Abstract
During prenatal and postnatal development of the mammalian brain, new neurons are generated by precursor cells that are located in the germinal zones. Subsequently newborn neurons migrate to their destined location in the brain. On the migrational route immature neurons interact via a series of recognition molecules with a plethora of extracellular cues. Stimuli that are conveyed by extracellular cues are translated into complex intracellular signaling networks that eventually enable neuronal migration. In this Focused Review we discuss signaling networks underlying neuronal migration emphasizing molecules and pathways that appear to be neuron-specific.
Keywords: neuroblasts, signaling networks, neuronal-specific pathways
Introduction
In mammals neuronal migration is a fundamental process in the development of nervous system. The peripheral and central nervous system (CNS) comprise numerous neurons that are born in various locations during development and migrate shorter or longer distances to their destination site. Precise coordination of neuronal migration is a prerequisite for the correct positioning and subsequent wiring of neurons into functional circuits. Impairment in migration results in structural defects that are accompanied by severe mental abnormalities.
In the peripheral nervous system, neuronal precursors originate from undifferentiated neural crest cells and migrate during embryonic development to their final locations (Francis and Landis, 1999; Glebova and Ginty, 2005). In the CNS, most neurons derive from precursor cells that reside in the ventricular zone. Projection neurons are born in the ventricular zone of the dorsal telencephalon and migrate radially toward the pia (Marin and Rubenstein, 2003). On the contrary, interneurons are born in the ventral telencephalon, i.e., in the ventricular zone of the medial, lateral, and caudal ganglionic eminences from where they migrate tangentially to the dorsal telencephalon (Corbin et al., 2001; Marin and Rubenstein, 2003). Postnatally new neurons continue to be generated in two brain regions, namely in the subgranular zone (SGZ) of the dentate gyrus in the hippocampus and the subventricular zone (SVZ) of the lateral ventricles (Lledo et al., 2006; Zhao et al., 2008). Neuroblasts (immature neurons) in the hippocampal SGZ migrate a short distance into the granule cell layer of the dentate gyrus and integrate into previously established neural circuits (Kempermann et al., 2004). Neuroblasts originating in the SVZ migrate over long distances via the rostral migratory stream to the olfactory bulb, where they mature into granule or periglomerular neurons (Lledo and Saghatelyan, 2005; Alonso et al., 2006). Under pathological conditions, neuroblasts generated in the SVZ migrate also into injured cortex and striatum (Parent et al., 2002; Kreuzberg et al., 2010). On the migratory route, neuroblasts receive different stimuli from extracellular cues that are paramount in migration guiding. As a result, a number of intracellular signaling molecules are activated. Ultimately, all signaling pathways are integrated in a “master network” that controls the final intracellular output in response to all extracellular inputs and modifies the cytoskeleton machinery accordingly resulting in neuroblast migration.
Specific Signaling for Neuronal Migration
An ever-growing number of studies on neuronal migration employing by and large anatomical techniques goes back to the 1960s. Starting in the 1980s, the function of particular genes/proteins in neuronal migration became the focus of numerous experimental investigations. Whilst these valuable studies have identified important players in migration, many of which were cytoskeleton proteins, during the last decade the interest grew in identifying whole pathways and understanding their complex interactions.
In a recent study (Khodosevich et al., 2009) using microarray gene expression analysis we identified several intracellular signaling pathways underlying neuroblast migration. The identified pathways were corroborated and further expanded using bioinformatics analysis derived from mining of public data that also included information obtained in other cell-types and species (see Data mining for signaling pathway analysis). This powerful approach allows filling in of gaps and proposing whole pathways that would be difficult to identify if one used experimental data from one system only. Needless to say, the signaling components resulting from bioinformatics analysis can and should be tested experimentally to provide functional proof for their involvement in a specific system (in our study it was murine neuroblasts migrating in the rostral migratory stream). For this Focused Review we used only data obtained in studies on mammalian neuronal migration to generate a signaling network whose components are discussed in more detail. The ultimate goal is to eventually arrive at a unique intracellular network underlying signaling in migrating neuroblasts. Here we collected published data for all subtypes of migrating neuroblasts regardless of their birth-place (prenatal ganglionic eminences, postnatal SVZ, precursors of peripheral nervous system, etc.), mode of migration (radial and tangential migration; migration in cell chains and individual cell migration). We also used data obtained in vitro including neurite outgrowth studies. Clearly, each subtype of migrating neuroblasts has its own signaling components that are “tuned” to the microenvironment (i.e., available extracellular stimuli) of migration. Furthermore, different modes of neuroblast migration might rely more on one type of molecular cues in microenvironment than the other, e.g., soluble factors, membrane-bound receptors, or extracellular matrix. However, subtypes of migrating neuroblasts share the majority of intracellular signaling components that integrate external stimuli and result in appropriate output. With the increasing availability of experimental data it will be eventually possible to perform a similar analysis focusing on distinct types of migration and investigate shared and specific molecules and their connections.
Hubs in a Signaling Network Controlling Neuronal Migration
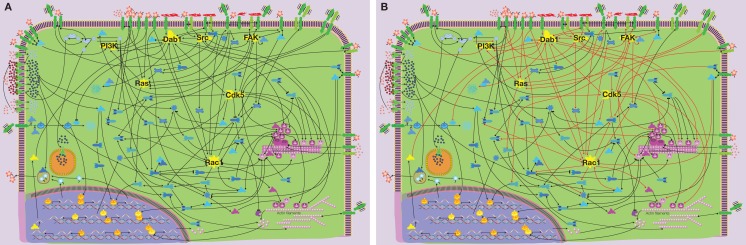
In Figure 1 we summarize the results derived from several hundred studies focusing on some aspects of signaling that control neuronal migration/neurite outgrowth (the names of the individual components are legible upon downloading of Figure S1 in Supplementary Material). The main feature of the signaling network is the uneven distribution of connections between individual molecules resulting in clustering of connections. Seven key “hubs” (shown in yellow) of intracellular signaling involve 2/3 of the connections within the network (Figures 1A,B). Such network clustering is typical for signaling networks and was shown in many proteomic studies (see, e.g., Giot et al., 2003; Pocklington et al., 2006). These signaling hubs control the input and output of the network: cell division protein kinase 5 (Cdk5), disabled homolog 1 (Dab1), ras-related C3 botulinum toxin substrate 1 (Rac1), focal adhesion kinase (FAK), rat sarcoma (Ras), Rous sarcoma oncogene (Src), and phosphatidylinositol 3 kinase (PI3K). Based on their connectivity resulting from our analysis, seven hubs can be further subdivided into two groups: Cdk5, Dab1, and Rac1 having each 13–14 connections, while FAK, Ras, Src, and PI3K having 7–9 connections.
