Abstract
Over the past two decades, the field of biosensors has been developing fast, portable, and convenient detection tools for various molecules of interest, both biological and environmental. While much attention is paid to the transduction portion of the sensor, the actual bioreceptor that binds the ligand is equally critical. Tight, specific binding by the bioreceptor is required to detect low levels of the relevant ligand, and the bioreceptor must be stable enough to survive immobilization, storage, and in ideal cases, regeneration on the biosensing device. Often, naturally-occurring bioreceptors or antibodies that are specific for a ligand either express affinities which may be too low to detect useful levels, or the proteins are too unstable to be used and re-used as a biosensor. Further engineering of these receptors can improve their utility. Here, we describe in detail the use of yeast surface display techniques to carry out directed evolution of bioreceptors to increase both the stability of the molecules and their affinity for the ligands. This powerful technique has enabled the production of stabilized, single-chain antibodies, T cell receptors, and other binding molecules that exhibit affinity increases for their ligands of up to 1 million-fold and expression of stable molecules in E. coli.
Keywords: Yeast Surface Display, Directed Evolution, Affinity Maturation, Thermal Stability
1. Introduction
The design of rapid, accurate, transportable, and inexpensive detection tools for a variety of biological and environmental molecules is an extremely desirable goal. One way that this may be achieved is using biosensors, which most often combine a bioreceptor that can unambiguously detect low levels of the target molecule, and a transducer that can sense and transmit binding events (reviewed in (1, 2)). Current research seeks to design biosensors that can sensitively and specifically detect infectious agents, toxins, allergens, hormones, illicit drugs, and even environmental contaminants (reviewed in (3–5)). A successful biosensor should achieve multiple goals: it must be able to clearly detect the molecule of interest at a level of practical consequence (e.g. infectious or toxic levels); it must be able to provide reproducible results after medium- to long-term storage; and it must be able to achieve a sufficiently low cost per test through ease of production, durability, and ability to be regenerated and reused. As the critical discriminating element of a biosensor, the bioreceptor molecule must exhibit exquisite stability, sensitivity, and specificity of binding (5).
While specific receptors such as monoclonal antibodies can be generated against many target molecules, further engineering of these antibodies can make them more suitable for biosensing applications (6). Yeast surface display is a powerful technique that can be applied toward increasing the desirable properties of antibodies and other biomolecules ((7), and reviewed in (8)). This method includes several advantages over other display and engineering systems (9). The ability to screen bioreceptor libraries by fluorescence activated cell sorting (FACS) allows quantitative comparisons between mutants to help guide the selection. In addition, ligand binding constants to soluble target molecules may be directly measured on the yeast cell surface; these values are typically in qualitative agreement with Biacore measurements of the interaction (10). Furthermore, yeast display incorporates adaptability for alternative sorting protocols for ligands that are difficult to produce or fluorescently label in soluble form, including magnetic bead separation (11) and cell-cell conjugates (12, 13). Yeast surface display has been successfully applied to the engineering of many bioreceptor proteins including antibodies (14–16), T-cell receptors (17–19), major histocompatibility complex proteins (20–23), NK receptors (24), growth factors (25), superantigen antagonists (26), fibronectin domains (27, 28) and cytokines (29, 30).
In this chapter, we describe detailed protocols for expression, mutation, and selection of optimal bioreceptor proteins by yeast surface display. Through this process, bioreceptor mutants can be engineered with greater stability for storage and regeneration conditions, as well as higher affinities for the detection of very low ligand concentrations.
2. Materials
2.1. Yeast strain
Saccharomyces cerevisiae yeast display strain EBY100 (a GAL1-AGA1∷URA3 ura3–52 trp1 leu2Δ1 his3Δ200 pep4∷HIS2 prb1Δ1.6R can1 GAL) is commercially available from Invitrogen (cat. no. C839-00).
2.2. Yeast display plasmid
Yeast display plasmid pCT302 (or a variant thereof) is commercially available from Invitrogen as pYD1 (cat. no. V835-01).
2.3. Electrocompetent E. coli strain for plasmid propagation
DH10B™ (Invitrogen cat. no. 18290-015).
2.3. Restriction Enzymes
NheI (Invitrogen cat. no.15444-011), BglII (Invitrogen cat. no. 15213-010), XhoI (Invitrogen cat. no. 15231-012), and DpnI (Invitrogen cat. no. 15242-019).
2.4. Polymerase chain reaction (PCR)
100 mM dNTPs (Invitrogen cat. no. 10297-018)
Taq DNA polymerase (Invitrogen cat. no. 18038-042)
High-fidelity Pfu Turbo® DNA polymerase (Stratagene cat. no. 600250)
Bovine serum albumin (BSA) (Sigma cat. no. A4503)
2.5. DNA purification
Plasmid rescue and purification from E. coli: QIAprep spin miniprep kit (QIAGEN cat. no. 27104)
Plasmid rescue from yeast: Zymoprep kit I or II (Zymo research cat. no. D2001 (I) or D2004 (II))
QIAquick PCR purification kit (QIAGEN cat. no. 28104)
2.6. Yeast media
YPD media for propagation of EBY100. To prepare 500 mL YPD, dissolve 5 g yeast extract (BD Bacto cat. no. 212750), 10 g peptone (BD Bacto cat. no. 211677), and 10 g dextrose (EMD cat. no. 346351) to 500 mL in ddH20. Autoclave. Store at room temperature no longer than 1 month. To make 1 sleeve of YPD plates, add 7.5 g of agar to the above recipe. Autoclave. Pour plates when cool enough to handle.
SD-citrate-CAA media for propagation of EBY100 transformed with pCT302. To prepare 500 mL SD-citrate-CAA, dissolve 7.4 g sodium citrate (Fisher cat. no. S279–500), 2.1 g citric acid monohydrate (Sigma cat. no. C1909), 2.5 g casamino acids (BD/Bacto cat. no. 223120), 10 g dextrose, and 3.35 g yeast nitrogen base without amino acids (Sigma cat. no. Y0626) to 500 mL in ddH20. Filter sterilize. Store at 4°C for up to 6 months. For SD-citrate-CAA plates, dissolve 91 g sorbitol (Sigma cat. no. S1876), 7.5 g agar, 7.4 g sodium citrate, and 2.1 g citric acid monohydrate to 400 mL in ddH20. Autoclave. In a separate container, dissolve 2.5 g casamino acids, 10 g dextrose, and 3.35 g yeast nitrogen base without amino acids to 100 mL in ddH20; add to the autoclaved solution when it is cool. Mix gently. Pour plates. Store plates at 4°C for up to several months.
SG-citrate-CAA media for induction of yeast display construct. Follow the recipe for 500 mL SD-citrate-CAA above substituting an equal mass of D-(+)-galactose (Sigma cat. no. G-0625) for dextrose.
Antibiotic for SD-citrate-CAA and SG-citrate-CAA cultures. Use 50 µg/mL kanamycin. Prepare 50 mg/mL stock by dissolving 500 mg kanamycin sulfate (Sigma cat. no. K4000) in 10 mL ddH20. Filter sterilize. Aliquot and store at −20°C.
2.7. Solutions for high-efficiency yeast transformation protocol
10 × TE buffer pH 7.5: To make 250 mL, add 25 mL of a 1M Tris base stock (pH 7.5) and 5 mL 0.5 M EDTA pH 8.0 stock to 200 mL ddH20, pH to 7.5 using HCl. Fill to 250 mL with ddH20. Filter sterilize. Store at room temperature for up to several months.
1 M lithium acetate: dissolve 25.5 g lithium acetate dehydrate to 250 mL in ddH20. Filter sterilize. Store at room temperature for up to several months.
1M DTT. Dissolve 1.545 g DTT in 10 mL. Filter sterilize. For best results, make fresh each time.
1 M sorbitol: Dissolve 45.55 g sorbitol to 250 mL in ddH20. Filter sterilize or autoclave. Store at 4°C for up to several months.
2.8. Flow cytometry/FACS reagents
Phosphate-buffered saline (PBS) containing 0.5% BSA (PBS/0.5% BSA). Prepare a 10X PBS pH 7.4 stock: For 100 mL, dissolve 0.257 g NaH2PO4 · H20, 2.249 g Na2HPO4 · 7H20, and 8.765 g NaCl to 100 mL in ddH20 (pH to 7.4 using NaOH or HCl). Filter sterilize. Store at room temperature for up to several months. Add 50 mL 10X PBS stock to 450 mL ddH20. Dissolve 2.5 g BSA into this solution. Filter sterilize. Store at 4°C for up to several months.
Monoclonal antibodies: anti-HA mouse antibody HA.11 (Covance MMS-101P), anti-c-myc chicken antibody (Molecular Probes cat. no. A-21281), Alexa488-labeled goat-anti-mouse secondary antibody (Invitrogen cat. no. A-11017), Alexa647-labeled goat-anti-chicken secondary antibody (Molecular Probes cat. no. A-21449), and streptavidin:phycoerythrin (SA:PE) (BD cat. no. 554061).
2.9. Equipment
Thermocycler
Two temperature controlled incubator shakers.
