Abstract
In mammals, the three classical ras genes encode four highly homologous proteins, N-Ras, H-Ras, and the isoforms K-Ras 4A and 4B. Previous studies have shown that K-ras is essential for mouse development and that while K-ras 4A and 4B are expressed during development, K-ras 4A expression is regulated temporally and spatially and occurs in adult kidney, intestine, stomach, and liver. In the present study, the pattern of K-ras 4A expression was examined in a wide range of wild-type adult mouse tissues, and gene targeting was used to generate K-ras 4A-deficient mice to examine its role in development. It was found that K-ras 4A is also expressed in uterus, lung, pancreas, salivary glands, seminal vesicles, bone marrow cells, and cecum, where it was the major K-Ras isoform expressed. Mating between K-rastmΔ4A/+ mice produced viable K-rastmΔ4A/tmΔ4A offspring with the expected Mendelian ratios of inheritance, and these mice expressed the K-ras 4B splice variant only. K-rastmΔ4A/tmΔ4A mice were fertile and showed no histopathological abnormalities on inbred (129/Ola) or crossbred (129/Ola × C57BL/6) genetic backgrounds. The results demonstrate that K-Ras 4A, like H- and N-Ras, is dispensable for normal mouse development, at least in the presence of functional K-Ras 4B.
The three classical mammalian ras genes, K-, N-, and H-ras, encode proteins that are members of the guanine nucleotide binding protein superfamily (8). Ras proteins are small (≈21 kDa) GTPases which cycle between inactive (GDP-bound) and active (GTP-bound) conformations at the plasma membrane, by interaction with a variety of guanine nucleotide exchange factors and GTPase activating proteins in response to stimulation by a diverse array of cell surface receptors, including the epidermal growth factor receptor and cytokine receptors such as interleukin-2 (14). Following activation, Ras proteins bind and activate a plethora of downstream effector proteins, including Raf kinases and phosphatidylinositol 3-kinases, and by this means control many cellular functions, including proliferation, differentiation, migration, and apoptosis (4, 11, 28).
Activating point mutations leading to constitutive activation of the Ras proteins are associated with some 30% of all human malignancies (1), and these mutations render Ras insensitive to the regulatory action of GTPase activating proteins, leading to excessive and inappropriate signaling, resulting in the promotion of cellular transformation. However, while mutationally active ras genes are generally believed to act as dominant oncogenes, recent reports indicate that wild-type K- and N-Ras proteins have tumor suppressor activity (7, 32). Thus, K- and N-ras appear to exert a dual function in that they promote cancer development as gain-of-function oncogenes when mutated and inhibit cancer by tumor suppressor activity when wild type (proto-oncogene).
The high degree of homology between Ras proteins suggests that functional redundancy may exist among these proteins, yet mounting evidence exists for unique roles for the ras gene family members. For example, most human cancers with ras activating mutations are associated exclusively with a particular ras gene, suggesting tissue-specific activity: K-ras mutations are prevalent in lung (≥30%), colon (≥40%), and pancreatic (≥90%) cancers, while H-ras and N-ras activating mutations are associated with bladder cancer and myeloid leukemia, respectively (1). Furthermore, while N-ras and H-ras are dispensable for development both individually and in combination (10, 26), mice harboring a homozygous K-ras null mutation (K-ras−/−) are not viable and die between day 12.5 post coitum and term (depending on the genetic background) due to cardiac, liver, neurological, and hematopoietic defects (18, 19). Thus, of the classical ras gene family members (K-, N-, and H-ras), only K-ras is necessary for embryonic development. However, the findings that (i) N-ras−/− mice are healthy but N-ras−/−, K-ras+/− mice die in utero (18), (ii) K-ras−/− N-ras−/− mice exhibit a more severe phenotype than K-ras−/− mice (18), and (iii) fewer than expected H-ras−/− N-ras−/− mice survive embryogenesis (10) underline the essential role for K-Ras in development and imply partial functional overlap between different Ras proteins.
Understanding the role(s) of K-Ras in development and tissue function is an important route to gain insight as to how K-ras activating mutations promote neoplastic change. However, this is complicated by the fact that the K-ras gene encodes two protein isoforms, K-Ras 4A and K-Ras 4B, of 189 and 188 residues, respectively, by alternative splicing of the fourth coding exons 4A and 4B, and K-ras activating mutations that usually affect codons 12, 13, and 61 jointly affect both isoforms (5, 9, 21). The isoforms differ significantly at their C termini after residue 165, including the hypervariable domains and CAAX motifs. These regions are involved in membrane association, which is essential for Ras function (16) via a series of posttranslational modifications, which include isoprenylation, endoproteolysis, and methylation (25). However, these modifications differ between the isoforms due to the sequence differences at the hypervariable domain: K-Ras 4A, like N- and H-Ras, is palmitoylated at cysteine residues upstream of the CAAX motif, which are replaced with a polylysine domain in K-Ras 4B.
