Abstract
Like the vertebrate enteric nervous system (ENS), the insect ENS consists of interconnected ganglia and nerve plexuses that control gut motility. However, the insect ENS lies superficially on the gut musculature, and its component cells can be individually imaged and manipulated within cultured embryos. Enteric neurons and glial precursors arise via epithelial-to-mesenchymal transitions that resemble the generation of neural crest cells and sensory placodes in vertebrates; most cells then migrate extensive distances before differentiating. A balance of proneural and neurogenic genes regulates the morphogenetic programs that produce distinct structures within the insect ENS. In vivo studies have also begun to decipher the mechanisms by which enteric neurons integrate multiple guidance cues to select their pathways. Despite important differences between the ENS of vertebrates and invertebrates, common features in their programs of neurogenesis, migration, and differentiation suggest that these relatively simple preparations may provide insights into similar developmental processes in more complex systems.
Keywords: stomatogastric, ENS, placode, delamination, invagination, epithelial-to-mesenchymal transition, EMT, neurogenesis, gliogenesis, migration
Introduction
Analogous to the enteric nervous system (ENS) of vertebrates, the ENS of higher invertebrates comprises a distinct division of the peripheral nervous system (PNS), providing innervation to the alimentary tract and related secretory organs. As in vertebrates, the ENS of invertebrates includes interconnected networks of ganglia and nerve plexuses that span different domains of the gut. Depending on their locations, these components contribute to regulation of feeding and swallowing, gut peristalsis, metabolism, and some endocrine functions (Penzlin, 1985; Harris-Warrick et al., 1992; Bestman and Booker, 2003; Ayali, 2004). In contrast to vertebrates, however, the ENS of invertebrates typically consists of a relatively small number of enteric ganglia and nerves that occupy stereotyped positions on the most superficial layers of visceral musculature. Consequently, the simplicity and accessibility of these preparations has been exploited for a variety of developmental and functional studies. Seminal work using the ENS of crustaceans (specifically the stomatogastric nervous system of the foregut and stomach) has advanced our understanding of how neural networks produce modulated motor patterns (reviewed in Nusbaum and Beenhakker, 2002; Hooper and DiCaprio, 2004; Marder et al., 2005). In contrast, the ENS of insects has proven particularly valuable for developmental studies.
The formation of the insect ENS involves many of the same types of cellular and molecular interactions that have been documented in vertebrate neurodevelopment. Whereas the vertebrate ENS is derived from neural crest cells that delaminate from the neural plate and then migrate onto the gut (Gershon, 1981; Le Douarin and Kalcheim, 1999), the insect ENS is formed primarily by cells that emerge from neurogenic zones within the foregut epithelium itself. Like neural crest cells, most neural and glial precursors in the insect ENS subsequently migrate substantial distances before assembling into the ganglia and nerve plexuses that innervate the gut. Both lineage-dependent and environmental factors affect the phenotypes that insect enteric neurons ultimately express once they complete their migration, as has been suggested for neural crest cells that populate the vertebrate ENS (Burns, 2005). Malleability in the neurogenic program of the insect ENS has apparently accommodated the structural diversity seen in the alimentary tracts of different species. Since the excellent summary by Hartenstein (Hartenstein, 1997), a number of studies have provided additional evidence for convergent aspects of ENS development in insects and vertebrates. This review is intended to provide a brief overview of these themes, and a discussion of potential areas in which the insect ENS might be exploited to investigate developmental mechanisms that also regulate neurogenesis, migration, and neuronal differentiation in vertebrate systems.
1. Species-specific differences in the morphology of the insect ENS are common
The insect ENS has been a focus of study since the 17th century, when the Dutch microscopist Jan Swammerdam first detected nerves on the insect gut (Swammerdam, 1752; as cited in Penzlin, 1985). Other investigators subsequently applied a variety of names to the insect ENS, including the stomatogastric, visceral, and sympathetic nervous system (Brandt, 1836; Janet, 1905; Snodgrass, 1935; Bickley, 1942). While the components of the insect nervous system should not be considered directly homologous to vertebrate structures, both the general organization and functions of the ENS in both phyla are clearly analogous, justifying the applicability of this term (Furness, 2006). At first glance, the distributions of enteric ganglia and nerves found in different insects appear surprisingly diverse (Fig. 1; Copenhaver, 1993; Hartenstein et al., 1994; Ganfornina et al., 1996). Given evidence that the modern insect orders radiated from a common ancestor over 300 million years ago, around the time that reptiles diverged from birds (Gaunt and Miles, 2002; Nobrega and Pennacchio, 2004), it is not surprising that the insect ENS evolved to meet the feeding requirements of animals with radically different lifestyles. Nevertheless, a common organizational pattern can be readily discerned in the ENS of different insect species (see also Snodgrass, 1935; Bickley, 1942; Penzlin, 1985).
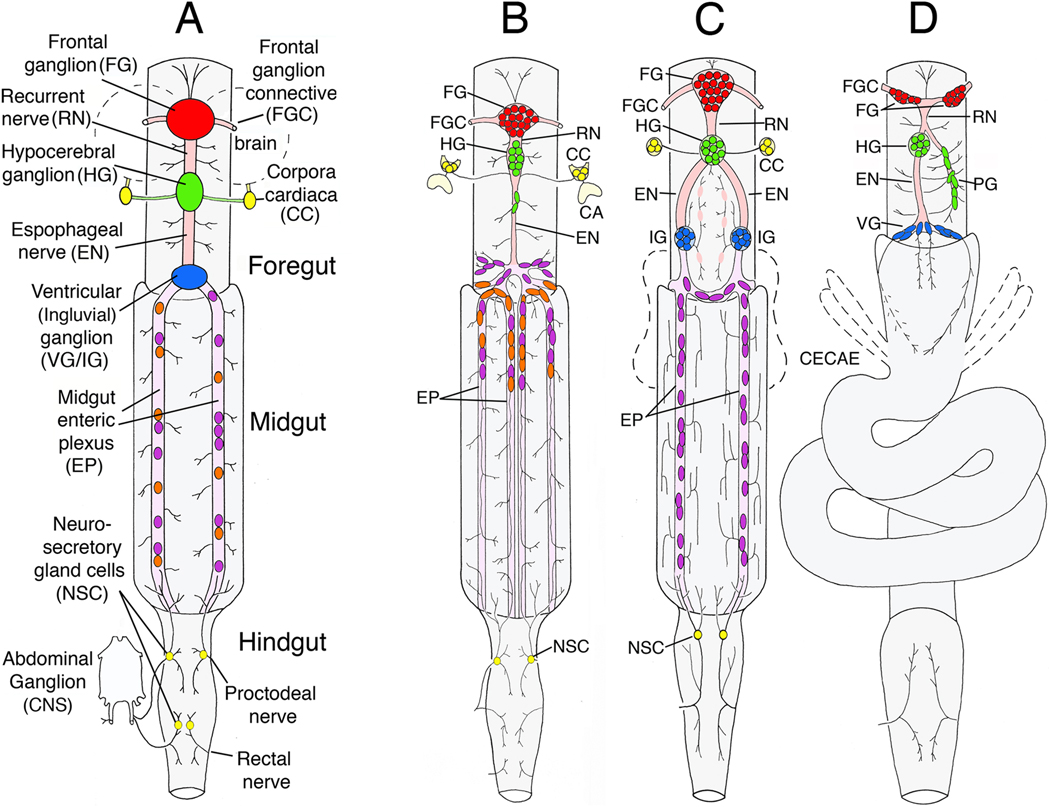
Figure 1. Schematic drawings of the ENS in different insect species.
A. Generalized organization of ganglia and nerves found in the insect ENS. The primary foregut ganglion is the frontal ganglion (FG; red), connected to the overlying brain (dotted outline) by paired frontal ganglion connectives (FGC). Several nerve branches extend anteriorly onto the pharynx, while the recurrent nerve (RN) extends posteriorly to the hypocerebral ganglion (HG; green), situated below the brain. The HG is also usually connected to the paired corpora cardiaca (CC), the primary neurosecretory organs of the brain. From the HG, the esophageal nerve (EN) extends to the ventricular ganglion (VG; blue; also called the ingluvial ganglion, or IG). The foregut ganglia also give rise to a diffuse plexus of nerves that innervate the musculature and may include peripheral stretch receptors. From the ventricular ganglion, a branching nerve plexus (the midgut enteric plexus, EP) extends along the superficial musculature of the midgut; typically, this plexus contains a distributed set of enteric neurons within its major branches. The hindgut is innervated by branches of the proctodeal and rectal nerves that originate in the terminal abdominal ganglion of the ventral nerve cord (CNS); branches of the proctodeal nerve also extend onto the posterior midgut. Several peripheral neurosecretory cells are often found within these nerves (yellow). B. Diagram of the ENS in the tobacco hornworm Manduca sexta (larval stage). The FG is closely apposed to a diminutive HG, but no VG forms in this species. The esophageal nerve connects with the enteric plexus that spans the foregut-midgut boundary and contains dispersed populations of neurons. Several distinct neuronal phenotypes are intermingled within the anterior portion of the midgut nerves (orange and purple cells). C. Diagram of the ENS in the grasshopper Schistocerca americana (after Ganfornina et al., 1996). Posterior to the HG, two esophageal nerves extend to paired IG near the foregut-midgut boundary; nerves from these ganglia connect with the midgut enteric plexus, which contains distributed neurons throughout its length in this species. D. Diagram of the ENS in the fruit fly Drosophila melanogaster (after Skaer, 1993 and Hartenstein et al., 1994). The FG consists of an asymmetric pair of hemi-ganglia on either side of the foregut. Branches of the recurrent nerve extend to the HG on the left and to the paraesophageal ganglion (PG) on the right. From the HG, the esophageal nerve extends to a single VG, consisting of a small number of neurons near the foregut-midgut boundary. Two sets of nerves extend from the VG onto the anterior portion of the midgut, but these nerves do not contain enteric neurons.
As in vertebrates, the insect gut consists of foregut, midgut, and hindgut divisions with corresponding specializations in their innervation. Most prominent is the ENS of the foregut, which consists of interconnected ganglia, nerves, and stretch receptors that are distributed across the gut musculature (Fig. 1A). In contrast, the midgut ENS generally lacks ganglia but is formed by diffuse nerve plexuses that extend across the midgut musculature and often contain distributed populations of enteric neurons. The hindgut ENS is primarily supplied by the proctodeal and rectal nerves originating from the terminal ganglia of the CNS, which give rise to a diffuse plexus that spans the pyloric region (between midgut and hindgut). In many species, these nerves also contain a small number of neurosecretory cells (or “epiproctodeal gland” cells; Evans, 1980; McCormick and Nichols, 1993; Lengyel and Iwaki, 2002). Peptides released from these cells have myoinhibitory effects on the gut musculature but may also regulate hormonally driven behaviors (Davis et al., 2003), one of several ways that the insect ENS contributes to endocrine functions. Input from the CNS plays an important role in regulating feeding behavior and defecation (Chapman, 1982; Eaton, 1985). Nevertheless, like the ENS of vertebrates, the insect ENS controls ingestion and rhythmic peristalsis of the gut in a largely autonomous fashion (Penzlin, 1985; Miles and Booker, 1994; Ayali, 2004; Zilberstein et al., 2004).
The primary ganglion on the foregut is the frontal ganglion (FG), positioned on the pharynx just anterior to the brain (Fig. 1, red). This ganglion is usually unpaired, although in Drosophila, its cells spread to form two hemi-ganglia (Fig. 1D). The frontal ganglion is joined to the brain via the bilaterally paired frontal ganglion connectives (FGC), through which a small number of CNS neurons project to the ENS (Penzlin, 1985). Anteriorly, the frontal ganglion gives rise to nerves that innervate the pharynx, while posteriorly, the recurrent nerve (RN) extends to the hypocerebral ganglion (HG, green), generally positioned below the brain. In many species, the hypocerebral ganglion is also connected to the corpora cardiaca (CC, yellow), the paired neurosecretory organs of the brain (part of the ring gland in Drosophila). Like the adenohypophysis of vertebrates, these organs often arise from the foregut epithelium, as described below.
From the hypocerebral ganglion, esophageal nerves traverse the foregut and connect with additional ganglia whose organization varies markedly among different species. In the tobacco hornworm Manduca sexta (Fig. 1B), a single esophageal nerve (EN) extends from the hypocerebral ganglion to a branching nerve plexus that spans the foregut-midgut boundary (Copenhaver and Taghert, 1989a). In the grasshopper Schistocerca americana (Fig. 1C), two esophageal nerves extend to a pair of “ingluvial” ganglia (IG; blue) that then connect with the midgut plexus (Ganfornina et al., 1996). An unusual example of asymmetry is found in the fruitfly Drosophila melanogaster (Fig. 1D), where the recurrent nerve branches before the hypocerebral ganglion. One branch connects with a diffuse paraesophageal ganglion (PG; or “esophageal ganglion 1”) on the right, while the other extends to the hypocerebral ganglion on the left (Hartenstein et al., 1994; González-Gaitán and Jäckle, 1995). An esophageal nerve then extends from the hypocerebral ganglion to the ventricular ganglion (VG; also called “esophageal ganglion 2”), while a second nerve from the paraesophageal ganglion innervates the opposite side of the foregut. Despite this structural diversity, the components of the foregut ENS in different species have similar developmental origins, providing an intriguing example of evolutionary plasticity in the refinement of neurogenic programs (summarized in the following section).
Unlike the insect foregut, the innervation of the midgut typically consists of gastric nerve plexuses that extend across the visceral musculature. In Drosophila, this innervation is surprisingly sparse: six nerves extend a short distance onto the anterior midgut before terminally branching, leaving much of the midgut without direct innervation (Fig. 1D; González-Gaitán and Jäckle, 1995). In contrast, the midgut of Manduca is innervated by an elaborate nerve plexus (originally termed the “enteric plexus”; Copenhaver and Taghert, 1989a), containing several hundred neurons in its anterior regions that extend long axons to innervate the rest of the midgut musculature (Fig. 1B). In grasshopper, the midgut enteric neurons as well as their processes are distributed throughout the entire length of the midgut plexus (Ganfornina et al., 1996). As summarized below, these populations differentiate via a sequence of placode invagination, delamination, and migration that resembles the programs of neurogenesis giving rise to the vertebrate PNS.
2. Experimental strategies for studying ENS development in insects
Despite radical differences in the gross morphologies of invertebrates and vertebrate nervous systems, it is now well established that many fundamental aspects of neural development have remained evolutionarily conserved at both the cellular and molecular level (Goodman, 1996; Tessier-Lavigne and Goodman, 1996; Buss and Oppenheim, 2004; Pearson and Doe, 2004; Charron and Tessier-Lavigne, 2005). With respect to the developing ENS, insect preparations offer several experimental advantages for investigating specific aspects of neurogenesis, migration, and differentiation within an intact nervous system, permitting mechanistic questions to be addressed in an in vivo context. In the case of Drosophila, the most obvious advantage is the growing repertoire of genetic tools for analyzing the functions of particular genes in specific cellular contexts. In particular, this approach has been used to delineate the genetic basis of how the insect ENS is first patterned. Studies of this type have also revealed an intriguing interplay between transcriptional regulatory genes and signal transduction pathways that may regulate important aspects of vertebrate neurogenesis, as well.
However, due to its small size and convoluted morphology, the ENS of Drosophila is not particularly amenable to acute experimental manipulations within developing embryos. For these types of studies, several larger species have proven valuable for investigating the embryonic development of the ENS. In particular, the tobacco hornworm (Manduca sexta) and grasshopper species (Schistocerca americana and Locusta migratoria) have been used to delineate the cellular origins of the ENS, and to test the role of specific guidance cues, receptors, and intracellular signaling molecules in controlling the migration and differentiation of enteric neurons and glia. While in post-embryonic stages, these larger insects rival juvenile mice in size (Fig. 2A), even as embryos they exceed the size of fully grown Drosophila larvae (> 5 mm), facilitating investigations into the morphogenesis of the ENS. Most notably, these larger preparations allow individual migratory cells within the developing ENS to be labeled, imaged, and experimentally challenged within their normal developmental context.
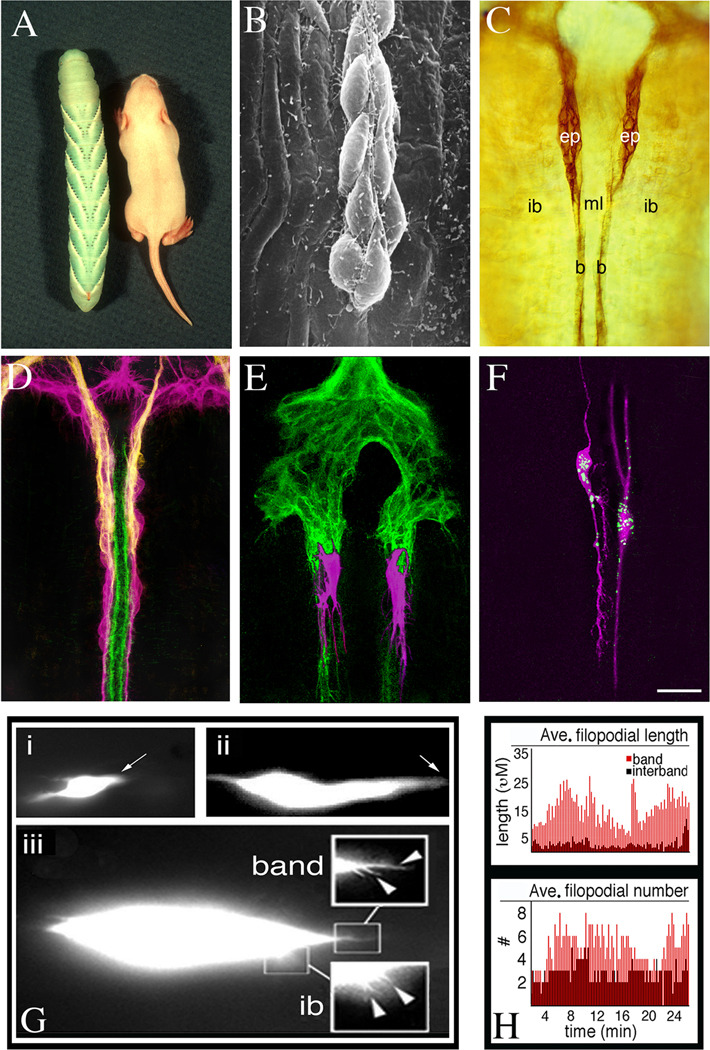
Figure 2. Manduca as an experimental model offers the advantages of simplicity and accessibility, permitting a variety of manipulations to be performed in the intact ENS.