Electroporator (Gene pulser, BioRad cat. no. 1652076)
Flow cytometer and FACS apparatus.
3. Methods
This section will describe a robust method for engineering a binding protein of interest using yeast surface display technology. Similar methods for gene engineering via yeast display have been reported (9, 31, 32), and slight variations may be used for particular applications to attain optimal results.
3.1. Gene Construction of Yeast Surface Displayed Bioreceptor
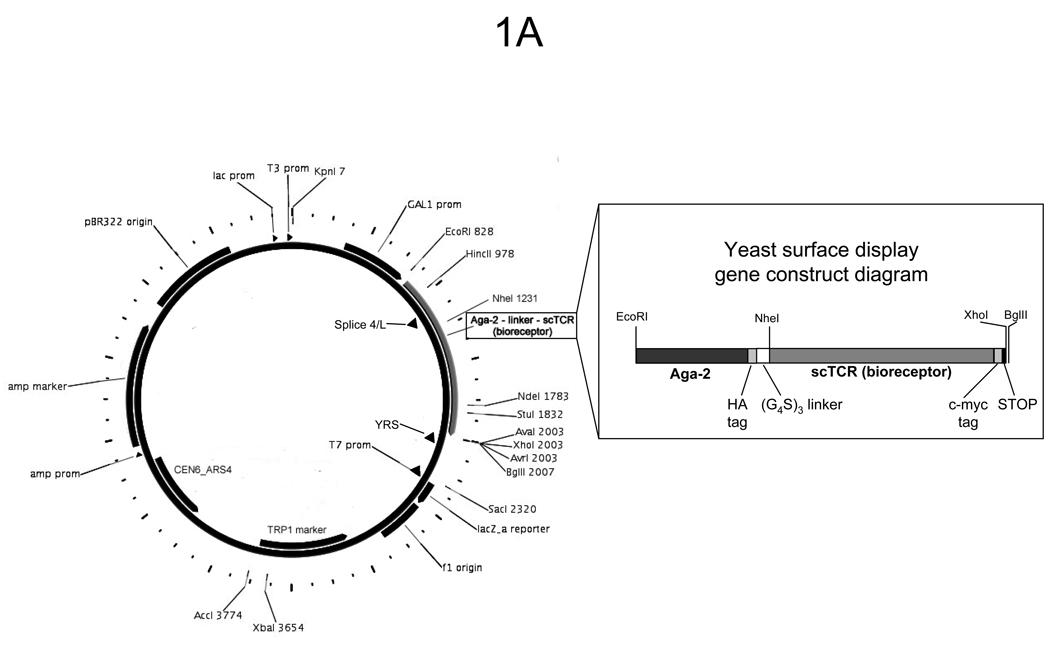
In order to engineer a more stable, higher affinity bioreceptor for biosensing applications, the protein is expressed as a fusion protein to the mating agglutinin protein Aga2 displayed on the surface of yeast. To achieve this, the bioreceptor gene can be inserted into the pCT302 vector using the in-frame NheI restriction site, and a downstream unique BglII or XhoI restriction site (See Fig. 1A)(7, 9). This plasmid allows galactose-inducible overexpression of the Aga2-gene fusion. Since the Aga2 protein is displayed on the surface of yeast linked to the Aga1 protein by a pair of disulfide bonds, a yeast strain containing a stably-integrated Aga1 gene under control of the GAL1 promoter should be used, such as EBY100 (See Note 1).
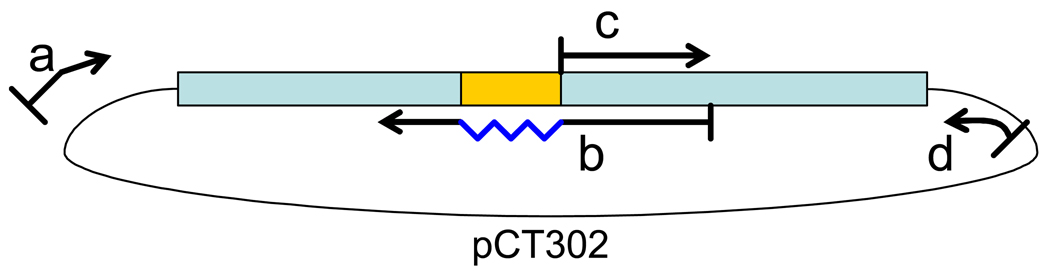
Fig. 1.
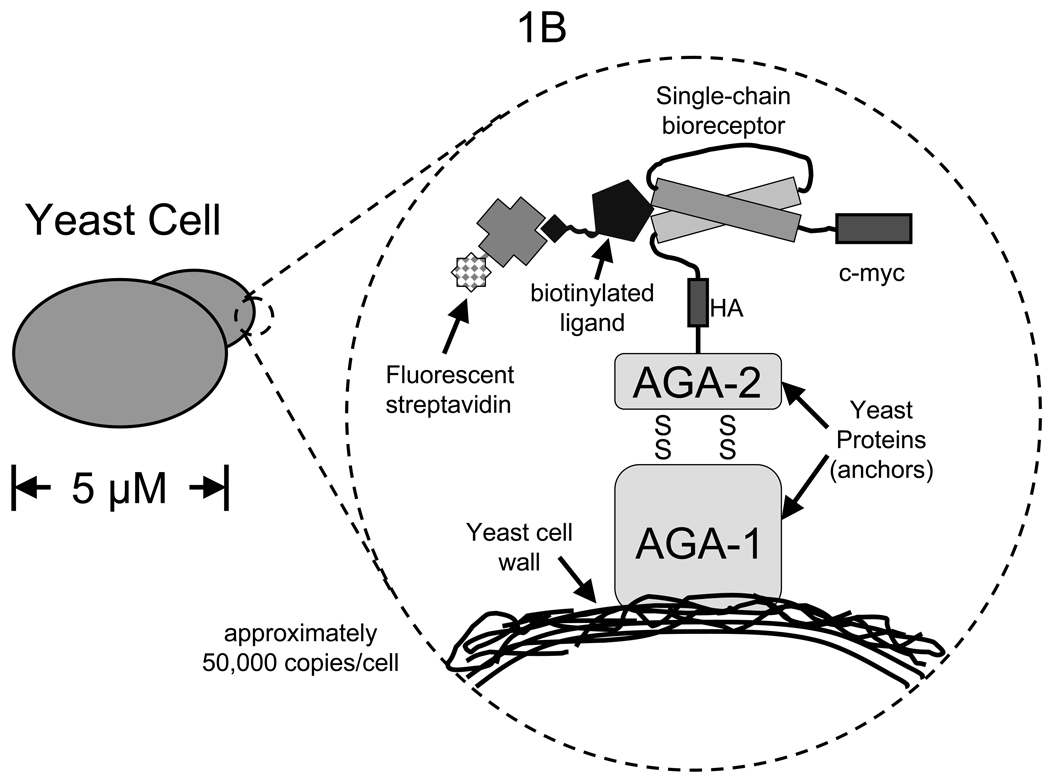
Schematic diagrams of yeast surface display elements. (A) Plasmid map of the yeast surface display vector pCT302 with a single-chain T cell receptor (scTCR) cloned in as a fusion with the yeast mating agglutinin protein Aga-2. Map was prepared with the help of PlasMapper (48). The open reading frame containing Aga-2 and the scTCR is expanded to show the important elements and restriction sites. (B) Diagram of a bioreceptor displayed on the surface of yeast fused to Aga-2. A detection scheme involving biotinylated ligand bound to the bioreceptor and fluorescent streptavidin may be used to analyze bioreceptor libraries on the surface of yeast.
Most often, the bioreceptor of interest is expressed as a single-chain construct fused to Aga2 (See Fig. 1B). For example, the binding domain of a monoclonal antibody, excluding the constant domains, may be conveniently expressed as a single-chain Fv (scFv) comprising the VH and VL genes connected by a ~16 amino acid long flexible linker. Usually a sequence like (G3S)4 or (G4S)3 will perform well as a flexible linker that is not very proteolytically sensitive. Similarly, a T cell receptor may be expressed as a single-chain construct containing the linked Vα and Vβ chains. The pCT302 vector includes the gene expressing Aga2, followed by the affinity tag HA (sequence: YPYDVPDYA), which can be used as an expression marker. Incorporation of a C-terminal expression tag is also useful to monitor full-length expression of the gene on the yeast surface. We use the c-myc affinity tag (sequence: EQKLISEEDL) at the C-terminus. Upon induction of protein over-expression in the yeast, antibodies against these epitopes (HA and c-myc) can be used to monitor expression of the fusion by flow cytometry.
Conventional cloning techniques are used to ligate the wild-type gene of interest into the pCT302 vector using a subcloning-competent E. coli strain, such as DH10B, DH5α, or XL1-blue. The pCT302 vector confers ampicillin resistance (100 µg/mL) to the bacterial cells. The sequence of fused gene in the vector can be confirmed using primers that flank the insert (See Fig. 1A): “Splice 4/L” (forward: GGC AGC CCC ATA AAC ACA CAG TAT), “YRS” (reverse: CGA GCT AAA AGT ACA GTG GG), or “T7” (reverse: TAA TAC GAC TCA CTA TAG).