Figure 1.
Signaling network controlling neuronal migration – the scheme is based on experimental data derived from several hundred publications. The only legible names denote network components that constitute “signaling hubs.” The scheme is legible upon downloading Figure S1 in Supplementary Material. (A) Seven hubs in the neuronal migration signaling network (shown in yellow). (B) More than 2/3 of the network connections (shown by red lines) involve hubs. Color code for molecules: yellow – signaling hubs, red – extracellular ligands/matrix components, green – transmembrane receptors/channels/transporters, etc., blue – intracellular signaling molecules, magenta – microtubule/actin-associated proteins, orange – cell nucleus components.
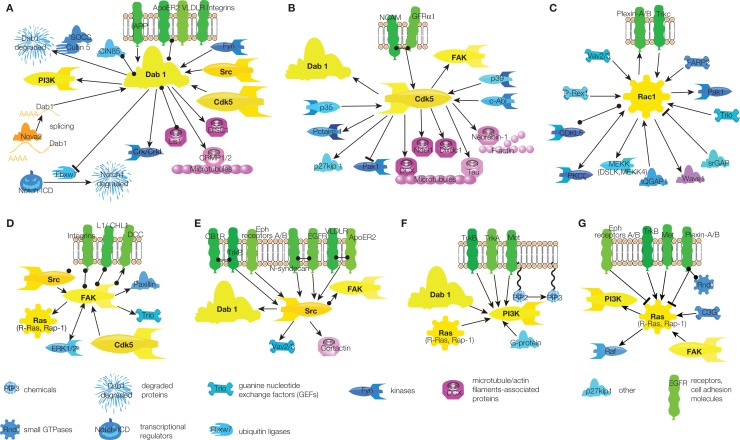
While the small GTPase Rac1 and to a much lesser extent Cdk5 kinase are involved in migration of non-neuronal cell-types, Dab1 is a specific component of neuronal migration signaling (Bielas et al., 2004; Ayala et al., 2007). Dab1 is a cytoplasmic adaptor molecule that was first described as a binding partner of the Src family kinases Src and Fyn (Howell et al., 1997). Later its action was also linked to Reelin signaling (Bielas et al., 2004). However, Dab1 is involved in neuronal migration not only as a target in Reelin signaling, but also in amyloid precursor protein (APP; Young-Pearse et al., 2010) and integrin signaling (Dulabon et al., 2000; Figure 2A). Furthermore, Dab1 is connected to other pathways via Cdk5 and Src kinases (Keshvara et al., 2002; Bock et al., 2003; Kuo et al., 2005; Figure 2A). Since Dab1 also directly binds to several microtubule-associated proteins – Lis1, DISC1, and CRMP (Assadi et al., 2003; Yamashita et al., 2006; Young-Pearse et al., 2010) – it is a strong candidate that “tunes” common mechanisms of cell migration to neuron-specific migration (Figure 2A).
Figure 2.
Signaling hubs in migrating neurons. (A) Dab1 signaling. (B) Cdk5 signaling. (C) Rac1 signaling. (D) FAK signaling. (E) Src signaling. (F) PI3K signaling. (G) Ras signaling. Arrow – activation, circle – interaction, T-shaped end – inhibition. Color code of molecules is the same as in Figure 1. Note that some links are indirect, for full scheme see Figure S1 in Supplementary Material.
Other central hubs in the signaling network of neuronal migration are Cdk5 and Rac1 (Figures 2B,C, respectively). Although Cdk5 was mentioned also in the context of non-neuronal cell migration, most evidence for its role in migration has been derived from studies focusing on neuronal migration. Thus, this signaling hub can be considered by and large neuron-specific (Gupta and Tsai, 2003; Nikolic, 2004). Conversely, Rac1 is a signaling component of many migratory cell-types (Fukata et al., 2003). In neurons almost all extracellular migratory stimuli, such as Slit, integrins, laminins, semaphorins, Reelin, GDNF, EGF, neurotrophin-3 (NT-3), neurotrophin-4 (NT-4), brain-derived neurotrophic factor (BDNF), netrin, calcium, etc. (see Figure S1 in Supplementary Material), eventually converge on these two intracellular proteins. Both hubs are directly connected to microtubules and actin filaments (Figures 2B,C). Thus, the convergence of activating and inhibitory signals onto Cdk5 and Rac1 confers them a pivotal role in determining the direction and speed of neuronal migration. Cdk5/Rac1 are also key players in controlling the initiation and stop of migration.
Cdk5 and Rac1 activation is modulated by other signaling molecules. Cdk5 is phosphorylated and thus activated by p35, p39, and c-Abl (Zukerberg et al., 2000; Keshvara et al., 2002; Beffert et al., 2004; Zhao et al., 2009). However, the regulation of these proteins that modulate Cdk5 activity in migrating neurons has not been explored much so far. Whilst it was shown that PKCδ and Pctaire-1 regulate p35 activity (Cheng et al., 2002; Zhao et al., 2009), there is no information regarding the identity of molecules operating upstream of p39. c-Abl tyrosine kinase, known to be involved in migration of other cell-types as well, exerts its action in neuronal migration downstream of Slit/Robo signaling (Rhee et al., 2002). Thus, modulation of Cdk5 in migrating neurons remains to be explored.
Rac1 is a small GTPase whose activity is triggered by guanine nucleotide exchange factors (GEFs) and inhibited by GTPase-activating proteins (GAPs; Figure 2C). Four GEFs activate Rac1 in migrating neurons – Vav2/3, FARP2, Trio, and P-Rex1 (Schmid et al., 2004; Toyofuku et al., 2005; Yoshizawa et al., 2005; Khodosevich et al., 2009; Peng et al., 2010), but only one GAP inactivates it – Slit–Robo GAP (srGAP; Wong et al., 2001). The discrepancy in inhibitory and activating inputs can be explained to some extent that activating signaling is easier to detect. However, it is very likely that there are other, still to be identified GAPs that inactivate Rac1 in migrating neuroblasts. Interestingly, Rac1 activity is also modulated by PKCζ, a kinase critically involved in the neuronal polarization (Khodosevich et al., 2009; Khodosevich and Monyer, 2010).