A. Photograph of a fully grown Manduca larva (fifth instar) and a juvenile mouse (courtesy of Dr. Rita Balice-Gordon). B. Scanning electron micrograph of EP cells that are migrating on the surface of the gut musculature. C. Whole-mount preparation of a Manduca embryo immunostained with an antibody against the cell adhesion receptor Fas II (at 58% of development; 1% of development = 1 hr). Both the EP cells (ep) and their muscle band pathways (b) express Fas II at this stage of development. EP cells migrate exclusively on the midgut bands but not onto adjacent interband muscles (ib), nor across the midline interband cells (ml). Only the dorsal pair of eight midgut bands is shown. D. Whole-mount preparation of the embryonic ENS (at 65% of development) immunostained with anti-MsEphrin (magenta; to label the EP ells and their processes); anti-GPI-Fas II (yellow; to label the glial cells ensheathing the midgut EP cells), and anti-Neuroglian (green; to label the muscle band pathways). E. Whole-mount preparation of the embryonic ENS (at 58% of development) in which two migratory EP cells were injected with DiI (magenta) prior to immunostaining the preparation with anti-TM-Fas II (green). As in panel C, the muscle band pathways are also stained by anti-Fas II antibodies. Each neuron extends an array of filopodial processes in advance of its cell body; filopodia that extend along the muscle band pathways are longer and are often become incorporated into the leading process, while filopodia that extend onto the adjacent interband muscles remain shorter and usually are rapidly retracted. F. Embryo in which two EP cells were injected prior to migration onset (at 50% of development) with mRNA encoding monomeric DsRed and Alexa Fluor 488 hydrazide (green). After 20 hr in culture, the preparation was immunostained with anti-DsRed antibodies (magenta). G. Single frames taken from QuickTime movies of EP cells that were injected with Alexa Fluor-488 dextran and monitored by time-lapse imaging. Panel “i” shows an EP cell migrating along one of the midgut muscle bands (the band is not labeled); arrow indicates the position of the leading process (see Supplementary movie #S1). Panel “ii” shows an EP cell transitioning from migration to axonal outgrowth (see Supplementary movie #S2). Arrow indicates the position of the leading process that will form the axon. Panel “iii” shows a higher magnification image of a migrating EP cell to visualize the filopodia associated with its leading process (see Supplementary movie #S3). Boxed insets indicate filopodia extending onto its muscle band pathway (band) and onto adjacent interband musculature (ib) that are quantified in panel H. H. Filopodial dynamics of a migrating EP cell over the course of 30 min (data collected from Supplementary movie #S3); upper panel indicates the average length of filopodia extending along the band pathway (red histograms) or adjacent interband musculature (shaded histograms). Lower panel shows the average number of filopodia on the band versus interband regions. Scale = 0.75 cm in A; 10 µm in B; 40 µm in C–D; 20 µm in E–F. The average somal length of the migrating EP cells in panel G is ~15 µm.
Morphological and histochemical analyses of the ENS can be performed in whole-mount
Since all of the components of the insect ENS arise superficially on the embryonic gut (Fig. 2B–F), a variety of methods can be used to visualize their development in minimally dissected, whole-mount preparations, mitigating the need for more extreme dissections or histological sectioning and reconstruction. By simply opening the dorsal body wall of embryos before fixation, both mRNA distributions (via in situ hybridization histochemistry) and protein expression patterns (via immunostaining) can be visualized in the intact ENS. For example, Figure 2C shows a whole-mount preparation of the Manduca gut immunostained for the adhesion receptor fasciclin II, revealing that both the migrating enteric neurons (“ep”) and their muscle band pathways (“b”) on the gut surface express this homophilic adhesion receptor (discussed in more detail below).
Manipulations of the developing ENS can be performed in cultured embryos
The robust nature of insect preparations has also permitted the establishment of intact and semi-intact culture protocols for directly accessing the ENS within developing embryos (Horgan et al., 1994; Ganfornina et al., 1996; Wright and Copenhaver, 2001; Haase and Bicker, 2003). Because insect embryogenesis is both precisely regulated and temperature-dependent, timetables of unambiguous markers can be used to stage embryos within 1% of development, greatly facilitating the generation of synchronous groups of animals (Copenhaver, 1993; Ganfornina et al., 1996; Horgan and Copenhaver, 1998; Haase and Bicker, 2003). Epitope-tagged ligands and antibodies against cell surface receptors can be used to label embryos prior to fixation (Fig. 2D), providing accurate maps of bioavailable proteins in the ENS at specific times during embryogenesis. Reagents designed to target specific protein interactions (such as competing peptides and blocking antibodies) or pharmacological compounds directed against intracellular signaling pathways can also be applied directly to cells within the developing ENS (Horgan et al., 1994; Wright et al., 1998; Wright et al., 1999; Haase and Bicker, 2003). In addition, cell-impermeable compounds (including bioactive toxins and antisense oligonucleotides) have been successfully introduced with selective permeabilizing agents to test the role of particular gene products in the differentiation of enteric neurons and glia (Horgan et al., 1994; Wright and Copenhaver, 2000).
For more sophisticated analyses, intracellular injection techniques have been established that permit compounds to be directly introduced into individual cells within the embryonic ENS, while neighboring cells within the same preparation provide an ideal set of matched internal controls. Labeled molecules and fluorescent dyes can be injected into enteric neurons or glial precursors at specific developmental stages, followed by immunohistochemical counterstaining to delineate their complete morphology (Fig. 2E). For functional assays of candidate signaling pathways, cells can be injected in vivo with activated or inactivated forms of a candidate molecule (or appropriate toxins) to test their effects on specific aspects of neuronal migration and differentiation behavior (Horgan et al., 1995; Copenhaver et al., 1996; Horgan and Copenhaver, 1998). Alternatively, a variety of recording techniques can be used to monitor the electrophysiological activity and intracellular calcium dynamics of individual neurons in vivo (e.g. Horgan and Copenhaver, 1998). To inhibit the expression of a particular gene of interest, antisense oligonucleotides and double-stranded RNAs can be injected into the different cell types associated with the developing ENS (Knittel et al., 2005), while intracellular injections of synthetic RNAs can be used to induce ectopic gene expression in a cell-specific manner (Fig. 2F). The unique features of the insect ENS thus permit manipulations of a particular gene to be performed within individual cells at specific phases of development, thereby avoiding the types of pleiotropic effects that often result from more global expression methods.
Neuronal migration can be imaged within the developing ENS
Several groups have established methods for examining the behavior of insect enteric neurons and glia in vivo. Live-cell staining with the membrane-permeable dye DiO has been used to label the ENS of dissected grasshopper embryos, allowing the migration of particular groups of neurons to be followed by time-lapse imaging (Haase and Bicker, 2003). We also recently adapted our intracellular injection protocols using fluorescent dextrans to monitor the behavior of individual neurons in the ENS of Manduca, including cells undergoing active migration (Fig. 2G panel i; Supplementary movie #S1), and neurons transitioning from migration to axonal outgrowth (Fig. 2G, panel ii; Supplementary movie #S2). Faster image-capture rates have also been used to visualize the filopodial dynamics of migrating neurons (Fig. 2G, panel iii; Supplementary movie #S3). Besides revealing the intrinsic behavior of enteric neurons at different times and locations within the ENS, these protocols also provide a sensitive bioassay for quantifying the effects of a particular manipulation on specific aspects of their motility and guidance—again, at the level of single cells (Fig. 2H).
In summary, the accessibility of the insect ENS permits a range of methodologies to be directly applied to neurons in semi-intact embryos, eliminating the need for more reduced cell culture preparations. These approaches have been used to provide comprehensive views of the normal sequence of development in these systems, and they can be exploited to investigate how different components of the ENS are assembled. As summarized in the following sections, many of the morphogenetic processes associated with the vertebrate PNS also occur in the insect ENS, highlighting the potential value of these simpler preparations for deciphering the underlying molecular mechanisms that regulate them.
3. Neurogenesis in the insect ENS
Unlike the generation of neurons in the insect CNS, where most precursors delaminate as individual cells and produce stereotyped lineages (Doe et al., 1985; Pearson and Doe, 2004; Karcavich, 2005), the neurons and glia of the insect ENS arise via developmental programs that resemble the epithelial-to-mesenchymal transitions associated with the vertebrate neural crest and cranial sensory placodes. Early in embryogenesis, the embryonic foregut and hindgut of insects originate as ectodermal invaginations named the stomodeum and proctodeum, by direct analogy to the vertebrate gut (Poulson, 1950; Campos-Ortega and Hartenstein, 1997). Anterior and posterior midgut primordia associated with the foregut and hindgut then fuse to form the endodermal layer of the midgut, while visceral mesodermal cells invest the gut with appropriate layers of circular and longitudinal muscle (Skaer, 1993; Campos-Ortega and Hartenstein, 1997). As the foregut invaginates, midline cells within its dorsal epithelium thicken and become columnar, forming the ENS anlage (Schoeller, 1964; Hartenstein et al., 1994; Ganfornina et al., 1996). Three neurogenic zones then differentiate within this primordium, which will give rise to the enteric ganglia and nerves of the foregut (Fig. 3).
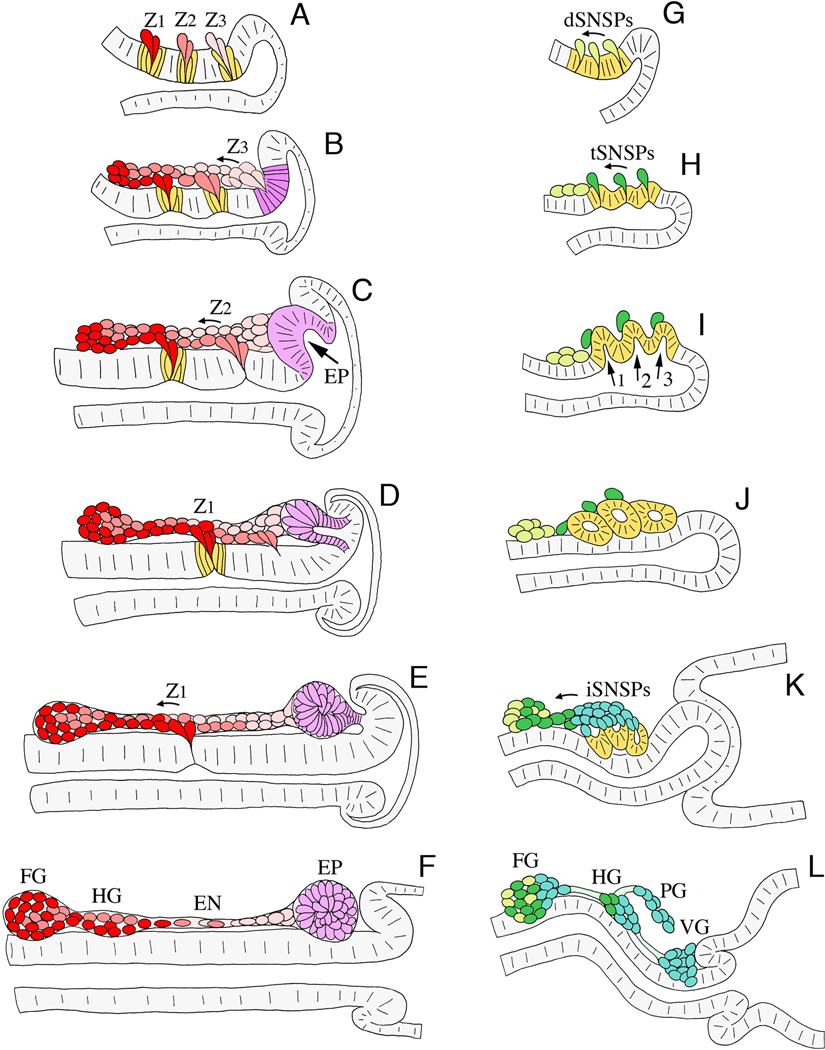
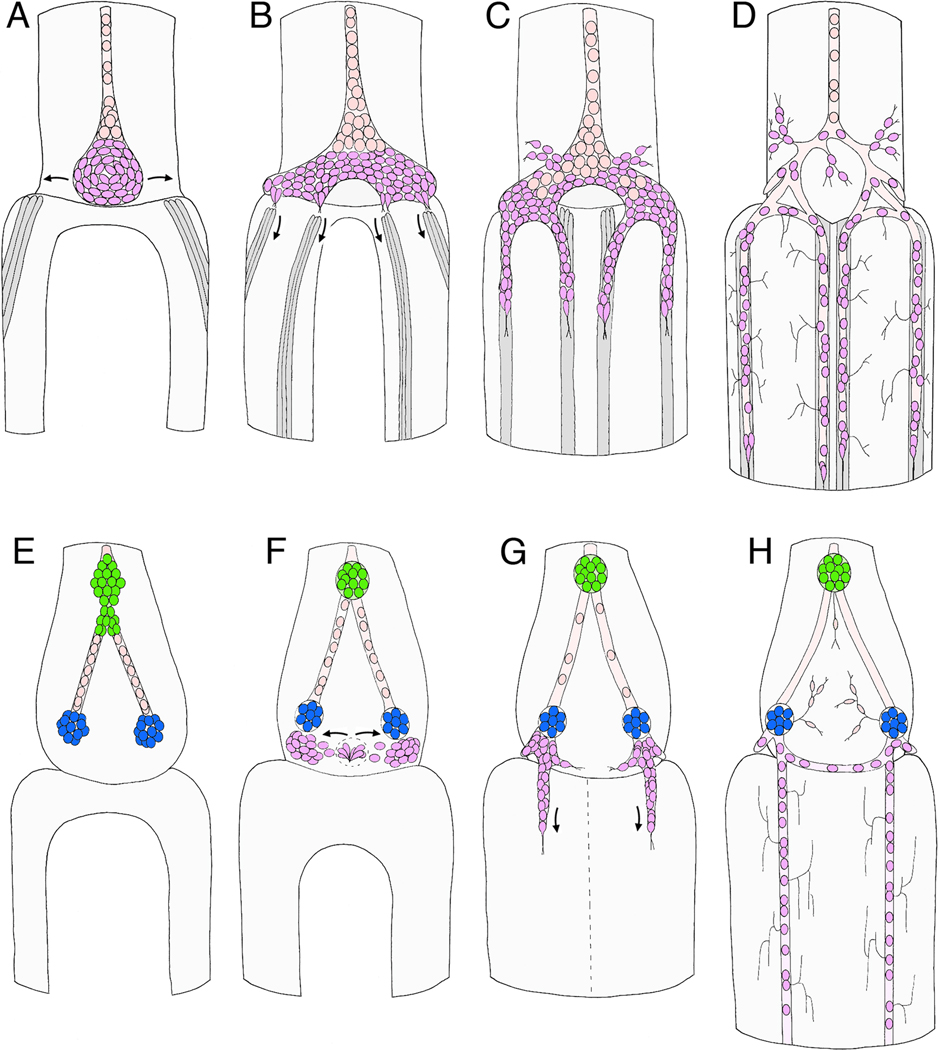
Figure 3. Schematic illustration of neurogenesis in Manduca (A–F; after Copenhaver and Taghert, 1991) and Drosophila (G–L; after Hartenstein, 1997).
Each panel represents a sagittal view of the foregut midline; anterior is to the left, dorsal is to the top. A. At ~24% of development in Manduca, three neurogenic zones have formed in the dorsal foregut epithelium (Z1, Z2, & Z3), from which a series of individual precursors will delaminate; each precursor cell divides once or twice after emerging. B. By 28% of development, streams of zone-derived cells have begun to migrate anteriorly along the foregut, while all of the remaining zone 3 cells delaminate. The epithelium surrounding the original position of zone 3 subsequently differentiates into a distinct placode (purple). C. By 33% of development, migrating zone cells have begun to form the frontal ganglion anteriorly, while all of the remaining zone 2 cells delaminate. The EP cell placode has also begun to invaginate. D. At 36% of development, zone-derived cells continue to migrate along the pathways that will form the recurrent and esophageal nerves, while the EP cell placode invaginates. E. By 39% of development, the last of the zone 1 cells delaminate, and the EP cell placode has invaginated onto the foregut surface. F. By 42% of development, the frontal and hypocerebral ganglia have formed; subsets of the residual zone cells will proliferate to form glial populations that ensheath the nerves and ganglia of the ENS. At this stage, the invaginated EP cells form a condensed packet of post-mitotic neurons adjacent to the foregut-midgut boundary. G. In Drosophila at stage 10 (~24% of development), three neurogenic centers form within the ENS anlage (yellow) and give rise to a set of delaminating precursors (dSNSPs; light green). H. At stage 11 (~30% of development), the dSNSPs have migrated anteriorly, where they will help form the frontal ganglion and its nerves. A second wave of precursors (tSNSPs, dark green) delaminates from the neurogenic centers, marking the positions where three invaginations will form. I. By stage 12 (~34% of development), the three invaginations form distinct pouches (1, 2, & 3) that protrude onto the foregut surface. J. By stage 13 (~38% of development), the three pouches have pinched off to form a set of neurogenic vesicles, while the tSNSPs migrate anteriorly to help form the frontal and hypocerebral ganglia. K. By stage 14 (~42% of development), invaginated cells from the three vesicles begin to dissociate (iSNSPs) and migrate anteriorly. L. By stage 16 (~60% of development), the SNSPs have assembled into the enteric ganglia of the foregut: frontal ganglion (FG), hypocerebral ganglion (HG), paraesophageal ganglion (PG), and ventricular ganglion (VG). Processes from these neurons also pioneer the interganglionic nerves.
Two distinct “programs of neurogenesis” can be distinguished in the developing ENS that correspond to the organization of the neurons and glia they produce. Many of the cells that form the enteric ganglia emerge from the neurogenic zones via sequential delamination, while placode invagination generates groups of neurons that can rapidly disperse to form more diffusely organized nerve plexuses. Species-specific differences in the utilization of these two modes of neurogenesis give rise to the morphological variations seen in the ENS. However, the control of these two neurogenic programs involves closely similar genetic mechanisms, suggesting that subtle variations in the regulation of epithelial-to-mesenchymal transitions underlie the evolutionary plasticity manifested in the development of the insect ENS.
In the larger embryos of Lepidopteran and Orthopteran species, the two neurogenic programs are separated both temporally and spatially (Copenhaver and Taghert, 1991; Ganfornina et al., 1996). Sequential delamination of neuronal precursors from three neurogenic zones (Z1, Z2, & Z3) first gives rise to the ganglia and nerves of the foregut (Figs. 3A–E, 4A–C). Cells within each zone expand apically while contracting basally (Figs. 3A, 4A), forming rosettes of cells that correspond to the “clear zones” originally described by Baden (1936). A series of individual precursors then delaminate from each zone, emerging as streams of cells that migrate anteriorly to form the frontal and hypocerebral ganglia (Figs. 3B–F, 4B–D). Each zone-derived cell becomes mitotically quiescent as it undergoes an epithelial-to-mesenchymal transition, but then typically undergoes one or two rounds of additional divisions as it begins to migrate, giving rise to post-mitotic neurons. In this regard, these zone-derived precursors resemble the midline precursors of the insect CNS, which also typically divide once after they delaminate before differentiating (Klämbt and Goodman, 1991). At the same time, the primary neurites of these cells begin to pioneer the tracts that will eventually form the nerves of the foregut (Fig. 4D). Towards the end of this neurogenic sequence, the remaining cells of each zone emerge as a group; first zone 3, then zone 2, and then zone 1 cells ingress onto the foregut as clusters (Figs. 3, 4). Interestingly, these last waves of cells provide glial precursors that continue to proliferate as they migrate along the pathways formed by the enteric neurons, ensheathing the nerves and ganglia of the ENS (Copenhaver, 1993).