3.2. Engineering of Bioreceptor for Improved Stability
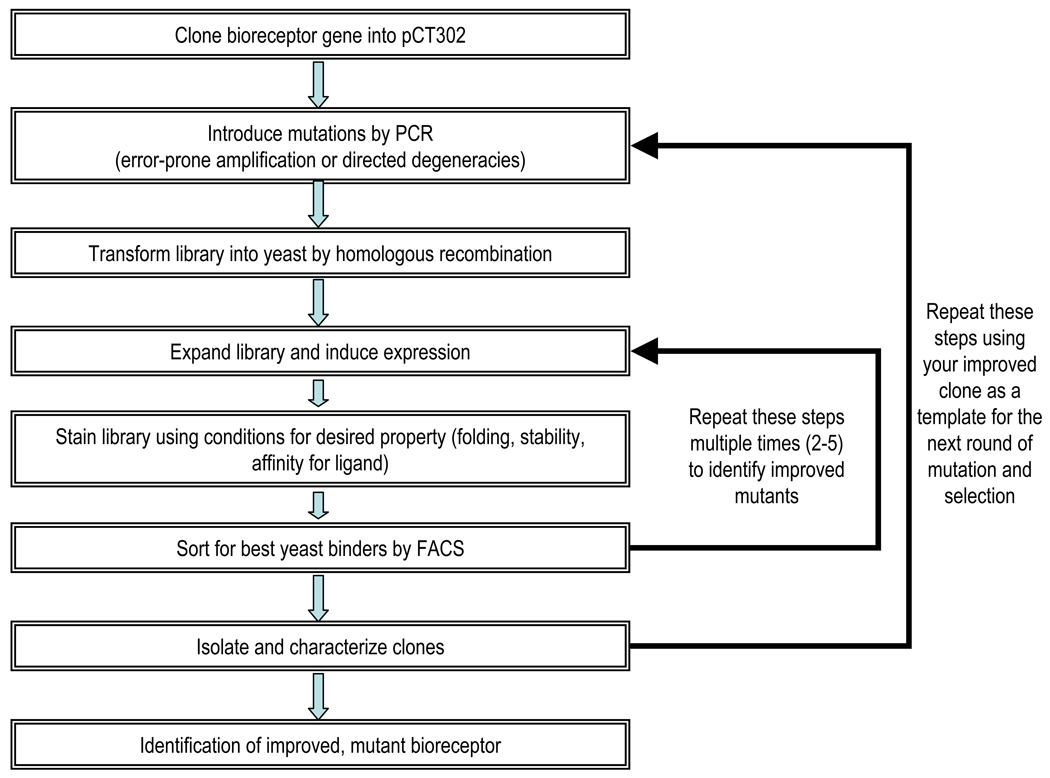
An overall flow chart of this step of the protocol is shown in Fig. 2. The method presumes that the protein of interest is not already optimally stable and thus it is expressed on the surface of yeast at sub-optimal levels, or it unfolds irreversibly under conditions of extreme pH or high temperature.
Fig. 2.
Flow diagram for bioreceptor engineering by yeast surface display.
3.2.1 Introduction of Random Mutations Into Bioreceptor Using Error-Prone PCR
The method for random mutagenesis of genes using error-prone PCR was adapted from previous reports (33, 34). An error rate of approximately 0.5% should be expected using this protocol. For a single yeast library of random mutants of approximately 105 clones, you should prepare enough reactions to yield 50–80 µg error-prone amplified insert (between 3–8 reactions).
-
Prepare a 100 µL solution in a PCR tube for each reaction. The following components should be present in the solution (final concentrations listed):
-
1.1.
220 µM dATP
-
1.2.
200 µM dCTP
-
1.3.
340 µM dGTP
-
1.4.
2.4 mM dTTP
-
1.5.
0.3 ng/µL template DNA (gene of interest in pCT302 vector)
-
1.6.
250 nM Splice 4/L (forward primer)
-
1.7.
250 nM T7 (reverse primer)
-
1.8.
5 ng/µL Bovine Serum Albumin (BSA)
-
1.9.
3.325 mM MgCl2
-
1.10.
0.5 mM MnCl2
-
1.11.
1:10 dilution of Taq Polymerase Reaction Buffer (10 µL per 100 µL reaction)
-
1.12.
1 µL Taq Polymerase
-
1.1.
-
Place PCR tube in thermocycler and run through the following program:
-
2.1.
94°C for 3 minutes
-
2.2.
30 cycles of:
-
2.2.1
94°C for 1 minute
-
2.2.2
50°C for 2 minutes
-
2.2.3
72°C for 3 minutes
-
2.2.1
-
2.3.
72°C for 5 minutes
-
2.4.
4°C forever
-
2.1.
Check for amplification of the gene on a 1% agarose gel.
To reduce insert background from remaining template, digest the PCR reaction with 1 µL DpnI for 1 hour at 37°C, which will fragment the template plasmid, but not the PCR product.
Clean up the reaction using the PCR Clean-up Kit from Qiagen. Elute each reaction in 30 µL from the Qiagen column.
3.2.2 Preparation of pCT302 Vector
This protocol will result in a linearized, dephosphorylated vector with a sequence overlap with your insert of at least 50 base pairs. No ligation with the insert will be necessary, as the yeast will recombine the vector and insert through homologous recombination. (See Note 1 for tips on choosing an appropriate vector.)
Starting from 50–80 µg full pCT302 vector obtained from Qiagen mini-prep or a protocol yielding similar purity, digest with NheI in appropriate buffer overnight at 37°C, while reserving a small aliquot (1 µL) uncut for later comparison on a gel.
Clean up the reaction after the digestion using the PCR Clean-up Kit from Qiagen. Save another 1 µL sample for gel analysis.
Next, digest with XhoI in appropriate buffer for at least 1 hour at 37°C. Clean up the reaction using PCR Clean-up Kit, and save another 1 µL sample.
Digest next with BglII in appropriate buffer for at least 1 hour at 37°C. Digesting with all three enzymes decreases the possibility of remaining full-length vector with the potential to re-circularize. Clean up the reaction using PCR Clean-up Kit, and save another 1 µL sample.
Finally, dephosphorylate the linearized vector using Calf Intestine Phosphatase (CIP) for 1 hour at 37°C. This should reduce vector background when transforming your yeast library. Clean up the reaction using PCR Clean-up Kit, and take another 1 µL sample for your gel.
Run all of your samples (uncut pCT302, NheI digested pCT302, NheI+XhoI digested pCT302, NheI+XhoI+BglII digested pCT302, and fully cut, dephosphorylated pCT302) on a 1% agarose gel to make sure you have not lost vector at some step. If your vector has disappeared, go back to the step before you lost the plasmid and trouble-shoot that reaction, and try again.
Run an additional gel comparing several dilutions of your cut, dephosphorylated vector along with your error-prone insert to determine their absolute and relative concentrations. (See Note 2).
3.2.3 Preparation of Electrocompetent Yeast
This protocol has been adapted from previously reported methods (35, 36).
Two days before preparation of your library, inoculate 2 or 3, 3 mL cultures of sterile YPD media with a colony of EBY100 cells from a freshly-streaked YPD plate. Grow to stationary phase at 30°C (for ~ 48 hours), shaking at 220 rpm. If required, these cultures may be prepared further in advance and stored at 4°C.
The night before your library transformation, inoculate a 2 L sterile flask containing 500 mL YPD medium with 5–9 mL of starter EBY100 culture. Grow overnight at 30°C with vigorous shaking until the cells reach an OD600 of 1.3–1.5. (See Note 3).
Harvest the culture in sterile centrifuge bottles that can hold 500 mL, spinning at 4000 × g, 4°C, and resuspend vigorously in 80mL sterile ddH2O.
-
To increase electrocompetence:
-
4.1.
Add 10 mL of sterile, 10x TE buffer, pH 7.5 to the yeast pellet. Swirl to mix. If multiple centrifuge bottles were used, combine the cultures into a single sterile centrifuge bottle.
-
4.2.
Add 10 mL of 10x (1M) Lithium Acetate stock solution. Swirl to mix, and shake in the sterile centrifuge bottle gently (100–150 rpm) for 45 minutes at 30°C.
-
4.3.
Freshly prepare and then add 2.5 mL of 1M DTT while swirling. Shake gently 15 minutes at 30°C.
-
4.1.
Dilute yeast suspension to 500mL with sterile water.
-
Wash and concentrate the cells three times by centrifuging at 4000–6000 × g, and resuspend the pellets each time as follows:
-
6.1.
First pellet—250 mL ice-cold water
-
6.2.
Second pellet—20–30 mL ice-cold 1M sorbitol
-
6.3.
Third pellet—0.5 mL ice-cold 1M sorbitol (See Note 4)
-
6.1.
The final volume of resuspended, electrocompetent yeast should be 1–1.5 mL. (See Note 5).
3.2.4 Transformation and Characterization of Randomized Bioreceptor Library
In this method, you will transform the overlapping error-prone amplified insert and vector into the yeast, and allow them to recombine the sequences through homologous recombination.
