki, K., Y. Li, T. Xu, and K. L. Guan. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17:1829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaeschke, A., J. Hartkamp, M. Saitoh, W. Roworth, T. Nobukuni, A. Hodges, J. Sampson, G. Thomas, and R. Lamb. 2002. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J. Cell Biol. 159:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozma, S. C., and G. Thomas. 2002. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays 24:65-71. [DOI] [PubMed] [Google Scholar]
- 24.Kwon, C. H., X. Zhu, J. Zhang, L. L. Knoop, R. Tharp, R. J. Smeyne, C. G. Eberhart, P. C. Burger, and S. J. Baker. 2001. PTEN regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat. Genet. 29:404-411. [DOI] [PubMed] [Google Scholar]
- 25.Lachance, P. E., M. Miron, B. Raught, N. Sonenberg, and P. Lasko. 2002. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol. Cell. Biol. 22:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawlor, M. A., A. Mora, P. R. Ashby, M. R. Williams, V. Murray-Tait, L. Malone, A. R. Prescott, J. M. Lucocq, and D. R. Alessi. 2002. Essential role of PDK1 in regulating cell size and development in mice. EMBO J. 21:3728-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, T. A., and J. C. Lawrence, Jr. 1996. Control of the translational regulators PHAS-I and PHAS-II by insulin and cAMP in 3T3-L1 adipocytes. J. Biol. Chem. 271:30199-30204. [DOI] [PubMed] [Google Scholar]
- 28.Lizcano, J. M., S. Alrubaie, A. Kieloch, M. Deak, S. J. Leevers, and D. R. Alessi. 2003. Insulin-induced Drosophila S6 kinase activation requires phosphoinositide 3-kinase and protein kinase B. Biochem. J. 374:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15:4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcotrigiano, J., A. C. Gingras, N. Sonenberg, and S. K. Burley. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3:707-716. [DOI] [PubMed] [Google Scholar]
- 31.Marygold, S. J., and S. J. Leevers. 2002. Growth signaling: TSC takes its place. Curr. Biol. 12:R785-R787. [DOI] [PubMed] [Google Scholar]
- 32.McManus, E. J., and D. R. Alessi. 2002. TSC1-TSC2: a complex tale of PKB-mediated S6K regulation. Nat. Cell Biol. 4:E214-E216. [DOI] [PubMed] [Google Scholar]
- 33.Miron, M., J. Verdu, P. E. Lachance, M. J. Birnbaum, P. F. Lasko, and N. Sonenberg. 2001. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat. Cell Biol. 3:596-601. [DOI] [PubMed] [Google Scholar]
- 34.Nojima, H., C. Tokunaga, S. Eguchi, N. Oshiro, S. Hidayat, K. Yoshino, K. Hara, N. Tanaka, J. Avruch, and K. Yonezawa. 2003. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278:15461-15464. [DOI] [PubMed] [Google Scholar]
- 35.Oldham, S., H. Stocker, M. Laffargue, F. Wittwer, M. Wymann, and E. Hafen. 2002. The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP(3) levels. Development 129:4103-4109. [DOI] [PubMed] [Google Scholar]
- 36.Patel, P. H., N. Thapar, L. Guo, M. Martinez, J. Maris, C. L. Gau, J. A. Lengyel, and F. Tamanoi. 2003. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J. Cell Sci. 116:3601-3610. [DOI] [PubMed] [Google Scholar]
- 37.Pause, A., G. J. Belsham, A.-C. Gingras, O. Donzé, T. A. Lin, J. C. Lawrence, Jr., and N. Sonenberg. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762-767. [DOI] [PubMed] [Google Scholar]
- 38.Peng, X.-D., P.-Z. Xu, M.-L. Chen, A. Hahn-Windgassen, J. Skeen, J. Jacobs, D. Sundararajan, W. S. Chen, S. E. Crawford, K. G. Coleman, and N. Hay. 2003. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 17:1352-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potter, C. J., L. G. Pedraza, and T. Xu. 2002. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4:658-665. [DOI] [PubMed] [Google Scholar]
- 40.Poulin, F., A.-C. Gingras, H. Olsen, S. Chevalier, and N. Sonenberg. 1998. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem. 273:14002-14007. [DOI] [PubMed] [Google Scholar]