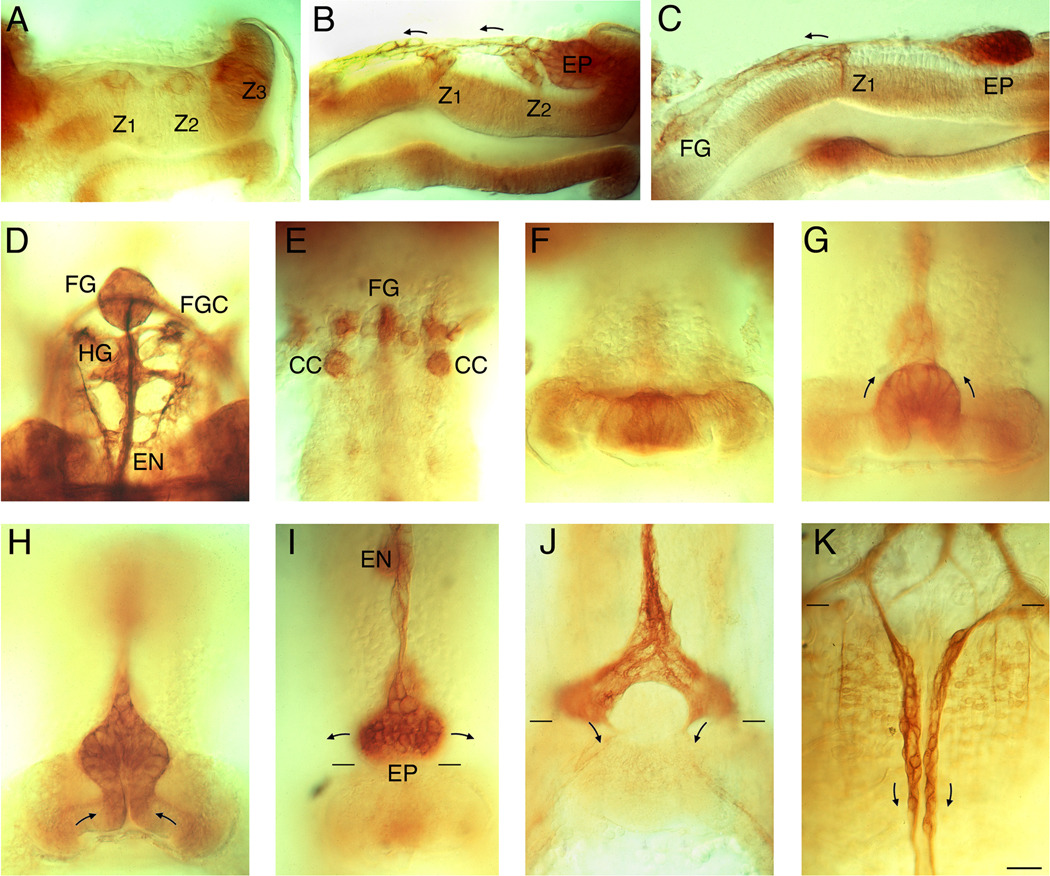
Figure 4. Photomicrographs of the developing ENS in Manduca (immunostained with anti-Fas II antibodies; after Copenhaver and Taghert, 1990, 1991).
A–C. Lateral views of the foregut at 24%, 33%, and 39% of development. Z1, Z2, and Z3 indicate the three neurogenic zones of the foregut; EP = the invaginating placode giving rise to the migratory neurons that populate the midgut. D. Dorsal view of the frontal ganglion (FG) and hypocerebral ganglion (HG) at ~60% of development; the frontal ganglion connectives to the brain (FGC) and posterior esophageal nerve (EN) are also visible. E Dorsal view of a younger embryo (~35% of development) shows the emergence of cells that will migrate off the foregut to form the intrinsic neurons of the corpora cardiaca (CC). F–H: dorsal views of the posterior lip of the foregut at 30%, 34%, and 38% of development, showing the invagination of the EP cell placode. I. By 42% of development, the EP cells have invaginated to form a packet of post-mitotic neurons adjacent to the foregut-midgut boundary (paired black lines). Anteriorly, the EP cell packet is in continuity with the residual zone 3 cells and the developing esophageal nerve (EN). Arrows indicate the directions the EP cells will subsequently follow as they spread bilaterally around the foregut. J. By 55% of development, the EP cells have spread almost completely around the foregut and have begun to align with eight longitudinal muscle bands that form on the midgut as it closes dorsally. Arrows indicate the direction that subsets of EP cells will follow once migration onto the midgut commences (the dorsal pair of muscle bands can be faintly seen below the arrows). K. By 58%, subsets of EP cells have begun to migrate rapidly along the muscle band pathways on the midgut; only the dorsal pair of eight parallel bands are shown. Scale = 40 µm.
As this period of sequential delamination is ending in Manduca, a second program of neurogenesis commences via placode invagination (Copenhaver and Taghert, 1990). Once all of the zone 3 cells have delaminated, a new neurogenic placode forms in the posterior foregut epithelium, adjacent to the foregut-midgut boundary (EP; Figs. 3C, 4B, 4F). The cells of this placode then invaginate en masse to form a tight packet of immature neurons on the foregut surface (Figs. 3D–F, 4G–I). Unlike the earlier waves of precursors that arise by delamination, these placode-derived neurons (“EP” cells) become post-mitotic as they lose their epithelial organization (Copenhaver and Taghert, 1990). As described below, the EP cells subsequently disperse via two phases of migration, first spreading bilaterally around the foregut and then migrating rapidly onto the midgut (Fig. 4J–K). Small contingents of these neurons also occupy scattered positions across the posterior foregut, perhaps functionally replacing the ventricular/ingluvial ganglia found in other species (Fig. 1B).
A variation on this dual program of neurogenesis occurs in the grasshopper ENS (Ganfornina et al., 1996). As in Manduca, three neurogenic zones within the foregut epithelium give rise to streams of precursors that migrate anteriorly to form the frontal and hypocerebral ganglia, while a smaller subset migrates bilaterally to form the paired ingluvial ganglia (Fig. 1C). Towards the end of this period of sequential delamination, a second phase of neurogenesis commences with an expansion of the third neurogenic zone. However, rather than invaginating as a coherent placode, the cells of this region undergo a rapid period of ingression, giving rise to a distinct population of precursors, which become post-mitotic soon after they emerge onto the foregut. Like the EP cells of Manduca, this new population subsequently undergoes two phases of migration, ultimately forming the midgut enteric plexus (summarized below; see Fig. 6).
Figure 6. Schematic illustrations of the sequence of migration that forms the midgut enteric plexus in the insect ENS.
A–D represents Manduca (after Copenhaver and Taghert, 1989b; Copenhaver, 1993); E–H represents Schistocerca (after Ganfornina et al., 1996). All panels show dorsal views of the developing ENS at the foregut-midgut boundary. A. At 40% of development, the EP cells have invaginated onto the dorsal foregut surface and begin to spread bilaterally around the foregut-midgut boundary. Concurrently, subsets of longitudinal muscles on the midgut (dark grey cells) begin to coalesce as dorsal closure of the midgut proceeds. Anteriorly, the EP cell packet is in continuity with the residual zone 3 cells that help form the esophageal nerve; subsets of these zone-derived cells will subsequently proliferate to form glial cells that ensheath the enteric plexus. B. By 55% of development, the EP cells have almost completely surrounded the foregut, and subsets of the cells have begun to align with each of the eight midgut muscle bands (only the dorsal four are shown). C. By 58% of development, the EP cells have begun to migrate posteriorly along the muscle bands on the midgut; a small number of neurons also migrate laterally onto radial muscles of the foregut (foregut muscles not shown). D. By 80%, the EP cells have completed their migration, forming the enteric plexus that spans the foregut-midgut boundary; they have also extended axons along the posterior midgut (not shown) and short terminal branches onto the adjacent interband musculature. Glial cells (pink) derived from the residual zone 3 cells have also migrated over the major branches of the enteric plexus to ensheath them. E. By 40% in the Grasshopper ENS, the neurogenic zones of the foregut have given rise to the cells of frontal ganglion (not shown) and hypocerebral ganglion (green), while posteriorly, cells derived from the third neurogenic zone have migrated bilaterally to form the incipient ingluvial ganglia (blue). F. By ~50% of development, a second wave of neurogenesis from the vicinity of zone 3 has begun to produce a new population of ingressing cells (purple); these cells then migrate bilaterally and aggregate adjacent to the ingluvial ganglia. G. By ~60% of development, four streams of cells have begun to migrate posteriorly from the ingluvial ganglia onto the midgut (only the dorsal pair is shown). Unlike Manduca, distinct muscle bands on the midgut have not been detected in grasshopper. H. By 80% of development, the migratory populations of neurons have become distributed along the entire length of the midgut and have extended terminal branches onto the adjacent musculature. An additional set of cells derived from the neurogenic zones (presumably sensory neurons) also contributes to an extensive foregut plexus (not detected in Manduca).
In Drosophila, the formation of the ENS is compressed both temporally and spatially, with a concomitant overlap in the two programs of neurogenesis (Fig. 3G–L; after Hartenstein et al., 1996). As in larger species, the formation of the Drosophila ENS commences with the appearance of a neurogenic primordium in the foregut epithelium (Schoeller, 1964; Campos-Ortega and Hartenstein, 1997). Three neurogenic centers then coalesce within this primordium, equivalent to the distinct neurogenic zones of other species. Once formed, these centers generate an initial wave of delaminating neural precursors named “dSNSPs” (for delaminating stomatogastric nervous system precursors; Hartenstein et al., 1994). The newly delaminated dSNSPs then migrate anteriorly to help form the frontal ganglion and its nerves (Fig. 3G–J), and the cycle is repeated. However, as the second wave of cells begins to delaminate (called “tSNSPs” for tip cell SNSPs; Fig. 3H), the three neurogenic centers also begin to invaginate, each with a tSNSP at its apical tip (Fig. 3I). These three invaginations then rapidly bud off to form epithelial vesicles on the foregut surface (Fig. 3J), akin to the invagination of the EP cell packet in Manduca. Besides foreshadowing the positions at which these vesicles emerge, the initial waves of dSNSPs and tSNSPs may actively define the invagination centers by altering the mechanical stability of the epithelium and by releasing signals that promote these morphogenetic movements (Hartenstein, 1997; González-Gaitán and Jäckle, 2000; and discussed below). Only subsequently do the three epithelial vesicles disperse, extruding streams of cells called “iSNSPs” (for invaginating SNSPs) that form the rest of the ganglia and nerves of the ENS (Fig. 3K–L).
As seen in larger insects, the precursors that arise by sequential delamination in Drosophila (dSNSPs and tSNSPs) may divide once or twice during their initial migration before they differentiate. In contrast, neurons derived by placode invagination become post-mitotic as they lose their epithelial organization and tend to assume more variable distributions (Hartenstein et al., 1994; Hartenstein, 1997). These two neurogenic programs may therefore affect both the developmental potential of the cells that they produce and the morphogenesis of different structural features in the ENS. Precursors that delaminate individually tend to produce neurons that are recruited into the enteric ganglia in an orderly sequence, where many assume unique fates (Copenhaver, 1993; Miles and Booker, 1994; Miles and Booker, 1998). As in the CNS, the identity of some of the neurons derived by this process may be specified entirely by their lineage, but variability in the patterns of migration and assembly suggests that local interactions may also regulate their ultimate fates (Copenhaver and Taghert, 1991; Hartenstein et al., 1994; Ganfornina et al., 1996). In contrast, placode invagination tends to produce large groups of migratory cells that form more diffuse plexuses, and the differentiation of these populations appears to be regulated by environmental as well as lineage-derived cues. This second strategy may therefore provide a means of rapidly assembling simple neural networks (Ganfornina et al., 1996; Hartenstein, 1997).
Although not functionally part of the ENS, the intrinsic cells of the corpora cardiaca also emerge from neurogenic placodes in the lateral stomodeum of larger insects (Fig. 4E), whereupon they migrate off the foregut and become associated with nerves from the brain (Baden, 1936; Schoeller, 1964; Copenhaver and Taghert, 1991). The position and neurosecretory function of these organs is clearly analogous to that of the adenohypophysis in mammals (Rathke, 1838; Clynen et al., 2001; Kouki et al., 2001), and it is intriguing that both structures originate from placodes in the stomodeal ectoderm. By contrast, corpora cardiaca cells in Drosophila (which form part of the ring gland of the brain) segregate from the ectoderm before the foregut invaginates (De Velasco et al., 2004), similar to the segregation of adenohypophyseal precursors in zebrafish before invagination of the stomodeum (Herzog et al., 2003). Given recent evidence that similar regulatory genes control the development of the insect corpora cardiaca and vertebrate pituitary, these parallel variations in their morphogenesis support the proposal that the elements of a primordial neuroendocrine system were established early in the evolution of a common bilateral ancestor (De Velasco et al., 2004).
4. Genetic control of neurogenesis in the insect ENS
A cascade of pattern organizer genes establish the positions of neurogenic zones
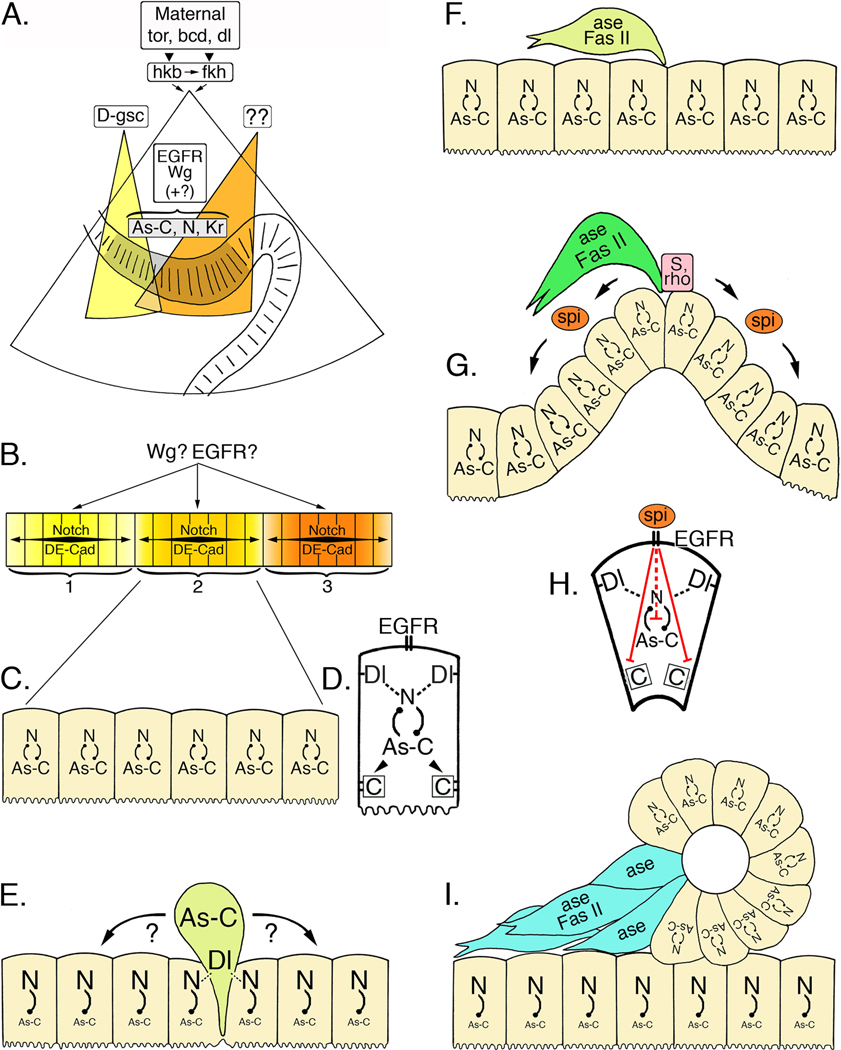
Meticulous studies by the Hartenstein and Jäckle laboratories have characterized the transcription factors and signaling molecules that establish the positions of the neurogenic zones within the stomodeum of Drosophila (González-Gaitán and Jäckle, 1995; Hartenstein et al., 1996; Dumstrei et al., 2002). As summarized in Hartenstein (1997), maternally derived torso, bicoid, and dorsal combine to drive the expression of huckebein and fork head (related to the FOX transcription factor family) throughout the stomodeum (Fig. 5A). Subsequently, the expression of several evolutionarily conserved regulatory genes, including Krüppel and members of the proneural Achaete-scute complex (As-C), delineate the ENS primordium within the stomodeum (shaded epithelium, Fig. 5A).
Figure 5. A cascade of regulatory genes controls neurogenesis in the ENS of Drosophila.
A. Maternally expressed torso (tor), bicoid (bcd), and dorsal (dl) control the expression of huckebein (hkb) and fork head (fkh) in the invaginating stomodeum (after Hartenstein, 1997). The homeobox gene D-goosecoid (D-gsc; yellow field) is required for the differentiation of the anterior stomodeum, including the anterior neurogenic center (zone 1) of the ENS. Other patterning genes (as yet unidentified; orange field) may specify the formation of the more posterior zones. As the stomodeum invaginates, the ENS anlage (grey shaded epithelium) becomes morphologically distinguishable and begins to express a combination of proneural genes in the Achaete-scute Complex (As-C), neurogenic genes (including Notch; N), and several other transcription factors, including Krüppel (Kr). Both Wingless (Wg) and EGFR signaling (plus other identified regulatory genes) play essential but poorly defined roles in this initial phase of ENS development. B. Wg and EGFR signaling may also help delineate the three neurogenic centers (1, 2, & 3) that subsequently form within the ENS anlage, possibly by limiting the range of Notch signaling within each zone and by modulating cell adhesive interactions mediated by Drosophila E-Cadherin (DE-Cad). C. All of the cells within each zone initially express intermediate levels of both proneural genes (As-C) and neurogenic genes (N). D. Enlarged view of a single zone cell at this stage. Notch (N)-Delta (Dl) interactions between adjacent cells are regulated in part by inhibitory feedback with the proneural genes (As-C); proneural genes may also regulate DE-Cadherin expression (boxed C). E. During the sequential delamination of individual precursors, lateral inhibition by the neurogenic genes promotes enhanced expression of the proneural genes (As-C) in a single zone cell, which then emerges from the foregut epithelium (light green cell represents a dSNSP). Cadherin-mediated adhesive interactions must also be down-regulated at this time. Concurrently, elevated levels of Notch signaling in the remaining zone cells help maintain their epithelial organization. F. Once delaminated, the precursor cell expresses the proneural gene Asense (ase), which may restrict further mitotic divisions, and the cell adhesion receptor Fas II, which may promote directed migration. The remaining zone cells re-acquire a balanced expression of both proneural genes and neurogenic genes. G. A second cycle of delamination gives rise to another set of precursors that emerge onto the foregut (dark green cell represents a tSNSP). As these precursors emerge, elevated levels of Star (S) and Rhomboid (rho) result in the localized release of the EGFR ligand, Spitz (spi), which in turn promotes the invagination of the remaining zone cells. H. Enlarged view of a single invaginating cell; EGFR signaling induced by Spitz interrupts the normal inhibitory feedback between neurogenic and proneural genes, permitting their continued co-expression. EGFR signaling also down-regulates DE-Cadherin-mediated adhesion, thereby promoting the morphological reorganization of the invaginating cells. I. Invagination of the neurogenic zone produces a discrete epithelial vesicle (see Fig. 3J), in which all of the cells continue to express both proneural genes (maintaining their potential to become neurons) and neurogenic genes (which help maintain their epithelial organization). By contrast, proneural gene expression is inhibited in the underlying epithelial layer. Individual cells from the vesicle then down-regulate neurogenic gene expression and disperse, while they upregulate Asense and Fas II (blue cells represent iSNSPs).
But how are discrete neurogenic zones within the ENS primordium actually specified? Work by González-Gaitán and Jäckle (1995) showed that the morphogen Wingless (Wg; the fly Wnt homologue) may play a role in this process. Mutations affecting the Wg signaling pathway resulted in the formation of only a single invagination center rather than three, suggesting that Wg signaling may control the range of Notch-dependent interactions that delimit the neurogenic zones (Fig. 5B). Alternatively, Wg signaling might only be required for the formation of two of the three zones, while one of the zones arises in a Wg-independent manner. This effect was originally proposed to be due to Wg-dependent regulation of armadillo/β-catenin, which in turn would modulate cadherin-dependent cell adhesion within the developing ENS (González-Gaitán and Jäckle, 1995). Subsequent studies argued against such a direct effect of Wg signaling on cell adhesion, due in part to the functional separation of β-catenin pools involved in Wg signaling versus cadherin-mediated interactions (Peifer et al., 1994; Dumstrei et al., 2002). Nevertheless, Wg/Wnt signaling can regulate cadherin-dependent adhesion by a variety of indirect pathways (Willert and Jones, 2006); conversely, Notch may inhibit Wg signaling via association with either disheveled or Axin, both of which regulate β-catenin levels (Axelrod et al., 1996; Hayward et al., 2006). How these types of interactions between the Wg and Notch signaling pathways help define neurogenic zones in the developing ENS remains enigmatic.
