Abstract
Telomerase-deficient mutants of Saccharomyces cerevisiae can survive death by senescence by using one of two homologous recombination pathways. The Rad51 pathway amplifies the subtelomeric Y′ sequences, while the Rad50 pathway amplifies the telomeric TG1-3 repeats. Here we show that telomerase-negative cells require Clb2 (the major B-type cyclin in this organism), in association with Cdc28 (Cdk1), to generate postsenescence survivors at a normal rate. The Rad50 pathway was more sensitive to the absence of Clb2 than the Rad51 pathway. We also report that telomerase RAD50 RAD51 triple mutants still generated postsenescence survivors. This novel Rad50- and Rad51-independent pathway of telomeric recombination also appeared to be controlled by Clb2. In telomerase-positive cells, a synthetic growth defect between mutations in CLB2 and RAD50 or in its partners in the conserved MRX complex, MRE11 and XRS2, was observed. This genetic interaction was independent of Mre11 nuclease activity but was dependent on a DNA repair function. The present data reveal an unexpected role of Cdc28/Clb2 in telomeric recombination during telomerase-independent maintenance of telomeres. They also uncover a functional interaction between Cdc28/Clb2 and MRX during the control of the mitotic cell cycle.
Telomeres, which represent specialized structures at the ends of linear chromosomes in most eukaryotic organisms, are composed of G-rich repetitive tracts of DNA sequences and associated specialized proteins (for a recent review, see reference 2). One of the major functions of telomeres is to provide a shield against nucleolytic attack and subsequent degradation of the linear chromosomes. Such inappropriate interventions of nucleases include those by DNA repair enzymes and checkpoint proteins that are susceptible to treating telomeres as DNA double-strand breaks in the absence of adequate protection (reviewed in references 9 and 16). Another major function of telomeres is to provide mechanisms ensuring the full replication of the telomeres and to overcome the end-replication problem, which inexorably leads to sequence loss as cells divide, followed by telomere shortening and, ultimately, chromosome degradation (reviewed in reference 2). These problems are counteracted by telomerase-dependent replication, during which telomerase, a ribonucleoprotein enzyme that synthesizes DNA by using its own RNA moiety as a template, is recruited at telomere ends (reviewed in references 13, 20, and 36). Both the activity of telomerase and its accessibility to telomeres are regulated during telomere replication (reviewed in references 2, 18, 20, 36, and 53).
In the yeast Saccharomyces cerevisiae, the ∼300 bp of TG1-3 repeats become eroded after 50 to 100 cell divisions in cells lacking the telomerase RNA component TLC1 or the reverse transcriptase component Est2, as well as in mutants with mutations in telomerase regulatory components, such as est1Δ, est3Δ, and est4-1/cdc13-2 mutants (40, 44). In yeast, these events are known as senescence (44). Replicative senescence, the finite replicative potential of many cultured human cell types (29), appears to result from the natural shortening of telomeres in aging cells deprived of telomerase activity (3, 62). In yeast, aging and senescence are two distinct processes, with telomere lengthening rather than telomere shortening being associated with aging (reviewed in reference 33). In fact, proliferating cultures of yeast cells, in which aging cells are naturally replaced by newly born cells, contain a mixture of young and old cells at all stages. Nevertheless, senescence in yeast has in common with senescence in higher eukaryotes induction by inactivation of telomerase with resulting telomere erosion and chromosome instability (reviewed in reference 42).
Although the vast majority of yeast senescing cells die once they have accumulated a lethal amount of DNA damage, rare survivors succeed in maintaining functional telomeres in the absence of telomerase, thus relieving the damage (43). These survivors employ homologous recombination mechanisms to restore telomere function (43), a strategy that also seems to be employed by immortalized mammalian cell lines and tumors to impose proliferation despite inactivation of telomerase (reviewed in reference 30). In S. cerevisiae, two telomerase-independent telomere maintenance pathways have been identified (reviewed in references 35 and 42). The Rad51 pathway, which relies on Rad52, Rad54, Rad55, and Rad57, yields telomeres that display recombination events within the subtelomeric Y′ sequences, referred to as type I recombination (39, 43, 56). The Rad50/Rad59 pathway, which amplifies the TG1-3 sequences (so-called type II recombination), relies on Rad52, Rad50, Mre11, Xrs2, Rad59, and Sgs1 (11, 12, 31, 34, 39, 43, 55, 56).
Here, we implicate Clb2, the major B-type cyclin in S. cerevisiae, in association with Cdc28 (Cdk1), in the survival mechanisms that operate in telomerase negative cells. More specifically, Clb2 controlled the Rad50/Rad59 pathway of homologous recombination that functions in parallel with the Rad51 pathway. Clb2 also appeared to control a third, novel, Rad52-dependent recombination program that could also generate postsenescence survivors in parallel with, and in the absence of, the Rad50 and Rad51 pathways.
MATERIALS AND METHODS
Strains and plasmids.
The strain background was 15Daub, a ura3Δns derivative of BF264-15D which was also leu2-3,112 trp1-1a ade1 his2 (51). The mre11::LEU2 disruptant, as well as the YCp33-MRE11 and YCp33-mre11(ts) plasmids, have been described previously (10). YEp-ADH1-MRE11 and YEp-ADH1-mre11D16A were from Kunihiro Ohta's laboratory. The cdc28-4 and cdc28-13 alleles (51) and clb1::URA3, clb2::LEU2, clb3::TRP1, and clb4::HIS2 (52) have been described previously. The xrs2::KanMX4, dnl4::KanMX4, rad59::KanMX4, and clb5::KanMX4 strains were from Euroscarf (Frankfurt, Germany). The origins of the rad50::URA3, yku70::URA3, rad52::LEU2, and tel1::KanMX4 strains and the tlc1::LEU2 plasmid are provided in reference 23. All strains were backcrossed at least five times against the genetic background used in our lab prior to experimentation.
Analysis of telomere structure.
Genomic DNAs were prepared as described previously and digested with XhoI, and the resulting Southern blots were analyzed with a 270-bp TG1-3 32P-labeled probe (24). To analyze recombination events in the so-called streak assays (see below), a single colony was picked out of the agar plate and grown overnight in liquid yeast extract-peptone-dextrose (YEPD) in order to obtain enough material for genomic DNA preparation and Southern blot analysis.
Analysis of the kinetics of senescence and survival.
Streak assays for analysis of telomeric recombination events, performed on agar-based plates (see Fig. 2), were based on protocols described previously (24, 56). Briefly, restreaking of single colonies on a YEPD plate was repeated every 48 h (typically, cells underwent ∼25 generations per streakout or passage at 29°C) to allow loss of viability and appearance of survivors. Measurements of cell viability in liquid cultures were done as described previously (11, 39). Briefly, cells were grown to saturation (108 cells/ml) in liquid YEPD and then, every 24 h, were diluted to 105 cells/ml with fresh medium after they were counted with a Neubauer hematocytometer (for Fig. 1A and 5) or were diluted to 5 × 106 cells/ml after spotting of 5 μl on to a fresh YEPD plate (for Fig. 4). In some experiments, cells were withdrawn from liquid cultures at various intervals during senescence and fixed with formaldehyde. Cells were then observed in a BX50 Olympus light microscope, using a ×40 lens and Nomarski optics, and photographs were taken with an Olympus numerical camera.
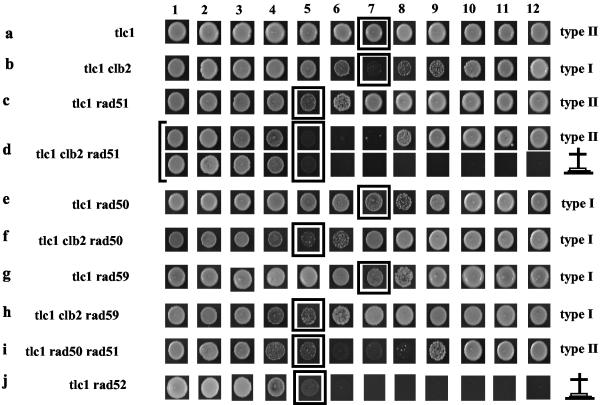
FIG. 2.