Prepare a mixture of error-prone insert and linearized vector DNA at approximately a 6:1 molar ratio, or roughly a 1:1 mass ratio. The DNA concentration in the mixture should be 10–100 ng in no more than 5 µL. (See Note 2).
-
In addition to your error-prone library mixture, set up the following additional controls to transform into the yeast:
-
2.1.
Linearized vector alone—use the same final concentration as in the DNA mixture
-
2.2.
Error-prone insert alone—use the same final concentration as in the DNA mixture
-
2.3.
Your unmutated gene in pCT302—these cells will serve as a control for later experiments
-
2.4.
No DNA in the transformation
-
2.1.
In a sterile, ice-cold eppendorf tube, mix 40 µL concentrated, electrocompetent yeast with 10–100 ng of transforming DNA (≤ 5 µL). (See Note 6).
Transfer to an ice-cold, 0.2-cm-gap disposable electroporation cuvette.
Pulse at 1.5 kV, 25 µF, 200 Ω. The time constant reported will vary between 4.2–4.9 msec. Time constants that are too low (<4 msec) or a current arc indicates that the conductance of the yeast/DNA mixture is too high. (See Note 7).
Add 1 mL ice-cold sorbitol to the cuvette to recover the yeast with gentle pipetting. Mix the recovered yeast from your error-prone DNA mixture electroporations into a single, sterile conical tube, but keep your controls in separate tubes.
Plate 10 µL of each control transformation onto a labeled SD-citrate-CAA plate, making sure that the yeast have not settled to the bottom of your tubes. Grow the plates at 30°C for approximately 48 hours.
-
Resuspend your library-transformed yeast well in the sorbitol, and plate the following dilutions onto SD-citrate-CAA plates to determine the size of your library. Grow the plates at 30°C for approximately 48 hours. (See Note 8).
-
8.1.
10 µL undiluted library
-
8.2.
10 µL of a 1:100 dilution of your library
-
8.3.
10 µL of a 1:500 dilution of your library
-
8.1.
Grow the positive control transformation (unmutated pCT302 plasmid) in several 3 mL culture tubes of SD-citrate-CAA media with 50µg/mL kanamycin, shaking at 220 rpm at 30°C for 48 hours.
Allow the library to recover and culture in 500 mL SD-citrate-CAA media with 50µg/mL Kanamycin, shaking at 220 rpm at 30°C for 48 hours.
After 48 hours of library growth, seed 10–50 mL of the expanded library into a sterile flask with 1000 mL SD-citrate-CAA media with 50µg/mL kanamycin, shaking at 220 rpm at 30°C for another 24 hours to reduce untransformed or unhealthy cells.
After the second passage of the library, freeze 10–30 aliquots of the library containing 1 mL of dense culture (approximately 108 cells/mL) with 70 µL sterile DMSO. Store these at −80°C.
Store the remaining passaged library in 3 mL aliquots in sterile culture tubes at 4°C.
To analyze your library for diversity, take 0.5 mL of dense yeast from the library and isolate plasmid from the yeast using the Zymoprep II kit. Use DNA isolated from the yeast to transform into E. coli, culture at least 8–10 colonies, and isolate the plasmids from these cells. Sequence the plasmids using the YRS reverse primer and ensure that you have obtained a reasonable level of mutations through your error-prone library protocol.
3.2.5 Sorting for High Expression and Proper Folding of Bioreceptor
Once you have generated and expanded your library, it is possible to induce it to produce your protein of interest and monitor expression on the surface using flow cytometry. Since affinity tags have been incorporated at the N-terminus (HA tag) and the C-terminus (c-myc tag) of the gene product, it is possible to use antibodies against these epitopes to probe for protein expression.
Some proteins do not fold properly on the surface of yeast without some stabilizing mutations which may be isolated from your error-prone library. While many scFv fragments will fold properly, other proteins such as scTCRs are not stably expressed. To isolate the yeast clones expressing properly folded mutants from your error-prone library, it is useful to probe your library with an additional ligand that detects the properly folded protein. This probe could be the biotinylated ligand, if the affinity of your bioreceptor is already sufficiently high to detect binding and thus staining (KD ~ 100 nM or less). Alternatively, the probe could be a conformationally sensitive antibody to the bioreceptor that will only detect properly folded protein. (See Note 9).
While the main goal of this protocol is to use fluorescence activated cell sorting (FACS) to separate the most stable, highly expressing yeast clones, it is essential to perform small scale test stains to analyze by flow cytometry before and after each round of sorting to monitor for affinity tag expression and ligand or conformational antibody binding.
To sort a library, include at least 10-fold more yeast in your sample than the theoretical size of your library (if possible). For example, if your library size is 107 unique yeast, make sure you induce and stain at least 108 yeast. To perform an analytical flow cytometry experiment, smaller samples may be used, around 106 yeast per staining.
When carrying out staining for subsequent sorting, be sure to use 0.2 µm filter-sterilized media, buffers, and stain solutions, as well as sterile (autoclaved) tubes, since you will be expanding the cells after the experiment. This is less of a concern for analytical samples, since you will not be collecting the cells.
Take 1 mL of freshly passaged, dense library culture in SD-citrate-CAA media (about 108 yeast) and spin it down in a sterile eppendorf tube for 3 minutes at 3000 rpm in a tabletop microcentrifuge. Aspirate the supernatant.
Resuspend the yeast in SG-citrate-CAA media (substituting galactose for glucose). Spin down again for 3 minutes at 3000 rpm and aspirate the supernatant.
Resuspend the yeast again in SG-citrate-CAA media containing 50 µg/mL kanamycin. Incubate this culture at 20°C for 24–48 hours, shaking at 220 rpm. This will induce the yeast to express the Aga2-bioreceptor fusion (See Note 10). After the induction, the yeast may be stored at 4°C in SG-citrate-CAA media for up to two weeks.
-
Stain your cells for flow cytometry or sorting.
-
4.1.
Spin down the appropriate number of cells in SG-citrate-CAA media (see above) at 3000 rpm for 3 minutes. Aspirate the supernatant, and rinse one time with PBS-BSA. Spin the cells down again and aspirate the supernatant.
-
4.2.
Resuspend the cells in an appropriate volume of PBS/0.5% BSA containing the desired primary staining reagent(s). Large samples for sorting by FACS should be stained in a large enough volume to stay in suspension—often 0.5–1.0 mL at least for 108 cells. Smaller, test samples (0.5–1.0 × 106 cells) can be stained in 20–50 µL. Useful primary stains include mouse anti-HA antibody (1:25 dilution), chicken anti-c-myc (1:100 dilution), a conformation-sensitive antibody against the bioreceptor (if available, titrate for dilution where antibody is in excess – typically 10 µg/ml), or biotinylated ligand for the bioreceptor (saturating concentration). It is useful to keep some samples of unstained yeast, as well as stained, non-expressing yeast, as controls.
-
4.3.
Incubate the yeast in the stain solution at room temperature for 30 minutes or on ice for 45 minutes. Make sure the yeast remain in suspension during the incubation with gentle agitation if required.
-
4.4.
Spin down the cells at 3000 rpm for 3 minutes. Wash with a large excess (e.g. 20 volumes) ice-cold PBS/0.5%BSA and spin down again. Perform all the steps from this point forward on ice.
-
4.5.
Resuspend cells in the appropriate volume of ice-cold secondary staining solution (fluorescently labeled reagents in PBS/0.5% BSA). While multiple detection schemes may be used, we recommend detection of chicken anti-c-myc primary antibody by using goat anti-chicken alexa 488 labeled antibody at a 1:100 dilution, and detection of mouse anti-HA primary antibody using goat anti-mouse F(ab’)2 Alexa 488 at 1:100 dilution. Incubate these samples on ice in the dark for 30–45 minutes. Be sure to include control samples that have been incubated with secondary, but not primary stain to determine the level of non-specific binding of the secondary reagent.
-
4.6.
Spin down the cells at 3000 rpm for 3 minutes. Wash with 1 mL ice-cold PBS/0.5% BSA and spin down again. Resuspend to an appropriate volume of ice-cold PBS/0.5% BSA for flow cytometry analysis or sorting immediately before analysis. (See Note 11).
-
4.1.