The difference in C-terminal modifications of Ras proteins leads to alternative trafficking pathways to the plasma membrane (3) and ultimately localization to different plasma membrane microdomains (23, 24). Indeed, reports that H-, N-, and K-Ras differentially affect Raf-1, phosphatidylinositol 3-kinase, and Rac (28, 29, 31) suggest that they elicit divergent biological responses by interacting with different subsets of downstream effectors. Thus, the posttranslational differences between K-Ras 4A and K-Ras 4B could affect their membrane localization and, therefore, interaction with different membrane targets. The specific association between K-Ras 4B and the guanine nucleotide exchange factor Smg GDS (27) and reports that the oncogenic mutant (G12V) K-Ras 4A and 4B differ in their ability to activate Raf-1, induce transformed foci, enable anchorage-independent growth, and promote cell migration in vitro (28) and that K-ras 4A and 4B are expressed differentially during mouse development and in adult tissues (22, 30) further suggest that they have distinct biological actions. Thus, the cooperative effects of both isoforms could account, at least in part, for the high frequency of K-ras activating mutations in human cancers.
To examine the role of the individual K-ras splice variants in development, gene targeting was used to delete exon 4A and generate K-ras 4A-deficient mice. It was found that K-rastmΔ4A/tmΔ4A mice are healthy and fertile. The result demonstrates that expression of K-ras 4A is dispensable for normal development, at least in the presence of functional K-Ras 4B.
MATERIALS AND METHODS
Design of the K-ras 4A targeting vector.
A nonisogenic targeting vector (pPTKiNKiΔ4A) was used to delete exon 4A of K-ras to generate a mutant allele designated K-rastmΔ4A. The 2.8-kb 5′ and the 1.3-kb 3′ arms of homology were isolated by HindIII and XbaI-EcoRI digests, respectively, of an 11.5-kb EcoR1 fragment (from BALB/c mice) stretching from upstream of exon 3 to upstream of exon 4B (13). The neomycin resistance cassette (containing a phosphoglycerate kinase I promoter) and the herpes simplex virus thymidine kinase gene cassette were employed for positive and negative selection, respectively (Fig. 1A).
FIG. 1.
Targeted deletion of the K-ras 4A splice variant in embryonic stem cells. (A) Schematic representation of the mouse K-ras locus and the targeting vector used to replace exon 4A. The exons are indicated by open boxes. The neomycin resistance cassette and the herpes simplex virus thymidine kinase gene cassette were employed as selectable markers. In the targeting vector, the thin line denotes plasmid DNA. The positions of the 5′ external and 3′ internal probes (solid boxes) and PCR primers (arrows) used for genotyping are indicated. Restriction enzyme sites: E, EcoRI; P, PvuII; H, HindIII. (B) Embryonic stem cells were genotyped by PCR with the primer set neo22 and Px4BA, which amplify a 2.1-kb product in clones that harbor the targeted allele (clones 173, 187, 194, 196, 221, 226, and 248). +/+, wild-type embryonic stem cells; −, PCR negative control; M, 1-kb DNA ladder showing the 2.0- and 2.1-kb size markers. (C) Confirmation of homologous recombination in embryonic stem cells by Southern blotting. Following digestion with HindIII, a wild-type 2-kb and a targeted 6-kb fragment were detected with the 3′ internal probe (upper panel). Following digestion with PvuII, a wild-type 5-kb and a targeted 4-kb band were detected with the 5′ external probe (lower panel). +/+, wild-type embryonic stem cells.
Production of K-rastmΔ4A/+ embryonic stem cells.