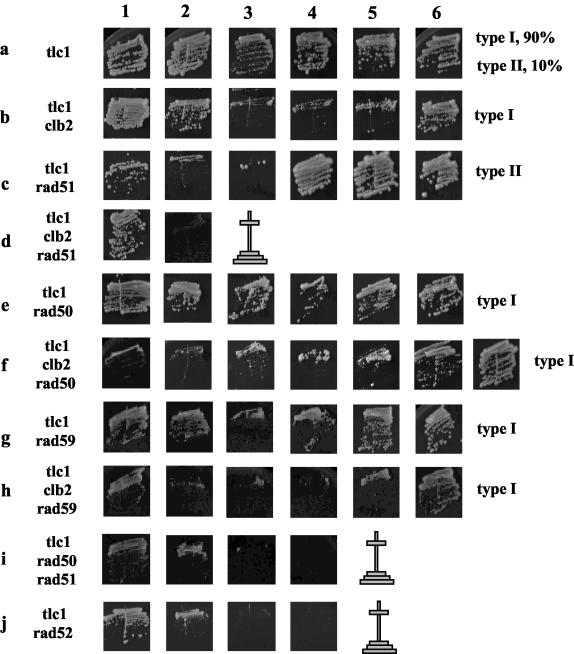
Clb2 controls the Rad50/Rad59-dependent type II survival pathway of telomeric recombination. Strains of the indicated relevant genotypes were assessed for their kinetics of senescence in streak assays. Each row represents the fate of a given strain at each restreak, indicated by numbers at the top (every 48 h, at 29°C). Because of the time for selection of the mutations (restreak from the sporulation plate onto the appropriate selective medium plates), an estimated ∼80 generations elapsed between sporulation and photography of the first streakout (first column). On the right of each row is indicated the survivor type or percentage of each type for each strain, i.e., type I (recombination of the subtelomeric Y′ sequences) or type II (recombination of the TG1-3 sequences), assessed after Southern analysis of 10 colonies for each strain (not shown). An absence of an indication of the survivor type indicates that the strain died without generating survivors; the cross indicates the time of death.
FIG. 1.
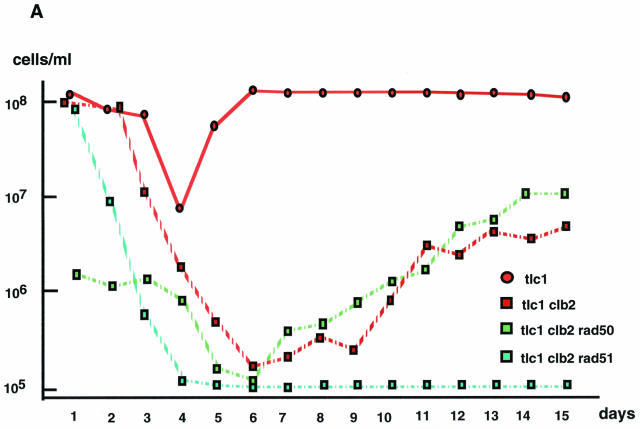
Clb2 is required for the generation of type II postsenescence survivors. (A) Cell viability in various telomerase-deficient strains of the indicated relevant genotypes during and after senescence. The trough of each viability curve indicates crisis, after which viability readily reincreases due to the onset of homologous recombination-based mechanisms that repair the eroded telomeres. A noticeable exception is the tlc1 rad51 clb2 triple mutant, which, under these conditions, could not recover from senescence. Cells were grown in liquid cultures at 29°C and diluted every 24 h to 105 cells/ml with fresh medium (see Materials and Methods). Two additional experiments gave similar results. (B) Clb2 inhibits the generation of type II survivors in streak assays (see Materials and Methods). Colonies emerging from senescing cultures of tlc1Δ clb2Δ or tlc1Δ CLB2+ strains grown on agar plates at 29°C were picked out, individually grown overnight in liquid, and harvested. In such samples, type I recombination can be conveniently distinguished from type II recombination by analyzing Southern blots from genomic DNA previously cut with XhoI. Classical criteria were used to determine formation of type I versus type II survivors (56, 58). Thus, type I survivors are characterized by the erosion of telomeres compared to wild-type (wt) cells (compare lanes 1 and 2; the mean size of the terminal telomere tracts in the wild type corresponds to the broad band at ∼1.3 kb), with the XhoI site within telomeres being located more distal than the Y′ sequences, while type II survivors exhibit very long and heterogeneous TG1-3 sequences (located more distal than the XhoI site) (lanes 12, 30, and 31). The disappearance of the four bands migrating at ∼2.1, 2.3, 3.4, and 4.2 kb in the nonrecombining cells (lane 1), representing non-Y′ fragments from the non-Y′ chromosomes, attests to the fact that these type I survivors have indeed undergone recombination. Exclusive type I survivors from tlc1Δ clb2Δ rad50Δ (lanes 26 and 27) and tlc1Δ rad50Δ (lanes 28 and 29) mutants and type II survivors from a tlc1Δ rad51Δ mutant (lanes 30 and 31) are shown for comparison. Southern blots were revealed with a 270-bp TG1-3 32P-labeled probe. (C) Clb2 is required for the transition from type I to type II that naturally occurs in liquid cultures, due to the better growth of type II survivors (56). Eight individual tlc1Δ and tlc1Δ clb2Δ survivors each were propagated for ∼100 generations as exponentially growing liquid cultures (at 29°C) that were diluted to 105 cells/ml every day, and their survivor type was determined as described above. XhoI cutting and a TG1-3 32P-labeled probe were used. (D) Full function of Cdc28 is required for the transition from type I to type II, under the same conditions as described above for tlc1Δ clb2Δ cells (daily dilutions of 105 cells/ml, ∼100 generations of growth), but here at either 25, 30, or 32°C (cdc28-4 is a temperature-sensitive allele of CDC28).
FIG. 5.
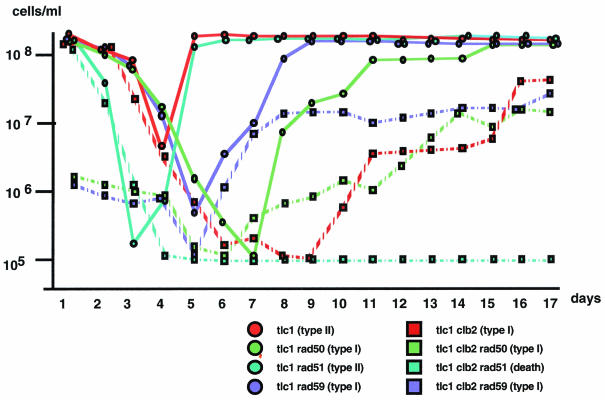
Impact of the absence of Clb2 on cell viability, expressed as the cell density, during senescence and acquisition of survival in various recombination mutants. Cells were grown in liquid cultures at 29°C and diluted every 24 h to 105 cells/ml with fresh medium (see Materials and Methods); this represents the standard conditions, while in the experiments shown in Fig. 4 the daily dilutions had been deliberately lowered.
FIG. 4.
Extremely rare survivors emerge from senescing tlc1 rad50 rad51 and tlc1 clb2 rad51 mutants when the culture dilution is lowered. Each row represents the kinetics of senescence for a strain of the indicated relevant genotype at 29°C. Successive dilutions of the liquid cultures, numbered at the top, consisted of bringing the final concentration down to 5 × 106 cells/ml every 24 h. Note that the standard condition for liquid culture assays (Fig. 1A) is 105 cells/ml. At the daily dilution of 5 × 106 cells/ml, the crisis (time at which the culture reaches the highest rate of death, indicated by the boxed picture for each strain) was barely visible in tlc1Δ cells (row a). These conditions highlight the existence of a novel pathway of recombination (of type II) operating in tlc1Δ rad50Δ rad51Δ cells (row i), which is thus independent of Rad50 and Rad51 but dependent on Rad52 (row j). See the text for interpretation of other events or phenotypes.
RESULTS
Cdc28/Clb2 regulates telomeric recombination in survivors from telomerase-negative cells.
Cells in which telomerase has been inactivated, for instance, cells of tlc1Δ, est2Δ, est1, or est3Δ mutants of S. cerevisiae, die of senescence (44). However, a very small percentage of senescing cells survive, owing to telomerase-independent pathways (43). Incidentally, we noticed that postsenescence survivors from a tlc1Δ clb2Δ double mutant strain exhibited poorer growth on agar-based plates than survivors from a tlc1Δ CLB2+ strain (data not shown).