- 41.Radimerski, T., J. Montagne, M. Hemmings-Mieszczak, and G. Thomas. 2002. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 16:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radimerski, T., J. Montagne, F. Rintelen, H. Stocker, J. van Der Kaay, C. P. Downes, E. Hafen, and G. Thomas. 2002. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat. Cell Biol. 4:251-255. [DOI] [PubMed] [Google Scholar]
- 43.Raught, B., A. C. Gingras, and N. Sonenberg. 2000. Regulation of ribosomal recruitment in eukaryotes, p. 245-293. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Rintelen, F., H. Stocker, G. Thomas, and E. Hafen. 2001. PDK1 regulates growth through Akt and S6K in Drosophila. Proc. Natl. Acad. Sci. USA 98:15020-15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saucedo, L., and B. Edgar. 2002. Why size matters: altering cell size. Curr. Opin. Genet. Dev. 12:565-571. [DOI] [PubMed] [Google Scholar]
- 46.Saucedo, L. J., X. Gao, D. A. Chiarelli, L. Li, D. Pan, and B. A. Edgar. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5:566-571. [DOI] [PubMed] [Google Scholar]
- 47.Schalm, S. S., and J. Blenis. 2002. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12:632-639. [DOI] [PubMed] [Google Scholar]
- 48.Schalm, S. S., D. C. Fingar, D. M. Sabatini, and J. Blenis. 2003. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13:797-806. [DOI] [PubMed] [Google Scholar]
- 49.Shioi, T., P. M. Kang, P. S. Douglas, J. Hampe, C. M. Yballe, J. Lawitts, L. C. Cantley, and S. Izumo. 2000. The conserved phosphoinositide 3-kinase pathway determines heart size in mice. EMBO J. 19:2537-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigrist, S. J., P. R. Thiel, D. F. Reiff, P. E. D. Lachance, P. Lasko, and C. F. Schuster. 2000. Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature 405:1062-1065. [DOI] [PubMed] [Google Scholar]
- 51.Stocker, H., M. Andjelkovic, S. Oldham, M. Laffargue, M. P. Wymann, B. A. Hemmings, and E. Hafen. 2002. Living with lethal PIP3 levels: viability of flies lacking PTEN restored by a PH domain mutation in Akt/PKB. Science 295:2088-2091. [DOI] [PubMed] [Google Scholar]
- 52.Stocker, H., T. Radimerski, B. Schindelholz, F. Wittwer, P. Belawat, P. Daram, S. Breuer, G. Thomas, and E. Hafen. 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5:559-565. [DOI] [PubMed] [Google Scholar]
- 53.Tee, A. R., and C. G. Proud. 2002. Caspase cleavage of initiation factor 4E-binding protein 1 yields a dominant inhibitor of cap-dependent translation and reveals a novel regulatory motif. Mol. Cell. Biol. 22:1674-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuttle, R. L., N. S. Gill, W. Pugh, J. P. Lee, B. Koeberlein, E. E. Furth, K. S. Polonsky, A. Naji, and M. J. Birnbaum. 2001. Regulation of pancreatic beta-cell growth and survival by the serine/threonine protein kinase Akt1/PKBα. Nat. Med. 7:1133-1137. [DOI] [PubMed] [Google Scholar]
- 55.Wang, X., W. Li, J. L. Parra, A. Beugnet, and C. G. Proud. 2003. The C terminus of initiation factor 4E-binding protein 1 contains multiple regulatory features that influence its function and phosphorylation. Mol. Cell. Biol. 23:1546-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, X., W. Li, M. Williams, N. Terada, D. R. Alessi, and C. G. Proud. 2001. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 20:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinkove, D., and S. J. Leevers. 2000. The genetic control of organ growth: insights from Drosophila. Curr. Opin. Genet. Dev. 10:75-80. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y., X. Gao, L. J. Saucedo, B. Ru, B. A. Edgar, and D. Pan. 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5:578-581. [DOI] [PubMed] [Google Scholar]