The targeting vector was linearized with EcoRI, and 150 μg of DNA was electroporated (800 V and 3 μF, Gene Pulser, Bio-Rad) into strain 129/Ola-derived HM-1 male mouse embryonic stem cells, which harbor an inactivating deletion in the X-linked Hprt gene (20). Homologous recombination in colonies resistant to G418 (300 μg/ml; Invitrogen) and ganciclovir (2 μM; Sigma) was identified initially by PCR and confirmed by Southern analysis. Embryonic stem cell clones were screened with the primer set neo22 (5′-CGATAGAAGGCGATGCGCTGCGAAT-3′) and Px4BA (5′-ATAACTGTACACCTTGTCCTTGACT-3′), positioned in the neomycin cassette and exon 4B, respectively (Fig. 1A), which amplify a 2.1-kb product. The PCR conditions were 1.0 μM each primer (Sigma), 1× PCR buffer (Invitrogen), 1.5 mM MgCl2 (Invitrogen), 200 μM each deoxynucleoside triphosphate (Amersham), 1 unit of Taq DNA polymerase (Invitrogen), and ≈100 ng of genomic DNA per reaction. Following denaturing for 4 min at 94°C, DNA was amplified for 45 cycles (94°C for 1 min, 58°C for 1 min, and 72°C for 2.5 min).
Homologous recombination was confirmed by Southern blotting. Genomic DNA (20 μg) was digested with HindIII or PvuII and, following electrophoresis, was transferred to a nylon membrane (GeneScreen) in denaturing buffer (0.5 M NaOH, 1.5 M NaCl). The membrane was neutralized and prehybridized at 65°C for 2 h (10% dextran sulfate, 1%sodium dodecyl sulfate, 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 200 μg of herring sperm DNA per ml). The 3′ internal and 5′ external probes (Fig. 1A) were labeled with [32P]dCTP by random priming (Stratagene) and hybridized to the membrane at 65°C overnight. Membranes were washed at room temperature with 2× SSC (twice for 5 min), at 65°C with 2× SSC-1% sodium dodecyl sulfate (twice for 30 min), then at room temperature with 0.1× SSC (twice for 30 min). Membranes were exposed to film overnight at −70°C.
Production of K-Ras 4A-deficient mice.
Embryonic stem cells harboring the heterozygous deletion of exon 4A (K-rastmΔ4A/+) were injected into day 3.5 post coitum C57BL/6 blastocysts, which were implanted into the uteri of day 2.5 post coitum CD1 pseudopregnant recipients. Male chimeras were mated with 129/Ola or C57BL/6 females to generate inbred and crossbred lines, and germ line transmission of embryonic stem cell markers was identified by coat color (light yellow and agouti, respectively).
Since long-range PCR proved unreliable for analyzing tail biopsy DNA, mice were genotyped with two different PCRs. In reaction 1, the primer set neo52 (5′-GATGCCTGCTTGCCGAATATCATGG-3′) and neo22 (5′-CGATAGAAGGCGATGCGCTGCGAAT-3′), positioned in the neomycin cassette (Fig. 1A), amplify a 206-bp product in K-rastmΔ4A/+ and K-rastmΔ4A/tmΔ4A mice. In reaction 2, the primer set Px4AS (5′-CATTGGTGAGAGAGATCCGACAGTAC-3′) and Px4AA (5′-TCACACAGCCAGGAGTCTTTTCTTC-3′), positioned in exon 4A (Fig. 1A), amplify a 72-bp product in wild-type and K-rastmΔ4A/+ mice. In both reactions, DNA was denatured for 4 min at 94°C and amplified for 30 cycles at 94°C for 30 s and 58°C for 30 s. The genotypes were subsequently confirmed by Southern blotting with internal and external probes as described above.
In the inbred colony, first-generation K-rastmΔ4A/+ males (which do not harbor the Hprt null mutation) were mated with wild-type 129/Ola mice to generate an inbred stock that was wild type for Hprt. In the crossbred colony, the Hprt mutation was allowed to segregate. All animal work was carried out under Home Office license, with ethically approved methods as set out by the UK Coordinating Committee on Cancer Research in their guidelines on the Welfare of Animals in Experimental Neoplasia.
RNA analysis.
K-ras 4A and 4B expression was determined in tissues from adult crossbred (129/Ola × C57BL/6) mice by reverse transcription-PCR (RT-PCR) with primers positioned within exons 1 and 4B, which amplify both isoforms in the same reaction (22). RNA was extracted with Trizol reagent (Invitrogen), and first-strand cDNA synthesis was performed with the SuperScript preamplification system, with 1 to 5 μg of RNA (Invitrogen). For each sample, controls without reverse transcriptase were included and were negative in all cases.
Histology.
Mice were sacrificed by cervical dislocation, and tissues were fixed overnight in 10% neutral buffered formalin except for the large and small intestines, which were fixed in Methacarn (methanol, chloroform, and glacial acetic acid, 4:2:1 by volume) overnight. Tissues were wax embedded, and 5-μm sections were cut and stained with hematoxylin and eosin.