To have a more accurate account of these events, cell viability in liquid cultures was compared in both strains. Immediately after sporulation, cell viability was similar in tlc1Δ clb2Δ and tlc1Δ CLB2+ cells (Fig. 1A ). As cells progressed towards senescence, cell viability decreased at the same rate in both strains (Fig. 1A). In other words, there was no accelerated senescence of tlc1Δ clb2Δ compared with tlc1Δ CLB2+ cells. After crisis, which refers to the peak of maximal mortality, after which the so-called postsenescence survivors take over the culture of senescent cells and start to proliferate again at a normal rate, the tlc1Δ clb2Δ strain generated survivors, albeit less efficiently than the tlc1Δ CLB2+ strain. Thus, the emergence of tlc1Δ clb2Δ survivors was preceded by a more prolonged period of poor growth than with the tlc1Δ CLB2+ strain (Fig. 1A). Moreover, although the slopes of growth for both strains were initially parallel immediately after crisis, recovery was eventually much smaller in tlc1Δ clb2Δ than in tlc1Δ CLB2+ cells (Fig. 1A). Similar events were observed in est1Δ clb2Δ, est2Δ clb2Δ, and est3Δ clb2Δ mutants (data not shown).
These observations suggested that Clb2 might be involved in the initiation of homologous recombination. Indeed, homologous recombination is absolutely required for postsenescence survival, as tlc1Δ rad52Δ cells, in which all types of recombination are basically impaired due to the absence of RAD52, do not generate any survivors (43). In fact, there are two distinct pathways, both dependent on Rad52, by which such postsenescence survivors can arise. In one pathway, the subtelomeric Y′ sequences are amplified (type I survival), while the second pathway (type II survival) amplifies the terminal TG1-3 sequences (43, 56). In our strain background, as in most other strain backgrounds studied to date, tlc1Δ cells or other telomerase-negative cells generate a large majority (70 to 95% [90 to 95% in our strain background]) of type I survivors (11, 24, 39, 55, 56). Strikingly, in streak assays, the percentage of type I survivors was shifted from ∼90% in tlc1Δ CLB2+ cells to 100% in tlc1Δ clb2Δ cells (Fig. 1B). Most probably, the failure of tlc1Δ clb2Δ cells to generate type II survivors did not result from aggravated growth prior to recombination (Fig. 1A), as cdc13-1 tlc1Δ, cdc13-1 yku70Δ, yku70Δ tlc1Δ, and sgs1Δ tlc1Δ cells, all of which displayed poorer growth than tlc1Δ cells before and during crisis, nevertheless readily generated type II survivors (22, 34).
To document our finding that Clb2 played a role in the type II survivor pathway, we exploited the previous observation that conversion from type I to type II is readily seen in liquid culture, where the more rapidly growing type II survivors overtake the population (56). All of the tlc1Δ liquid cultures had adopted the type II character after ∼100 generations, while in contrast, all of the tlc1Δ clb2Δ cultures had maintained a clear type I character (Fig. 1C). These results confirm that Clb2 is involved in the determination of the type II survivor pathway and indicate that Clb2 is important for the transition from type I to type II.
Of the mitotic cyclin genes CLB1, -2, -3, and -4, clb2 deletion alone results in a significant cell cycle delay before mitosis. Clb2 levels represent around 40% of the total mitotic Clb level (14). The amount of kinase activity associated with the Cdc28/Clb2 complex alone is even larger than that associated with the other three complexes (25). To test the possibility that the defect of the tlc1Δ clb2Δ strain in survivor type selection, described above, was due to a loss of kinase activity associated with the Cdc28/Clb2 complex, we constructed the tlc1Δ cdc28-4 and tlc1Δ cdc28-13 double mutants. cdc28-4 and cdc28-13 are two temperature-sensitive mutant alleles of CDC28 that both cause defects in kinase activity but to different extents. cdc28-13 cells exhibit a strong kinase activity at 25°C and an average activity at 37°C, while cdc28-4 cells exhibit an average kinase activity at 25°C and a very weak kinase activity at 37°C (7, 51). Interestingly, at 32°C, but not at 25 and 30°C, tlc1Δ cdc28-4 cells grown in liquid culture maintained a type I character after ∼100 generations (Fig. 1D), similarly to tlc1Δ clb2Δ cells but in contrast to tlc1Δ cells (Fig. 1C). Meanwhile, tlc1Δ cdc28-13 cells maintained a clear type II character under the exact same conditions of growth, at either 25, 30, 32, or 34°C (tlc1Δ cdc28-4 cells could not grow at 34°C) (data not shown). Since the cdc28-4 allele does not affect Cdc28 and Clb2 abundance (1), these data suggest that even in the presence of normal levels of Clb2, the loss of associated Cdc28 kinase activity down to a certain level (attained in tlc1Δ cdc28-4 cells but not in tlc1Δ cdc28-13 cells) confers a defect in the selection of the survivor type in telomerase-negative cells.
Clb2 controls the Rad50/Rad59-dependent survivor pathway but is not essential for amplification of the telomeric TG1-3 sequences.
In telomerase-negative, otherwise wild-type cells, type I recombination relies on (besides Rad52) Rad51, Rad54, Rad55, and Rad57, while type II recombination requires (in addition to Rad52), Mre11, Rad50, Xrs2 (the MRX complex), and Sgs1 (reviewed in references 35 and 42). Thus, tlc1Δ rad51Δ mutants exclusively generate type II survivors, while tlc1Δ rad50Δ mutants exclusively generate type I survivors (39) (Fig. 2). To confirm the role of Clb2 in the selection of the recombination pathway in cells lacking telomerase, we constructed the tlc1Δ clb2Δ rad50Δ and tlc1Δ clb2Δ rad51Δ mutants and analyzed their survival rate in liquid cultures as well as their mode of postsenescence recombination following growth on agar plates. Interestingly, the tlc1Δ clb2Δ rad51Δ strain did not generate any survivor at all (Fig. 1A and 2, row d), while type I survivors arose from the tlc1Δ clb2Δ rad50Δ strain (Fig. 2, row f). The former situation was reminiscent of that in the tlc1Δ rad50Δ rad51Δ strain (39, 55) (Fig. 2, row i). Therefore, deleting CLB2 in telomerase-negative cells has the same effect on the selection of the telomeric recombination pathway as deleting RAD50. In both cases, the generation of type II survivors from senescing cells grown on plates is totally impaired.
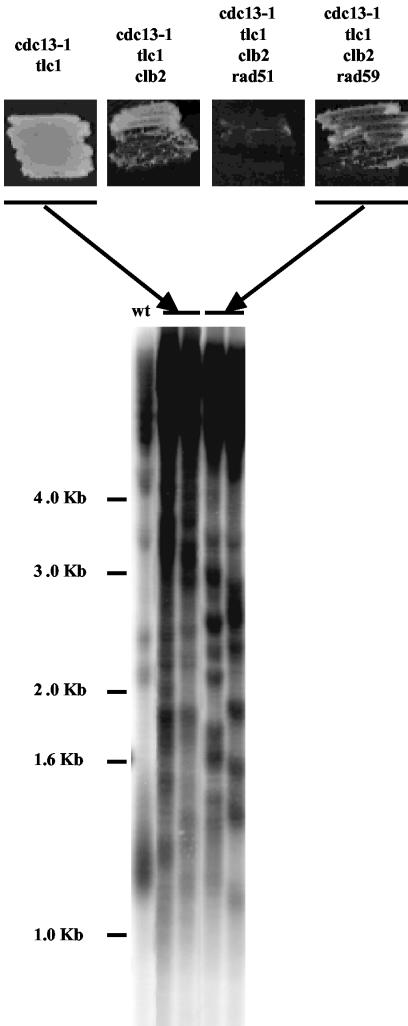
The data above indicated that, in either streak assays (Fig. 2) or standard liquid assays (Fig. 1A), telomeric homologous recombination could take place even in the absence of Clb2, but only on the subtelomeric Y′ sequences (type I survival). It was therefore possible that Clb2 was specifically required for recombination on the TG1-3 sequences. Alternatively, Clb2 could be necessary for operation of the Rad50/Rad59 pathway, independently of recombination per se. To distinguish between these two possibilities, we took advantage of the property that cdc13-1 tlc1Δ mutant cells have to generate 100% type II recombination (on TG1-3 sequences), which can be achieved either through the Rad50/Rad59-dependent pathway or, atypically, through the Rad51-dependent pathway (22). To find out whether clb2Δ mutants could amplify TG1-3 sequences by the Rad51 pathway, we constructed the cdc13-1 tlc1Δ clb2Δ rad59Δ mutant. This strain did generate survivors that exclusively amplified TG1-3 sequences (Fig. 3). Since in this strain the Rad50/Rad59 pathway had been genetically inactivated, these type II survivors presumably relied on the Rad51 pathway. This interpretation was confirmed by the finding that a cdc13-1 tlc1Δ clb2Δ rad51Δ quadruple mutant did not generate survivors at all (Fig. 3).
FIG. 3.