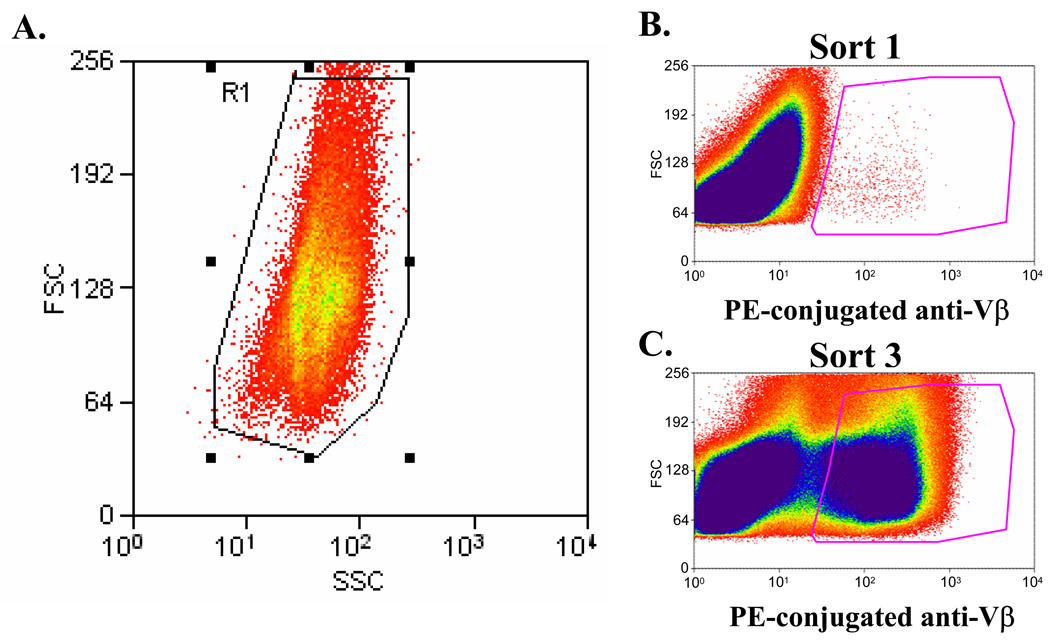
Run your samples on the flow cytometer or FACS instrument, using a gate on the forward scatter vs. side scatter plot to focus only on the live, healthy cells (See Fig. 3A). Normally, this will be the vast majority of the events that are detected.
-
Within the live population, observe the fluorescent probes with which you stained the cells.
-
6.1.
When sorting your error-prone library, it may be advantageous to stain either with ligand or conformation-sensitive antibody only, or to co-stain with c-myc to ensure full-length protein expression. To minimize compensation complications using two different fluorophores, we recommend a detection scheme that utilizes either fluorescein, Alexa 488, or phycoerythrin, combined with a higher wavelength fluorescent probe such as allophycocyanin or Alexa 647 when co-staining your cells. (See Note 12)
-
6.2.
To sort, set up a second gate based on fluorescence that will collect the yeast with the highest expression of your fluorescent probe of interest, such as ligand binding, expression of a bioreceptor-specific conformational epitope, and/or c-myc expression (See Fig. 3B). In the first round of sorting, it may be useful to collect around 5% of the most positive yeast, but smaller fractions of the yeast may be collected on subsequent sorts, down to 0.1–1%.
-
6.1.
Collect the yeast inside of both gates (scatter and fluorescence) into a sterile eppendorf tube on ice. You should sample enough events to exceed your library size by 10-fold, if possible. For practical purposes, you should plan to collect at least 50,000 cells.
Dilute the collected yeast into SD-citrate-CAA media and expand in a culture tube at 30°C, shaking at 220 rpm.
Repeat steps 1–8, sorting until you have a distinctly positive population in your re-expanded library (See Fig. 3C).
Once the library has been substantially enriched for positive cells, plate an aliquot of the sorted positive population onto SD-citrate-CAA plates to isolate single colonies.
Pick at least 10 colonies to inoculate small monoclonal test cultures (3 mL SD-citrate-CAA + 50 µg/mL kanamycin per colony). Grow cultures to stationary phase (approximately 48 hours in a 30 °C shaker incubator).
Induce an aliquot (e.g. 0.5 mL) of the culture by pelleting the cells and resuspending in SG-citrate-CAA + kanamycin. Incubate in a 20°C shaker for approximately 48 hours.
Characterize the individual colonies by flow cytometry, following the procedure in Step 4 for smaller test samples.
Based on the flow cytometric characterization of individual yeast colonies, isolate plasmids from approximately 0.5 mL of each monoclonal test SD-citrate-CAA culture using the Zymoprep I or II kit, transform back into E. coli to propagate the plasmid, and isolate plasmids for sequence analysis.
Fig. 3.
Light scatter and fluorescence properties of yeast populations stained for FACS. (A) Typical forward light scatter (FSC) versus side scatter (SSC) profile of a yeast population. A gate (R1) can be drawn to exclude any events that do not fall within a given range of FSC and SSC properties. (B) A large library of yeast expressing a single-chain T cell receptor (scTCR) was stained with a fluorescently-labeled anti-TCR Vβ domain antibody, and the most fluorescent cells were collected using FACS. (C) After two rounds of sorting, the yeast population was stained with the anti-Vβ antibody for a third sort, revealing that the population had been enriched for Vβ+ scTCR mutants. The most fluorescent cells were again collected using FACS.
If your desired stability or expression level has not been achieved through a single round of error-prone mutations, it is possible to select one of your best yeast clones and use the sequence of its mutated bioreceptor as the template for an additional round of error-prone amplification (See Section 3.2.1).
3.2.6 Sorting for Thermal Stability of Bioreceptor Mutants
Increased thermal stability of proteins used for biosensors is desirable to allow for robust devices that can withstand a variety of storage, assay, and regeneration conditions. In addition, some evidence suggests that starting from the most stable version of a bioreceptor by yeast surface display will aid in later affinity maturation efforts (37–39). In addition, thermally-stable mutants can enable higher expression levels as soluble proteins from yeast or E. coli (19, 20, 37). Thermal stability selection rounds have been carried out on many of the proteins that were later mutated for high affinity (17, 19–21, 37–41).
To select for increased thermal stability in an error-prone library, follow the protocol in Section 3.2.5, Steps 1–3. Next, proceed as follows:
-
3.1.
Separate the induced yeast into several samples. Spin cells down at 3000 rpm for 3 minutes and aspirate the supernatants. Resuspend each sample in an appropriate volume of PBS/0.5% BSA (See Section 3.2.5, Step 4b).
-
3.2.
Incubate the different samples each for 30 minutes at a different, elevated temperature, e.g. 37°C, 42°C, 50°C, and 60°C. After the incubation, return the cells to ice and dilute with 1 mL ice-cold PBS-BSA. (See Note 13)
-
3.3.
Return to Step 4 in Section 3.2.5 for staining and analysis of the yeast.
3.2.7 Sorting for pH Stability of Bioreceptor Mutants
Increased pH stability of a bioreceptor for the particular application of a biosensor can also be advantageous. For example, to keep device costs down, the ability to regenerate a single biosensor would be useful, and many convenient regeneration protocols may include a pH shift to release the ligand from the bioreceptor. It is important, in this case, that the bioreceptor itself survives the treatment and can be re-used to detect ligand in a subsequent assay. Data from our lab suggests that yeast survive 30 minute, room temperature treatments with solutions of pH 3–11 with no significant decrease in viability (unpublished results), making directed evolution on the yeast surface attractive to achieve bioreceptors that are stable to regeneration conditions.
To select for increased pH stability in an error-prone library, follow the protocol in Section 3.2.5, Steps 1–3. Next, proceed as follows:
-
3.1.
Separate the induced yeast into several samples. Spin cells down at 3000 rpm for 3 minutes and aspirate the supernatants. Resuspend each sample in an appropriate volume of a pre-prepared solution of known pH (See Section 3.2.5, Step 4b). If desired regeneration conditions are known, make sure you test the conditions that will be used. If not, you may use solutions of 100 mM buffer, pH adjusted, with 100 mM NaCl to protect against osmotic shock.
-
3.2.
Incubate the different samples each for 30 minutes at room temperature. After the incubation, return the cells to ice and dilute with 1 mL ice-cold PBS/0.5% BSA.
-
3.3.
Spin down the cells and aspirate the supernatant. Resuspend the cells in ice-cold PBS-BSA
-
3.4.
Return to Step 4 in Section 3.2.5 for staining and analysis of the yeast.
3.3 Engineering of Bioreceptor for Improved Ligand Affinity
3.3.1 Mutagenesis of bioreceptor
Several different mutagenesis strategies have proven successful in the construction of libraries from which higher-affinity mutants have been selected (See Note 14). These include error-prone PCR (described in 3.2.1) and site-directed randomization. The latter technique is described below.
-
Use splice by overlap extension (SOE) PCR (42) to randomize up to a five amino acid stretch of the bioreceptor (e.g. a ligand-binding loop of an antibody). A schematic for this technique is shown in Figure 4.
Prepare two “pre-SOE” PCRs
-
1.1.
Pre-SOE 1 (Amplify the 5’ half of the bioreceptor gene, including randomized codons)
-
1.1.1.
70 µL ddH20
-
1.1.2.
16 µL 5 mM dNTPs
-
1.1.3.
10 µL polymerase buffer (provided with polymerase)
-
1.1.4.
1 µL 20 µM Splice4L (forward flanking primer)
-
1.1.5.
1 µL 20 µM reverse overlap primer (5’ 50 nucleotides SOE primer overlap-(SNN)5-30 nucleotides uninterrupted template complementarity 3’ (N=any nucleotide, S= G or C)
-
1.1.6.
1 µL 10 ng/µL yeast display plasmid containing bioreceptor gene insert
-
1.1.7.
1 µL Pfu Turbo (Stratagene)
-
1.1.1.
-
1.1.
-
1.2.
Pre-SOE 2
-
1.2.1.