Other investigations have shown that the Drosophila homologue of the Epidermal Growth Factor Receptor (EGFR) is also essential for the initial formation of the ENS primordium (Fig. 5A). During gastrulation, normal EGFR signaling is required to prevent widespread apoptosis in the ENS anlage, similar to its actions in other developing midline structures within the head (Dumstrei et al., 1998). Studies on the differentiation of midline glia have shown that locally released EGFR ligands such as the TGFα-related protein Spitz suppress the activity of pro-apoptotic genes, providing a model for how trophic signals may regulate developmental cell death (Bergmann et al., 2002). EGFR signaling has also been shown to antagonize Notch-dependent transcription via several distinct mechanisms (Rohrbaugh et al., 2002; Voas and Rebay, 2004; Hasson et al., 2005), an interaction that plays an important role in regulating epithelial-mesenchymal transitions during later stages of neurogenesis in the ENS (summarized below). However, both Wg signaling and EGFR signaling can be detected uniformly throughout the ENS anlage during its initial formation (González-Gaitán and Jäckle, 1995; Dumstrei et al., 1998), so other patterning genes must restrict their effects to particular subdomains within the foregut epithelium. One of these genes is the homeobox gene D-goosecoid (Fig. 5A, yellow field), which is required for the formation of the most anterior neurogenic zone in the ENS (Hahn and Jäckle, 1996). Whether additional homeobox genes play similar roles in defining the more posterior zones remains to be determined.
A balance between proneural genes and neurogenic genes regulates the neurogenic programs of the ENS
As illustrated in Figure 5, all of the cells within the ENS primordium initially express a combination of proneural genes and neurogenic genes (González-Gaitán and Jäckle, 1995; Hartenstein et al., 1996). Given the conserved role of these two gene classes in regulating neurogenesis in both invertebrates and vertebrates (Anderson, 1999; Cau et al., 2002; Cornell and Eisen, 2005), it is not surprising that they are also essential to the differentiation of insect ENS. More unusual is the discovery that modulation of the normal inhibitory feedback interactions between proneural and neurogenic genes helps delineate the three neurogenic zones in the foregut epithelium, and also helps control the different neurogenic programs by which enteric neurons and glia are generated.
In the insect CNS, proneural gene expression becomes restricted to small clusters of cells within the neurectoderm, rendering these cells competent to form neuroblasts. Lateral inhibition by the neurogenic genes then rapidly restricts proneural gene expression to a single cell within each cluster, which subsequently delaminates and assumes a unique stem cell fate (Artavanis-Tsakonas et al., 1995; Wodarz and Huttner, 2003; Lai, 2004). A similar mechanism helps regulate the neurogenic program of sequential delamination in the developing ENS (Fig. 5C, D; after González-Gaitán and Jäckle, 1995; Hartenstein et al., 1996). Initially, all of the cells within each neurogenic zone simultaneously express a combination of neurogenic genes and members of the Achaete-scute Complex (As-C) of proneural genes, including achaete (ac), scute (sc), and lethal of scute (l’sc). Soon thereafter, lateral inhibition mediated by the neurogenic genes Notch (N), Delta (Dl), and Enhancer of split (E(spl)) down-regulates the As-C proneural genes in all but one of the cells in each zone, which in turn upregulates a subset of As-C genes as it delaminates (Fig. 5E).
Each delaminated cell subsequently expresses the proneural gene Asense (ase; Fig. 5F), which may help restrict its subsequent mitotic activity and promote its differentiation (Wallace et al., 2000). Newly delaminated cells also begin to express cell adhesion receptors like fasciclin II (Fas II; an orthologue of vertebrate NCAM), which play a prominent role in their subsequent migration (discussed below). Concurrently, the remaining epithelial cells within each zone re-acquire a balance of proneural and neurogenic gene expression (Fig. 5F), and the cycle of delamination is repeated. As noted above, the first wave of delaminated cells in Drosophila gives rise to the dSNSPs, while the second wave generates the tSNSPs (Fig. 3G–J). Loss-of-function mutations in the proneural genes reduce or eliminate the delamination of these precursors, as do gain-of-function mutations in the neurogenic genes. In contrast, loss-of-function mutations in the neurogenic genes results in the persistent expression of the proneural genes throughout the zones and a dramatic increase in the number of delaminating precursors. Thus, the neurogenic program of sequential delamination in the ENS resembles neurogenesis in the CNS: proneural genes render all of the zone cells competent to become neural precursors, while lateral inhibition by the neurogenic genes restricts the number of cells that can actually delaminate at any particular time (Hartenstein et al., 1996).
Still unanswered is how a single delaminating precursor can modulate proneural gene expression throughout the entire zone from which it emerges (Fig. 5E). One possible mechanism might involve Scabrous, a secreted glycoprotein that may help stabilize Notch in the membrane or prevent its inactivation, thereby extending the range of Notch signaling (Powell et al., 2001; Li et al., 2003). Scabrous-Notch interactions have been shown to sharpen the boundaries between proneural clusters in the developing eye (Powell et al., 2001) and regulate the spacing of sensory organ precursors (Renaud and Simpson, 2002). The scabrous gene also appears to be expressed within the newly formed ENS anlage (Mlodzik et al., 1990; Graba et al., 1992), but its potential role in establishing the neurogenic zones during subsequent development has yet to be investigated.
However, a markedly different relationship between proneural and neurogenic genes is required for placode invagination. While the tSNSPs increase their expression of ac as they delaminate onto the foregut, the remaining cells within each zone continue to express other proneural genes (including l’sc and sc) as well as the neurogenic genes (Fig. 5G). This pattern of coordinated gene expression persists within the zones for the next several hours, during which the cells invaginate as a group to form epithelial vesicles (Fig. 3J; González-Gaitán and Jäckle, 1995; Hartenstein et al., 1996). A loss of neurogenic gene function results in the premature, unorganized dispersal of the vesicles, while gain-of-function mutations (or a loss of proneural gene function) inhibits both the invagination of the vesicles and their subsequent dissociation. In this context, the normal inhibitory feedback by the neurogenic genes on proneural gene expression must be pre-empted, allowing all of the cells in each vesicle to retain their neurogenic potential while also maintaining their epithelial organization. This process, described as mutual inhibition (Hartenstein et al., 1996), is perhaps related to other instances where Notch signaling promotes the formation of sharply defined groups of cells with similar fates, which has been called lateral induction (de Celis and Bray, 1997; Portin, 2002; Daudet and Lewis, 2005). Only after the vesicles have invaginated do they begin to disperse as post-mitotic cells, producing iSNSPs (Fig. 5I). As with zone-derived precursors that delaminate individually, cells that have emerged via placode invagination also commence the expression of ase and Fas II as they lose their epithelial organization, consistent with their acquisition of a post-mitotic neuronal identity.
Thus, the interplay between proneural and neurogenic genes regulates whether cells within the ENS primordium assume an epithelial or mesenchymal organization, while subtle modifications in the balance between the two gene classes can produce remarkably different morphological rearrangements: delamination of individual cells, invagination of epithelial placodes, or their dissociation into populations of cells with similar fates. The same modulated pattern of gene regulation has been demonstrated in progenitor clusters giving rise to medial parts of the brain, optic lobes, and the larval eye (Dumstrei et al., 1998), and in the generation of clustered chordotonal sense organs (zur Lage and Jarman, 1999). In all of these regions, the prolonged co-expression of proneural and neurogenic genes maintains invaginating cell groups in an epithelial state for many hours before they dissociate, a sequence that is critical to their subsequent differentiation. Indeed, the regulation of epithelial-to-mesenchymal transitions by the neurogenic genes has been proposed to be a basic determinant of cell fate in many ectodermal and endodermal tissues (Hartenstein et al., 1992; Hartenstein et al., 1996; Dumstrei et al., 1998). Slight alterations in the balance between proneural and neurogenic genes might therefore provide a finely tuned mechanism for producing the diverse morphological features found in the developing ENS of different insect species. Vertebrate homologues of the neurogenic genes have similarly been implicated in the control of epithelial-to-mesenchymal transitions within the neural crest and cranial sensory placodes (Lewis, 1998; Anderson, 1999; Streit, 2004; Daudet and Lewis, 2005), although different experimental models have yielded disparate results regarding the contribution of these genes to neuronal differentiation (reviewed in Cornell and Eisen, 2005).
EGF receptor signaling plays a critical role in the morphogenesis of the ENS
Besides dynamic interactions between the proneural and neurogenic genes, EGFR signaling is also essential for the normal sequence of neurogenesis in the developing ENS (Dumstrei et al., 1998; González-Gaitán and Jäckle, 2000). Elegant work by Hartenstein et al. showed that local activation of the EGFR ligand Spitz by the rhomboid gene occurs specifically at the positions of the three neurogenic zones (Fig. 5G). EGFR signaling subsequently induces the invagination of these cell groups as neurogenic placodes, in part by down-regulating cadherin-mediated adhesion (Fig. 5H; Tepass et al., 1996; Dumstrei et al., 1998). Overactivation of EGFR signaling increases the number of invaginating cells and results in ENS hyperplasia, while inhibition of EGFR signaling eliminates placode invagination and results in a substantial increase in cell death. Localized EGFR signaling may be further enhanced by the secretion of Spitz from the tSNSPs after they delaminate from the neurogenic zones (Fig. 5G), helping to specify the sites of vesicle invagination (González-Gaitán and Jäckle, 2000). This pattern of EGFR signaling also regulates the morphogenesis of other midline components within the fly nervous system in a similar manner. In the developing optic placode, for example, direct interactions between the EGFR and cadherin-catenin complexes may modulate adhesion via the phosphorylation of armadillo/β-catenin, promoting the epithelial rearrangements required for normal eye development (Dumstrei et al., 2002).
The regulation of cadherin-mediated adhesion by EGFR signaling may therefore be a common mechanism for controlling epithelial-to-mesenchymal transitions by groups of cells that differentiate in a coherent manner. A similar mechanism has been demonstrated in vertebrate cell culture, where EGFR activation was shown both to phosphorylate β-catenin and to modulate cadherin expression (Huber et al., 2005). In the developing neural crest, regulation of cadherin expression by Snail family transcription factors plays an important role in initiating epithelial-to-mesenchymal transitions (Huang and Saint-Jeannet, 2004; Morales et al., 2005), while a variety of growth factors has also been shown to regulate cadherin-mediated adhesion (Pla et al., 2001). Whether receptor tyrosine kinases like the EGFR also induce more rapid alterations in cadher-independent adhesion remains to be determined.
How does EGFR signaling affect the balance of proneural and neurogenic genes to promote delamination versus invagination? Besides altering local adhesive interactions, EGFR signaling may also interrupt the normal inhibitory feedback between neurogenic genes and proneural genes, allowing all of the cells within a neurogenic group to remain neurally competent while undergoing morphogenetic rearrangements (Fig. 5H). In other contexts, the Notch and EGFR pathways have been shown to interact both antagonistically and synergistically. For example, Notch signaling can inhibit the production of EGFR ligands, while EGFR activation can stimulate Delta expression and promote Notch signaling (reviewed in Voas and Rebay, 2004; Doroquez and Rebay, 2006). During the segregation of chordotonal organ clusters in Drosophila, EGFR activation can also apparently countermand the repression of proneural gene expression by E(spl), thereby allowing proneural and neurogenic gene expression to be maintained in the same cell (zur Lage and Jarman, 1999). More recently, EGFR signaling has been shown to antagonize E(Spl) by compromising the activity of an essential co-repressor, Groucho (Hasson et al., 2005). Both of these effects would preclude the down-regulation of proneural genes by Notch activation without inhibiting neurogenic gene expression per se, as originally postulated by Dumstrei et al. (1998).
Within the neurogenic zones of the insect ENS, EGFR signaling may therefore modulate both cadherin-dependent aspects of epithelial morphogenesis and the process of lateral inhibition, allowing all of the cells within each placode to differentiate as neurons (Fig. 5H). At the same time, persistent levels of Notch signaling may restrict the expression of EGFR ligands like Spitz to the neurogenic zones. This model is also consistent with the proposal that EGFR signaling may adjust the range of Notch-dependent lateral inhibition within a neurogenic zone, possibly acting in conjunction with Wg signaling to regulate local adhesive interactions (Fig. 5B; González-Gaitán and Jäckle, 1995; González-Gaitán and Jäckle, 2000). In this manner, interactions between EGFR and Notch signaling may simultaneously regulate cell fate decisions within the neurogenic placodes and the morphogenetic rearrangements required for their differentiation (Dumstrei et al., 2002).
5. Cell migration is critical to the formation of the insect ENS
Whereas directed cell migration plays a prominent role in the developing nervous systems of vertebrates, most newly generated neurons and glial cells in insects undergo only limited movements and typically begin to differentiate soon after they are born (Heathcote, 1981; Doe et al., 1986; Klämbt et al., 1991; Edenfeld et al., 2005). In contrast, the development of the insect ENS involves extensive phases of neuronal and glial migration that are critical for the innervation of the gut, and many of these cells delay their differentiation until after migration is complete. As already noted, the foregut components of the ENS are formed by the migration and assembly of cells that emerge from the neurogenic zones. However, the most dramatic examples of neuronal migration occur during the formation of the midgut enteric plexuses of larger insects, providing useful preparations for investigating the mechanisms by which neurons select and follow particular migratory pathways in vivo.
In Manduca, the migratory sequence giving rise to the midgut enteric plexus begins with the en masse invagination of the EP cells from the foregut epithelium, as noted above (Figs. 3, 4F–I). This neurogenic sequence results in a tight packet of post-mitotic but undifferentiated neurons on the dorsal surface of the foregut (Fig. 4F–I), which then disperses via two distinct phases of migration (Copenhaver and Taghert, 1989b). Initially, the packet spreads bilaterally to form a ring of cells around the foregut (Figs. 4J, 6A–B); at the same time, subsets of longitudinal muscle cells on the midgut coalesce into eight distinct muscle bands (shaded cells in Fig. 6A–D). Most of the EP cells then disperse rapidly along these muscle bands to form the midgut plexus, while smaller subsets travel laterally onto radial muscles on the foregut (Figs. 4K, 6C–D; see Supplementary movie #S1). While the positions of the eight band pathways on the midgut is highly predicable, individual neurons select their migratory route in a probabilistic manner, based on their proximity to a particular band and by interactions among neighboring neurons (Copenhaver and Taghert, 1989a, b).
The EP cells travel up to 250 µm in 6–8 hours (~30–40 µm/hr) and then transition from migration to axonal outgrowth (Supplementary movie #S2). Consequently, the neurons occupy only the anterior portion of the midgut plexus, while their axons and terminal branches provide diffuse innervation to the rest of the midgut musculature (Figs. 1B, 6D). The EP cells also delay their differentiation until their migration is complete, and then acquire a variety of phenotypes that are regulated in part by their final positions on the gut (Copenhaver and Taghert, 1989a; Copenhaver et al., 1996). This developmental sequence of directed migration and delayed differentiation shares a number of general features with the formation of the vertebrate ENS, in which enteric neural crest-derived cells (ENCCs) migrate substantial distances to form the nerve plexuses of the gut while delaying their terminal differentiation until migration is largely complete (Gershon et al., 1993; Gershon, 1997; Burns, 2005). Unlike the ENCCs, which continue to proliferate throughout much of their migration, the EP cells in Manduca migrate as post-mitotic but undifferentiated neural precursors. In contrast, a trailing set of proliferating glial cells then migrates down the newly formed branches of the enteric plexus, ensheathing the newly dispersed populations of neurons (Copenhaver, 1993; pink cells in Fig. 6A–D). As noted above, these glial precursors are among the last cells to emerge from the neurogenic zones of the foregut, but they delay their subsequent proliferation and differentiation until after the EP cells have begun to migrate.
A similar developmental sequence gives rise to the midgut enteric plexus in grasshoppers, although with interesting variations (Ganfornina et al., 1996). As noted above, after the three neurogenic zones have produced the ganglia and nerves of the foregut, a second round of neurogenesis generates a distinct population of neurons that will populate the midgut (Fig. 6E–H; purple cells). Like the EP cells of Manduca, the cells produced during this second neurogenic phase become post-mitotic as they leave the foregut epithelium but remain undifferentiated as they spread bilaterally, accumulating near the ingluvial ganglia on either side of the foregut (Fig. 6E–F). Subsequently, they commence an extended phase of migration, forming four columns of neurons that eventually span the entire length of the midgut (Figs. 1C, 6G–H). During this process, they establish the major branches of the midgut enteric plexus, while their terminal processes innervate the lateral musculature. As in Manduca, these migratory neurons follow predictable trajectories along the gut; however, a distinct set of muscle bands do not form in this species, and the cues that establish migratory pathways on the grasshopper midgut are less well understood. Although these migratory neurons are generated by ingression rather than by the coherent invagination of a placode, they otherwise resemble the EP cells of Manduca, suggesting that a common developmental program underlies the formation of the midgut plexuses in both species. Surprisingly, Drosophila lacks this additional phase of neurogenesis and migration and never forms a midgut plexus (Fig. 1D; Hartenstein et al., 1994; González-Gaitán and Jäckle, 1995), perhaps reflecting the relatively small size and accelerated development of this species compared to larger insects.
Neuronal migration is precisely guided in the insect ENS
Using cultured embryos, we have investigated the control of EP cell behavior in vivo by a variety of methods (Fig. 2; Horgan et al., 1995; Copenhaver et al., 1996; Horgan and Copenhaver, 1998). During their initial phase of circumferential migration around the foregut, the neurons extend short, exploratory filopodia but remain closely aligned with the foregut-midgut boundary, and their dispersal onto the midgut is delayed until the muscle bands have condensed. Once they begin to migrate onto the midgut, the neurons actively continue to select their migratory route: filopodia that they extend onto the adjacent interband muscles are rapidly retracted, while those extended along their chosen band pathway are retained, helping to form their leading processes (Fig. 2E & G, and Supplementary movies S1 & S3). At the onset of migration, the leading groups of EP cells resemble neurons undergoing chain migration (Lois et al., 1996; Wichterle et al., 1997; Anderson et al., 2006), during which individual neurons crawl over one another in an apparently stochastic manner. As they continue to disperse, however, the cells become increasingly scattered along the length of the gut, during which individual cells (or small clusters) can migrate substantial distances in advance of the trailing population.
Using surgical manipulations of the developing ENS, we showed that the eight midgut muscle bands are both sufficient and necessary to promote neuronal migration (Copenhaver et al., 1996). When a particular band was removed before the onset of migration, neurons that had aligned with the ablated band could course-correct onto a nearby band but never migrated onto the adjacent interband musculature. Conversely, when EP cells were transplanted onto the posterior regions of a host midgut, the neurons grew out along the muscle bands but again avoided the interband regions. Thus, the eight muscle bands act as equivalent pathways that promote migration, while strong repellent cues associated with the interband muscles inhibit migration onto these inappropriate domains. What are the guidance cues that specify the muscle bands as pathways for neuronal migration? How does the distribution of a particular cue alter migratory behavior? And how does the control of migration in the insect ENS pertain to similar events in vertebrates? Although these questions have been only partially explored, a variety of studies have shown the control of migration in these simple preparations involves evolutionarily conserved families of guidance factors and signal transduction molecules that also help regulate neuronal guidance in more complex systems, as summarized below.