In type II survival, Clb2 is not required for recombination of TG1-3 sequences but rather is required for operation of the Rad50/Rad59 pathway of recombination. Cells of the relevant indicated genotype were restreaked on a YEPD plate immediately after sporulation, grown for 3 days at 25°C, and photographed. In all three viable strains, fast-growing colonies appeared on the first day following restreaking (not shown). The telomere structure (XhoI digestion, TG1-3 32P-labeled probe) is shown for two fast-growing colonies, representative of a total of 12, for two strains only, picked out individually at day 1. All of were type II recombinants (amplification of the TG1-3 sequences), as explained in the legend to Fig. 1B. On the plates shownhere, streaking approximately equal amounts of cells for all four strains allowed for the visualization of the much higher viability of the cdc13-1 tlc1Δ strain compared to the two viable clb2Δ strains.
These data demonstrate the possibility of recombination on TG1-3 sequences in the absence of Clb2. In such a situation, however, type II recombination (on the TG1-3 sequences) is accomplished by the Rad51 pathway rather than by the usual Rad50/Rad59 pathway. These data also strongly argue that the absence of survivors in tlc1Δ clb2Δ rad51Δ cells is due to the impairment of the Rad50 pathway in the absence of Clb2, rather than to an impairment of recombination per se.
Extremely rare survivors emerge from a tlc1Δ clb2Δ rad51Δ strain, but also from a tlc1Δ rad50Δ rad51Δ strain, when cell dilution during passages is reduced.
As seen above, no survivors were observed from cultures of the tlc1Δ clb2Δ rad51Δ or tlc1Δ rad50Δ rad51Δ strain grown on agar plates (Fig. 2) or from liquid cultures diluted every day during senescence under standard conditions (Fig. 1A). Standard dilutions used by several laboratories to analyze postsenescence survivors in liquid cultures consist of diluting cells to 105 per ml every day (11, 22, 39, 56). In four separate experiments, including the one shown in Fig. 1A, daily dilutions of 105 cells/ml did not allow survival of a single cell of the tlc1Δ clb2Δ rad51Δ strain. Incidentally, we noted that when the cell dilution was decreased (to 5 × 106 instead of 1 × 105 cells/ml, every 24 h), postsenescence survivors did emerge from the senescing tlc1Δ clb2Δ rad51Δ strain (Fig. 4, row d). Interestingly, postsenescence survivors also emerged from the senescing tlc1Δ rad50Δ rad51Δ strain (Fig. 4, row i). This was in contrast with previous data showing that, under the standard conditions used before, simultaneous inactivation of the Rad50/Rad59 and Rad51 pathways prevented the emergence of survivors in telomerase-negative cells (22, 39, 55) (Fig. 2).
From the detailed analysis of the growth characteristics of survivors from various strains illustrated in Fig. 4, we also noticed that postsenescence survivors were rarer in the tlc1Δ clb2Δ rad51Δ strain than in the tlc1Δ rad50Δ rad51Δ strain (Fig. 4, compare rows d and i). This suggested that Clb2 controlled both the Rad50 pathway and a third putative pathway. This overlapping function of Clb2 was confirmed by the finding that senescence took place more rapidly in tlc1Δ clb2Δ rad50Δ cells than in tlc1Δ rad50Δ cells (Fig. 4, compare rows e and g). A similar situation was found in the rad59Δ background (Fig. 4, compare rows g and h). In fact, experiments examining the viability of liquid cultures diluted under standard conditions (1 × 105 cells/ml every 24 h versus 5 × 106 cells/ml when the dilution is reduced, as in Fig. 4) also confirmed that Clb2 controlled a recombination pathway distinct from the Rad50 pathway (Fig. 5). In addition, under these conditions, the presumed accelerated senescence of the tlc1Δ clb2Δ rad50Δ and tlc1Δ clb2Δ rad59Δ cells appeared to correspond to a synthetic growth defect existing from the beginning (Fig. 5). In contrast, in actual accelerated senescence, observed in clb2Δ rad51Δ cells, for instance, as described previously (39), cell viability is high just after sporulation, at day 1 (Fig. 5). Incidentally, we note that clb2Δ rad50Δ but not clb2Δ rad59Δ cells are very sick (see below), thus explaining the low viability of the tlc1Δ clb2Δ rad50Δ strain but leaving unexplained that of the tlc1Δ clb2Δ rad59Δ strain (Fig. 5).
The third survival mechanism which functions in parallel with the Rad50 and Rad51 pathways is probably also based on homologous recombination, as tlc1Δ rad52Δ mutants did not generate any survivors at all even under these conditions of lower culture dilutions (Fig. 4, row j). Finally, the rare survivors emerging from the tlc1Δ rad50Δ rad51Δ and tlc1Δ clb2Δ rad51Δ strains were 100% type II (data not shown). Given that these rare survivors cannot be propagated on plates, it is not possible to know whether type I survivors exist at some time and are eventually overcome by type II survivors, as happens in liquid cultures (56). Alternatively, this third mechanism of recombination might, like the Rad50 pathway, exclusively generate type II survivors. Finally, but most importantly, we also noted that although the determination of type I was not affected by clb2Δ, as shown above, the efficiency of the Rad51-based recombination was nevertheless severely affected in the absence of Clb2 (Fig. 4, compare rows e and f and rows g and h).
Mus81, which potentially physically interacts with Clb2, is not involved in telomere elongation in the absence of telomerase.
Sgs1, a conserved DNA helicase of the RecQ family, has been recently implicated in the Rad50-dependent telomerase-independent pathway of telomere maintenance (12, 31, 34). Other studies have attributed to Sgs1 a potential role in the resolution of Holiday junctions that participate in the repair of damaged or stalled replication forks (reviewed in reference 28). Sgs1 and Mus81, a potential HJ resolvase, have been proposed to cooperate in these processes, and sgs1Δ mus81Δ double mutants are inviable (28, 47). Since Mus81 and Clb2 were found to physically interact by two-hybrid analysis (60) and since both Sgs1 and Clb2 have been implicated in the Rad50-dependent postsenescence survival pathway, as seen above, we wondered whether Mus81 might represent a link between Sgs1 and Clb2 in these processes. To answer this question, we constructed the tlc1Δ rad51Δ mus81Δ triple mutant and analyzed its behavior during and after senescence. Cells of that strain entered senescence at times similar to those for the tlc1Δ single and tlc1Δ rad51Δ double mutants. Moreover, the tlc1Δ rad51Δ mus81Δ strain generated normal quantities of postsenescence survivors on agar plates (data not shown). This is in contrast with the tlc1Δ clb2Δ rad51Δ strain, which did not generate survivors at all when grown on agar plates, as described above.
We conclude that the potential physical interaction between Clb2 and Mus81 (60) is probably not relevant to the role of Clb2 in senescence survival. Likewise, the functional interaction existing between Sgs1 and Mus81 (reviewed in reference 28) is probably not relevant to the participation of Sgs1 in telomerase-independent telomere maintenance.
In telomerase-positive cells, a clb2 null mutation aggravates the slow-growth phenotype associated with an mrx (mre11, rad50, or xrs2) null mutation, a characteristic linked to loss of Cdc28-associated kinase activity.
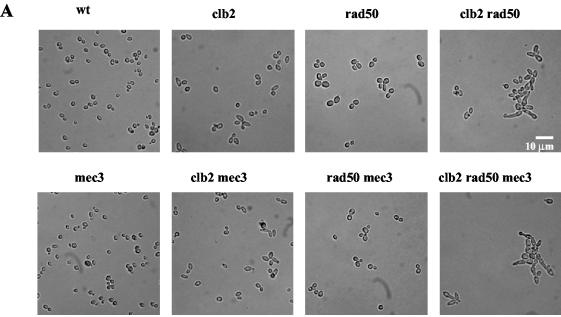
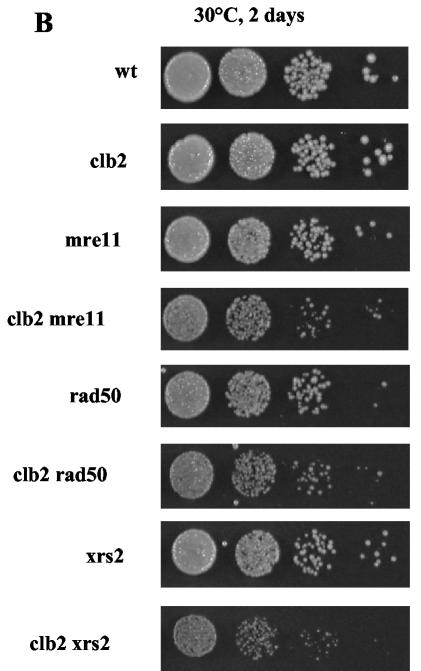
While performing the experiments described above, we noted that the tlc1Δ clb2Δ rad50Δ strain, unlike the other strains used in parallel, exhibited a growth defect well before the onset of senescence (Fig. 1A). Both the clb2Δ and rad50Δ single mutants exhibited morphological defects (Fig. 6A), as previously reported. Noticeably, rad50Δ-induced morphological defects were suppressed following inactivation of the DNA damage checkpoint (in the mec3Δ background), while those associated with the clb2Δ mutation were not (Fig. 6A). Despite its morphological defects, the clb2Δ strain was not affected in terms of colony formation compared with the wild type (Fig. 6B). On the other hand, the mre11Δ, rad50Δ, and xrs2Δ strains were slightly impaired in colony formation in comparison with a wild-type isogenic strain (Fig. 6B). When an mrxΔ mutation was combined with the clb2Δ mutation (Fig. 6B), growth was impaired in comparison with that in either single mutant.