Amongst the less connected hubs, FAK and Src are noteworthy (Figures 2D,E, respectively) since they receive input from many receptors, such as DCC, L1, integrins, Reelin receptors ApoER2 and VLDLR, EGFR, TrkB, etc. (see Figure S1 in Supplementary Material). Independent of their individual activation, these kinases can interact with each other resulting in FAK activation by Src (Kuo et al., 2005). FAK and Src operate upstream of the major cytoskeleton-linked hubs, Cdk5 and Rac1. There is only limited evidence for a direct connection between Src and the cytoskeleton in migrating neurons. It involves the phosphorylation of cortactin, an actin-binding protein, by Src (Hienola et al., 2006).
Interestingly, two other hubs, namely PI3K and Ras, that are importantly involved in migration of other cell-types are not that heavily connected within the signaling network in migrating neurons (Figures 2F,G, respectively). PI3K is a kinase involved in numerous cell processes. However, in neuronal migration signaling PI3K is activated only by growth factor receptors and it is also a downstream effector of Dab1 (Figure 2F). The same holds true for Ras, with an addition of semaphorin signaling (Figure 2G), in particular if one takes into account that in our scheme Ras stands for the whole protein family comprising about 30 members. Direct involvement in neuronal migration was demonstrated for two Ras family members, R-Ras and Rap1 (Oinuma et al., 2004; Toyofuku et al., 2005; Voss et al., 2008). Such limited involvement of PI3K and Ras activity in migrating neurons can hardly be explained by a scarcity of study – these are well-known signaling molecules, which have been studied for decades. Most likely, PI3K and Ras have major roles in growth factor and semaphorin signaling while in other signaling cascades they play a minor role (Figures 2F,G, see also Figure S1 in Supplementary Material for full scheme).
Neuronal-Specific Signaling Pathways
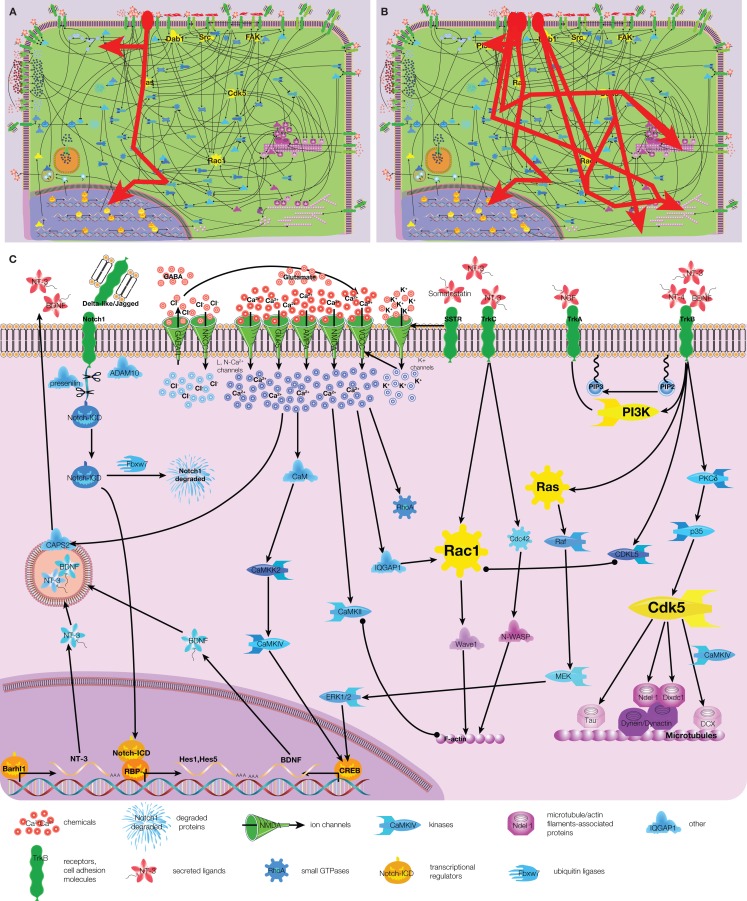
Signaling network in migrating neuroblasts is complex and consists of hundreds interconnected molecules that have been identified already and yet more to be added to this list (Figure 1 and Figure S1 in Supplementary Material). However, the complexity can be reduced when focusing on a specific stimulus (e.g., HGF or hepatocyte growth factor; Figure 3A) or a group of similar stimuli (e.g., all four members of neurotrophin protein family; Figure 3B). Distinct extracellular stimuli activate only one or a few subsets/pathways of the signaling network in migrating neuroblasts and some signaling pathways are specific for migrating neurons. Most notable neuronal-specific examples are those that involve calcium, neurotrophin, and Notch signaling (Figure 3C).
Figure 3.
Examples of signaling pathways in migrating neurons. (A) Scheme of HGF signaling (highlighted by red). (B) Scheme of signaling for neurotrophin protein family (NT-3, NT-4, NGF, BDNF; highlighted in red). (C) Calcium, neurotrophin, and Notch signaling are shown together. Arrow – activation, circle – interaction, T-shaped end – inhibition. Color code of molecules is the same as in Figure 1. Note that some links are indirect, for full scheme see Figure S1 in Supplementary Material.