RESULTS
Generation of K-ras 4A-deficient mice.
The targeting vector pPTKiNKiΔ4A was designed to replace exon 4A of K-ras with a neomycin resistance cassette (Fig. 1A). Of 324 embryonic stem clones screened by PCR, 17 were positive for homologous recombination, amplifying the correct 2.1-kb PCR product (Fig. 1B). The genotype of seven of these clones was confirmed by Southern analysis with a 5′ external probe that identified a 5-kb wild-type and a 4-kb targeted band and a 3′ internal probe that identified a 2-kb wild-type and a 6-kb targeted band (Fig. 1C). Of nine chimeras (generated from three different embryonic stem cell clones), five transmitted embryonic stem cell-derived coat color markers through the germ line.
Since the effects of mutations, including the K-ras null mutation (18), can vary greatly with the genetic background, the consequence of deleting K-ras 4A expression was examined in inbred (129/Ola) and crossbred (129/Ola × C57BL/6) strains of mice. The embryonic stem cell-derived offspring were genotyped at weaning by PCR (with the primer sets neo22/neo52 and Px4AS/Px4AA, which amplify the neomycin cassette and exon 4A, respectively; Fig. 2A) and by Southern blotting (with the 5′ external and 3′ internal probes; Fig. 2B), and it was found that K-rastmΔ4A/+ and wild-type mice were present in the expected Mendelian ratio (54 and 61, respectively; χ21 = 0.426, P = 0.514).
FIG. 2.
Genotyping of mice. (A) The upper panel shows amplification of K-ras exon 4A by PCR. The 72-bp product is present in wild-type (+/+, lanes 9 and 10) and K-rastmΔ4A/+ (+/−,lanes 14 and 15) mice, but not in K-rastmΔ4A/tmΔ4A mice (−/−, lanes 5 and 11). The lower panel shows amplification of the neomycin cassette by PCR. The 206-bp product is present in K-rastmΔ4A/+ and K-rastmΔ4A/tmΔ4A mice but not in wild-type mice. (B) Confirmation of genotypes by Southern blotting. Tail DNA digested with HindIII and hybridized with the 3′ internal probe (upper panel) gave a 2-kb fragment for the wild-type allele and a 6-kb fragment from the targeted allele. Only the 6-kb fragment is present in K-rastmΔ4A/tmΔ4A mice. Digestion with PvuII and hybridization with the 5′ external probe (lower panel) gave a 5-kb band for the wild-type allele and a 4-kb fragment for the targeted allele. Only the 4-kb fragment is present in K-rastmΔ4A/tmΔ4A mice.
K-ras 4A-deficient mice are healthy and fertile.
Offspring from crosses between K-rastmΔ4A/+ mice were genotyped by PCR (Fig. 2A) and Southern blotting (Fig. 2B). Wild-type, K-rastmΔ4A/+, and K-rastmΔ4A/tmΔ4A offspring were present in the expected Mendelian ratios on both crossbred (48, 101, and 58, respectively; χ22 = 1.087, P = 0.581) and inbred (11, 20, and 10, respectively; χ22 = 0.073, P = 0.964) genetic backgrounds, indicating that all K-rastmΔ4A/tmΔ4A mice develop normally. In addition, further breeding studies found that male and female K-rastmΔ4A/tmΔ4A offspring were present in the expected Mendelian ratios in both crossbred (113 and 113, respectively) and inbred (20 and 25, respectively; χ21 = 0.556, P = 0.456) stocks. Male and female K-rastmΔ4A/tmΔ4A mice were fertile, and females successfully weaned their young.
Detailed histopathological analysis was undertaken of 24 crossbred mice at 3 months, including 12 wild-type and 12 K-rastmΔ4A/tmΔ4A mice, with six males and six females in each cohort. No abnormalities were detected at necropsy, and examination of liver, kidney, large and small intestine, stomach, pancreas, spleen, thymus, heart, lung, brain, ovary, uterus, seminal vesicles, and testis found no difference between tissues from wild-type and K-rastmΔ4A/tmΔ4A mice. Inbred K-rastmΔ4A/tmΔ4A mice were also fertile and outwardly healthy, and histopathological analysis of a single animal at 3 months found no abnormalities. The oldest K-rastmΔ4A/tmΔ4A mice in the crossbred colony are currently 8 months old, and these too are outwardly healthy.