FIG. 6.
In telomerase-positive cells, the slow-growth phenotype of mre11Δ, rad50Δ, and xrs2Δ mutants is aggravated by deletion of CLB2. (A) Morphology of the clb2Δ, rad50Δ, and clb2Δ rad50Δ strains compared with that of an isogenic wild-type (wt) strain in the presence (top row) or absence, in the mec3Δ background (bottom row), of a functional DNA damage checkpoint. (B) Growth characteristics of various mutants combining mutations in the CLB2 and MRX genes (see text for explanations). Tenfold serial dilutions (from left to right in each row) of cultures were grown under the indicated conditions and photographed.
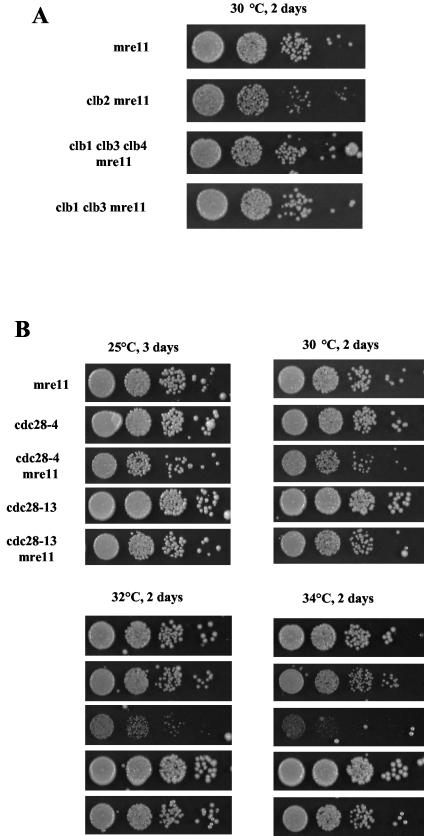
S. cerevisiae cells possess six CLB genes which function in pairs, i.e., Clb1-Clb2, Clb3-Clb4, and Clb5-Clb6, with each pair representing duplicated genes. The CLB genes assume specific, but overlapping and sometimes redundant, functions. The Clb1-Clb2 and Clb3-Clb4 pairs have been implicated in the triggering and control of mitosis, while the Clb5-Clb6 pair has been implicated in triggering DNA replication (see references in reference 14). A clb5Δ mre11Δ double mutant strain exhibited growth properties similar to those of the mre11Δ strain (data not shown). To determine whether loss of function of Clb2 was specifically responsible for the severe growth defect of the clb2Δ mrxΔ strains, we set out to inactivate mitotic CLB genes other than CLB2 in the mre11Δ background. Previous analyses of the consequences of the deletion of the different CLB genes indicate that CLB2 is the more important one, as all multiple-deletion combinations that are lethal or semilethal include clb2Δ. Therefore, we chose to simultaneously introduce into an mre11Δ strain mutations in two CLB genes other than CLB2. The clb1Δ clb3Δ mre11Δ and clb1Δ clb3Δ clb4Δ mre11Δ mutant strains were slightly more impaired in their growth characteristics than the mre11Δ strain but were less impaired than the clb2Δ mre11Δ strain (Fig. 7A).
FIG. 7.
In mre11Δ cells, simultaneous loss of CLB1, CLB3, and CLB4 (A), as well as loss of Cdc28 kinase activity, mimics loss of CLB2 (B). Tenfold serial dilutions (from left to right in each row) of cultures of the mutant strains of the indicated relevant genotypes were grown on YEPD plates under the indicated conditions and photographed.
To test the possibility that the severe growth defect of the clb2Δ mrxΔ strains was due to a loss of kinase activity associated with the Cdc28/Clb2 complex, we constructed the cdc28-4 mre11Δ and cdc28-13 mre11Δ double mutants. As seen above, both the temperature-sensitive cdc28-4 and cdc28-13 mutant alleles are defective in kinase activity but to different extents. cdc28-13 cells exhibit a strong kinase activity at 25°C and an average activity at 37°C, while cdc28-4 cells exhibit an average kinase activity at 25°C and a very weak kinase activity at 37°C (7, 51). Interestingly, we found that cdc28-13 mre11Δ cells exhibited a slightly more severe growth defect than mre11Δ cells at either 25, 30, 32, or 34°C, while cdc28-4 mre11Δ cells clearly exhibited a much more severe growth defect than mre11Δ cells even at 30°C (Fig. 7B). Since the cdc28-4 allele does not affect Cdc28 and Clb2 abundance (1), the present result suggests that even in the presence of normal levels of Clb2, the loss of associated Cdc28 kinase activity confers a growth defect when combined with the mre11 null mutation.
The defect in clb2Δ mre11Δ is independent of Mre11 nuclease activity but depends on the telomere maintenance and DNA repair functions of Mre11.
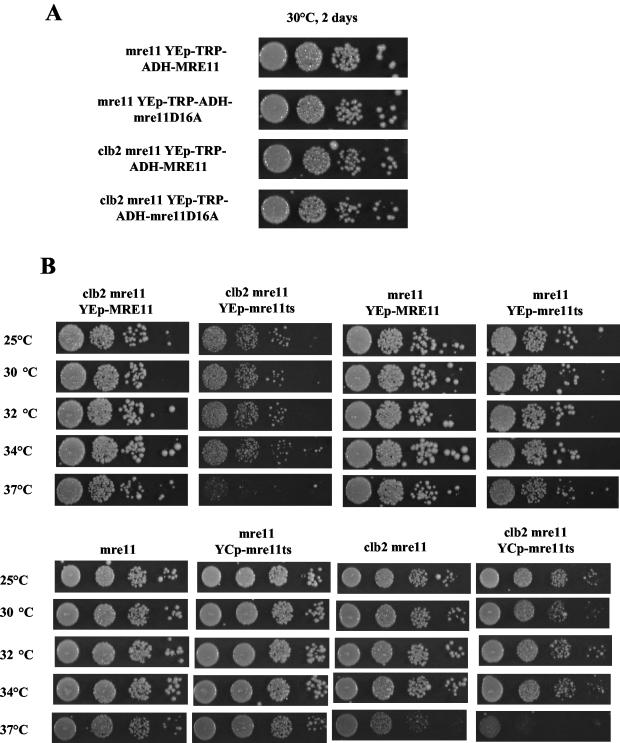
clb2Δ mre11Δ cells expressing the mre11D16A mutant allele, which is completely defective in nuclease activity (21), were not impaired in growth in comparison with clb2Δ mre11Δ cells expressing wild-type MRE11 (Fig. 8A). In contrast, clb2Δ mre11Δ cells expressing the mre11(ts) allele, which confers a defect in telomere maintenance at 25 to 37°C and a defect in DNA repair at 34 or 37°C only (10), still exhibited a growth defect at 25 or 30°C (Fig. 8B). This defect was much aggravated at 34 and 37°C (Fig. 8B). Therefore, defects in both the telomeric and DNA repair functions of the MRX complex appear to be responsible for the synthetic interaction between clb2Δ and mre11Δ. Noticeably, at 25 to 37°C the severity of the mre11(ts)-induced effect, in the clb2Δ background, depended on its level of expression (Fig. 8B). This stronger effect of an episomal over a centromeric plasmid can be called a dominant negative effect. This effect was specific for the interaction with clb2Δ, however, as mre11Δ YEp-mre11(ts) cells were only slightly impaired in growth compared with mre11Δ YEp-MRE11 cells (Fig. 8B).
FIG. 8.