Studies describing intracellular signaling mediating calcium responses in migrating neurons are by and large limited to the effect of calcium per se and calcium-dependent kinases (Figure 3C). Calcium was shown to activate RhoA (Guan et al., 2007) and the Rac1-activating protein IQGAP1 (Kholmanskikh et al., 2006), most likely also through some intermediate partners. Interestingly, while RhoA inhibits neuronal migration, Rac1 stimulates it, clearly showing that, depending on the upstream stimulus, calcium-signaling can have opposite effects. The first calcium-signaling intermediate protein is calmodulin (CaM) that is activated directly by calcium binding to it. CaM in turn activates CaMKII (Ca/calmodulin dependent kinase II) and CaMKK2 (Ca/calmodulin dependent kinase kinase 2). As CaMKII can bind F-actin filaments, it constitutes the link between calcium-signaling and cytoskeleton remodeling (Fink et al., 2003). CaMKK2 activates another Ca/calmodulin dependent kinase – CaMKIV that upon activation translocates into the nucleus and upregulates BDNF expression (Kokubo et al., 2009). Indeed, we could recently demonstrate the involvement of CaMKIV in the migration of neuroblasts in the rostral migratory stream (Khodosevich et al., 2009). These are the only intracellular calcium-signaling components for which there is experimental evidence that they are involved in neuronal migration. Missing players in the calcium-signaling network can be proposed, but such an educated guess would rely on data from other cell-types or model organisms. Of the signaling hubs only Rac1 has been linked to calcium-signaling via IQGAP1 (Kholmanskikh et al., 2006).
The neurotrophin protein family comprises four members, NT-3, NT-4, BDNF, and nerve growth factor (NGF), which regulate neuronal migration via tropomyosin-receptor-kinases (Trk) A, B, and C (Figure 3C). All neurotrophins activate hubs of intracellular signaling, such as PI3K, Ras, Rac1, and Cdk5 (Yamauchi et al., 2003; Berghuis et al., 2005; Yoshizawa et al., 2005; Chiaramello et al., 2007), that, as discussed above, will eventually lead to cytoskeleton remodeling. Interestingly, already the expression and release of the neurotrophins themselves is tightly controlled during neuronal migration. As we mentioned above, calcium-signaling can activate BDNF expression via CaMKIV (Kokubo et al., 2009). The expression of neurotrophin NT-3 on the other hand is activated by the homeobox protein Barhl1 (Li et al., 2004). On the next stage, the release of neurotrophin-containing secretory granules is mediated by Ca-dependent activator protein for secretion 2 (CAPS2), a secretory granule-associated protein that constitutes a further link between calcium and neurotrophin signaling (Sadakata et al., 2007).
In addition to its major and well-studied function in determining cell fate choice and survival, Notch1 also plays a role in neuronal migration (Figure 3C). Upon ligand binding and stimulation, the Notch receptor gets cleaved by ADAM10 and subsequently by the presenilin 1-containing γ-secretase complex (Louvi et al., 2004; Jorissen et al., 2010). Notch-ICD (intracellular domain) translocates into the nucleus to interact with RBP-J and activates Notch-dependent transcription of Hes1 and Hes5 (Hashimoto-Torii et al., 2008). It is of note that the signaling hub Dab1 controls Notch-ICD degradation by inhibiting Fbxw7, a component of the ubiquitin protein ligase complex (Hashimoto-Torii et al., 2008).
Transcription Activation in Migrating Neuroblasts
Although migrating neuroblasts are still able to proliferate, a major switch in the transcription program between precursor cells and neuroblasts must take place to account for proliferation and migration. Thus, it stands to reason that genes involved in precursor cell proliferation are downregulated with a concomitant upregulation of genes responsible for migration, leading to a dramatic change in the gene expression profile. This is supported by our previous results demonstrating that around 1000 genes changed their expression in neuroblasts that had migrated some distance from the site of origin, the postnatal SVZ (Khodosevich et al., 2009).
One of the most prominent transcription factors involved in the switch from precursor cells to neuroblasts is distal-less homeobox protein Dlx1/2 (see Figure S1 in Supplementary Material for full scheme of transcriptional control). Dlx1/2 knockout animals exhibit strong neuronal migration defects (Cobos et al., 2007). Regulation by Dlx1/2 include expression of a number of noteworthy genes that are known to mediate several fundamental processes in neuronal migration, including repulsion/attraction, stop signal for migration, motility, and cytoskeleton reorganization (Ghashghaei et al., 2007; Tran et al., 2007). Whilst expression of Sema3A, Reelin, and Arx is upregulated by Dlx1/2, that of Pak3, MAP2, Tau, Robo1, GAP43, and Npn2 is downregulated (Cobos et al., 2007; Le et al., 2007).
Another transcription factor, neuregulin 2 (Ngn2), downregulates the expression of the small GTPase RhoA and upregulates the expression of Dcx, p35, and small GTPase Rnd2 (Ge et al., 2006; Heng et al., 2008). Dcx (doublecortin) is a neuronal-specific microtubule-associated protein that, together with Dcx-like kinase (Dcxl), is required for proper microtubule remodeling in response to migratory stimuli (Friocourt et al., 2007). Rnd2 and p35 activate RhoA and Cdk5, respectively (Keshvara et al., 2002; Beffert et al., 2004; Tanaka et al., 2006; Heng et al., 2008). Since RhoA and Cdk5 have opposite functions in the regulation of neuronal migration, precise control of their signaling by Ngn2 is very important (also taking into account that RhoA expression per se is regulated by Ngn2). Which other factors may interact with Ngn2 and specify stimulation of either RhoA or Cdk5 signaling remains to be determined.
Cooperation between transcription factor E2F3 and retinoblastoma (Rb) results in the inhibited expression of several genes, including Reelin receptor VLDLR (McClellan et al., 2007). Reelin is one of the major extracellular ligands controlling several stages of neuronal migration. Knockout of Reelin or any of its receptors cause various defects in brain formation due to impaired neuronal migration during embryonic development (Assadi et al., 2003; Hack et al., 2007).
Finally, other mechanisms than conventional transcription factors that regulate gene expression in migrating neuroblasts should be mentioned. Thus, actin per se controls its own expression via a negative feedback loop. There are two types of actin in migrating neurons – polymerized, F-actin, and depolymerized or free, G-actin. When in excess, free G-actin translocates into the nucleus and downregulates the expression of actin by inhibiting the expression of the transcription factor MRTF (myocardin family transcription factor) that together with serum response factor (SRF) controls actin expression (Stern et al., 2009).
Posttranscriptional RNA processing is also subject to regulation in migrating neuroblasts. For instance, alternative splicing of the key signaling hub Dab1 is regulated by the RNA-binding protein Nova2 preventing the inclusion of two additional exons (Yano et al., 2010), which is necessary for correct migration of cortical neurons during development.
Last but not least, as already mentioned above, calcium and Notch signaling are involved in regulation of transcription in neuroblasts.