K-ras 4A-deficient mice express the K-ras 4B splice variant only.
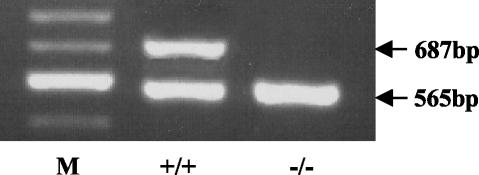
To confirm that the modification of the K-ras gene, by the introduction of the targeting vector, resulted in the deletion of the K-ras 4A splice variant, RT-PCR was used to examine K-ras expression in the large intestine of wild-type and K-rastmΔ4A/tmΔ4A mice. Only the K-ras 4B splice variant was expressed by K-rastmΔ4A/tmΔ4A mice (Fig. 3).
FIG. 3.
Expression of K-ras in the large intestine from wild-type and K-rastmΔ4A/tmΔ4A mice. RT-PCR analysis of K-ras 4A and 4B mRNAs with primers positioned in exons 1 and 4B that amplify both splice variants in the same reaction (22). The 687-bp band corresponds to K-ras 4A, and the 565-bp band corresponds to K-ras 4B. Wild-type (+/+) but not K-rastmΔ4A/tmΔ4A (−/−) mice express K-ras 4A. Lane M, 100-bp size markers; the major band shown is 600 bp.
K-ras 4A expression in wild-type adult mice.
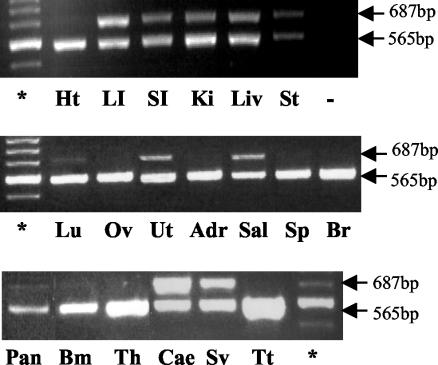
To date, the expression of K-ras 4A has been examined in only a limited number of adult mouse tissues (22, 30). In the present study, the analysis was extended further to gain a comprehensive view of the overall pattern of K-ras 4A expression in wild-type adult mice. RT-PCR analysis found that K-ras 4A was expressed in the liver, large and small intestine, stomach, cecum, kidney, uterus, salivary gland, and seminal vesicles and only at low levels in the lung, pancreas, and bone marrow cells (Fig. 4). In contrast, K-ras 4B was expressed ubiquitously and was the only isoform expressed in brain, spleen, heart, adrenal gland, thymus, testis, and ovary. While both isoforms were expressed at similar levels in stomach and large intestine, K-ras 4A was the major isoform expressed in the cecum.
FIG. 4.
K-ras 4A and 4B expression in adult mouse tissues. The PCR primers used amplify both K-ras splice variants in the same reaction (22). The K-ras 4A and 4B mRNAs generate PCR products of 687 and 565 bp, respectively. Tissues: Ht, heart; LI, large intestine; SI, small intestine; Ki, kidney; Liv, liver; St, stomach; Lu, lung; Ov, ovary; Ut, uterus; Adr, adrenal gland; Sg, salivary gland; Sp, spleen; Br, brain; Pan, pancreas; Bm, bone marrow; Th, thymus; Cae, cecum; Sv, seminal vesicles; Tt, testis. −, PCR negative control. *, 100-bp ladder; the major band shown is 600 bp.
DISCUSSION
To gain further understanding concerning the role(s) of K-Ras in development and tissue function, we examined the pattern of K-ras 4A expression in wild-type adult mouse tissues and used gene targeting to delete K-ras 4A expression. While previous studies have established that expression of the K-ras gene is essential for mouse development (18, 19), here we found that, even though K-ras 4A is expressed during development and in adult tissues (22, 30; this study), all K-rastmΔ4A/tmΔ4A mice on both inbred and crossbred genetic backgrounds developed normally, and the adult mice are fertile and healthy.