Cell adhesion receptors define the migratory pathways
Among the guidance cues identified in the ENS of Manduca, members of the immunoglobulin (Ig) superfamily of adhesion receptors play a prominent role in controlling EP cell migration and outgrowth. In particular, the coordinated expression of Fas II by both the neurons and their muscle band pathways (Fig. 2C) is required for normal migration to occur. Like its vertebrate orthologues NCAM and OCAM, Fas II acts as a homophilic receptor that is expressed as multiple isoforms, including a glycosylphosphatidylinositol (GPI)-linked form and at least two transmembrane (TM) isoforms (Snow et al., 1988; Grenningloh et al., 1991; Wright et al., 1999). Notably, different Fas II isoforms are expressed by the EP cells at specific times during development and regulate distinct aspects of their behavior. Initially. during placode invagination, the EP cells express GPI-Fas II, suggesting that this isoform may participate in the segregation of neurons from the adjacent epithelium (Fig. 4F–H). Once emerged onto the foregut, the EP cells continue to express GPI-Fas II as they migrate circumferentially around the foregut (Fig. 4I–J). This pattern of expression is unusual, as most growing neurons express only transmembrane forms of Fas II (Wright and Copenhaver, 2001). When GPI-Fas II expression was inhibited in the EP cells during this period, they were still able to spread around the gut, but they became increasingly disorganized and failed to align with the muscle bands on the midgut. GPI-Fas II-dependent interactions therefore help maintain cell-cell contacts within the packet of undifferentiated neurons, which in turn helps position them correctly for their next phase of development (Wright et al., 1999; Wright and Copenhaver, 2000).
Shortly before the onset of migration onto the midgut, the EP cells rapidly eliminate GPI-Fas II expression and begin to express TM-Fas II. At the same time, the muscle bands also begin to express TM-Fas II, a pattern that is maintained throughout the subsequent migratory period (Figs. 2C, 2E, 4K; Wright and Copenhaver, 2000). When we used blocking antibodies or synthetic fragments of Fas II to interfere with Fas II-dependent interactions, EP cell migration onto the muscle bands was consistently inhibited, although these treatments did not induce ectopic migration onto the adjacent interband regions (Wright et al., 1999). A similar effect was seen when we blocked TM-Fas II expression in the developing ENS with antisense oligonucleotides (Wright and Copenhaver, 2000). Recently, we extended this analysis to the single cell level, using intracellular injections of morpholino antisense probes against Fas II. Down-regulating Fas II expression in individual EP cells inhibited their migration without affecting the behavior of neighboring neurons. Conversely, injecting Fas II-specific morpholinos into muscle band cells caused neurons that had aligned with that band to stall and defasciculate, while neurons on adjacent muscle bands were unaffected (Knittel et al., 2005; and unpublished data). These results indicate that Fas II-dependent interactions are indeed necessary for the correct migration of individual neurons in the ENS. Our results also provide an example of how isoform switching by a specific Ig-family adhesion receptor may regulate distinct aspects of neuronal development, as has also recently been proposed for NCAM (Polo-Prada et al., 2004). With the establishment of methods to induce ectopic gene expression in individual cells within the developing ENS (Fig. 2F), we can now test whether Fas II is sufficient to promote migration into regions that normally repel the EP cells, such as the interband domains of the midgut (Fig. 2C).
Several other aspects of Fas II-dependent migration still need to be determined. Too much adhesion can be just as inhibitory to outgrowth as insufficient adhesion. Both receptor turnover and antagonism by other guidance cues have been shown to regulate Fas II-dependent interactions in other contexts (Lin et al., 1994; Davis et al., 1997; Winberg et al., 1998; Yu et al., 2000), but whether these mechanisms also modulate the behavior of the EP cells remains to be explored. In the vertebrate ENS, NCAM is expressed both by migrating ENCCs and by the enteric mesenchyme through which they travel, and recent studies have shown that transient polysialylation of NCAM (a modification known to decrease NCAM-dependent adhesion) promotes the remodeling of migratory cell groups into presumptive ganglia (Faure et al., 2007). Insect adhesion receptors like Fas II may be preferentially fucosylated in the nervous system (Rendic´ et al., 2006), but the role of post-translational modifications in controlling Fas II-dependent aspects of neuronal motility have yet to be explored. Equally important are the signal transduction pathways that may be activated by Fas II. During the growth of neuromuscular junctions, both cAMP-mediated and Ras1/MAP kinase-mediated signaling can modulate Fas II-dependent adhesion (Davis et al., 1996; Schuster et al., 1996), but pharmacological manipulations of these pathways in the developing ENS of Manduca did not perturb EP cell migration (unpublished data). The PDZ protein Discs-Large (Dlg) also mediates interactions between Fas II and other proteins at developing synapses (Thomas et al., 1997; Zito et al., 1997), but Dlg is not expressed by the EP cells until after migration is complete. In contrast, we recently identified Manduca Rasputin as a candidate RhoGAP-binding protein that interacts with Fas II and is expressed by the EP cells during their migration (Copenhaver et al., 2006; and unpublished data), suggesting that Fas II may associate with distinct adapter and signaling cassettes at different stages and locations within developing neurons.
Lastly, Fas II is certainly not the only guidance receptor in the insect ENS. In Manduca, Neuroglian (the insect orthologue of vertebrate L1CAM; Bieber et al., 1989) is also expressed by the muscle bands but only at the end of the migratory period, when it may guide axonal outgrowth (Fig. 2D; Wright et al., 1999). By comparison, adhesive interactions mediated by L1CAM have been shown to promote the chain migration of ENCCs in the developing mouse ENS (Anderson et al., 2006). In grasshoppers, migratory enteric neurons have also been shown to express other candidate guidance cues, including Fasciclin I, Semaphorin 1, and Lazarillo (Ganfornina et al., 1996), although their contribution to the migratory process have not been examined. Most challenging will be to determine how individual neurons interpret input from multiple guidance receptors in choosing a correct migratory pathway, an issue that pertains to the guidance of migrating cells in the vertebrate ENS, as well (Newgreen and Young, 2002a; Heuckeroth, 2003; Young et al., 2004a).
Ephrin-Eph receptor interactions prevent inappropriate midline crossing
Recently, we discovered that Ephrin-Eph receptor interactions play an important role in establishing midline boundaries within the developing ENS. As in other insects, Manduca expresses only a single Ephrin ligand (MsEphrin, a GPI-linked Ephrin) and one corresponding Eph receptor (MsEph; Kaneko and Nighorn, 2003). Like many neurons in the CNS, the EP cells express MsEphrin throughout their motile phases of development (Fig. 2D). In contrast, MsEph expression is confined to discrete populations of midline interband cells on the gut, where the migrating neurons normally never travel (Fig. 2C–E; Coate et al., in press). When we applied Fc-MsEphrin fusion proteins to the developing ENS as a means of blocking endogenous ligand-receptor interactions, a substantial number of EP cells migrated and extended processes across these inhibitory midline regions (unpublished data). While Ephrin-dependent activation of neuronally expressed Eph receptors has been shown to modulate neuronal guidance in a variety of contexts (Santiago-Garcia and Erickson, 2002; Young et al., 2004a; Poliakov et al., 2004), our results in Manduca suggest that reverse signaling through a GPI-linked Ephrin can also regulate the motile behavior of developing neurons. We are currently using this preparation to decipher the signaling mechanisms by which this type of reverse signaling may regulate neuronal migration in developing embryos.
Nitric oxide (NO) signaling may serve multiple functions in the ENS
Many developing neurons in both vertebrate and invertebrate systems express NO-dependent soluble guanylate cyclases (sGCs; Bruning and Mayer, 1996; Truman et al., 1996; Hindley et al., 1997), and the ENS is no exception: most of the neurons that form the enteric ganglia on the foregut and all of the cells that populate the midgut plexuses exhibit robust levels of sGCs (Wright et al., 1998; Haase and Bicker, 2003). More unusual is the discovery that the enteric neurons continue to express NO-sensitive sGC activity throughout their differentiation, whereas most CNS neurons express sGCs only transiently (Dawson et al., 1991; Roskams et al., 1994; Ward et al., 1994). In vertebrates, different isoforms of NOS have been detected in enteric neurons at various stages of development (Balaskas et al., 1995; Van Ginneken et al., 1998; Arnhold et al., 2004), but a role for NO-dependent sGC activity in the formation of the ENS has not been systematically explored.
In Manduca, the EP cells begin to express sGCs coincident with the onset of their migration, but no detectable source of nitric oxide synthase (NOS) appears in the gut until after the neurons have stopped migrating and begin to form terminal synapses. Consistent with these observations, manipulations of NO-dependent sGC activity had no effect on EP cell migration but did perturb synapse formation, indicating that NO signaling plays a delayed role in regulating the differentiation of these neurons (Wright et al., 1998). In contrast, Haase et al. showed that the expression of sGCs by migratory neurons in the grasshopper ENS coincides with the appearance of NOS in the underlying midgut cells, while inhibitors of either sGC or NOS reduced neuronal migration in the midgut plexus (Haase and Bicker, 2003). NO signaling may therefore play a more immediate role during ENS development in this species, promoting the migration of enteric neurons along their pathways. Because both of the foregoing studies were based on pharmacological manipulations, additional experiments that directly target sGC and NOS expression will be needed to verify these results. Nevertheless, they provide an interesting view of how the same signal transduction pathway may be recruited to control different phases of enteric neuron development in a species-dependent manner.
A role for an Amyloid Precursor Protein-like protein in migration
The Amyloid Precursor Protein (APP) has been extensively studied as the source of Aβ fragments associated with amyloid plaque formation in Alzheimer’s disease, but the role of this protein during normal brain development remains controversial (De Strooper and Annaert, 2000; Selkoe, 2000). Our recent studies suggest that the Manduca orthologue of APP (Amyloid Precursor Protein-Like protein, or APPL) may help confine the migrating EP cells to their normal band pathways. This work was motivated by reports that human APP can interact with Goα, a heterotrimeric G protein that has been proposed to modulate APP-dependent aspects of neuronal growth and survival (Okamoto et al., 1995; Brouillet et al., 1999). We previously showed that activating Goα in the EP cells inhibits their migration via the induction of calcium spiking activity (Horgan et al., 1995; Horgan and Copenhaver, 1998), suggesting that Goα-coupled receptors might mediate the response of the migrating neurons to repulsive guidance cues. However, candidate Goα-coupled receptors expressed by the EP cells and ligands within the ENS that might activate them remained unidentified.
Using whole-mount labeling methods, we discovered that APPL is strongly expressed within the leading processes of the EP cells, where it co-localizes with Goα, and we showed that APPL interacts with Goα during ENS development (Swanson et al., 2005). In addition, interfering with APPL expression caused the EP cells to wander inappropriately off their band pathways and onto the lateral interband muscles, suggesting that APPL may interact with inhibitory cues in these regions (Copenhaver et al., 2004; and unpublished data). Intriguingly, a recent study using transgenic mice showed that the combined deletion of APP plus two related proteins (APP-like proteins APLP1 & 2) also caused excessive, inappropriate migration of neurons in the developing brain (Herms et al., 2004), although whether this effect involved the misregulation of Goα was not explored. This convergence of effects on neuronal migration in mice and Manduca supports the hypothesis that the APP family of proteins is evolutionarily ancient and may regulate conserved functions in the developing nervous system (Price et al., 1998; Coulson et al., 2000).
6. Perspectives: what can the insect ENS tell us about neuronal development in vertebrates?
Although much of this review has emphasized common features that can be identified in the ENS of insects and vertebrates, there are also important differences in the developmental programs that underlie the formation of these systems. Whereas enteric neurons and glial cells undergo extensive phases of migration before differentiating in both classes of organisms, all of the cells that form the vertebrate ENS are derived from the neural crest, a structure that first appeared in evolution with the emergence of the chordates (Baker and Bronner-Fraser, 1997; Northcutt, 2005). The importance of this process has been highlighted by the identification of human disorders that occur when ENCCs fail to migrate correctly, resulting in segments of the gut that never receive innervation (reviewed in Newgreen and Young, 2002a; Furness, 2006). Both the phenomenology and the control of ENCC development have now been extensively investigated; not surprisingly, these studies have identified a number of regulatory processes that may uniquely affect neural crest cells, and hence can only be tested in vertebrate model systems. Elegant work has been performed using avian, murine, and amphibian preparations to explore the mechanisms by which ENCCs populate the gut (reviewed in Newgreen and Young, 2002a; Burns, 2005; Heuckeroth and Pachnis, 2006), and a variety of sophisticated genetic approaches are now being used in zebrafish to identify additional genes that regulate ENS development and function (Pietsch et al., 2006; Kuhlman and Eisen, 2007). While a comprehensive discussion of the vertebrate ENS is obviously beyond the scope of this review, several key aspects should be considered when comparing this process to the formation of the ENS in insects.
Dynamic patterns of migratory and proliferative behavior drive the formation of the vertebrate ENS
The migration of ENCCs from the neural crest has been the focus of many studies over the past 30 years (Gershon, 1997; Le Douarin and Kalcheim, 1999), but only recently have new methods been developed that permit fluorescently labeled ENCCs to be monitored in vivo (Young et al., 2004b; Druckenbrod and Epstein, 2007). From these studies has emerged an intriguing view of how the ganglia and nerves of the vertebrate ENS are formed. As precursor cells travel from the vagal and lumbrosacral regions of the neural crest onto the embryonic gut, they establish migratory “wavefronts” of ENCCs that continue to proliferate as they spread through the gut mesenchyme. Time-lapse imaging has revealed that the behavior of the ENCCs is both dynamic and complex. Most ENCCs tend to migrate in strands that wind across the gut surface. In some regions, these columns resemble the patterns of “chain migration” of neural precursors in the developing olfactory bulb (Lois et al., 1996; Kornack and Rakic, 2001): columns of ENCCs stay closely associated as they migrate, crawling over one another as they advance. The behavior of individual cells (acting as leaders or followers) depends in part on their position relative to the migratory wavefront, reflecting a balance between factors that support intercellular adhesion and those that promote cell-cell dissociation (Young et al., 2004b; Druckenbrod & Epstein, 2007). The continued proliferation of undifferentiated ENCCs also promotes their continued dispersal, while perturbations that restrict ENCC numbers or induce their premature differentiation can lead to structural deficits in the ENS (Vohra et al., Flynn et al., 2007; and discussed below). This effect may be due in part to “population pressure” that drives ENCCs away from regions of high cell density (Hearn et al., 1998), although recent studies suggest that more subtle aspects of cell-cell interactions may determine when ENCCs disperse or aggregate (Natarajen et al., 1999; Druckenbrod and Epstein, 2007).
Unlike chain migration in the brain, however, where the columns of cells are confined by surrounding “tubes” of ensheathing astrocytes (Lois et al. 1996) and are guided by a variety of secreted molecules (Chen et al., 2001; Alberti et al., 2005), the factors that determine the specific directions taken by migratory ENCC columns are not well understood. Many of the same guidance cues that regulate neuronal migration and outgrowth in other contexts have been implicated in the control of ENCC migration, including components in the extracellular matrix, cell adhesion receptors, and diffusible guidance cues (reviewed in Young et al., 2004a; Burns, 2005; Faure et al., 2007). Local heterogeneity in the distributions of these cues within the gut mesenchyme may therefore help define the trajectories and termination sites of migratory ENCCs, although methods for visualizing these “microdomains” have yet to be developed (Young et al., 2004b). Also mysterious is the role of a subset of ENCCs (“advance” cells) that occasionally migrate in front of the interconnected ENCC strands, which might act as pioneers or might alter the distribution of local cues available to the trailing columns (Druckenbrod & Epstein, 2007). Lastly, in vitro studies have implicated both netrins (Jiang et al., 2003) and bone morphogenetic proteins (Chalazonitis et al., 2004) in the aggregation of enteric neurons into ganglia, but the molecular mechanisms regulating the formation and dissolution of migratory chains within the developing ENS remain largely unexplored (Young et al., 2004b).
Although these innovative approaches have revealed the dynamics of ENCC migration in unprecedented detail, manipulations of individual migratory ENCCs still cannot be readily performed within the developing ENS. By comparison, the simplified morphology and superficial position of the insect ENS has permitted the behavior of single cells to be monitored and manipulated in vivo, which has revealed some interesting parallels with the migratory behaviors of the ENCCs in vertebrates. As noted previously, neurons and glial precursors in the insect ENS also migrate in columns that resemble chain migration (Fig. 2), during which individual cells travel over one another in an apparently stochastic manner as they advance along the gut (Copenhaver and Taghert, 1989b; Horgan and Copenhaver, 1998). The speed at which cells travel within a column are also similar: in the Manduca ENS, average rates of migration are 30–40 µm/hr (at 28°C), while rates ranging from 35–50 µm/hr have been reported in the developing mouse ENS, although cells at the leading edge of the wavefront can occasionally move much faster (~85–90 µm/hr; Young et al., 2004b; Druckenbrod & Epstein, 2007). Transplantation and ablation studies in Manduca have shown that isolated columns and individual cells can continue to migrate normally when isolated from the rest of the developing ENS (Copenhaver et al., 1996; and unpublished observations), although repulsive interactions among neighboring cells may contribute to their dispersal along a pathway (similar to the population pressure model described above). Given these parallels, it would be interesting to test the role of cell-cell interactions within chains of migrating enteric neurons by selectively ablating individual cells (e.g. leaders or followers) within a motile column, or by manipulating the expression of candidate receptor-ligand complexes within individual cells to test their effects on chain migratory behavior.
Whereas the ongoing proliferation of undifferentiated ENCCs is required for their continued migration along the gut, the relationship between proliferation and migration tends to be slightly different in the insect ENS, where most neural precursors stop dividing shortly after they emerge from the neurogenic zones. As a result, most insect enteric neurons migrate as post-mitotic but undifferentiated cells, although like ENCCs in vertebrates, they typically delay their terminal differentiation until after their migration is complete. Thus, the continued proliferation of neural precursors is not required for the normal sequence of migration in the insect ENS (perhaps because of their smaller size and fewer cell numbers), but local cues encountered in the post-migratory environment appear to help regulate the terminal differentiation of enteric neurons, as in the vertebrate ENS. In contrast, glial precursors in the insect ENS do continue to proliferate throughout the course of their migration, and it has been presumed (but not proven) that this mitotic activity is required for the proper ensheathment of the enteric nerves (Copenhaver, 1993). It would be interesting to compare the effects of blocking glial proliferation in the insect ENS with the well-documented effects of premature ENCC differentiation on the innervation of the vertebrate gut.
Diffusible trophic factors play a prominent role in the developing vertebrate ENS
Investigations into the genes that cause developmental defects in humans and in rodent models have identified two interacting signaling systems that play critical roles in controlling ENS development. Migrating ENCCs express the RET receptor tyrosine kinase and the co-receptor GFRα1, which together mediate responses to glial-derived growth factor (GDNF) that is produced by embryonic gut mesenchyme. Concurrently, ENCCs also express the Endothelin-B receptor (ENDR-B), which mediates responses to Endothelin-3 (ET-3) derived from gut mesenchyme. Both of these diffusible trophic factors are required for normal ENS development: mutations in either signaling pathway cause serious abnormalities in the human ENS, including familial Hirschsprung's disease and intestinal neuronal displasia (Newgreen and Young, 2002a; Hofstra et al., 2002; Vohra et al., 2007). However, the effects these signaling systems on ENS development are complex, and the molecular mechanisms by which they interact are still being deciphered. GDNF can promote ENCC proliferation, differentiation, chemoattraction, and survival in different assays, while ET-3 appears to be particularly important during the early phases of ENS development, when the ENCCs first begin to migrate onto the gut (reviewed in Newgreen & Young, 2002a; Burns, 2005). ET-3 has also been shown to inhibit the induction of neuronal differentiation by GDNF without affecting its mitogenic or trophic effects (Hearn et al., 1998). Given the evidence that continued ENCC proliferation may play a critical role in ensuring their migratory dispersal, an attractive model is that ET-3-dependent signaling modulates the effects of GDNF on ENCC development, thereby regulating the timing and position of neuronal differentiation within the ENS (Hearn et al., 1998; Wu et al., 1999; Flynn et al., 2007). Recent studies have also begun to explore how specific transcription factors regulate these signaling pathways may contribute to the production of different cell types within the ENS in a spatially distinct manner along the gut (Newgreen and Young, 2002a; Hendershot et al., 2007). As recently noted, however, the mechanisms underlying the generation of phenotypic diversity within the ENS remain poorly understood (Hendershot et al., 2007).