Conclusion
This study and others (see, e.g., Lee and Megeney, 2005; Pocklington et al., 2006) using similar approaches clearly highlight how complex signaling in different systems (e.g., migrating neurons, synapse signaling, yeast kinase signaling) can be reduced to major components eventually leading to the identification of general network principles.
The morphological features of migrating neuroblasts differ from those of many other types of migratory cells. During migrational “step” neuroblast extends leading process in the direction controlled by external stimuli, then nucleus moves into the leading process and finally the trailing process is retracted. The majority of cytoskeleton-associated components are concentrated at the very tip of the leading process – the growth cone. Hence neuronal migration differs from the classical one known in fibroblasts. It is not surprising that the unique neuroblast morphology is also reflected in idiosyncratic intracellular signaling. Thus, although migrating neuroblasts share a significant number of hubs and pathways involved in the migration of other cell-types, they are equipped with neuronal-specific actors in migration that most likely adjust the common “migratory machinery” to the specific neuronal needs. Furthermore the weights between certain components or even whole pathways within a network exhibit neuronal-specificity. It may be that these neuronal-specific pathways in migration are not only critical for neuroblasts getting to the right place but are already a prerequisite for neuronal connectivity.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Neuroscience/10.3389/fnins.2011.00028/abstract
Full scheme of signaling network controlling neuronal migration. The scheme was generated by using published data regarding neuronal migration/neurite outgrowth in mammals only (i.e., excluding other cell-types and model organisms). See full list of literature in the main manuscript and supplementary references. Note that we excluded some molecules that were shown to be involved in neuronal migration/neurite outgrowth, but their intracellular partners are not known in migrating neuroblasts. Color code of molecules: yellow – signaling hubs, red – extracellular ligands/matrix components, green – transmembrane receptors/channels/transporters, etc., blue – intracellular signaling molecules, magenta – microtubule/actin-associated proteins, orange – cell nucleus components. Arrow – activation, circle – interaction, T-shaped end – inhibition. Some links are indirect due to scarcity of published data.
Acknowledgments
Images were made with the use of vector graphics from DragonArtz Designs (www.dragonartz.net). The work was supported in part by the Schilling Foundation, DFG (SFB488 grant), and BMBF (RUS 09/B38).
Key Concepts
- Subventricular zone
Germinal zone in the postnatal brain in many mammals. Spans a few intermixed cell layers located in the lateral wall of the paired lateral ventricles. Contains slow-dividing neural stem cells, fast-dividing transit-amplifying precursors, neuroblasts, and a few other support cell-types.
- Neuroblasts
Immature neurons with a small cell body and usually one leading neurite. Neuroblasts migrate by protruding neurite and then pulling cell body in the direction of migration.
- Rostral migratory stream
Stream of migrating neuroblasts from the subventricular zone to the olfactory bulb. Identified in many mammals including rodents, sheep, and primates.
- Small GTPases
GTP/GDP-binding proteins. They are active when bound to GTP and inactive when bound to GDP. Exchange of GDP with GTP (=activation) is catalyzed by proteins called “guanine nucleotide exchange factors” (GEFs). GTP hydrolysis (=inactivation) is catalyzed by “GTPase-activating proteins” (GAPs).
- Kinases
Enzymes that add phosphate groups (=phosphorylate) to other proteins or to themselves. Phosphorylation can result in activation or inhibition of the recipient protein.
- Data mining for signaling pathway analysis
Analysis of published data to connect many molecules, usually obtained after microarray search, in signaling pathways. Data mining is performed with the help of bioinformatics software searching in published literature for pairs of genes/proteins in the same article/abstract/sentence, with/without linking words, such as activation/inhibition, etc.
Biography
 Hannah Monyer got her MD at the University of Heidelberg, Germany. She was a postdoctoral fellow at Stanford University, USA and ZMBH, Germany. Since 1999, she is Professor and Head of the Department of Clinical Neurobiology at the Medical Center Heidelberg University, and since 2010 also Professor at the German Cancer Research Center, Heidelberg. The studies of Hannah Monyer's lab aim at understanding molecular and cellular mechanisms underlying synchronous network oscillations, learning, and memory with particular focus on the functional characterization of GABAergic interneurons.
Hannah Monyer got her MD at the University of Heidelberg, Germany. She was a postdoctoral fellow at Stanford University, USA and ZMBH, Germany. Since 1999, she is Professor and Head of the Department of Clinical Neurobiology at the Medical Center Heidelberg University, and since 2010 also Professor at the German Cancer Research Center, Heidelberg. The studies of Hannah Monyer's lab aim at understanding molecular and cellular mechanisms underlying synchronous network oscillations, learning, and memory with particular focus on the functional characterization of GABAergic interneurons.
 Hannah Monyer got her MD at the University of Heidelberg, Germany. She was a postdoctoral fellow at Stanford University, USA and ZMBH, Germany. Since 1999, she is Professor and Head of the Department of Clinical Neurobiology at the Medical Center Heidelberg University, and since 2010 also Professor at the German Cancer Research Center, Heidelberg. The studies of Hannah Monyer's lab aim at understanding molecular and cellular mechanisms underlying synchronous network oscillations, learning, and memory with particular focus on the functional characterization of GABAergic interneurons.
Hannah Monyer got her MD at the University of Heidelberg, Germany. She was a postdoctoral fellow at Stanford University, USA and ZMBH, Germany. Since 1999, she is Professor and Head of the Department of Clinical Neurobiology at the Medical Center Heidelberg University, and since 2010 also Professor at the German Cancer Research Center, Heidelberg. The studies of Hannah Monyer's lab aim at understanding molecular and cellular mechanisms underlying synchronous network oscillations, learning, and memory with particular focus on the functional characterization of GABAergic interneurons.