The finding that K-Ras 4A is dispensable for development suggests that the embryonic lethality of K-ras−/− mice (which do not express either isoform) may result solely from failure of expression of the K-Ras 4B isoform. However, the very fact that K-ras 4A is expressed during development, albeit in a spatially and temporally regulated manner (22), raises the possibility that the lethal K-ras−/− phenotype could result from loss of synergistic function of the two protein isoforms or else from loss of a critical function(s) that can be performed by either isoform, such that one or the other is necessary but in the absence of both development cannot occur. While the latter possibility is less likely, given that only the K-Ras 4B isoform is expressed ubiquitously throughout development (22), questions concerning whether expression of the K-Ras 4B isoform is essential for normal development and whether K-Ras 4A influences its action wait to be addressed by comparing the phenotypes of K-ras−/− and K-ras 4B-deficient mice on the same inbred genetic background. It is nevertheless important to stress that whatever the result of these future studies, our conclusion that K-Ras 4A is dispensable for normal mouse development in the presence of functional K-Ras 4B remains valid.
The C-terminal modifications of Ras proteins are essential for protein function (16), and functional differences between Ras proteins may reflect, at least in part, these different modifications. Therefore, the findings by Hancock et al. that N-, H-, and K-Ras 4A undergo similar C-terminal modifications which involve palmitoylation of cysteine residues upstream of the common CAAX motif (14) are significant because these proteins are dispensable for normal development. Furthermore, the viability of K-rastmΔ4A/tmΔ4A mice is unlikely to reflect a compensatory upregulation of these closely related Ras proteins, since the simultaneous deletion of both K-ras splice variants does not result in the upregulation of H-ras or N-ras expression in either tissues or fibroblast cultures from K-ras−/− embryos (18). Likewise, H-ras−/− N-ras−/− mutant mice show no change in K-ras expression (10).
Importantly, since N-ras−/−, H-ras−/−, and K-rastmΔ4A/tmΔ4A mice are fertile, it is now possible, by crossing these mice, to formally test whether the viability of K-rastmΔ4A/tmΔ4A mice is indeed independent of N- and H-ras expression. Also, since K-Ras 4B, unlike K-Ras 4A, associates with the plasma membrane by virtue of a polybasic domain and, thereby, may localize to different membrane microdomains, it could selectively activate Ras effectors required for embryonic development. Indeed, Ras proteins, including the K-Ras isoforms, differentially activate particular downstream effectors (28, 29, 31). Since K-rastmΔ4A/tmΔ4A mice are viable, their analysis can also address whether both K-Ras isoforms differentially affect Ras effector signal transduction pathways in vivo. This information is crucial in view of current research interest in anti-Ras agents, some of which target posttranslational modifications that affect one isoform and not the other, in cancer therapy (2).
Previous studies based on RT-PCR and Northern blotting reported that K-ras 4A is expressed in adult liver, stomach, small intestine, colon, and kidney but disagree concerning its expression in the lung (22, 30). Furthermore, these studies reported that K-ras 4A is invariably the minor isoform expressed. The present findings that K-ras 4A is expressed in a wide range of adult tissues, including lung and in some cases at levels equal to or greater than that of K-ras 4B (Fig. 4) imply it has an important role in tissue homeostasis in general, even though it is dispensable for normal development. In view of recent reports that N-ras−/− and H-ras−/− mice exhibit phenotypes but only after challenge with exogenous agents (6, 15), a similar approach with K-rastmΔ4A/tmΔ4A mice, combined with examination of the long-term consequence of K-Ras 4A-deficiency, may yield insight concerning the role(s) of K-Ras 4A in tissue function.
The finding that K-Ras 4A is dispensable for development also presents an opportunity to examine the role of the K-Ras isoforms in tumorigenesis in vivo. While expression of a mutationally activated K-ras 4B transgene can promote development of lung and intestinal tumors (12, 17), evidence suggests that mutant K-Ras 4A could also be implicated in tumorigenesis: (i) expression of a K-ras 4A transgene encoding an activating mutation can transform cells in vitro and with greater efficiency than K-ras 4B (28); (ii) mutationally activated K-ras 4A is expressed by the SW480 human colorectal carcinoma cell line (5); (iii) the level of K-ras 4A expression in lung correlates with lung tumor susceptibility among inbred mouse strains (30); and (iv) here we found that K-ras 4A is expressed in the colon, lung. and pancreas, where tumors with K-ras activating mutations generally arise in human malignancy (1). Since mutant K-Ras isoforms differ in their ability to promote cell transformation and cell migration (28), it is conceivable that synergistic interactions between activated K-Ras 4A and K-Ras 4B could play a crucial role in driving tumor development. In view of recent evidence that wild-type K-Ras exhibits tumor suppressor activity (32), K-rastmΔ4A/tmΔ4A mice can be used to address not only the role of K-Ras 4A in neoplastic progression in vivo, but also whether both isoforms exhibit tumor suppressor activity.