Growth defects in clb2Δ cells expressing mre11D16A or mre11(ts). (A) Loss of Mre11 nuclease activity (conferred by the mre11D16A allele [21] expressed under the control of the conditional GAL1 promoter) in mre11Δ cells is not responsible for the synthetic interaction with clb2Δ. Tenfold serial dilutions (from left to right in each row) of mutants with the indicated relevant genotypes were grown for the indicated period of time at the indicated temperature. (B) Expression of the mre11(ts) allele, which confers a telomeric defect at 25 to 37°C and a DNA repair defect at 34 to 37°C only (10), confers a growth defect in the clb2Δ background at 25 to 37°C. The defect is aggravated at 34 to 37°C (see text for explanations). The mre11(ts) allele was expressed either from an episomal (high-copy-number) (YEp) or centromeric (one or two copies) (YCp) plasmid in an mre11 null background. Growth was for 2 days at the indicated temperatures.
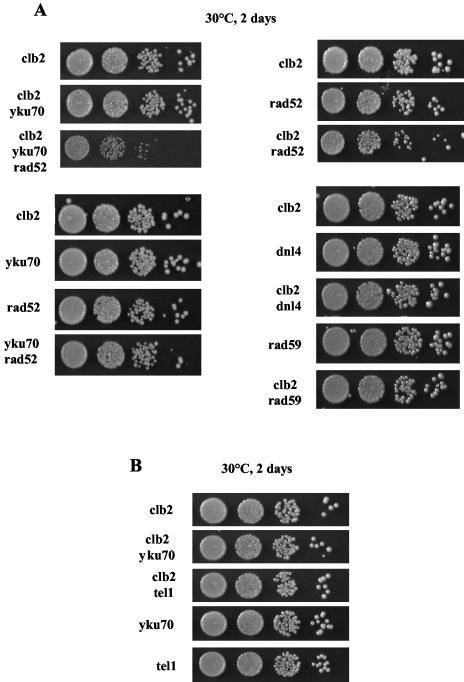
In S. cerevisiae, the DNA repair functions of the MRX complex are in both the nonhomologous end-joining (NHEJ) and homologous recombination pathways (reviewed in reference 26). If loss of the MRX complex's DNA repair functions was responsible for the synthetic growth defect observed in clb2Δ mrxΔ mutants, then this should be mimicked by inhibiting NHEJ or homologous recombination. Since a null mutation in YKU70, a gene essential for NHEJ, also interacts with telomeric functions, we also used dnl4Δ to inhibit NHEJ. Indeed, DNL4, which codes for a DNA ligase, ligase IV, which is essential for NHEJ (57), has no known role in telomere maintenance. On the other hand, Rad52 (which has no known role in telomere maintenance in telomerase-positive cells) is essential for all types of homologous recombination (reviewed in reference 50). Interestingly, we found that clb2Δ dnl4Δ cells had no aggravated phenotype compared with clb2Δ cells (Fig. 9A), thus confirming, along with the growth characteristics of clb2Δ yku70Δ (Fig. 9A), that loss of NHEJ is not responsible for the growth defect in clb2Δ mrxΔ cells. In contrast, growth of clb2Δ rad52Δ cells was affected compared with that of each single mutant (Fig. 9A); however, the defect was weaker than that resulting from the association of the clb2Δ and mrxΔ mutations (Fig. 6B). Interestingly, growth of the clb2Δ yku70Δ rad52Δ triple mutant was more affected than that of the clb2Δ rad52Δ double mutant (Fig. 9A), thus highlighting a latent defect conferred by the yku70Δ mutation in the clb2Δ background. Rad59 is in the same pathway of telomeric recombination as Rad50 but has no major role in nontelomeric DNA repair (reviewed in reference 50). Interestingly, a clb2Δ rad59Δ strain exhibited no aggravated phenotype compared with either single mutant (Fig. 9A).
FIG. 9.
(A) In telomerase-positive cells, the growth phenotype of clb2 null cells is aggravated by loss of DNA repair functions (see text for explanations). The growth characteristics of various mutants with the indicated relevant genotypes are shown as 10-fold serial dilutions (from left to right in each row). (B) Comparison of growth characteristics in clb2Δ cells also bearing a telomere-shortening mutation, yku70Δ or tel1Δ. The mode of representation is as in panel A.
Like mre11Δ, rad50Δ, and xrs2Δ mutant cells, yku70Δ and tel1Δ mutant cells exhibit telomere shortening (5, 37, 45). Unlike clb2Δ mre11Δ, clb2Δ rad50Δ, and clb2Δ xrs2Δ cells (Fig. 6B), however, clb2Δ yku70Δ and clb2Δ tel1Δ cells exhibited no aggravated growth defect (Fig. 9B). Therefore, the defect of the MRX complex in telomere maintenance that is responsible for the genetic interaction with clb2Δ does not implicate telomere length regulation per se.
DISCUSSION
A novel pathway of telomeric recombination functions in parallel with the Rad50 and Rad51 pathways.
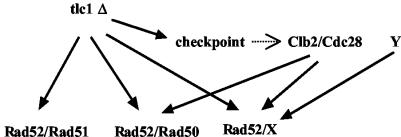
tlc1Δ rad50Δ rad51Δ cells have been previously reported to die without generating postsenescence survivors in several genetic backgrounds (39, 55), including ours (24). Those experiments were based either on streak assays, in which senescing cells are grown on agar-based media, or on liquid culture assays, in which cells were diluted down to 105 cells/ml every day (11, 39, 55, 56). We find here that liquid cultures of tlc1Δ rad50Δ rad51Δ cells can generate survivors, albeit at an extremely low rate, when diluted down to 5 × 106 cells/ml every day. The finding that postsenescence survivors could emerge from a tlc1Δ clb2Δ rad51Δ strain at an even lower rate strongly suggests that Clb2 functions in this Rad50-, Rad51-independent pathway, as well as in the Rad50 pathway (Fig. 10). This third pathway must also be controlled by an additional, unknown, factor distinct from Clb2, which is responsible for the emergence of extremely rare survivors of tlc1Δ clb2Δ rad51Δ strain under decreased culture dilutions (Fig. 4). This novel pathway, also based on homologous recombination (there were no survivors in a tlc1Δ rad52Δ strain), could operate recombination on the TG1-3 sequences, independently of the Rad50 pathway. However, the conditions of the experiments, which necessitated cultivation of the cells in liquid cultures, cannot rule out the existence, at earlier stages, of type I recombinants that would eventually be overcome by type II recombinants. Loss of Cdc28 kinase activity mimicked loss of CLB2 function in determining the choice for the survival type in cells lacking telomerase (Fig. 1D). This was expected, since Cdk1 (Cdc28) is known to be active only when in complex with a cyclin, and consequently, any function of Clb2, including here in telomeric recombination, most probably implicates Cdc28 kinase activity.
FIG. 10.
Postsenescence survival in telomerase-deficient (tlc1Δ) budding yeast cells is controlled by three distinct Rad52-based homologous recombination pathways. When both the Rad50 pathway (which amplifies the telomeric TG1-3 sequences) and the Rad51 pathway (which amplifies the subtelomeric Y′ sequences) have been genetically inactivated, there remains a third pathway, operating with a much lower efficiency than the other two, which exclusively amplifies the TG1-3 sequences. The lower number of survivors in tlc1Δ clb2Δ rad51Δ cells than in tlc1Δ rad50Δ rad51Δ cells suggests that Clb2 controls both the Rad50 pathway and the third, unknown (X) pathway. The fact that there are still survivors in tlc1Δ clb2Δ rad51Δ cells but none in tlc1Δ rad52Δ cells suggests that another component, Y (unknown), controls pathway X. On the other hand, it is also clear that the Rad50 pathway is more sensitive than the Rad51 pathway to the absence of Clb2, as indicated, for instance, by the absence of survivors in the tlc1Δ clb2Δ rad51Δ strain grown in liquid cultures under standard conditions (Fig. 1A) or on plates (Fig. 2). However, the Rad51 pathway, although not absolutely requiring Clb2 for telomeric recombination (tlc1Δ clb2Δ rad50Δ cells can generate survivors under standard conditions [Fig. 1A and 2]), functioned less efficiently in the absence of Clb2 (Fig. 1A and 5). The DNA damage checkpoint, which is activated in response to telomerase loss (see references in the text), represents a possible link with Cdc28/Clb2.