References
- Alonso M., Viollet C., Gabellec M. M., Meas-Yedid V., Olivo-Marin J. C., Lledo P. M. (2006). Olfactory discrimination learning increases the survival of adult-born neurons in the olfactory bulb. J. Neurosci. 26, 10508–10513 10.1523/JNEUROSCI.2633-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assadi A. H., Zhang G., Beffert U., McNeil R. S., Renfro A. L., Niu S., Quattrocchi C. C., Antalffy B. A., Sheldon M., Armstrong D. D., Wynshaw-Boris A., Herz J., D'Arcangelo G., Clark G. D. (2003). Interaction of reelin signaling and Lis1 in brain development. Nat. Genet. 35, 270–276 [DOI] [PubMed] [Google Scholar]
- Ayala R., Shu T., Tsai L. H. (2007). Trekking across the brain: the journey of neuronal migration. Cell 128, 29–43 10.1016/j.cell.2006.12.021 [DOI] [PubMed] [Google Scholar]
- Beffert U., Weeber E. J., Morfini G., Ko J., Brady S. T., Tsai L. H., Sweatt J. D., Herz J. (2004). Reelin and cyclin-dependent kinase 5-dependent signals cooperate in regulating neuronal migration and synaptic transmission. J. Neurosci. 24, 1897–1906 10.1523/JNEUROSCI.4084-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghuis P., Dobszay M. B., Wang X., Spano S., Ledda F., Sousa K. M., Schulte G., Ernfors P., Mackie K., Paratcha G., Hurd Y. L., Harkany T. (2005). Endocannabinoids regulate interneuron migration and morphogenesis by transactivating the TrkB receptor. Proc. Natl. Acad. Sci. U.S.A. 102, 19115–19120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas S., Higginbotham H., Koizumi H., Tanaka T., Gleeson J. G. (2004). Cortical neuronal migration mutants suggest separate but intersecting pathways. Annu. Rev. Cell Dev. Biol. 20, 593–618 [DOI] [PubMed] [Google Scholar]
- Bock H. H., Jossin Y., Liu P., Forster E., May P., Goffinet A. M., Herz J. (2003). Phosphatidylinositol 3-kinase interacts with the adaptor protein Dab1 in response to reelin signaling and is required for normal cortical lamination. J. Biol. Chem. 278, 38772–38779 [DOI] [PubMed] [Google Scholar]
- Cheng K., Li Z., Fu W. Y., Wang J. H., Fu A. K., Ip N. Y. (2002). Pctaire1 interacts with p35 and is a novel substrate for Cdk5/p35. J. Biol. Chem. 277, 31988–31993 [DOI] [PubMed] [Google Scholar]
- Chiaramello S., Dalmasso G., Bezin L., Marcel D., Jourdan F., Peretto P., Fasolo A., De Marchis S. (2007). BDNF/TrkB interaction regulates migration of SVZ precursor cells via PI3-K and MAP-K signalling pathways. Eur. J. Neurosci. 26, 1780–1790 [DOI] [PubMed] [Google Scholar]
- Cobos I., Borello U., Rubenstein J. L. (2007). Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron 54, 873–888 10.1016/j.neuron.2007.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin J. G., Nery S., Fishell G. (2001). Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat. Neurosci. 4(Suppl.), 1177–1182 [DOI] [PubMed] [Google Scholar]
- Dulabon L., Olson E. C., Taglienti M. G., Eisenhuth S., McGrath B., Walsh C. A., Kreidberg J. A., Anton E. S. (2000). Reelin binds alpha3beta1 integrin and inhibits neuronal migration. Neuron 27, 33–44 10.1016/S0896-6273(00)00007-6 [DOI] [PubMed] [Google Scholar]
- Fink C. C., Bayer K. U., Myers J. W., Ferrell J. E., Jr., Schulman H., Meyer T. (2003). Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron 39, 283–297 10.1016/S0896-6273(03)00428-8 [DOI] [PubMed] [Google Scholar]
- Francis N. J., Landis S. C. (1999). Cellular and molecular determinants of sympathetic neuron development. Annu. Rev. Neurosci. 22, 541–566 [DOI] [PubMed] [Google Scholar]
- Friocourt G., Liu J. S., Antypa M., Rakic S., Walsh C. A., Parnavelas J. G. (2007). Both doublecortin and doublecortin-like kinase play a role in cortical interneuron migration. J. Neurosci. 27, 3875–3883 10.1523/JNEUROSCI.4530-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M., Nakagawa M., Kaibuchi K. (2003). Roles of Rho-family GTPases in cell polarisation and directional migration. Curr. Opin. Cell Biol. 15, 590–597 [DOI] [PubMed] [Google Scholar]
- Ge W., He F., Kim K. J., Blanchi B., Coskun V., Nguyen L., Wu X., Zhao J., Heng J. I., Martinowich K., Tao J., Wu H., Castro D., Sobeih M. M., Corfas G., Gleeson J. G., Greenberg M. E., Guillemot F., Sun Y. E. (2006). Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. U.S.A. 103, 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghashghaei H. T., Lai C., Anton E. S. (2007). Neuronal migration in the adult brain: are we there yet? Nat. Rev. Neurosci. 8, 141–151 [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., Vijayadamodar G., Pochart P., Machineni H., Welsh M., Kong Y., Zerhusen B., Malcolm R., Varrone Z., Collis A., Minto M., Burgess S., McDaniel L., Stimpson E., Spriggs F., Williams J., Neurath K., Ioime N., Agee M., Voss E., Furtak K., Renzulli R., Aanensen N., Carrolla S., Bickelhaupt E., Lazovatsky Y., DaSilva A., Zhong J., Stanyon C. A., Finley R. L., Jr., White K. P., Braverman M., Jarvie T., Gold S., Leach M., Knight J., Shimkets R. A., McKenna M. P., Chant J., Rothberg J. M. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 10.1126/science.1090289 [DOI] [PubMed] [Google Scholar]
- Glebova N. O., Ginty D. D. (2005). Growth and survival signals controlling sympathetic nervous system development. Annu. Rev. Neurosci. 28, 191–222 [DOI] [PubMed] [Google Scholar]
- Guan C. B., Xu H. T., Jin M., Yuan X. B., Poo M. M. (2007). Long-range Ca2+ signaling from growth cone to soma mediates reversal of neuronal migration induced by slit-2. Cell 129, 385–395 10.1016/j.cell.2007.01.051 [DOI] [PubMed] [Google Scholar]
- Gupta A., Tsai L. H. (2003). Cyclin-dependent kinase 5 and neuronal migration in the neocortex. Neurosignals 12, 173–179 10.1159/000074618 [DOI] [PubMed] [Google Scholar]
- Hack I., Hellwig S., Junghans D., Brunne B., Bock H. H., Zhao S., Frotscher M. (2007). Divergent roles of ApoER2 and Vldlr in the migration of cortical neurons. Development 134, 3883–3891 10.1242/dev.005447 [DOI] [PubMed] [Google Scholar]