Examination of the role of wild-type K-Ras in adult tissues, where tumors with oncogenic K-ras mutations normally arise, is one approach to gain valuable insight concerning how these mutations promote tumorigenesis. While this has been hindered by the embryonic lethality of the K-ras null mutation (18, 19), we envisage that future studies with K-rastmΔ4A/tmΔ4A mice will contribute significantly to understanding why K-ras activating mutations are so prevalent in human malignancies and why they are linked with particular types of epithelial cell-derived tumors.
Acknowledgments
This study was supported by a Medical Research Council studentship and grants from the Cancer Research Campaign (CRC, now Cancer Research UK), the Melville Trust for the Care and Cure of Cancer, Action Research for the Crippled Child (new Action Medical Research), and the Row Fogo Charitable Trust.
REFERENCES
- 1.Adjei, A. A. 2001. Blocking oncogenic Ras signalling for cancer therapy. J. Natl. Cancer Inst. 93:1062-1074. [DOI] [PubMed] [Google Scholar]
- 2.Ahmadian, M. R. 2002. Prospects for anti-ras drugs. Br. J. Hematol. 116:511-518. [DOI] [PubMed] [Google Scholar]
- 3.Apolloni, A. I., A. Prior, M. Lindsay, R. G. Parton, and J. F. Hancock. 2000. H-Ras but not K-Ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20:2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, D. J., R. M. James, C. E. Patek, J. Williamson, and M. J. Arends. 2001. Mutant K-ras enhances apoptosis in embryonic stem cells in combination with DNA damage and is associated with increased levels of p19 (ARF). Oncogene 20:2144-2152. [DOI] [PubMed] [Google Scholar]
- 5.Capon, D. J., P. H. Seeburg, J. P. McGrath, J. S. Hayflick, U. Edman, A. D. Levinson, and D. V. Goeddel. 1983. Activation of Ki-ras2 gene in human colon and lung carcinomas by two different point mutations. Nature 304:507-513. [DOI] [PubMed] [Google Scholar]
- 6.de Castro, I. P., R. Diaz, M. Malumbres, M-I. Hernandez, J. Jagirdar, M. Jimenez, D. Ahn, and A. Pellicer. 2003. Mice deficient for N-ras: Impaired antiviral immune response and T-cell function. Cancer Res. 63:1615-1622. [PubMed] [Google Scholar]
- 7.Diaz, R., D. Ahn, L. Lopez-Barcons, M. Malumbres, I. P. de Castro, J. Lue, N. Ferrer-Miralles, R., Mangues, J. Tsong, R. Garcia, R. Perez-Soler, and A. Pellicer. 2002. The N-ras proto-oncogene can suppress the malignant phenotype in the presence or absence of its oncogene. Cancer Res. 62:4514-4518. [PubMed] [Google Scholar]
- 8.Ehrhardt, A., G. R. A. Ehrhardt, X. Guo, and J. W. Schrader. 2002. Ras and relatives-job sharing and networking keep an old family together. Exp. Hematol. 30:1089-1106. [DOI] [PubMed] [Google Scholar]
- 9.Ellis, C. A., and G. Clark. 2000. The importance of being K-Ras. Cell. Signalling 12:425-434. [DOI] [PubMed] [Google Scholar]
- 10.Esteban, L. M., C. Vicario-Abejon, P. Fernandez-Salguero, A. Fernandez-Medarde, N. Swaminathan, K. Yienger, E. Lopez, M. Malumbres, R. McKay, J. M. Ward, A. Pellicer, and E. Santos. 2001. Targeted disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol. Cell. Biol. 21:1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, J., and J. R. Bertino. 1997. K-ras modulates the cell cycle via both positive and negative regulatory pathways. Oncogene 14:2595-2607. [DOI] [PubMed] [Google Scholar]
- 12.Fisher, G. H., S. L. Wellen, D. Klimstra, J. M. Lenczowski, J. W. Tichelaar, M. J. Lizak, J. A. Whitsett, A. Koretsky, and H. E. Varmus. 2001. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumour suppressor genes. Genes Dev. 15:3249-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.George, D. L., A. F. Scott, S. Trusko, B. Glick, E. Ford, and D. J. Dorney. 1985. Structure and expression of amplified cKi-ras gene sequences in Y1 mouse adrenal tumour cells. EMBO J. 4:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock, J. F. 2003. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell. Biol. 4:273-284. [DOI] [PubMed] [Google Scholar]