70 µL ddH20
-
1.2.2.
16 µL 5 mM dNTPs
-
1.2.3.
10 µL polymerase buffer (provided with polymerase)
-
1.2.4.
1 µL 20 µM forward overlap primer (5’ 50 nucleotides primer overlap/template complementarity 3’)
-
1.2.5.
1 µL 20 µM T7 (See section 3.1) (reverse flanking primer)
-
1.2.6.
1 µL 10 ng/µL yeast display plasmid containing bioreceptor gene insert
-
1.2.7.
1 µL Pfu Turbo (Stratagene)
-
1.2.1.
-
1.3.
Place PCR tubes in thermocycler and run the following program:
-
1.3.1.
94 °C for 3 minutes
-
1.3.2.
35 cycles of
-
1.3.2.1.
94 °C for 1.5 minutes
-
1.3.2.2.
55 °C for 2 minutes
-
1.3.2.3.
72 °C for 3 minutes
-
1.3.2.1.
-
1.3.3.
72 °C for 7 minutes
-
1.3.4.
4 °C forever
-
1.3.1.
Check for PCR products on a 1% agarose gel
Purify and concentrate PCR products using QIAGEN PCR clean-up kit, following manufacturer’s instructions, and eluting in 32 µL ddH20.
-
Set up 4 identical SOE PCR reactions:
-
4.1.
66.5 µL ddH20
-
4.2.
16 µL 5 mM dNTPs
-
4.3.
10 µL polymerase buffer (provided with polymerase)
-
4.4.
2 µL 20 µM Splice4L
-
4.5.
2 µL 20 µM T7
-
4.6.
1.5 µL eluted 5’ pre-SOE 1 product (from 3.)
-
4.7.
1.5 µL eluted 3’ pre-SOE 2 product (from 3.)
-
4.8.
1 µL Pfu Turbo
-
4.1.
-
Place PCR tubes in thermocycler and run the following program:
-
5.1.
94 °C for 3 minutes
-
5.2.
40 cycles of
-
5.1.1.
94 °C for 1.5 minutes
-
5.1.2.
55°C for 2 minutes
-
5.1.3.
72 °C for 3 minutes
-
5.1.1.
-
5.3.
72 °C for 7 minutes
-
5.4.
4 °C forever
-
5.1.
Check for PCR product, clean, and concentrate as described in steps 2. and 3.
Store PCR product at 4 °C.
Fig. 4.
Schematic representation of pre-SOE PCR primers. Forward flanking primer a and reverse overlap primer b are used in pre-SOE 1 to amplify the 5’ portion of the construct. Primer b contains the degenerate codons (saw-tooth), a ~50 nucleotide region of complementarity to forward overlap primer (c) at its 5’ end, and ~30 nucleotides of template complementarity to direct annealing for elongation at its 3’ end. Forward overlap primer c and reverse flanking primer d are used in pre-SOE 2 to amplify the 3’ portion of the construct, with a ~50 nucleotide region at its 5’ end that is complementary to the 3’ end of pre-SOE 1 product. This complementarity or “overlap” mediates the subsequent SOE PCR.
See Note 15 for brief discussion of SOE PCR troubleshooting.
3.3.2 Transformation of EBY100 with cut vector and mutagenized insert containing flanking homologous ends
Follow high-efficiency transformation protocol described in 3.2.3 and 3.2.4 to create the yeast library.
3.3.3 Selecting higher-affinity mutants: equilibrium staining conditions
The large library of bioreceptor mutants is incubated with fluorescently labeled ligand under conditions in which higher ligand binding affinity will result in a detectable increase in fluorescence. As indicated in 3.2.5, the following steps should be carried out using sterile technique.
Pellet sufficient library yeast cells to 10-fold oversample the library diversity. Resuspend in several mL ice cold PBS/0.5% BSA. Pellet the cells again and aspirate the supernatant.
-
Primary stain: Resuspend cells in ice-cold 100 µL PBS/0.5% BSA. Transfer the cells, noting the volume, to a fresh sterile tube. To this tube, add a dilution of biotinylated ligand (in ice-cold PBS/0.5% BSA) to the cells to yield a predetermined appropriate concentration of biotinylated ligand, typically 0.05–0.1 × KD, diluted in ice-cold PBS.0.5% BSA. See Note 16 for further discussion on optimal equilibrium ligand staining conditions for the first sort. The concentration of biotinylated ligand used to stain the population can be progressively decreased with each round of sorting to increase the stringency of labeling conditions.
An equilibrium staining procedure should also include a counterstain with an antibody to an epitope tag (e.g. chicken anti-c-myc (1:100 dilution)) to normalize the ligand staining for differences in expression. Incubate primary stain at room temperature long enough for the bioreceptor:ligand interaction to reach equilibrium (e.g. ~ 30 minutes). (See Note 16)
Wash away unbound ligand/antibody by adding a large excess (≥ 20-fold) of ice-cold PBS/0.5% BSA. Then pellet and aspirate the supernatant.
Resuspend cells in dilution of compatible fluorescent secondary reagents. (See Note 12). Secondary reagents should be in excess, and should be titrated ahead of time to determine the optimal concentration (i.e. a concentration that yields saturated signal but is not so high as to encourage non-specific binding). A common dilution is Alexa647-conjugated anti-chicken 1:100, streptavidin:phycoerythrin (SA:PE) 1:200. Cells should be shielded from light during this and subsequent steps to avoid photobleaching of the fluorophores. Incubate on ice for 30 minutes.
Wash away unbound reagent by adding a large excess ice-cold PBS/0.5% BSA. Pellet cells, aspirate supernatant.
-
Resuspend in ice-cold PBS/0.5% BSA to ~107 cells per mL.
Keep cells on ice. Also prepare a small sample (104-105 cells) cells resuspended in just PBS/0.5% BSA (unstained cells), and a sample of cells stained individually with a single fluorophore if compensation is needed (e.g. for FITC and PE) (See Note 12 for further discussion of fluorescently-labeled secondary reagents).
3.3.4 Staining conditions for selection of mutants with longer t1/2
Library cells can be sorted not only for higher equilibrium binding affinity but also specifically for longer half-life of ligand binding. This method is especially useful when the protein of interest already has a relatively high equilibrium binding affinity (KD value <1 nM) and equilibrium labeling poses a challenge (See Note 16).
Surface polypeptide expression is induced as described previously.
Pellet enough induced library yeast cells to 10-fold oversample the library diversity. Resuspend in at least 20 volumes of ice cold PBS/0.5% BSA. Pellet the cells again and aspirate the supernatant.
Incubate cells with a saturating concentration of biotinylated ligand in 500 µL until equilibrium (30 minutes).
Wash with a large excess of ice-cold PBS/0.5% BSA
Competition step: Incubate with a large molar excess (e.g. 10-fold) of unbiotinylated ligand at room temperature or 37 °C. The optimal competition time to achieve maximum fluorescence discrimination depends on the koff. This time can be determined mathematically (43) but can be estimated as ~5/koff.
Cells are then treated as in step 3.3.3 steps 4–5.
3.3.5 Select for high-affinity mutants by FACS
Analyze (i.e. acquire data but do not sort) ~50,000 unstained cells to create a gate containing yeast cells based on forward scatter and side scatter properties, as well as to control for autofluorescence. (If compensation is needed, analyze the singly-stained cells and compensate for emission overlap (See Note 12))
Analyze ~50,000 labeled library cells to set the gate, i.e. the fluorescence threshold for collection. In order to avoid excluding any desirable mutants during the first sort, set the gates liberally to collect the most fluorescent ~1% of the library.
Sort the cells into a sterile tube (sorting directly into SD-citrate-CAA + 50 µg/mL kanamycin may improve viability). As indicated previously, sample at least 10 times the library size. For example, for a library size of 107, during the first sort, sample 108 cells, collecting the top 1% (106 cells). During subsequent sorts, the gates may be set more stringently to collect a smaller percentage (e.g. the most fluorescent 0.25-0.5%) of cells.
Culture the collected cells in SD-citrate-CAA + 50 µg/mL kanamycin to stationary phase.
Freeze down an aliquot of saturated culture collected from the first sort.
Induce the cells to stain for the second sort.
The staining and sorting procedure is typically repeated several times until the population is substantially enriched for higher-affinity mutants (See Fig. 2).
Isolate individual yeast clones as described in 3.2.5 step 10.
3.3.6 Characterization of mutants using flow cytometry
One significant advantage of yeast display over other in vitro engineering methods is the ability to asses KD directly on the isolated yeast clones. On induced yeast, titrate the ligand and measure ligand binding at various concentrations by flow cytometry. Plot fraction bound vs. ligand concentration to approximate KD. The entire process (including growth, induction, and staining) should be repeated at least 2 additional times.
Aliquot 20 µL of induced cells (~2 × 106 cells)
Add ~500 µL ice cold PBS/0.5%BSA
Pellet cells
Aspirate supernatant.