- Hashimoto-Torii K., Torii M., Sarkisian M. R., Bartley C. M., Shen J., Radtke F., Gridley T., Sestan N., Rakic P. (2008). Interaction between reelin and notch signaling regulates neuronal migration in the cerebral cortex. Neuron 60, 273–284 10.1016/j.neuron.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng J. I., Nguyen L., Castro D. S., Zimmer C., Wildner H., Armant O., Skowronska-Krawczyk D., Bedogni F., Matter J. M., Hevner R., Guillemot F. (2008). Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 455, 114–118 10.1038/nature07198 [DOI] [PubMed] [Google Scholar]
- Hienola A., Tumova S., Kulesskiy E., Rauvala H. (2006). N-syndecan deficiency impairs neural migration in brain. J. Cell Biol. 174, 569–580 10.1083/jcb.200602043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. W., Gertler F. B., Cooper J. A. (1997). Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO J. 16, 121–132 10.1093/emboj/16.1.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorissen E., Prox J., Bernreuther C., Weber S., Schwanbeck R., Serneels L., Snellinx A., Craessaerts K., Thathiah A., Tesseur I., Bartsch U., Weskamp G., Blobel C. P., Glatzel M., De Strooper B., Saftig P. (2010). The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J. Neurosci. 30, 4833–4844 10.1523/JNEUROSCI.5221-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G., Wiskott L., Gage F. H. (2004). Functional significance of adult neurogenesis. Curr. Opin. Neurobiol. 14, 186–191 [DOI] [PubMed] [Google Scholar]
- Keshvara L., Magdaleno S., Benhayon D., Curran T. (2002). Cyclin-dependent kinase 5 phosphorylates disabled 1 independently of reelin signaling. J. Neurosci. 22, 4869–4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodosevich K., Monyer H. (2010). Signaling involved in neurite outgrowth of postnatally born subventricular zone neurons in vitro. BMC Neurosci. 11, 18. 10.1186/1471-2202-11-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodosevich K., Seeburg P. H., Monyer H. (2009). Major signaling pathways in migrating neuroblasts. Front. Mol. Neurosci. 2:7. 10.3389/neuro.02.007.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholmanskikh S. S., Koeller H. B., Wynshaw-Boris A., Gomez T., Letourneau P. C., Ross M. E. (2006). Calcium-dependent interaction of Lis1 with IQGAP1 and Cdc42 promotes neuronal motility. Nat. Neurosci. 9, 50–57 [DOI] [PubMed] [Google Scholar]
- Kokubo M., Nishio M., Ribar T. J., Anderson K. A., West A. E., Means A. R. (2009). BDNF-mediated cerebellar granule cell development is impaired in mice null for CaMKK2 or CaMKIV. J. Neurosci. 29, 8901–8913 10.1523/JNEUROSCI.0040-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzberg M., Kanov E., Timofeev O., Schwaninger M., Monyer H., Khodosevich K. (2010). Increased subventricular zone-derived cortical neurogenesis after ischemic lesion. Exp. Neurol. 226, 90–99 [DOI] [PubMed] [Google Scholar]
- Kuo G., Arnaud L., Kronstad-O'Brien P., Cooper J. A. (2005). Absence of Fyn and Src causes a reeler-like phenotype. J. Neurosci. 25, 8578–8586 10.1523/JNEUROSCI.1656-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T. N., Du G., Fonseca M., Zhou Q. P., Wigle J. T., Eisenstat D. D. (2007). Dlx homeobox genes promote cortical interneuron migration from the basal forebrain by direct repression of the semaphorin receptor neuropilin-2. J. Biol. Chem. 282, 19071–19081 [DOI] [PubMed] [Google Scholar]
- Lee R. E., Megeney L. A. (2005). The yeast kinome displays scale free topology with functional hub clusters. BMC Bioinformatics 6, 271. 10.1186/1471-2105-6-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Qiu F., Xu A., Price S. M., Xiang M. (2004). Barhl1 regulates migration and survival of cerebellar granule cells by controlling expression of the neurotrophin-3 gene. J. Neurosci. 24, 3104–3114 10.1523/JNEUROSCI.4444-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lledo P. M., Saghatelyan A. (2005). Integrating new neurons into the adult olfactory bulb: joining the network, life-death decisions, and the effects of sensory experience. Trends Neurosci. 28, 248–254 10.1016/j.tins.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Lledo P. M., Alonso M., Grubb M. S. (2006). Adult neurogenesis and functional plasticity in neuronal circuits. Nat. Rev. Neurosci. 7, 179–193 [DOI] [PubMed] [Google Scholar]
- Louvi A., Sisodia S. S., Grove E. A. (2004). Presenilin 1 in migration and morphogenesis in the central nervous system. Development 131, 3093–3105 10.1242/dev.01191 [DOI] [PubMed] [Google Scholar]
- Marin O., Rubenstein J. L. (2003). Cell migration in the forebrain. Annu. Rev. Neurosci. 26, 441–483 [DOI] [PubMed] [Google Scholar]
- McClellan K. A., Ruzhynsky V. A., Douda D. N., Vanderluit J. L., Ferguson K. L., Chen D., Bremner R., Park D. S., Leone G., Slack R. S. (2007). Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol. Cell Biol. 27, 4825–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic M. (2004). The molecular mystery of neuronal migration: FAK and Cdk5. Trends Cell Biol. 14, 1–5 10.1016/j.tcb.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Oinuma I., Ishikawa Y., Katoh H., Negishi M. (2004). The semaphorin 4D receptor plexin-B1 is a GTPase activating protein for R-Ras. Science 305, 862–865 10.1126/science.1097545 [DOI] [PubMed] [Google Scholar]
- Parent J. M., Vexler Z. S., Gong C., Derugin N., Ferriero D. M. (2002). Rat forebrain neurogenesis and striatal neuron replacement after focal stroke. Ann. Neurol. 52, 802–813 [DOI] [PubMed] [Google Scholar]