The central defect of telomerase-deficient clb2Δ cells affected the emergence of postsenescence survivors, rather than the kinetics of senescence itself. Indeed, unlike tlc1Δ rad51Δ (39) (Fig. 1A), tlc1Δ cdc13-1 and tlc1Δ yku70Δ (49), and tlc1Δ sgs1Δ (34) mutants, tlc1Δ clb2Δ mutants did not exhibit accelerated senescence (Fig. 1A). Although the present experiments showed that the Rad50/Rad59 pathway, but not the Rad51 pathway, was blocked in the absence of Clb2, it is nevertheless clear that clb2Δ also affected Rad51-based recombination. Indeed, in all strains lacking Clb2, recovery was poorer than in the corresponding CLB2+ strains, presumably reflecting less efficient recombinational repair of damaged telomeres (Fig. 5). We also note that accelerated senescence was clearly associated with the clb2Δ mutation only in the rad51Δ background (Fig. 5) but that rad51Δ also induced accelerated senescence on its own (Fig. 5), as reported before (39). The apparent accelerated senescence in tlc1Δ clb2Δ rad50Δ and tlc1Δ clb2Δ rad59Δ mutants (Fig. 4) more likely corresponds to the low viability of these two strains prior to senescence (Fig. 5). However, the exact reason for the low viability of the tlc1Δ clb2Δ rad59Δ strain is unknown, while that of the tlc1Δ clb2Δ rad50Δ could be explained by the synthetic growth defect between clb2Δ and rad50Δ (see below). The exact reasons for the accelerated senescence of the tlc1Δ rad51Δ (39) and tlc1Δ sgs1Δ strains (34) remain obscure. On the other hand, inactivation of one of two telomere protection pathways complementary to the telomerase pathway, namely, Cdc13 and Yku, more readily explains the accelerated senescence of the tlc1Δ cdc13-1 and tlc1Δ yku70Δ strains (49). Our present limited knowledge about Rad50-based telomeric recombinational repair and about its peculiarities compared with the better-studied Rad51-based pathway (54) leaves huge gaps to be filled in.
Based on the impact of the clb2Δ mutation on the kinetics of senescence and recovery, discussed above, it is more probable that the absence of Clb2 directly affects the homologous recombination repair pathway rather than that Clb2 is involved in resolving structural changes associated with telomerase loss. Interestingly, clb2Δ cells were not deficient in recombination on the TG1-3 sequences but, rather, were deficient in operation of the Rad50-dependent pathway. Indeed, in the cdc13-1 tlc1Δ clb2Δ rad59Δ quadruple mutant, for instance, survivors could still amplify the TG1-3 sequences. Type II recombination in these cells relied on the Rad51 pathway, a recently demonstrated possibility (22). An interesting hypothesis is that during homologous recombination, Cdc28/Clb2-associated kinase activity is required for phosphorylation of one or several members of the MRX complex but not for that of a member of the Rad51 pathway. Even if in the absence of Clb2, Clb1-, Clb3-, and Clb4-associated Cdc28 activities take over, these might not be high enough or specific enough to perform the needed functions of phosphorylation of the Rad50 recombination complex. However, in view of recent results, Tel1 and Mec1 (two ATM-ATR-related kinases) rather than Cdc28 appear to be able to phosphorylate members of the MRX complex, at least during the intra-S-phase checkpoint (reviewed in reference 15). Interestingly, the survivors from tlc1Δ tel1Δ and tlc1Δ mec1Δ strains were recently found to be 100% type I, suggesting that Tel1 and Mec1 might mediate the generation of type II survivors, perhaps through direct control of the Rad50 pathway (58). It is now well established that telomerase loss activates a DNA damage response, which is globally similar to that induced by other types of damage but also seems to be specific for some of its components (48). Although the exact composition of the checkpoint response to telomerase loss is not known in detail yet, it nevertheless appears that Mec1, but not Tel1, is involved in activating the checkpoint machinery (19, 32). On the other hand, it has been known for a long time that one of the major functions of the checkpoint network is to affect the cell cycle machinery. We therefore propose that telomerase loss activates a DNA damage response that might directly affect recombination proteins, as suggested before (58), or might indirectly act on the selection of the recombinational pathway via the Cdc28/Clb2 complex, as shown here (Fig. 10).
Break-induced replication (BIR) represents the likely mechanism for telomere elongation in the absence of telomerase (4, 39). During BIR, only one end around a double-strand break can find homologies with sequences located at other places within the genome, thus resulting in copying of the invaded chromosome by replication all the way to its end (reviewed in reference 38). After invasion, the BIR intermediates resemble Holiday junctions, structures believed to reinitiate replication from stalled replication forks (reviewed in references 27 and 28). Mus81 appears to play a crucial role in these processes. On the basis of the two-hybrid interaction between Clb2 and Mus81 (60), the functional interaction between Mus81 and Sgs1 (47), and the implication of Sgs1 in type II recombination (12, 31, 34), it was tempting to speculate on a role for Clb2 in telomeric recombination in cooperation with Mus81 and Sgs1. However, our experiments with mus81 mutant cells render this possibility unlikely. In addition, Clb2-myc and Mus81-hemagglutinin did not coimmunoprecipitate in extracts made from cells in which they had been overexpressed (unpublished data).
Finally, it is interesting that in the yeast Schizosaccharomyces pombe, Cdc2/cyclin B activity has recently been implicated in indirectly regulating recombinational repair of radiation-induced double-strand breaks (8). It is therefore tempting to speculate that the homologues of S. cerevisiae Clb2 in humans, i.e., cyclins B1 and B2, play a similar role in ALT, the telomerase-independent pathway of telomere maintenance in tumor cells.
Cdc28/Clb2 cooperates with MRX for optimal functioning of a DNA repair event in telomerase-positive cells.
The mre11Δ, rad50Δ, and xrs2Δ mutants exhibit a slow-growth phenotype, the origin of which, due to the numerous functions of the MRX complex, is not known with certainty (reviewed in reference 15). rad50Δ mec3Δ cells, which are defective in the DNA damage checkpoint, recovered wild-type growth (Fig. 6A). We note that the growth defect in clb2Δ xrs2Δ cells was more severe than that in clb2Δ mre11Δ and clb2Δ rad50Δ cells (Fig. 6B). This was unexpected, as these three proteins function in the same complex and deletion of either one indeed conferred identical phenotypes (reviewed in references 15 and 26). However, it should be noted that Xrs2 possesses a forkhead-associated (FHA) domain that is not implicated in the interaction with Mre11 and that, in addition, Xrs2 does not appear to directly interact with Rad50 (reviewed in reference 15). This FHA domain could possibly mediate a physical interaction with a protein distinct from its usual partners in the MRX complex. In the clb2Δ xrs2Δ mutant, loss of the function of a protein binding the FHA domain of the Xrs2 protein not participating in the MRX complex would take place in addition to that of Clb2 and MRX. It should be noted that in this hypothesis, Cdc28/Clb2 is likely to also have functional interactions with the Xrs2 FHA domain-bound protein.
Based on previous data (10), our experiments using the mre11(ts) mutant allele suggest that the defect of clb2Δ mrxΔ cells has a telomeric origin. Diede and Gottschling (17) have recently provided evidence that in cells held in M phase by nocodazole treatment, Rad50 is required for telomere addition by telomerase. However, those authors also noted that even in the absence of Rad50, telomere addition still took place when cells were allowed to cycle (17). Although this was contradicted by other data (59), Diede and Gottschling suggested that the damage in rad50Δ cells might result from inefficient loading of Cdc13 onto ill-processed telomere ends that would in turn create unprotected DNA ends (17). Given the synthetic lethality between rad50Δ and clb2Δ observed here, it is probable that Cdc28/Clb2 is doing more than just sustaining cell cycle-dependent mechanisms leading to activation of Rad50 prior to, or during, mitosis. Since Clb2 also acts primarily during G2/M (14), it is probable that the target of Cdc28/Clb2 other than MRX, which is involved in the synthetic interaction observed here, also functions during G2/M. However, the multiplicity of targets of Cdc28/Clb2 makes the identification of this target improbable at this time.
Although our experiments with mre11(ts) suggest a telomeric origin for the synthetic defect in clb2Δ mrxΔ strains, it should be noted that the effect at 25 to 32°C was relatively slight (Fig. 8B). On the other hand, at 34 and 37°C the synthetic growth defect between the clb2Δ and mrxΔ mutations was much more severe, thus highlighting the implication of DNA repair functions of the MRX complex (10). Loss of homologous recombination repair functions was aggravated by loss of NHEJ, and loss of both functions mimicked the growth defect of clb2Δ mrxΔ cells (Fig. 9A). Therefore, some DNA damage must be created (or becomes apparent) when Clb2 is absent or Cdc28 kinase activity is lowered (Fig. 6B and 7B). We do not yet know whether the damaged DNA that failed to be repaired in clb2Δ yku70Δ rad52Δ cells was the same as the DNA that failed to be repaired in clb2Δ mrxΔ cells. That a DNA repair function of MRX is involved in the interaction with Clb2 is also based on the finding that the growth defect of clb2Δ mre11Δ cells could be rescued by overexpression of EXO1 (unpublished data). Interestingly, Lewis et al. (41) recently found that overexpression of EXO1 rescued the sensitivity of rad50, mre11, and xrs2 mutants to DNA damage. Although the exonuclease activity of Exo1 was required for this rescue (41), it is generally accepted that Mre11 nuclease activity is not required for its DNA repair function in mitotic cells (6, 46, 61). This would explain the fact that the defect we saw here in clb2Δ mrxΔ cells was clearly independent of Mre11 nuclease activity. Importantly, overexpression of TLC1 also rescued the DNA damage sensitivity of rad50, mre11, and xrs2 mutants (41). Those authors suggested that telomerase might have affinity for damaged DNA, competitively with MRX, independent of its catalytic activity (41).