What role, if any, might analogous signaling systems play in the formation of the insect ENS? By the most stringent criteria, the answer is “none”: insects do not express immediate homologues of either GDNF or the endothelins, suggesting that the developmental functions of these factors might have first been established during the emergence of the vertebrate lineage. However, there is intriguing evidence that components of these pathways evolved before the separation of protostomes and deuterostomes and may contribute to the differentiation of the ENS in simpler organisms, albeit via undefined mechanisms. For example, while insects lack a true GDNF homologue, they possess at least six other members of the TGFβ superfamily, many of which are expressed in the mesoderm and/or endoderm of the developing gut (Lo and Frash, 1999; Raftery and Sutherland, 1999; Nguyen et al., 2000; Lee and Frash, 2005). More striking is the identification of an authentic homologue of Ret in Drosophila (dRet), containing the same conserved residues required for ligand-mediated activation and signaling in vertebrate Ret proteins (Sugaya et al., 1994; Abrescia et al., 2005). Based on mutations in human Ret that cause multiple endocrine neoplasia type 2, gain-of-function mutations in dRet have been used to investigate the downstream signaling pathways by which these genetic defects cause aberrant cell proliferation, and to identify interacting genes that may contribute to the progression of Ret-dependent oncogenesis (Read et al., 2005). The developmental role of Ret homologues in insects has not been comprehensively explored. In Drosophila, however, dRet is strongly expressed by all neurons in the embryonic ENS as they emerge from the neurogenic zones and begin to migrate (Sugaya et al., 1994; Hahn and Bishop, 2001), a remarkable parallel to the expression of Ret by ENCCs as they enter the foregut (Newgreen and Young, 2002a).
As noted earlier, GDNF signaling is mediated by a receptor complex consisting of Ret and GFRα1, a member of the GFRα family of GPI-linked co-receptors (Airaksinen et al, 2006). Intriguingly, insects also express at least one GFRα gene (named munin in Drosophila; NCBI accession # NM 001014642), indicating that Ret-GFRα complexes may represent extremely ancient receptor components that arose early in bilateran evolution, along with the diversification of the TGFβ superfamily (Enomoto, 2005; Airaksinen et al., 2006). The developmental expression of Munin-related proteins and their potential role as Ret-associated co-receptors remains to be determined. However, given other parallels that can be drawn between ENS development in insects and vertebrates, it will be interesting to determine whether Ret-dependent signaling mediates the response of ENS precursors to TGFβ-related secreted factors that are derived from the embryonic gut, and whether this signaling pathway may affect the proliferation, migration, and differentiation of enteric neurons and glia in these relatively simple systems.
Might an analogue of the endothelin-endothelin receptor signaling system also contribute to the development of the insect ENS? Evidence for such a parallel is speculative at best. Insects certainly express G protein-coupled receptors that share structural similarities with the endothelin receptors in vertebrates (c.f. Kristiansen, 2004; Hauser et al., 2006). Insects also express neprilysin-family endopeptidases (Macours et al., 2003; Thomas et al., 2005) that align closely with the endothelin converting enzymes, proteases that are required for generating functional ET-3 (Hofstra et al., 1999; Vohra et al., 2007). These insect homologues undoubtedly regulate the activation of neuropeptides and peptide hormones in a tissue-specific manner; whether bioactive peptides derived from the insect gut also contribute to the differentiation of the ENS (analogous to the effects of ET-3 in vertebrates) remains unexplored.
7. Unresolved issues and areas for future research
As has been previously noted, the establishment of suitable animal models has played an essential role in characterizing the developmental origins of the vertebrate ENS and in deciphering the function of genes that are essential to this process (Newgren and Young, 2002a, b). Despite these advances, the mechanisms controlling many aspects of ENS development and their potential disruption in the context of congenital abnormalities remain poorly understood (Heuckeroth and Pachnis, 2006). In recent years, insect model systems have proven increasingly useful for delineating the signal transduction pathways that regulate key aspects of development and disease (Kango-Singh & Halder, 2004; Bilen & Bonini, 2005; Kretzschmar, 2005; Vidal, 2006); can they also contribute to our understanding of the developmental events underlying the formation of the ENS? Despite obvious differences in the embryonic origins and gross morphology of the ENS in vertebrates and invertebrates, common themes in their programs of neurogenesis, migration, and differentiation suggest that insect preparations can be exploited to investigate a number of issues that are relevant to more complex systems, as well. In particular, our ability to explore these issues at the level of individual cells within the developing insect ENS may provide a means of examining specific aspects of intercellular and intracellular signal integration in motile neurons.
How do similar gene cassettes regulate distinct morphogenic programs in different species?
As already noted, elegant work in Drosophila has shown that a delicate interplay between proneural genes and neurogenic genes can produce markedly different programs of neurogenesis in the ENS, and that both Wg and EGFR signaling influence the epithelial-to-mesenchymal transitions that distinguish sequential delamination from placode invagination. With the advent of tools that permit transgenic gene expression in larger insects such as Lepidoptera and Orthoptera (Moto et al., 2003; Shinmyo et al., 2004; Yamamoto et al., 2004), it should now be possible to investigate how species-specific differences in the regulation of homologous genes produce markedly different structures in the ENS. In turn, this approach might provide insight into the evolutionary changes in these developmental programs that allowed adaptations in the insect ENS to match the structural and functional diversity of the gut in different species (an “evo-devo” analysis; Minelli and Fusco, 2004; Breuker et al., 2006). Besides obvious similarities between the morphogenetic programs giving rise to the insect ENS and both the sensory placodes and neural crest cell populations in vertebrates, increasing evidence suggests that many of the same gene classes are involved in controlling epithelial-to-mesenchymal transitions in both phyla (e.g. Streit, 2004; Cornell and Eisen, 2005; Doroquez and Rebay, 2006). Thus, this type of comparative analysis might also provide important clues about the developmental mechanisms controlling the morphogenesis of structural diversity in more complex nervous systems, and how errors in this process may lead to morphological and functional defects.
There are also some apparent differences between the regulation of epithelial-to-mesenchymal transitions in vertebrate and invertebrate nervous systems. In both phyla, regulated changes in cadherin-mediated cell adhesion play an essential role in the de-epithelialization of neural precursors but may involve distinct mechanisms of control. In the insect ENS, neurogenesis most likely requires the direct down-regulation of cadherin-dependent adhesion by EGFR signaling (Dumstrei et al., 1998; Dumstrei et al., 2002), while alterations in cadherin expression by the Snail family of transcriptional repressors helps drive the differentiation of neural crest cells (Tucker, 2004; Barembaum and Bronner-Fraser, 2005). Given the prominent role of Snail genes in other aspects of insect development, including neuroblast proliferation (Cai et al., 2001), it will be interesting to assess whether they also contribute to ENS development. Likewise, the potential involvement of EGFR signaling in balancing proneural and neurogenic gene activity might add to our understanding of how Notch signaling is regulated during neural crest cell delamination (Cornell and Eisen, 2005).
How are the fates of individual neurons and glial cells regulated in the insect ENS?
Similar to the insect CNS, the ganglia of the insect ENS contain a substantial number of uniquely identifiable neurons (Zitnan et al., 1989; Copenhaver and Taghert, 1989a; Miles and Booker, 1998). Rather than arising from specific neuroblast precursors with defined lineages, however, the enteric neurons appear to differentiate via a more stochastic sequence of delamination, migration, and assembly (Figs. 3, 4), resembling the generation of peripheral neurons from the neural crest in vertebrates. An obvious question is whether neuronal fate in insect enteric ganglia is indeed regulated by epigenetic interactions or whether unrecognized lineage relationships within the neurogenic zones drive this process. One means of examining this issue would be to alter the expression of lineage-associated genes in the zone cells to test how they affect the differentiation of specific types of enteric neurons. Genetic manipulations to target single zone cells (Lee and Luo, 1999) or acute manipulations of gene expression via intracellular injections of mRNA and antisense constructs could be used to distinguish between cell-autonomous and non-cell autonomous aspects of their differentiation. The larger insect species also lend themselves to targeted ablation strategies that could be used to eliminate individual cells as they emerge from the zones; alternatively, supernumerary precursors could be transplanted into the migratory streams of neurons destined to coalesce into the enteric ganglia. These same methods could be used to explore the genetic regulation of neuronal versus glial fate in the ENS, an issue that is also of interest in the developing vertebrate ENS (Burns, 2005). Both cell types are generated from the neurogenic zones in the insect ENS but at distinct developmental phases, as summarized above. Manipulations of the different precursor classes that arise from each zone might provide clues about how lineage-derived versus cell-cell interactions contribute to the regulation of alternative cell fates.
How do migratory neurons respond to multiple guidance cues simultaneously?
As in other regions of the nervous system, the guidance of neuronal motility in the insect ENS is undoubtedly regulated by a combination of attractive and repulsive cues. To date, the roles of particular guidance molecules have only been examined individually, and only a subset of candidate guidance cues has been investigated in any detail. The potential contribution of soluble chemoattractants and chemorepellents to the formation of the insect ENS should be examined, including Slit (Kidd et al., 1999), the Semaphorins (Ayoob et al., 2006), the Netrins (Mitchell et al., 1996), and their receptors. Recent studies have shown that morphogenic molecules can also serve as guidance molecules for neurons, including Wg/Wnt family members, EGFs, and members of the TGF-β/BMP family (Lehmann, 2001; Zou, 2006). As noted above, the potential role of Ret-dependent responses to TGFβ-related factors during ENS development in insects deserves particular attention. Embryo culture preparations that are now available for Manduca and Locusta should facilitate the rapid screening of these candidate guidance factors to test their role on the migratory behavior of enteric neurons. These studies would also provide the framework for genetic manipulations of the developing ENS in Drosophila.
Perhaps the most important feature of the insect ENS is that it permits individual migratory neurons to be monitored by a variety of cell-labeling and time-lapse protocols in cultured embryos. These methods can now be used to quantify how particular guidance cues affect key aspects of migration, guidance, and outgrowth in vivo, and to examine how multiple cues (both positive and negative) simultaneously regulate the behavior of motile neurons. Similarly, the signal transduction pathways associated with particular guidance receptors can be manipulated to examine their role in the integrating migratory responses to multiple cues, in regulating the transition from migration to axon outgrowth, and in promoting the terminal differentiation of enteric neurons once their migration is complete. While investigations into this type of signal integration are being approached in a variety of preparations (Lehmann, 2001; Ghose and Van Vactor, 2002; Lee and Van Vactor, 2003; Gallo and Letourneau, 2004), the simplicity and experimental accessibility of the insect ENS provides an opportunity to explore the control of cell migration at high resolution within the developing nervous system.
Supplementary Material
Acknowledgements
I wish to thank Dr. Doris Kretzschmar, Dr. David Morton, Dr. Marcel Wehrli, and Mr. Thomas Coate for helpful comments about this review. I also thank Mr. Coate and Dr. Laura Knittel for providing figures from their unpublished work, and Dr. Knittel for generating the QuickTime movies of EP cell migration. Dr. Stephanie Kaech-Petrie in the Live Cell Imaging Facility, Center for Research on Occupational and Environmental Toxicology provided outstanding assistance and support for our confocal imaging studies. This summary of ENS development in different insects benefited greatly from the published discussions of Dr. Volker Hartenstein and Dr. Michael Bastiani; I apologize for any inadvertent misinterpretations or misrepresentations of their excellent work. Supported by NIH # R01 NS34439 and R01 AG025525 to PFC.
Literature Cited
- Airaksinen MS, Holm L, Hatinen T. Evolution of the GDNF family ligands and receptors. Brain Behav Evol. 2006;68:181–190. doi: 10.1159/000094087. [DOI] [PubMed] [Google Scholar]
- Alberti S, Krause SM, Kretz O, Philippar U, Lemberger T, Casanova E, Wiebel FF, Schwarz H, Frotscher M, Schutz G, Nordheim A. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc Natl Acad Sci U S A. 2005;102:6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ. Stem cells and transcription factors in the development of the mammalian neural crest. Faseb J. 1994;8:707–713. doi: 10.1096/fasebj.8.10.8050669. [DOI] [PubMed] [Google Scholar]
- Anderson DJ. Lineages and transcription factors in the specification of vertebrate primary sensory neurons. Curr Opin Neurobiol. 1999;9:517–524. doi: 10.1016/S0959-4388(99)00015-X. [DOI] [PubMed] [Google Scholar]
- Anderson RB, Turner KN, Nikonenko AG, Hemperly J, Schachner M, Young HM. The cell adhesion molecule L1 is required for chain migration of neural crest cells in the developing mouse gut. Gastroenterology. 2006;130:1221–1232. doi: 10.1053/j.gastro.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Arnhold S, When M, Labbe D, Andressen C, Addicks K. Transient expression of NOS-II during development of the murine enteric nervous system. J Mol Histol. 2004;35:741–748. doi: 10.1007/s10735-004-5675-8. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Axelrod JD, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Interaction between Wingless and Notch signaling pathways mediated by Dishevelled. Science. 1996;271:1826–1832. doi: 10.1126/science.271.5257.1826. [DOI] [PubMed] [Google Scholar]
- Ayali A. The insect frontal ganglion and stomatogastric pattern generator networks. Neurosignals. 2004;13:20–36. doi: 10.1159/000076156. [DOI] [PubMed] [Google Scholar]
- Ayoob JC, Terman JR, Kolodkin AL. Drosophila Plexin B is a Sema-2a receptor required for axon guidance. Development. 2006;133:2125–2135. doi: 10.1242/dev.02380. [DOI] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. The origins of the neural crest. Part II: an evolutionary perspective. Mech Dev. 1997;69:13–29. doi: 10.1016/s0925-4773(97)00129-9. [DOI] [PubMed] [Google Scholar]
- Baden V. Embryology of the nervous system in the grasshopper, Melanoplus differentialis (Acrididae: Orthoptera) Journal of Morphology. 1936;60:159–188. [Google Scholar]
- Balaskas C, Saffrey MJ, Burnstock G. Distribution and colocalization of NADPH-diaphorase activity, nitric oxide synthase immunoreactivity, and VIP immunoreactivity in the newly hatched chicken gut. Anat Rec. 1995;243:10–18. doi: 10.1002/ar.1092430103. [DOI] [PubMed] [Google Scholar]
- Barembaum M, Bronner-Fraser M. Early steps in neural crest specification. Semin Cell Dev Biol. 2005;16:642–646. doi: 10.1016/j.semcdb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Tugentman M, Shilo BZ, Steller H. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev Cell. 2002;2:159–170. doi: 10.1016/s1534-5807(02)00116-8. [DOI] [PubMed] [Google Scholar]
- Bestman JE, Booker R. Modulation of foregut synaptic activity controls resorption of molting fluid during larval molts of the moth Manduca sexta. J Exp Biol. 2003;206:1207–1220. doi: 10.1242/jeb.00237. [DOI] [PubMed] [Google Scholar]
- Bickley WE. On the stomodeal nervous system of insects. Annals of the Entomological Society of America. 1942;35:343–354. [Google Scholar]
- Bieber AJ, Snow PM, Hortsch M, Patel NH, Jacobs JR, Traquina ZR, Schilling J, Goodman CS. Drosophila Neuroglian: a member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 1989;59:447–460. doi: 10.1016/0092-8674(89)90029-9. [DOI] [PubMed] [Google Scholar]
- Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- Brandt JF. Remarqeus sur les nerfs stomato-gastriqiue on intesinaux (nervus sympatheticus sue nervi reproductorii). dans les animaux invertebres. Ann. Sci. Nat. Zool. Ser. 1836;2:81–110. 138–154. [Google Scholar]
- Breuker CJ, Debat V, Klingenberg CP. Functional evo-devo. Trends Ecol Evol. 2006;21:488–492. doi: 10.1016/j.tree.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Brouillet E, Trembleau A, Galanaud D, Volovitch M, Bouillot C, Valenza C, Prochiantz A, Allinquant B. The amyloid precursor protein interacts with Go heterotrimeric protein within a cell compartment specialized in signal transduction. J Neurosci. 1999;19:1717–1727. doi: 10.1523/JNEUROSCI.19-05-01717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning G, Mayer B. Prenatal development of nitric oxide synthase in the mouse spinal cord. Neurosci. Letts. 1996;202:189–192. doi: 10.1016/0304-3940(95)12239-7. [DOI] [PubMed] [Google Scholar]
- Burns AJ. Migration of neural crest-derived enteric nervous system precursor cells to and within the gastrointestinal tract. Int J Dev Biol. 2005;49:143–150. doi: 10.1387/ijdb.041935ab. [DOI] [PubMed] [Google Scholar]
- Buss RR, Oppenheim RW. Role of programmed cell death in normal neuronal development and function. Anat Sci Int. 2004;79:191–197. doi: 10.1111/j.1447-073x.2004.00088.x. [DOI] [PubMed] [Google Scholar]
- Cai Y, Chia W, Yang X. A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. Embo J. 2001;20:1704–1714. doi: 10.1093/emboj/20.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. 2nd edition. Heidelberg: Springer-Verlag; [Google Scholar]