- Peng Y. J., He W. Q., Tang J., Tao T., Chen C., Gao Y. Q., Zhang W. C., He X. Y., Dai Y. Y., Zhu N. C., Lv N., Zhang C. H., Qiao Y. N., Zhao L. P., Gao X., Zhu M. S. (2010). Trio is a key guanine nucleotide exchange factor coordinating regulation of the migration and morphogenesis of granule cells in the developing cerebellum. J. Biol. Chem. 285, 24834–24844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocklington A. J., Cumiskey M., Armstrong J. D., Grant S. G. (2006). The proteomes of neurotransmitter receptor complexes form modular networks with distributed functionality underlying plasticity and behaviour. Mol. Syst. Biol. 2, 2006 0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J., Mahfooz N. S., Arregui C., Lilien J., Balsamo J., VanBerkum M. F. (2002). Activation of the repulsive receptor roundabout inhibits N-cadherin-mediated cell adhesion. Nat. Cell Biol. 4, 798–805 [DOI] [PubMed] [Google Scholar]
- Sadakata T., Kakegawa W., Mizoguchi A., Washida M., Katoh-Semba R., Shutoh F., Okamoto T., Nakashima H., Kimura K., Tanaka M., Sekine Y., Itohara S., Yuzaki M., Nagao S., Furuichi T. (2007). Impaired cerebellar development and function in mice lacking CAPS2, a protein involved in neurotrophin release. J. Neurosci. 27, 2472–2482 10.1523/JNEUROSCI.2279-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid R. S., Midkiff B. R., Kedar V. P., Maness P. F. (2004). Adhesion molecule L1 stimulates neuronal migration through Vav2-Pak1 signaling. Neuroreport 15, 2791–2794 [PubMed] [Google Scholar]
- Stern S., Debre E., Stritt C., Berger J., Posern G., Knoll B. (2009). A nuclear actin function regulates neuronal motility by serum response factor-dependent gene transcription. J. Neurosci. 29, 4512–4518 10.1523/JNEUROSCI.0333-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Katoh H., Negishi M. (2006). Pragmin, a novel effector of Rnd2 GTPase, stimulates RhoA activity. J. Biol. Chem. 281, 10355–10364 [DOI] [PubMed] [Google Scholar]
- Toyofuku T., Yoshida J., Sugimoto T., Zhang H., Kumanogoh A., Hori M., Kikutani H. (2005). FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nat. Neurosci. 8, 1712–1719 [DOI] [PubMed] [Google Scholar]
- Tran T. S., Kolodkin A. L., Bharadwaj R. (2007). Semaphorin regulation of cellular morphology. Annu. Rev. Cell Dev. Biol. 23, 263–292 [DOI] [PubMed] [Google Scholar]
- Voss A. K., Britto J. M., Dixon M. P., Sheikh B. N., Collin C., Tan S. S., Thomas T. (2008). C3G regulates cortical neuron migration, preplate splitting and radial glial cell attachment. Development 135, 2139–2149 10.1242/dev.016725 [DOI] [PubMed] [Google Scholar]
- Wong K., Ren X. R., Huang Y. Z., Xie Y., Liu G., Saito H., Tang H., Wen L., Brady-Kalnay S. M., Mei L., Wu J. Y., Xiong W. C., Rao Y. (2001). Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the slit-robo pathway. Cell 107, 209–221 10.1016/S0092-8674(01)00530-X [DOI] [PubMed] [Google Scholar]
- Yamashita N., Uchida Y., Ohshima T., Hirai S., Nakamura F., Taniguchi M., Mikoshiba K., Honnorat J., Kolattukudy P., Thomasset N., Takei K., Takahashi T., Goshima Y. (2006). Collapsin response mediator protein 1 mediates reelin signaling in cortical neuronal migration. J. Neurosci. 26, 13357–13362 10.1523/JNEUROSCI.4276-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi J., Chan J. R., Shooter E. M. (2003). Neurotrophin 3 activation of TrkC induces Schwann cell migration through the c-Jun N-terminal kinase pathway. Proc. Natl. Acad. Sci. U.S.A. 100, 14421–14426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M., Hayakawa-Yano Y., Mele A., Darnell R. B. (2010). Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron 66, 848–858 10.1016/j.neuron.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M., Kawauchi T., Sone M., Nishimura Y. V., Terao M., Chihama K., Nabeshima Y., Hoshino M. (2005). Involvement of a Rac activator, P-Rex1, in neurotrophin-derived signaling and neuronal migration. J. Neurosci. 25, 4406–4419 10.1523/JNEUROSCI.4955-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young-Pearse T. L., Suth S., Luth E. S., Sawa A., Selkoe D. J. (2010). Biochemical and functional interaction of disrupted-in-schizophrenia 1 and amyloid precursor protein regulates neuronal migration during mammalian cortical development. J. Neurosci. 30, 10431–10440 10.1523/JNEUROSCI.1445-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Deng W., Gage F. H. (2008). Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660 10.1016/j.cell.2008.01.033 [DOI] [PubMed] [Google Scholar]
- Zhao C. T., Li K., Li J. T., Zheng W., Liang X. J., Geng A. Q., Li N., Yuan X. B. (2009). PKCdelta regulates cortical radial migration by stabilizing the Cdk5 activator p35. Proc. Natl. Acad. Sci. U.S.A. 106, 21353–21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukerberg L. R., Patrick G. N., Nikolic M., Humbert S., Wu C. L., Lanier L. M., Gertler F. B., Vidal M., Van Etten R. A., Tsai L. H. (2000). Cables links Cdk5 and c-Abl and facilitates Cdk5 tyrosine phosphorylation, kinase upregulation, and neurite outgrowth. Neuron 26, 633–646 10.1016/S0896-6273(00)81200-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full scheme of signaling network controlling neuronal migration. The scheme was generated by using published data regarding neuronal migration/neurite outgrowth in mammals only (i.e., excluding other cell-types and model organisms). See full list of literature in the main manuscript and supplementary references. Note that we excluded some molecules that were shown to be involved in neuronal migration/neurite outgrowth, but their intracellular partners are not known in migrating neuroblasts. Color code of molecules: yellow – signaling hubs, red – extracellular ligands/matrix components, green – transmembrane receptors/channels/transporters, etc., blue – intracellular signaling molecules, magenta – microtubule/actin-associated proteins, orange – cell nucleus components. Arrow – activation, circle – interaction, T-shaped end – inhibition. Some links are indirect due to scarcity of published data.