In conclusion, the present study allowed us to identify a third, novel, telomere maintenance pathway controlled by the yeast mitotic Cdk/cyclin complex, Cdc28/Clb2, in the absence of telomerase. Future experiments will aim at determining whether the DNA damage checkpoint represents an intermediate between telomerase loss and activation of Cdc28/Clb2-induced telomeric recombination, as well as the nature of the target of the Cdk/cyclin complex that regulates telomeric recombination. Meanwhile, it will also be important to determine the nature of the functional interactions, uncovered here, between the ubiquitous Mre11/Rad50/Xrs2 and Cdc28/Clb2 complexes, as the MRX complex plays an as-yet-unidentified role in telomerase loading.
FIG. 1—Continued.
Acknowledgments
We thank W. Xiao, K. Ohta, S. I. Reed, T. Petes, J. Haber, and D. Gottschling for gifts of strains and plasmids.
This work was supported by grants from the Association pour la Recherche contre le Cancer and the Comité Départemental de la Savoie de la Ligue Nationale contre le Cancer.
REFERENCES
- 1.Ahn, S. H., B. T. Tobe, J. N. FitzGerald, S. L. Anderson, A. Acurio, and S. J. Kron. 2001. Enhanced cell polarity in mutants of the budding yeast cyclin-dependent kinase Cdc28p. Mol. Biol. Cell. 12:3589-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 3.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 4.Bosco, G., and J. E. Haber. 1998. Chromosome break-induced DNA replication leads to non-reciprocal translocations and telomere capture. Genetics 150:1037-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent nonhomologous end-joining pathways are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bressan, D. A., B. K. Baxter, and J. H. Petrini. 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7681-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calzada, A., M. Sanchez, E. Sanchez, and A. Bueno. 2000. The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J. Biol. Chem. 275:9734-9741. [DOI] [PubMed] [Google Scholar]
- 8.Caspari, T., J. M. Murray, and A. M. Carr. 2002. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cervantes, R. B., and V. Lundblad. 2002. Mechanisms of chromosome-end protection. Curr. Opin. Cell Biol. 14:351-356. [DOI] [PubMed] [Google Scholar]
- 10.Chamankhah, M., T. Fontanie, and W. Xiao. 2000. The Saccharomyces cerevisiae mre11 (ts) allele confers a separation of DNA repair and telomere maintenance functions. Genetics 155:569-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, H., and D. A. Sinclair. 2001. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. USA 98:3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins, K., and J. R. Mitchell. 2002. Telomerase in the human organism. Oncogene 21:564-579. [DOI] [PubMed] [Google Scholar]
- 14.Cross, F. R., V. Archambault, M. Miller, and M. Klovstad. 2002. Testing a mathematical model of the yeast cell cycle. Mol. Biol. Cell 13:52-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Amours, D., and S. P. Jackson. 2002. The MRE11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 16.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 17.Diede, S. J., and D. E. Gottschling. 2001. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 11:1336-1340. [DOI] [PubMed] [Google Scholar]
- 18.Dubrana, K., S. Perrod, and S. M. Gasser. 2001. Turning telomeres on and off. Curr. Opin. Cell Biol. 13:281-289. [DOI] [PubMed] [Google Scholar]
- 19.Enomoto, S., L. Glowczewski, and J. Berman. 2002. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell. 13:2626-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, S. K., and V. Lundblad. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113:3357-3364. [DOI] [PubMed] [Google Scholar]
- 21.Furuse, M., Y. Nagase, H. Tsubouchi, K. Murakami-Murofushi, T. Shibata, and K. Ohta. 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17:6412-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grandin, N., and M. Charbonneau. 2003. The Rad51 pathway of telomerase-independent maintenance of telomeres can amplify TG1-3 sequences in yku and cdc13 mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 23:3721-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grandin, N., C. Damon, and M. Charbonneau. 2000. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 20:8397-8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grandin, N., C. Damon, and M. Charbonneau. 2001. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 20:6127-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandin, N., and S. I. Reed. 1993. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol. Cell. Biol. 13:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95:583-586. [DOI] [PubMed] [Google Scholar]
- 27.Haber, J. E. 2000. Partners and pathways: repairing a double-strand break. Trends Genet. 16:259-264. [DOI] [PubMed] [Google Scholar]
- 28.Haber, J. E., and W. D. Heyer. 2001. The fuss about Mus81. Cell 107:551-554. [DOI] [PubMed] [Google Scholar]
- 29.Hayflick, L. 1980. Cell aging. Annu. Rev. Geront. Geriatr. 1:26-67. [Google Scholar]
- 30.Henson, J. D., A. A. Neumann, T. R. Yeager, and R. R. Reddel. 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21:598-610. [DOI] [PubMed] [Google Scholar]
- 31.Huang, P. H., F. E. Pryde, D. Lester, R. L. Maddison, R. H. Borts, I. D. Hickson, and E. J. Louis. 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11:125-129. [DOI] [PubMed] [Google Scholar]
- 32.Ijpma, A. S., and C. W. Greider. 2003. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14:987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, F. B., R. A. Marciniak, and L. Guarente. 1998. Telomeres, the nucleolus and aging. Curr. Opin. Cell Biol. 10:332-338. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, F. B., R. A. Marciniak, M. McVey, S. A. Stewart, W. C. Hahn, and L. Guarente. 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20:905-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kass-Eisler, A., and C. W. Greider. 2000. Recombination in telomere-length maintenance. Trends Biol. Sci. 25:200-204. [DOI] [PubMed] [Google Scholar]
- 36.Kelleher, C., M. T. Teixeira, K. Förstemann, and J. Lingner. 2002. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27:572-579. [DOI] [PubMed] [Google Scholar]
- 37.Kironmai, K. M., and K. Muniyappa. 1997. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells 2:443-455. [DOI] [PubMed] [Google Scholar]
- 38.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lendvay, T. S., D. K. Morris, J. Sah, B. Balasubramanian, and V. Lundblad. 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis, L. K., G. Karthikeyan, J. W. Westmoreland, and M. A. Resnick. 2002. Differential suppression of DNA repair deficiencies of yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 160:49-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundblad, V. 2002. Telomere maintenance without telomerase. Oncogene 21:522-531. [DOI] [PubMed] [Google Scholar]
- 43.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 44.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 45.Lustig, A. J., and T. D. Petes. 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 83:1398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mullen, J. R., V. Kaliraman, S. S. Ibrahim, and S. J. Brill. 2001. Requirement for three novel protein complexes in the absence of Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157:103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nautiyal, S., J. L. DeRisi, and E. H. Blackburn. 2002. The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:9316-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger, J. K. Moore, J. E. Haber, and V. Lundblad. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8:657-660. [DOI] [PubMed] [Google Scholar]
- 50.PÂques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reed, S. I., J. A. Hadwiger, and A. T. Lorincz. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. USA 82:4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson, H. E., M. Henze, K. Sugimoto, and S. I. Reed. 1992. Cyclin B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 6:2021-2034. [DOI] [PubMed] [Google Scholar]
- 53.Shore, D. 2001. Telomeric chromatin: replicating and wrapping up chromosome ends. Curr. Opin. Genet. Develop. 11:189-198. [DOI] [PubMed] [Google Scholar]
- 54.Signon, L., A. Malkova, M. L. Naylor, H. Klein, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif1-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 56.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teo, S. H., and S. P. Jackson. 1997. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 2:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai, Y. L., S. F. Tseng, S. H. Chang, C. C. Lin, and S. C. Teng. 2002. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 22:5679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsukamoto, Y., A. K. P. Taggart, and V. A. Zakian. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11:1328-1335. [DOI] [PubMed] [Google Scholar]
- 60.Uetz, P., et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 61.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 62.Vaziri, H., and S. Benchimol. 1998. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol. 8:279-282. [DOI] [PubMed] [Google Scholar]