- Cau E, Casarosa S, Guillemo F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Chalazonitis A, D'Autreaux F, Guha U, Pham TD, Faure C, Chen JJ, Roman D, Kan L, Rothman TP, Kessler JA, Gershon MD. Bone morphogenetic protein-2 and -4 limit the number of enteric neurons but promote development of a TrkC-expressing neurotrophin-3-dependent subset. J Neurosci. 2004;24:4266–4282. doi: 10.1523/JNEUROSCI.3688-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RF. The Insects. Cambridge: Harvard U. Press; 1982. [Google Scholar]
- Charron F, Tessier-Lavigne M. Novel brain wiring functions for classical morphogens: a role as graded positional cues in axon guidance. Development. 2005;132:2251–2262. doi: 10.1242/dev.01830. [DOI] [PubMed] [Google Scholar]
- Chen JH, Wen L, Dupuis S, Wu JY, Rao Y. The N-terminal leucine-rich regions in Slit are sufficient to repel olfactory bulb axons and subventricular zone neurons. J Neurosci. 2001;21:1548–1556. doi: 10.1523/JNEUROSCI.21-05-01548.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynen E, Baggerman G, Veelaert D, Cerstiaens A, Van der Horst D, Harthoorn L, Derua R, Waelkens E, De Loof A, Schoofs L. Peptidomics of the pars intercerebralis-corpus cardiacum complex of the migratory locust, Locusta migratoria. Eur J Biochem. 2001;268:1929–1939. doi: 10.1046/j.1432-1327.2001.02067.x. [DOI] [PubMed] [Google Scholar]
- Coate TM, Swanson TL, Proctor TM, Nighorn AJ, Copenhaver PF. Eph receptor expression defines midline boundaries for ephrin-positive migratory neurons in the enteric nervous system of Manduca sexta. J. Comp. Neurol. doi: 10.1002/cne.21260. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver PF. Origins, migration, and differentiation of glial cells in the insect nervous system from a discrete set of glial precursors. Devt. 1993;117:59–74. [Google Scholar]
- Copenhaver PF, Horgan AM, Combes S. An identified set of visceral muscle bands is essential for the guidance of migratory neurons in the enteric nervous system of Manduca sexta. Dev. Biol. 1996;179:412–426. doi: 10.1006/dbio.1996.0271. [DOI] [PubMed] [Google Scholar]
- Copenhaver PF, Knittel LM, Swanson TL, Farley SM. Developmental regulation of APPL, the insect homologue of the amyloid precursor protein (APP), and its potential role in neuronal migration. Soc. Neurosci. 2004 abst. 492.2. [Google Scholar]
- Copenhaver PF, Taghert PH. Development of the enteric nervous system in the moth I. Diversity of cell types and the embryonic expression of FMRFamide-related neuropeptides. Developmental Biology. 1989a;131:70–84. doi: 10.1016/s0012-1606(89)80039-9. [DOI] [PubMed] [Google Scholar]
- Copenhaver PF, Taghert PH. Development of the enteric nervous system in the moth II. Stereotyped cell migration precedes the differentiation of embryonic neurons. Developmental Biology. 1989b;131:85–101. doi: 10.1016/s0012-1606(89)80040-5. [DOI] [PubMed] [Google Scholar]
- Copenhaver PF, Taghert PH. Neurogenesis in the insect enteric nervous system: generation of pre-migratory neurons from an epithelial placode. Development. 1990;109:17–28. doi: 10.1242/dev.109.1.17. [DOI] [PubMed] [Google Scholar]
- Copenhaver PF, Taghert PH. Origins of the insect enteric nervous system: differentiation of the enteric ganglia from a neurogenic epithelium. Development. 1991;113:1115–1132. doi: 10.1242/dev.113.4.1115. [DOI] [PubMed] [Google Scholar]
- Copenhaver PF, Vogt TM, Swanson TL, Coate TM. A function-independent screen for proteins that interact with fasciclin II in the control of neuronal migration. Soc. Neurosci. 2006 Abst. 514.19. [Google Scholar]
- Cornell RA, Eisen JS. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin Cell Dev Biol. 2005;16:663–672. doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Coulson EJ, Paliga K, Beyreuther K, Masters CL. What the evolution of the amyloid protein precursor supergene family tells us about its function. Neurochem Int. 2000;36:175–184. doi: 10.1016/s0197-0186(99)00125-4. [DOI] [PubMed] [Google Scholar]
- Daudet N, Lewis J. Two contrasting roles for Notch activity in chick inner ear development: specification of prosensory patches and lateral inhibition of hair-cell differentiation. Development. 2005;132:541–551. doi: 10.1242/dev.01589. [DOI] [PubMed] [Google Scholar]
- Davis GW, Schuster CM, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. III. CREB is necessary for presynaptic functional plasticity. Neuron. 1996;17:669–679. doi: 10.1016/s0896-6273(00)80199-3. [DOI] [PubMed] [Google Scholar]
- Davis GW, Schuster CM, Goodman CS. Genetic analysis of the mechanisms controlling target selection: target-derived fasciclin II regulates the pattern of synapse formation. Neuron. 1997;19:561–573. doi: 10.1016/s0896-6273(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Davis NT, Blackburn MB, Golubeva EG, Hildebrand JG. Localization of myoinhibitory peptide immunoreactivity in Manduca sexta and Bombyx mori, with indications that the peptide has a role in molting and ecdysis. J Exp Biol. 2003;206:1449–1460. doi: 10.1242/jeb.00234. [DOI] [PubMed] [Google Scholar]
- Dawson TM, Bredt DS, Fotuhi M, Hwang PM, Snyder SH. Nitric oxide synthase and neuronal NADPH-diaphorase are identical in brain and peripheral tissues. Proc Natl Acad Sci U S A. 1991;88:7797–7801. doi: 10.1073/pnas.88.17.7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, Bray S. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124:3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W. Proteolytic processing and cell biological functions of the amyloid precursor protein. J Cell Sci. 2000;113(Pt 11):1857–1870. doi: 10.1242/jcs.113.11.1857. [DOI] [PubMed] [Google Scholar]
- De Velasco B, Shen J, Go S, Hartenstein V. Embryonic development of the Drosophila corpus cardiacum, a neuroendocrine gland with similarity to the vertebrate pituitary, is controlled by sine oculisand glass. Dev Biol. 2004;274:280–294. doi: 10.1016/j.ydbio.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Bastiani MJ, Goodman CS. Guidance of neuronal growth cones in the grasshopper embryo. IV. Temporal delay experiments. Journal of Neuroscience. 1986;6:3552–3563. doi: 10.1523/JNEUROSCI.06-12-03552.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ, Kuwada JY, Goodman CS. From epithelium to neuroblasts to neurons: the role of cell interactions and cell lineage during insect neurogenesis. Phil. Trans. R. Soc. Lond. B. 1985;312:67–81. doi: 10.1098/rstb.1985.0178. [DOI] [PubMed] [Google Scholar]
- Doroquez DB, Rebay I. Signal integration during development: mechanisms of EGFR and Notch pathway function and cross-talk. Crit Rev Biochem Mol Biol. 2006;41:339–385. doi: 10.1080/10409230600914344. [DOI] [PubMed] [Google Scholar]
- Druckenbrod NR, Epstein ML. Behavior of enteric neural crest-derived cells varies with respect to the migratory wavefront. Dev Dyn. 2007;236:84–92. doi: 10.1002/dvdy.20974. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Nassif C, Abboud G, Aryai A, Hartenstein V. EGFR signaling is required for the differentiation and maintenance of neural progenitors along the dorsal midline of the Drosophila embryonic head. Development. 1998;125:3417–3426. doi: 10.1242/dev.125.17.3417. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Shy D, Tepass U, Hartenstein V. Interaction between EGFR signaling and DE-cadherin during nervous system morphogenesis. Development. 2002;129:3983–3994. doi: 10.1242/dev.129.17.3983. [DOI] [PubMed] [Google Scholar]
- Eaton JL. Nervous System: Functional Role. In: Blum MS, editor. Fundamentals of Insect Physiology. New York: Wiley; 1985. pp. 358–390. [Google Scholar]
- Edenfeld G, Stork T, Klämbt C. Neuron-glia interaction in the insect nervous system. Curr Opin Neurobiol. 2005;15:34–39. doi: 10.1016/j.conb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Enomoto H. Regulation of neural development by glial cell line-derived neurotrophic factor family ligands. Anat Sci Int. 2005;80:42–52. doi: 10.1111/j.1447-073x.2005.00099.x. [DOI] [PubMed] [Google Scholar]
- Evans PD. Biogenic amines in the insect nervous system. In: Berridge MJ, Treherne JE, Wigglesworth VB, editors. Advances in Insect Physiology. New York: Academic Press; 1980. pp. 318–473. [Google Scholar]
- Faure C, Chalazonitis A, Rheaume C, Bouchard G, Sampathkumar SG, Yarema KJ, Gershon MD. Gangliogenesis in the enteric nervous system: roles of the polysialylation of the neural cell adhesion molecule and its regulation by bone morphogenetic protein-4. Dev Dyn. 2007;236:44–59. doi: 10.1002/dvdy.20943. [DOI] [PubMed] [Google Scholar]
- Flynn B, Bergner AJ, Turner KN, Young HM, Anderson RB. Effect of GDNF haploinsufficiency on rate of migration and number of enteric neural crest-derived cells. Dev Dyn. 2007;236:134–141. doi: 10.1002/dvdy.21013. [DOI] [PubMed] [Google Scholar]
- Fu M, Lui VC, Sham MH, Pachnis V, Tam PK. Sonic hedgehog regulates the proliferation, differentiation, and migration of enteric neural crest cells in gut. J Cell Biol. 2004;166:673–684. doi: 10.1083/jcb.200401077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness JB. The Enteric Nervous System. Malden, MA: Blackwell Publishing, Inc.; 2006. p. 274. [Google Scholar]
- Gallo G, Letourneau PC. Regulation of growth cone actin filaments by guidance cues. J Neurobiol. 2004;58:92–102. doi: 10.1002/neu.10282. [DOI] [PubMed] [Google Scholar]
- Ganfornina MD, Sanchez D, Bastiani MJ. Embryonic development of the enteric nervous system of the grasshopper Schistocerca americana. J Comp Neurol. 1996;372:581–596. doi: 10.1002/(SICI)1096-9861(19960902)372:4<581::AID-CNE7>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Gershon MD. The enteric nervous system. Annual Review of Neuroscience. 1981;4:227–272. doi: 10.1146/annurev.ne.04.030181.001303. [DOI] [PubMed] [Google Scholar]
- Gershon MD. Genes and lineages in the formation of the enteric nervous system. Curr Opin Neurobiol. 1997;7:101–109. doi: 10.1016/s0959-4388(97)80127-4. [DOI] [PubMed] [Google Scholar]
- Gershon MD, Chalazonitis A, Rothman TP. From neural crest to bowel: development of the enteric nervous system. J Neurobiol. 1993;24:199–214. doi: 10.1002/neu.480240207. [DOI] [PubMed] [Google Scholar]
- Ghose A, Van Vactor D. GAPs in Slit-Robo signaling. Bioessays. 2002;24:401–404. doi: 10.1002/bies.10080. [DOI] [PubMed] [Google Scholar]
- González-Gaitán M, Jäckle H. Invagination centers within the Drosophila stomatogastric nervous system anlage are positioned by Notch-mediated signaling which is spatially controlled through wingless. Development. 1995;121:2313–2325. doi: 10.1242/dev.121.8.2313. [DOI] [PubMed] [Google Scholar]
- González-Gaitán M, Jäckle H. Tip cell-derived RTK signaling initiates cell movements in the Drosophila stomatogastric nervous system anlage. EMBO Rep. 2000;1:366–371. doi: 10.1093/embo-reports/kvd064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CS. Mechanisms and molecules that control growth cone guidance. Annu Rev Neurosci. 1996;19:341–377. doi: 10.1146/annurev.ne.19.030196.002013. [DOI] [PubMed] [Google Scholar]
- Graba Y, Aragnol D, Laurenti P, Garzino V, Charmot D, Berenger H, Pradel J. Homeotic control in Drosophila; the scabrous gene is an in vivo target of Ultrabithorax proteins. Embo J. 1992;11:3375–3384. doi: 10.1002/j.1460-2075.1992.tb05416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh G, Rehm EJ, Goodman CS. Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell. 1991;67:45–57. doi: 10.1016/0092-8674(91)90571-f. [DOI] [PubMed] [Google Scholar]
- Haase A, Bicker G. Nitric oxide and cyclic nucleotides are regulators of neuronal migration in an insect embryo. Development. 2003;130:3977–3987. doi: 10.1242/dev.00612. [DOI] [PubMed] [Google Scholar]
- Hahn M, Bishop J. Expression pattern of Drosophila ret suggests a common ancestral origin between the metamorphosis precursors in insect endoderm and the vertebrate enteric neurons. Proc Natl Acad Sci U S A. 2001;98:1053–1058. doi: 10.1073/pnas.98.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M, Jäckle H. Drosophila goosecoid participates in neural development but not in body axis formation. Embo J. 1996;15:3077–3084. [PMC free article] [PubMed] [Google Scholar]
- Harris-Warrick RM, Marder E, Selverston AI, Moulins M. The Stomatogastric Nervous System. Cambridge: MIT Press; 1992. [Google Scholar]
- Hartenstein AY, Rugendorff A, Tepass U, Hartenstein V. The function of the neurogenic genes during epithelial development in the Drosophila embryo. Development. 1992;116:1203–1220. doi: 10.1242/dev.116.4.1203. [DOI] [PubMed] [Google Scholar]
- Hartenstein V. Development of the insect stomatogastric nervous system. TINS. 1997;20:412–427. doi: 10.1016/s0166-2236(97)01066-7. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Tepass U, Gruszynski-Defeo E. Embryonic development of the stomatogastric nervous system in Drosophila. J. Comp. Neurol. 1994;350:367–381. doi: 10.1002/cne.903500304. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Tepass U, Gruszynski-deFeo E. Proneural and neurogenic genes control specification and Morphogenesis of stomatogastric nerve cell precursors in Drosophila. Dev Biol. 1996;173:213–227. doi: 10.1006/dbio.1996.0018. [DOI] [PubMed] [Google Scholar]
- Hasson P, Egoz N, Winkler C, Volohonsky G, Jia S, Dinur T, Volk T, Courey AJ, Paroush Z. EGFR signaling attenuates Groucho-dependent repression to antagonize Notch transcriptional output. Nat Genet. 2005;37:101–105. doi: 10.1038/ng1486. [DOI] [PubMed] [Google Scholar]
- Hauser F, Cazzamali G, Williamson M, Blenau W, Grimmelikhuijzen CJ. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog Neurobiol. 2006;80:1–19. doi: 10.1016/j.pneurobio.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Hayward P, Balayo T, Martinez Arias A. Notch synergizes with axin to regulate the activity of armadillo in Drosophila. Dev Dyn. 2006;235:2656–2666. doi: 10.1002/dvdy.20902. [DOI] [PubMed] [Google Scholar]
- Hearn CJ, Murphy M, Newgreen D. GDNF and ET-3 differentially modulate the numbers of avian enteric neural crest cells and enteric neurons in vitro. Dev Biol. 1998;197:93–105. doi: 10.1006/dbio.1998.8876. [DOI] [PubMed] [Google Scholar]
- Heathcote RD. Differentiation of an identified sensory neuron (SR) and associated structures (CTO) in grasshopper embryos. Journal of Comparative Neurology. 1981;202:1–18. doi: 10.1002/cne.902020103. [DOI] [PubMed] [Google Scholar]
- Hendershot TJ, Liu H, Sarkar AA, Giovannucci DR, Clouthier DE, Abe M, Howard MJ. Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev Dyn. 2007;236:93–105. doi: 10.1002/dvdy.20989. [DOI] [PubMed] [Google Scholar]
- Herms J, Anliker B, Heber S, Ring S, Fuhrmann M, Kretzschmar H, Sisodia S, Muller U. Cortical dysplasia resembling human type 2 lissencephaly in mice lacking all three APP family members. Embo J. 2004;23:4106–4115. doi: 10.1038/sj.emboj.7600390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog W, Zeng X, Lele Z, Sonntag C, Ting JW, Chang CY, Hammerschmidt M. Adenohypophysis formation in the zebrafish and its dependence on sonic hedgehog. Dev Biol. 2003;254:36–49. doi: 10.1016/s0012-1606(02)00124-0. [DOI] [PubMed] [Google Scholar]
- Heuckeroth RO, Pachnis V. Getting to the guts of enteric nervous system development. Development. 2006;133:2287–2290. doi: 10.1242/dev.02418. [DOI] [PubMed] [Google Scholar]
- Hindley S, Juurlink BH, Gysbers JW, Middlemiss PJ, Herman MA, Rathbone MP. Nitric oxide donors enhance neurotrophin-induced neurite outgrowth through a cGMP-dependent mechanism. J. Neurosci. Res. 1997;15:427–439. [PubMed] [Google Scholar]
- Hofstra RM, Elfferich P, Osinga J, Verlind E, Fransen E, Lopez Pison J, de Die-Smulders CE, Stolte-Dijkstra I, Buys CH. Hirschsprung disease and L1CAM: is the disturbed sex ratio caused by L1CAM mutations? J Med Genet. 2002;39:E11. doi: 10.1136/jmg.39.3.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstra RM, Valdenaire O, Arch E, Osinga J, Kroes H, Loffler BM, Hamosh A, Meijers C, Buys CH. A loss-of-function mutation in the endothelin-converting enzyme 1 (ECE-1) associated with Hirschsprung disease, cardiac defects, and autonomic dysfunction. Am J Hum Genet. 1999;64:304–308. doi: 10.1086/302184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SL, DiCaprio RA. Crustacean motor pattern generator networks. Neurosignals. 2004;13:50–69. doi: 10.1159/000076158. [DOI] [PubMed] [Google Scholar]
- Horgan AM, Copenhaver PF. G protein-mediated inhibition of neuronal migration requires calcium influx. J. Neurosci. 1998;18:4189–4200. doi: 10.1523/JNEUROSCI.18-11-04189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan AM, Lagrange MT, Copenhaver PF. Developmental expression of G proteins in a migratory population of embryonic neurons. Development. 1994;120:729–742. doi: 10.1242/dev.120.4.729. [DOI] [PubMed] [Google Scholar]
- Horgan AM, Lagrange MT, Copenhaver PF. A developmental role for the heterotrimeric G protein Goα in a migratory population of embryonic neurons. Dev. Biol. 1995;172:640–653. doi: 10.1006/dbio.1995.8042. [DOI] [PubMed] [Google Scholar]
- Huang X, Saint-Jeannet JP. Induction of the neural crest and the opportunities of life on the edge. Dev Biol. 2004;275:1–11. doi: 10.1016/j.ydbio.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Janet C. Anatomie de lat tete du Lasius niger. Limoges. 1905:1–40. [Google Scholar]
- Jiang Y, Liu MT, Gershon MD. Netrins and DCC in the guidance of migrating neural crest-derived cells in the developing bowel and pancreas. Dev Biol. 2003;258:364–384. doi: 10.1016/s0012-1606(03)00136-2. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Nighorn A. Interaxonal Eph-ephrin signaling may mediate sorting of olfactory sensory axons in Manduca sexta. J Neurosci. 2003;23:11523–11538. doi: 10.1523/JNEUROSCI.23-37-11523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M, Halder G. Drosophila as an emerging model to study metastasis. Genome Biol. 2004;5:216. doi: 10.1186/gb-2004-5-4-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcavich RE. Generating neuronal diversity in the Drosophila central nervous system: a view from the ganglion mother cells. Dev Dyn. 2005;232:609–616. doi: 10.1002/dvdy.20273. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Klämbt C, Goodman CS. Role of the midline glia and neurons in the formation of the axon commissures in the central nervous system of the Drosophila embryo. Ann N Y Acad Sci. 1991;633:142–159. doi: 10.1111/j.1749-6632.1991.tb15604.x. [DOI] [PubMed] [Google Scholar]
- Klämbt C, Jacobs JR, Goodman CS. The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- Knittel LM, Swanson TL, Copenhaver PF. Expression of the cell adhesion receptor fasciclin II mediates the guidance of individual migratory neurons along specific muscle cell pathways in vivo. Soc. Neurosci. Abst. 2005;480:13. [Google Scholar]
- Kornack DR, Rakic P. The generation, migration, and differentiation of olfactory neurons in the adult primate brain. Proc Natl Acad Sci U S A. 2001;98:4752–4757. doi: 10.1073/pnas.081074998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouki T, Imai H, Aoto K, Eto K, Shioda S, Kawamura K, Kikuyama S. Developmental origin of the rat adenohypophysis prior to the formation of Rathke's pouch. Development. 2001;128:959–963. doi: 10.1242/dev.128.6.959. [DOI] [PubMed] [Google Scholar]