Resuspend the cells in 100 µL PBS/0.5% BSA. Combine 50 µL resuspended cells with a range of biotinylated ligand dilutions (in PBS/0.5%BSA) and to an adequate final volume to maintain excess ligand compared to yeast displayed polypeptide. (If necessary, the number of cells can be reduced to 105 in order to achieve a reasonable staining volume)
Incubate for 1 hour
Wash unbound ligand by adding 20 volumes of ice-cold PBS/0.5% BSA. Pellet cells and aspirate supernatant.
Resuspend each tube in 1:100 SA:PE (cold) in 100 µL.
Incubate on ice 30 minutes
Wash unbound reagent by adding 500 µL ice-cold PBS/0.5% BSA. Pellet cells. Aspirate supernatant.
Resuspend cells in 500 µL ice-cold PBS/0.5% BSA.
Analyze using flow cytometry.
Plot percent of maximum ligand bound vs. ligand concentration, and determine the KD as ligand concentration yielding half-maximum bound using non-linear regression.
Flow cytometric analysis of mutants stained with a panel of non-cognate but practically relevant ligands should be performed to ensure that specificity of the mutants has been maintained throughout the affinity maturation process. A stain using just the secondary reagents should also be performed to ensure the mutant is specifically binding the ligand and not the secondary reagents.
If after characterization of isolated mutants it is determined that further affinity maturation is required, new libraries can be constructed, using plasmid DNA isolated from high-affinity mutants as a template. See Note 17.
Acknowledgements
The authors would like to thank members of our laboratory and K. Dane Wittrup and members of his lab for working out many of the conditions and methods described in this chapter. This work was supported by NIH grants AI064611 and GM55767 (to DMK), a postdoctoral fellowship from the Cancer Research Institute (to JDS), and pre-doctoral fellowship ES013571 (to SAR).
Footnotes
Despite several restriction enzyme digestions, some vector background is consistently observed. Consequently, if possible, choose a vector plasmid that does not express epitope(s) that will be used as part of the selection scheme. For example, if the library is to be sorted based on levels of the c-myc epitope tag expression, use a c-myc non-expressing vector to serve as the backbone for the library to avoid inadvertently selecting those rare yeast that were transformed with uncut or restored vector plasmid.
When transforming into yeast, you will make a mixture of insert and vector DNA at approximately a 6:1 molar ratio, which is often about a 1:1 mass ratio. In addition, your DNA mixture should contain 10–100 ng in no more than 5 µL. If your DNA is too dilute, you may concentrate using several techniques, such as a Qiagen kit, or by precipitating the DNA and re-dissolving in a smaller volume of ddH2O.
It is important that the overnight culture not be allowed to reach stationary phase, or your transformation efficiency will be greatly reduced. If you are unsure how much inoculum to use to obtain the proper density the next day, seed three independent flasks with varying amounts of inoculum to ensure one flask will have the appropriate cell density.
At this point the yeast suspension will be too viscous to easily manipulate with standard P200 or P1000 pipet tips. The day before transformation, prepare some clipped P200 and P1000 pipet tips by carefully removing the distal ¼” with a razor blade. The wider orifice will facilitate handling. Autoclave.
Electrocompetent yeast can be stored at −80°C for future use by adding glycerol to 15% final concentration and snap-freezing in a dry ice/ethanol bath. To use frozen cells, thaw slowly on ice, pellet the cells, and resuspended to the same volume in fresh, ice-cold 1M sorbitol. This will remove excess ions released by dead cells. The transformation efficiency using these frozen/thawed cells will drop >10-fold from fresh yeast.
Mixing the DNA with the electrocompetent yeast and allowing them to incubate together on ice for 30–45 minutes may improve transformation efficiency.
If the conductance is too high to electroporate yeast, make sure you have adequately washed away all free ions in the electrocompetent yeast, and make sure you have prepared the DNA in salt-free ddH2O. Do not use DNA in restriction digest or dephosphorylation reaction buffers for electroporation.
To determine library size, count the colonies that grow up on your dilution plates. For each plate, calculate a theoretical library size by multiplying the number of colonies on the plate by the dilution factor and the total volume of resuspended library in µL divided the number of µL on the plate. For example, an error-prone library of 1 × 107 clones where the final resuspended sorbitol volume was 30 mL would yield an expected 3333, 33, and 6 colonies on your 10 µL, 10 µL at 1:100 dilution, and 10 µL at 1:500 dilution plates, respectively. If you do not get the expected numbers of colonies on your plates after dilution, it is possible that you have not adequately mixed the sorbitol-dissolved library or the diluted culture before plating.
If your bioreceptor cannot be probed with biotinylated ligand at its wild-type affinity and there is not a conformation-specific antibody to the protein, then the level of c-myc staining may be used as a proxy for native expression. Although some full-length protein containing the c-myc tag may be expressed on yeast even if the protein is unfolded or partially folded (44), in some cases properly folded clones may be expressed at higher levels on the yeast surface (37–39, 41). If using this technique to isolate stable mutants, it is important to frequently isolate plasmid from the yeast and analyze the sequence to make sure you did not yield sequence artifacts during homologous recombination that give high c-myc staining for reasons other than stable, native folding.
Optimal induction temperatures and durations should be determined for each bioreceptor. We have found that 20°C is consistently the optimal induction temperature, and the optimal induction duration ranged from 24–72 hours, depending on the polypeptide.
The resuspension volume for flow cytometry will vary based on the number of cells in the sample and the instrument on which you will perform the flow. For analysis, usually a volume of 400–500 µL will give an acceptable sampling rate if 0.5–1.0 × 106 cells were included per sample. For samples that will be sorted, it may be advantageous to resuspend a sample of 1 × 108 cells to a larger volume of 5–10 mL to allow for slower sampling and more careful separation.
When designing your flow cytometry samples, take into consideration how many fluorescent labels will appear in a single sample. If the fluorescent probes included in a single sample have some overlap in excitation and/or emission spectra, such as fluorescein and phycoerythrin, it is important to include valid single-stained control samples in order to adequately compensate the data. In addition, take care not to use multiple primary antibodies that can be detected by the same secondary reagent, such as two different mouse antibodies in the same sample if you plan to detect even one of them with a polyclonal anti-mouse secondary reagent.
When planning to select for higher thermostability in the bioreceptor displayed on the surface of yeast, it is important to check the percent of viability of your cells after the incubation conditions. This may be carried out by plating dilutions of cells onto SD-citrate-CAA plates after incubation at the temperatures you will use to select mutants, compared to those kept at 4°C. If a significant fraction of cells are no longer viable after the treatment (yeast cells appear to begin to lose significant viability at about 50°C), increase the initial sample size to ensure you will still sufficiently sample your library with cells that can recover and expand.
One general approach for mutagenesis is to direct mutations to certain region(s) of the bioreceptor, usually those regions known or believed to contact ligand. These site-directed mutations are encoded by primers that anneal to the template and contain randomized codons at specified positions. These primers are used in a PCR-based amplification method, “splicing by overlap extension” (SOE) (42). Issues of theoretical diversity and maximum library size arise when considering how many codons to randomize. The degenerate codon NNS, where N represents any nucleotide A/T/G/C and S represents G or C, will cover all amino acids (eliminating two of three stop codons) with a theoretical diversity of 32 (4 × 4 × 2). Therefore, a library of yeast randomized at one codon using an NNS degenerate primer need only contain 32 mutants, theoretically, to fully sample all possible mutations. Yeast transformation efficiencies are such that 107-108 independent transformants can be obtained, providing sufficient library sizes to comprehensively sample all possible codon combinations in a 5-codon library (325). Other techniques that have been used include DNA shuffling (45) and “look-through” mutagenesis (46).
Conditions that affect the robustness of the SOE PCR include: annealing temperature, mole ratio of pre-SOE products, and competing undesired amplification. If the SOE product is in low abundance or appears contaminated by unwanted product, try a gradient of annealing temperatures in the SOE PCR program to optimize that parameter. Also, adjusting the ratio of pre-SOE product 1 to pre-SOE product 2 in the SOE reaction mixture may yield improved results. DpnI digestion of the pre-SOE reactions (which will fragment template DNA) prior to the SOE reaction (subjecting to a QIAgen PCR cleanup kit between digest and SOE) may also improve the PCR efficiency.
Staining conditions that yield maximum increase in fluorescence of a high-affinity mutant over the template/wild-type will provide optimal sorting results. A quantitative theoretical analysis of equilibrium labeling conditions of yeast display populations indicated the ligand concentration yielding maximized fluorescence discrimination on the first sort was equal to ~ 0.05–0.1 × the KD of the template or “wild-type” interaction (43). The optimal staining volume, in any equilibrium staining procedure, depends on the numbers of bioreceptors on the yeast surface and the KD of the bioreceptor:ligand interaction (and consequently the concentration of ligand in the staining mixture). The staining volume must be large enough to ensure that the number of bioreceptors does not exceed the moles of ligand, otherwise, the depletion of ligand from solution upon binding bioreceptor would have a non-negligible effect on free ligand concentration and thus staining. In cases where the template affinity is lower, 500 µL volume is likely sufficient. However, when the template affinity is already quite high ( KD < 10 nM) and the concentration of ligand used for staining is therefore low, a larger staining volume must be used so the moles of ligand at least equalize (but ideally exceed) the number of yeast surface displayed bioreceptors (assume ~5 × 105 bioreceptors per yeast cell) (7). If the kon and koff are known, the incubation time that allows approach to equilibrium can also be determined mathematically (31, 43).