- 15.Ise, K., K. Nakamura, K. Nakao, S. Shimizu, H. Harada, T. Ichise, J. Miyoshi, Y. Gondo, T. Ishikawa, A. Aiba, and M. Katsuki. 2000. Targeted deletion of the H-ras gene decreases tumour formation in mouse skin carcinogenesis. Oncogene 19:2951-2956. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, J. H., J. W. Li, J. E. Buss, C. J. Der, and C. G. Cochrane. 1994. Polylysine domain of K-ras 4B protein is crucial for malignant transformation. Proc. Natl. Acad. Sci. 91:12730-12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen, K. P., F. el Marjou, D. Pinto, X. Sastre, D. Rouillard, C. Fouquet, T. Soussi, D. Louvard, and S. Robine. 2002. Targeted expression of oncogenic K-ras in intestinal epithelium causes spontaneous tumorigenesis in mice. Gastroenterology 123:492-504. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, L., D. Greenbaum, K. Cichowski, K. Mercer, E. Murphy, E. Schmitt, R. T. Bronson, H. Umanoff, W. Edelmann, R. Kucherlapati, and T. Jacks. 1997. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 11:2468-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koera, K., K. Nakamura, K. Nakao, J. Miyoshi, K. Toyoshima, T. Hatta, H. Otani, A. Aiba, and M. Katsuki. 1997. K-ras is essential for the development of the mouse embryo. Oncogene 15:1151-1159. [DOI] [PubMed] [Google Scholar]
- 20.Magin, T. W., J. McWhir, and D. W. Melton. 1992. A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res. 20:3795-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGrath, J. P., D. J. Capon, D. H. Smith, E. Y. Chen, P. H. Seeburg, D. V. Goeddel, and A. D. Levinson. 1983. Structure and organization of the human Ki-ras proto-oncogene and a related processed pseudogene. Nature 304:501-506. [DOI] [PubMed] [Google Scholar]
- 22.Pells, S., M. Divjak, P. Romanowski, H. Impey, N. J. Hawkins, A. R. Clarke, M. L. Hooper, and D. J. Williamson. 1997. Developmentally regulated expression of murine K-ras isoforms. Oncogene 15:1781-1786. [DOI] [PubMed] [Google Scholar]
- 23.Prior, I. A., A. Harding, J. Yan, J. Sluimer, R. G. Parton, and J. F. Hancock. 2001. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell. Biol. 3:368-375. [DOI] [PubMed] [Google Scholar]
- 24.Roy, S., R. Luetterforst, A. Harding, A. Apolloni, M. Etheridge, E. Stang, B. Rolls, J. F. Hancock, and R. G. Parton. 1999. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell. Biol. 1:98-105. [DOI] [PubMed] [Google Scholar]
- 25.Silvius, J. R. 2002. Mechanisms of Ras protein targeting in mammalian cells. J. Membrane Biol. 190:83-92. [DOI] [PubMed] [Google Scholar]
- 26.Umanoff, H., W. Edelmann, A. Pellicer, and R. Kucherlapati. 1995. The murine N-ras gene is not essential for growth and development. Proc. Natl. Acad. Sci. 92:1709-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vikis, H. G., S. Stewart, and K. L. Guan. 2002. SmgGDS displays differential binding and exchange activity towards different Ras isoforms. Oncogene 21:2425-2432. [DOI] [PubMed] [Google Scholar]
- 28.Voice, J. K., R. L. Klemke, A. Le, and J. H. Jackson. 1999. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J. Biol. Chem. 274:17164-17170. [DOI] [PubMed] [Google Scholar]
- 29.Walsh, A. B., and D. Bar-Sagi. 2001. Differential activation of the Rac pathway by Ha-Ras and K-Ras. J. Biol. Chem. 276:15609-15615. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Y., M. You, and Y. Wang. 2001. Alternative splicing of the K-ras gene in mouse tissues and cell lines. Exp. Lung Res. 27:255-267. [DOI] [PubMed] [Google Scholar]
- 31.Yan, J., S. Roy, A. Apolloni, A. Lane, and J. F. Hancock. 1998. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J. Biol. Chem. 273:24052-24056. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Z., Y. Wang, H. G. Vikis, L. Johnson, G. Liu, J. Li, M. W. Anderson, R. C. Sills, H. L. Hong, T. R. Devereux, T. Jacks, K-L. Guan, and M. You. 2001. Wild-type Kras2 can inhibit lung carcinogenesis in mice. Nat. Genet. 29:25-33. [DOI] [PubMed] [Google Scholar]