- Kretzschmar D. Neurodegenerative mutants in Drosophila: a means to identify genes and mechanisms involved in human diseases? Invert Neurosci. 2005;5:97–109. doi: 10.1007/s10158-005-0005-8. [DOI] [PubMed] [Google Scholar]
- Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther. 2004;103:21–80. doi: 10.1016/j.pharmthera.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Kuhlman J, Eisen JS. Genetic screen for mutations affecting development and function of the enteric nervous system. Dev Dyn. 2007;236:118–127. doi: 10.1002/dvdy.21033. [DOI] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Kalcheim C. The Neural Crest. Cambridge, England: Cambridge U. Press; 1999. [Google Scholar]
- Lee HH, Frasch M. Nuclear integration of positive Dpp signals, antagonistic Wg inputs and mesodermal competence factors during Drosophila visceral mesoderm induction. Development. 2005;132:1429–1442. doi: 10.1242/dev.01687. [DOI] [PubMed] [Google Scholar]
- Lee H, Van Vactor D. Neurons take shape. Curr Biol. 2003;13:R152–R161. doi: 10.1016/s0960-9822(03)00080-0. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lehmann R. Cell migration in invertebrates: clues from border and distal tip cells. Curr Opin Genet Dev. 2001;11:457–463. doi: 10.1016/s0959-437x(00)00217-3. [DOI] [PubMed] [Google Scholar]
- Lengyel JA, Iwaki DD. It takes guts: the Drosophila hindgut as a model system for organogenesis. Dev Biol. 2002;243:1–19. doi: 10.1006/dbio.2002.0577. [DOI] [PubMed] [Google Scholar]
- Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- Li Y, Fetchko M, Lai ZC, Baker NE. Scabrous and Gp150 are endosomal proteins that regulate Notch activity. Development. 2003;130:2819–2827. doi: 10.1242/dev.00495. [DOI] [PubMed] [Google Scholar]
- Lin DM, Fetter RD, Kopczynski C, Grenningloh G, Goodman CS. Genetic analysis of fasciclin II in Drosophila: defasciculation, refasciculation, and altered fasciculation. Neuron. 1994;13:1055–1069. doi: 10.1016/0896-6273(94)90045-0. [DOI] [PubMed] [Google Scholar]
- Lo PC, Frasch M. Sequence and expression of myoglianin, a novel Drosophila gene of the TGF-beta superfamily. Mech Dev. 1999;86:171–175. doi: 10.1016/s0925-4773(99)00108-2. [DOI] [PubMed] [Google Scholar]
- Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271:978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- Macours N, Poels J, Hens K, Luciani N, De Loof A, Huybrechts R. An endothelin-converting enzyme homologue in the locust, Locusta migratoria: functional activity, molecular cloning and tissue distribution. Insect Mol Biol. 2003;12:233–240. doi: 10.1046/j.1365-2583.2003.00406.x. [DOI] [PubMed] [Google Scholar]
- Marder E, Bucher D, Schulz DJ, Taylor AL. Invertebrate central pattern generation moves along. Curr Biol. 2005;15:R685–R699. doi: 10.1016/j.cub.2005.08.022. [DOI] [PubMed] [Google Scholar]
- McCormick J, Nichols R. Spatial and Temporal Expression Identify Dromyosuppressin as a Brain-Gut Peptide in Drosophila-Melanogaster. Journal of Comparative Neurology. 1993;338:279–288. doi: 10.1002/cne.903380210. [DOI] [PubMed] [Google Scholar]
- Miles CI, Booker R. The Role of the Frontal Ganglion in Foregut Movements of the Moth, Manduca-Sexta. Journal of Comparative Physiology a-Sensory Neural and Behavioral Physiology. 1994;174:755–767. [Google Scholar]
- Miles CI, Booker R. The role of the frontal ganglion in the feeding and eclosion behavior of the moth Manduca sexta. J Exp Biol. 1998;201(Pt 11):1785–1798. doi: 10.1242/jeb.201.11.1785. [DOI] [PubMed] [Google Scholar]
- Minelli A, Fusco G. Evo-devo perspectives on segmentation: model organisms, and beyond. Trends Ecol Evol. 2004;19:423–429. doi: 10.1016/j.tree.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Doyle JL, Serafini T, Kennedy TE, Tessier-Lavigne M, Goodman CS, Dickson BJ. Genetic analysis of Netrin genes in Drosophila: Netrins guide CNS commissural axons and peripheral motor axons. Neuron. 1996;17:203–215. doi: 10.1016/s0896-6273(00)80153-1. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Baker NE, Rubin GM. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 1990;4:1848–1861. doi: 10.1101/gad.4.11.1848. [DOI] [PubMed] [Google Scholar]
- Morales AV, Barbas JA, Nieto MA. How to become neural crest: from segregation to delamination. Semin Cell Dev Biol. 2005;16:655–662. doi: 10.1016/j.semcdb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Moto K, Kojima H, Kurihara M, Iwami M, Matsumoto S. Cell-specific expression of enhanced green fluorescence protein under the control of neuropeptide gene promoters in the brain of the silkworm, Bombyx mori, using Bombyx mori nucleopolyhedrovirus-derived vectors. Insect Biochem Mol Biol. 2003;33:7–12. doi: 10.1016/s0965-1748(02)00185-6. [DOI] [PubMed] [Google Scholar]
- Newgreen D, Young HM. Enteric nervous system: development and developmental disturbances--part 1. Pediatr Dev Pathol. 2002a;5:224–247. doi: 10.1007/s10024-001-0142-y. [DOI] [PubMed] [Google Scholar]
- Newgreen D, Young HM. Enteric nervous system: development and developmental disturbances--part 2. Pediatr Dev Pathol. 2002b;5:329–349. doi: 10.1007/s10024-002-0002-4. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Parker L, Arora K. Identification of maverick, a novel member of the TGF-beta superfamily in Drosophila. Mech Dev. 2000;95:201–206. doi: 10.1016/s0925-4773(00)00338-5. [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Pennacchio LA. Comparative genomic analysis as a tool for biological discovery. J Physiol. 2004;554:31–39. doi: 10.1113/jphysiol.2003.050948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt RG. The new head hypothesis revisited. J Exp Zoolog B Mol Dev Evol. 2005;304:274–297. doi: 10.1002/jez.b.21063. [DOI] [PubMed] [Google Scholar]
- Nusbaum MP, Beenhakker MP. A small-systems approach to motor pattern generation. Nature. 2002;417:343–350. doi: 10.1038/417343a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Takeda S, Murayama Y, Ogata E, Nishimoto I. Ligand-dependent G protein coupling function of amyloid transmembrane precursor. J Biol Chem. 1995;270:4205–4208. doi: 10.1074/jbc.270.9.4205. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Peifer M, Pai LM, Casey M. Phosphorylation of the Drosophila adherens junction protein Armadillo: roles for wingless signal and zeste-white 3 kinase. Dev Biol. 1994;166:543–556. doi: 10.1006/dbio.1994.1336. [DOI] [PubMed] [Google Scholar]
- Penzlin H. Stomatogastric nervous system. In: Kerkut GA, Gilbert LI, editors. Comprehensive Insect Physiology, Biochemistry, and Pharmacology. Oxford: Pergamon Press; 1985. pp. 371–406. [Google Scholar]
- Pietsch J, Delalande JM, Jakaitis B, Stensby JD, Dohle S, Talbot WS, Raible DW, Shepherd IT. lessen encodes a zebrafish trap100 required for enteric nervous system development. Development. 2006;133:395–406. doi: 10.1242/dev.02215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla P, Moore R, Morali OG, Grille S, Martinozzi S, Delmas V, Larue L. Cadherins in neural crest cell development and transformation. J Cell Physiol. 2001;189:121–132. doi: 10.1002/jcp.10008. [DOI] [PubMed] [Google Scholar]
- Poliakov A, Cotrina M, Wilkinson DG. Diverse roles of Eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev Cell. 2004;7:465–480. doi: 10.1016/j.devcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Portin P. General outlines of the molecular genetics of the Notch signalling pathway in Drosophila melanogaster: a review. Hereditas. 2002;136:89–96. doi: 10.1034/j.1601-5223.2002.1360201.x. [DOI] [PubMed] [Google Scholar]
- Poulson DF. Histogenesis, organogenesis, and differentiation in the embryo of Drosophila melanogaster (Meigen) In: Demerec M, editor. Biology of Drosophila. New York: Wiley; 1950. pp. 168–274. [Google Scholar]
- Powell PA, Wesley C, Spencer S, Cagan RL. Scabrous complexes with Notch to mediate boundary formation. Nature. 2001;409:626–630. doi: 10.1038/35054566. [DOI] [PubMed] [Google Scholar]
- Price DL, Tanzi RE, Borchelt DR, Sisodia SS. Alzheimer's disease: genetic studies and transgenic models. Annu Rev Genet. 1998;32:461–493. doi: 10.1146/annurev.genet.32.1.461. [DOI] [PubMed] [Google Scholar]
- Raftery LA, Sutherland DJ. TGF-beta family signal transduction in Drosophila development: from Mad to Smads. Dev Biol. 1999;210:251–268. doi: 10.1006/dbio.1999.9282. [DOI] [PubMed] [Google Scholar]
- Rathke H. Ueber die Entstehung der Glandula pituitaria. Arch. Anat. Physiol. Wissen. Med. 1838:482–485. [Google Scholar]
- Renaud O, Simpson P. Movement of bristle precursors contributes to the spacing pattern in Drosophila. Mech Dev. 2002;119:201–211. doi: 10.1016/s0925-4773(02)00381-7. [DOI] [PubMed] [Google Scholar]
- Read RD, Goodfellow PJ, Mardis ER, Novak N, Armstrong JR, Cagan RL. A Drosophila model of multiple endocrine neoplasia type 2. Genetics. 2005;171:1057–1081. doi: 10.1534/genetics.104.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendic D, Linder A, Paschinger K, Borth N, Wilson IB, Fabini G. Modulation of neural carbohydrate epitope expression in Drosophila melanogaster cells. J Biol Chem. 2006;281:3343–3353. doi: 10.1074/jbc.M508334200. [DOI] [PubMed] [Google Scholar]
- Rohrbaugh M, Ramos E, Nguyen D, Price M, Wen Y, Lai ZC. Notch activation of yan expression is antagonized by RTK/pointed signaling in the Drosophila eye. Curr Biol. 2002;12:576–581. doi: 10.1016/s0960-9822(02)00743-1. [DOI] [PubMed] [Google Scholar]
- Roskams AJ, Bredt DS, Dawson TM, Ronnett GV. Nitric oxide mediates the formation of synaptic connections in developing and regenerating olfactory receptor neurons. 1994;13:289–299. doi: 10.1016/0896-6273(94)90347-6. 1994. [DOI] [PubMed] [Google Scholar]
- Santiago A, Erickson CA. Ephrin-B ligands play a dual role in the control of neural crest cell migration. Development. 2002;129:3621–3632. doi: 10.1242/dev.129.15.3621. [DOI] [PubMed] [Google Scholar]
- Santiago-Garcia J, Mas-Oliva J, Innerarity TL, Pitas RE. Secreted forms of the amyloid-beta precursor protein are ligands for the class A scavenger receptor. J Biol Chem. 2001;276:30655–30661. doi: 10.1074/jbc.M102879200. [DOI] [PubMed] [Google Scholar]
- Schoeller J. Recherches descriptives et experimentales sur la cephalogenese de Calliphora erythrocephala (Meigen), au cours des developpements embryonnaire et postembryonnaire. Arch. Zool. Exp. Gen. 1964;103:1–216. [Google Scholar]
- Schuster CM, Davis GD, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity II. Fasciclin II controls presynaptic structural plasticity. Neuron. 1996;17:655–667. doi: 10.1016/s0896-6273(00)80198-1. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Toward a comprehensive theory for Alzheimer's disease. Hypothesis: Alzheimer's disease is caused by the cerebral accumulation and cytotoxicity of amyloid beta-protein. Ann N Y Acad Sci. 2000;924:17–25. doi: 10.1111/j.1749-6632.2000.tb05554.x. [DOI] [PubMed] [Google Scholar]
- Shinmyo Y, Mito T, Matsushita T, Sarashina I, Miyawaki K, Ohuchi H, Noji S. piggyBac-mediated somatic transformation of the two-spotted cricket, Gryllus bimaculatus. Dev Growth Differ. 2004;46:343–349. doi: 10.1111/j.1440-169x.2004.00751.x. [DOI] [PubMed] [Google Scholar]
- Skaer H. The Alimentary Canal. In: Bate M, Martinez Arias A, editors. The Development of Drosophila. Plainview: Cold Spring Harbor Press; 1993. pp. 941–1012. [Google Scholar]
- Snodgrass RE. Principles of Insect Morphology. New York: McGraw-Hill; 1935. [Google Scholar]
- Snow PM, Zinn K, Harrelson AL, McAllister L, Schilling J, Bastiani MJ, Makk G, Goodman CS. Characterization and cloning of fasciclin I and fasciclin II glycoproteins in the grasshopper. Proceedings of the National Academy of Science (USA) 1988;85:5291–5295. doi: 10.1073/pnas.85.14.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A. Early development of the cranial sensory nervous system: from a common field to individual placodes. Dev Biol. 2004;276:1–15. doi: 10.1016/j.ydbio.2004.08.037. [DOI] [PubMed] [Google Scholar]
- Sugaya R, Ishimaru S, Hosoya T, Saigo K, Emori Y. A Drosophila homolog of human proto-oncogene ret transiently expressed in embryonic neuronal precursor cells including neuroblasts and CNS cells. Mech Dev. 1994;45:139–145. doi: 10.1016/0925-4773(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Swanson TL, Knittel LM, Coate TM, Farley SM, Snyder MA, Copenhaver PF. The insect amyloid precursor protein interacts with the heterotrimeric G protein Goα in an identified population of migratory neurons. Developmental Biology. 2005;288:160–178. doi: 10.1016/j.ydbio.2005.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Torok T, Hartenstein V. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neurectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10:672–685. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- Thomas JE, Rylett CM, Carhan A, Bland ND, Bingham RJ, Shirras AD, Turner AJ, Isaac RE. Drosophila melanogaster NEP2 is a new soluble member of the neprilysin family of endopeptidases with implications for reproduction and renal function. Biochem J. 2005;386:357–366. doi: 10.1042/BJ20041753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas U, Kim E, Kuhlendahl S, Koh YH, Gundelfinger ED, Sheng M, Garner CC, Budnik V. Synaptic clustering of the cell adhesion molecule fasciclin II by discs-large and its role in the regulation of presynaptic structure. Neuron. 1997;19:787–799. doi: 10.1016/s0896-6273(00)80961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, De Vente J, Ball EE. Nitric oxide-sensitive guanylate cyclase activity is associated with the maturational phase of neuronal development in insects. Devt. 1996;122:3949–3958. doi: 10.1242/dev.122.12.3949. [DOI] [PubMed] [Google Scholar]
- Tucker RP. Neural crest cells: a model for invasive behavior. Int J Biochem Cell Biol. 2004;36:173–177. doi: 10.1016/s1357-2725(03)00243-7. [DOI] [PubMed] [Google Scholar]
- Van Ginneken C, Van Meir F, Sommereyns G, Sys S, Weyns A. Nitric oxide synthase expression in enteric neurons during development in the pig duodenum. Anat Embryol (Berl) 1998;198:399–408. doi: 10.1007/s004290050192. [DOI] [PubMed] [Google Scholar]
- Vidal M, Cagan RL. Drosophila models for cancer research. Curr Opin Genet Dev. 2006;16:10–16. doi: 10.1016/j.gde.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Vohra BP, Planer W, Armon J, Fu M, Jain S, Heuckeroth RO. Reduced endothelin converting enzyme-1 and endothelin-3 mRNA in the developing bowel of male mice may increase expressivity and penetrance of Hirschsprung disease-like distal intestinal aganglionosis. Dev Dyn. 2007;236:106–117. doi: 10.1002/dvdy.21028. [DOI] [PubMed] [Google Scholar]
- Wallace K, Liu TH, Vaessin H. The pan-neural bHLH proteins DEADPAN and ASENSE regulate mitotic activity and cdk inhibitor dacapo expression in the Drosophila larval optic lobes. Genesis. 2000;26:77–85. doi: 10.1002/(sici)1526-968x(200001)26:1<77::aid-gene10>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Ward SM, Shuttleworth CWR, Kenyon JL. Dorsal root ganglion neurons of embryonic chick contain nitric oxide synthase and respond to nitric oxide. Brain Res. 1994;648:249–258. doi: 10.1016/0006-8993(94)91124-x. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Garcia-Verdugo JM, Alvarez-Buylla A. Direct evidence for homotypic, glia-independent neuronal migration. Neuron. 1997;18:779–791. doi: 10.1016/s0896-6273(00)80317-7. [DOI] [PubMed] [Google Scholar]
- Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- Winberg ML, Mitchell KJ, Goodman CS. Genetic analysis of the mechanisms controlling target selection: complementary and combinatorial functions of netrins, semaphorins, and IgCAMS. Cell. 1998;93:581–591. doi: 10.1016/s0092-8674(00)81187-3. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Huttner WB. Asymmetric cell division during neurogenesis in Drosophila and vertebrates. Mech Dev. 2003;120:1297–1309. doi: 10.1016/j.mod.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Wright JW, Copenhaver PF. Different isoforms of fasciclin II play distinct roles in the guidance of neuronal migration during insect embryogenesis. Dev Biol. 2000;225:59–78. doi: 10.1006/dbio.2000.9777. [DOI] [PubMed] [Google Scholar]
- Wright JW, Copenhaver PF. Cell type-specific expression of fasciclin II isoforms reveals neuronal-glial interactions during peripheral nerve growth. Dev Biol. 2001;234:24–41. doi: 10.1006/dbio.2001.0247. [DOI] [PubMed] [Google Scholar]
- Wright JW, Schwinof KM, Snyder MA, Copenhaver PF. A delayed role for nitric oxide-sensitive guanylate cyclases in a migratory population of embryonic neurons. Dev. Biol. 1998;203:15–33. doi: 10.1006/dbio.1998.9066. [DOI] [PubMed] [Google Scholar]
- Wright JW, Snyder MA, Schwinof KM, Combes S, Copenhaver PF. A role for fasciclin II in the guidance of neuronal migration. Development. 1999;126:3217–3228. doi: 10.1242/dev.126.14.3217. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Chen JX, Rothman TP, Gershon MD. Inhibition of in vitro enteric neuronal development by endothelin-3: mediation by endothelin B receptors. Development. 1999;126:1161–1173. doi: 10.1242/dev.126.6.1161. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Yamao M, Nishiyama H, Sugihara S, Nagaoka S, Tomita M, Yoshizato K, Tamura T, Mori H. New and highly efficient method for silkworm transgenesis using Autographa californica nucleopolyhedrovirus and piggyBac transposable elements. Biotechnol Bioeng. 2004;88:849–853. doi: 10.1002/bit.20296. [DOI] [PubMed] [Google Scholar]
- Young HM, Anderson RB, Anderson CR. Guidance cues involved in the development of the peripheral autonomic nervous system. Auton Neurosci. 2004a;112:1–14. doi: 10.1016/j.autneu.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Young HM, Bergner AJ, Anderson RB, Enomoto H, Milbrandt J, Newgreen DF, Whitington PM. Dynamics of neural crest-derived cell migration in the embryonic mouse gut. Dev Biol. 2004b;270:455–473. doi: 10.1016/j.ydbio.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Yu HH, Huang AS, Kolodkin AL. Semaphorin-1a acts in concert with the cell adhesion molecules fasciclin II and connectin to regulate axon fasciculation in Drosophila. Genetics. 2000;156:723–731. doi: 10.1093/genetics/156.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberstein Y, Fuchs E, Hershtik L, Ayali A. Neuromodulation for behavior in the locust frontal ganglion. Journal of Comparative Physiology a-Neuroethology Sensory Neural and Behavioral Physiology. 2004;190:301–309. doi: 10.1007/s00359-004-0496-5. [DOI] [PubMed] [Google Scholar]
- Zitnan D, Endo Y, Sehnal F. Stomatogastric nervous system of Galleria mellonella L. (Lepidoptera: Pyralidae): changes during metamorphosis with special reference to FMRFamide neurons. Int. J. Insect Morphol. & Embryol. 1989;18:227–237. [Google Scholar]
- Zito K, Fetter RD, Goodman CS, Isacoff EY. Synaptic clustering of fasciclin II and Shaker: essential targeting sequences and role of dlg. Neuron. 1997;19:1007–1016. doi: 10.1016/s0896-6273(00)80393-1. [DOI] [PubMed] [Google Scholar]
- Zou Y. Navigating the anterior-posterior axis with Wnts. Neuron. 2006;49:787–789. doi: 10.1016/j.neuron.2006.03.004. [DOI] [PubMed] [Google Scholar]
- zur Lage P, Jarman AP. Antagonism of EGFR and notch signalling in the reiterative recruitment of Drosophila adult chordotonal sense organ precursors. Development. 1999;126:3149–3157. doi: 10.1242/dev.126.14.3149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.