Although not the focus of this paper, an engineered high-affinity bioreceptor with broadened specificity (e.g. for multiple toxin subtypes) may be a desired feature of a biosensor. See Garcia-Rodriguez et. al. (47) for yeast display engineering of a scFv for high-affinity to two subtypes of botulinum neurotoxin.
References
- 1.Luppa PB, Sokoll LJ, Chan DW. Clin Chim Acta. 2001;314:1–26. doi: 10.1016/s0009-8981(01)00629-5. [DOI] [PubMed] [Google Scholar]
- 2.Pejcic B, De Marco R, Parkinson G. Analyst. 2006;131:1079–1090. doi: 10.1039/b603402k. [DOI] [PubMed] [Google Scholar]
- 3.Stefan RI, van Staden JF, Aboul-Enein HY. Fresenius J Anal Chem. 2000;366:659–668. doi: 10.1007/s002160051560. [DOI] [PubMed] [Google Scholar]
- 4.Warsinke A, Benkert A, Scheller FW. Fresenius J Anal Chem. 2000;366:622–634. doi: 10.1007/s002160051557. [DOI] [PubMed] [Google Scholar]
- 5.Peruski AH, Peruski LF., Jr Clin Diagn Lab Immunol. 2003;10:506–513. doi: 10.1128/CDLI.10.4.506-513.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodchild S, Love T, Hopkins N, Mayers C. Adv Appl Microbiol. 2006;58:185–226. [PubMed] [Google Scholar]
- 7.Boder ET, Wittrup KD. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 8.Kondo A, Ueda M. Appl Microbiol Biotechnol. 2004;64:28–40. doi: 10.1007/s00253-003-1492-3. [DOI] [PubMed] [Google Scholar]
- 9.Boder ET, Wittrup KD. Methods Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- 10.Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, Graff C, Wiley HS, Wittrup KD. Nat Biotechnol. 2003;21:163–170. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 11.Yeung YA, Wittrup KD. Biotechnol Prog. 2002;18:212–220. doi: 10.1021/bp010186l. [DOI] [PubMed] [Google Scholar]
- 12.Richman SA, Healan SJ, Weber KS, Donermeyer DL, Dossett ML, Greenberg PD, Allen PM, Kranz DM. Protein Eng Des Sel. 2006;19:255–264. doi: 10.1093/protein/gzl008. [DOI] [PubMed] [Google Scholar]
- 13.Wang XX, Shusta EV. J Immunol Methods. 2005;304:30–42. doi: 10.1016/j.jim.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Boder ET, Midelfort KS, Wittrup KD. Proc Natl Acad Sci U S A. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieke MC, Cho BK, Boder ET, Kranz DM, Wittrup KD. Protein Eng. 1997;10:1303–1310. doi: 10.1093/protein/10.11.1303. [DOI] [PubMed] [Google Scholar]
- 16.Feldhaus MJ, Siegel RW. J Immunol Methods. 2004;290:69–80. doi: 10.1016/j.jim.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Kieke MC, Shusta EV, Boder ET, Teyton L, Wittrup KD, Kranz DM. Proc Natl Acad Sci U S A. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richman SA, Kranz DM. Biomol Eng. 2007 doi: 10.1016/j.bioeng.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Weber KS, Donermeyer DL, Allen PM, Kranz DM. Proc Natl Acad Sci U S A. 2005;102:19033–19038. doi: 10.1073/pnas.0507554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones LL, Brophy SE, Bankovich AJ, Colf LA, Hanick NA, Garcia KC, Kranz DM. J Biol Chem. 2006;281:25734–25744. doi: 10.1074/jbc.M604343200. [DOI] [PubMed] [Google Scholar]
- 21.Starwalt SE, Masteller EL, Bluestone JA, Kranz DM. Protein Eng. 2003;16:147–156. doi: 10.1093/proeng/gzg018. [DOI] [PubMed] [Google Scholar]
- 22.Esteban O, Zhao H. J Mol Biol. 2004;340:81–95. doi: 10.1016/j.jmb.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 23.Boder ET, Bill JR, Nields AW, Marrack PC, Kappler JW. Biotechnol Bioeng. 2005;92:485–491. doi: 10.1002/bit.20616. [DOI] [PubMed] [Google Scholar]
- 24.Dam J, Guan R, Natarajan K, Dimasi N, Chlewicki LK, Kranz DM, Schuck P, Margulies DH, Mariuzza RA. Nat Immunol. 2003;4:1213–1222. doi: 10.1038/ni1006. [DOI] [PubMed] [Google Scholar]
- 25.Cochran JR, Kim YS, Lippow SM, Rao B, Wittrup KD. Protein Eng Des Sel. 2006;19:245–253. doi: 10.1093/protein/gzl006. [DOI] [PubMed] [Google Scholar]
- 26.Buonpane RA, Churchill HRO, Moza B, Sundberg EJ, Peterson ML, Schlievert PM, Kranz DM. Nat Med. 2007 doi: 10.1038/nm1584. (In Press) [DOI] [PubMed] [Google Scholar]
- 27.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. Proc Natl Acad Sci U S A. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipovsek D, Lippow SM, Hackel BJ, Gregson MW, Cheng P, Kapila A, Wittrup KD. J Mol Biol. 2007;368:1024–1041. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 29.Rao BM, Driver I, Lauffenburger DA, Wittrup KD. Mol Pharmacol. 2004;66:864–869. doi: 10.1124/mol.66.4.. [DOI] [PubMed] [Google Scholar]
- 30.Rao BM, Girvin AT, Ciardelli T, Lauffenburger DA, Wittrup KD. Protein Eng. 2003;16:1081–1087. doi: 10.1093/protein/gzg111. [DOI] [PubMed] [Google Scholar]
- 31.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 32.Colby DW, Kellogg BA, Graff CP, Yeung YA, Swers JS, Wittrup KD. Methods Enzymol. 2004;388:348–358. doi: 10.1016/S0076-6879(04)88027-3. [DOI] [PubMed] [Google Scholar]
- 33.Daugherty PS, Chen G, Iverson BL, Georgiou G. Proc Natl Acad Sci U S A. 2000;97:2029–2034. doi: 10.1073/pnas.030527597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fromant M, Blanquet S, Plateau P. Anal Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- 35.Meilhoc E, Masson JM, Teissie J. Biotechnology (N Y) 1990;8:223–227. doi: 10.1038/nbt0390-223. [DOI] [PubMed] [Google Scholar]
- 36.Becker DM, Lundblad V. In: Introduction of DNA into Yeast Cells, Transformation by Electroporation in "Current Protocols in Molecular Biology". Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Hoboken, NJ: John Wiley & Sons; 1996. [Google Scholar]
- 37.Shusta EV, Holler PD, Kieke MC, Kranz DM, Wittrup KD. Nat Biotechnol. 2000;18:754–759. doi: 10.1038/77325. [DOI] [PubMed] [Google Scholar]
- 38.Shusta EV, Kieke MC, Parke E, Kranz DM, Wittrup KD. J Mol Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 39.Kowalski JM, Parekh RN, Mao J, Wittrup KD. J Biol Chem. 1998;273:19453–19458. doi: 10.1074/jbc.273.31.19453. [DOI] [PubMed] [Google Scholar]
- 40.Kowalski JM, Parekh RN, Wittrup KD. Biochemistry. 1998;37:1264–1273. doi: 10.1021/bi9722397. [DOI] [PubMed] [Google Scholar]
- 41.Hagihara Y, Kim PS. Proc Natl Acad Sci U S A. 2002;99:6619–6624. doi: 10.1073/pnas.102172099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warrens AN, Jones MD, Lechler RI. Gene. 1997;186:29–35. doi: 10.1016/s0378-1119(96)00674-9. [DOI] [PubMed] [Google Scholar]
- 43.Boder ET, Wittrup KD. Biotechnol Prog. 1998;14:55–62. doi: 10.1021/bp970144q. [DOI] [PubMed] [Google Scholar]
- 44.Park S, Xu Y, Stowell XF, Gai F, Saven JG, Boder ET. Protein Eng Des Sel. 2006;19:211–217. doi: 10.1093/protein/gzl003. [DOI] [PubMed] [Google Scholar]
- 45.Stemmer WP. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 46.Rajpal A, Beyaz N, Haber L, Cappuccilli G, Yee H, Bhatt RR, Takeuchi T, Lerner RA, Crea R. Proc Natl Acad Sci U S A. 2005;102:8466–8471. doi: 10.1073/pnas.0503543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Rodriguez C, Levy R, Arndt JW, Forsyth CM, Razai A, Lou J, Geren I, Stevens RC, Marks JD. Nat Biotechnol. 2007;25:107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 48.Dong X, Stothard P, Forsythe IJ, Wishart DS. Nucleic Acids Res. 2004;32:W660–W664. doi: 10.1093/nar/gkh410. [DOI] [PMC free article] [PubMed] [Google Scholar]