Abstract
RapA, a prokaryotic member of the SWI/SNF protein superfamily, is an integral part of the RNA polymerase transcription complex. RapA’s function and catalytic mechanism have been linked to nucleic acid remodeling. In this work we show that mutations in the interface between RapA’s SWI/SNF and double-stranded nucleic acid-binding domains significantly alter ATP hydrolysis in purified RapA. The effects of individual mutations on ATP hydrolysis loosely correlated with RapA’s nucleic acid-remodeling activity, indicating that the interaction between these domains may be important for the RapA-mediated remodeling of nonproductive transcription complexes. In this study we introduced a model system for in vitro transcription of a full-length E. coli gene (slyD). To study the function of RapA, we fractionated and identified in vitro transcription reaction intermediates in the presence or absence of RapA. These experiments demonstrated that RapA contributes to the formation of free RNA species during in vitro transcription. This work further refines our models for RapA function in vivo and establishes a new role in RNA management for a representative of the SWI/SNF protein superfamily.
Keywords: RapA, RNA polymerase, SWI/SNF, transcription, DNA-RNA, triplex
Escherichia coli (E. coli) has served as a primary model system for the study of enzymes involved in RNA synthesis. A textbook example of polymerase function, the E. coli core RNA polymerase formed by the α2ββ’ω subunits is capable of transcription elongation and termination (1). Proteins called sigma factors (1–3) associate with the core enzyme and allow the resulting complex, referred to as the RNA polymerase holoenzyme, to bind DNA in a sequence-specific manner and initiate transcription from loosely homologous promoter sequences on the E. coli chromosome. Each sigma factor controls multiple groups of promoters; e.g, the key growth-related and housekeeping genes in E. coli are under control of σ70 (4), the rpoD gene product. Approximately one-third of the core E. coli RNA polymerase molecules in a given cell form a high-affinity, stoichiometric complex with σ70 under normal growth conditions; the α2ββ’ω and α2ββ’ωσ70 enzyme species display a number of distinct properties and can be chromatographically separated (5, 6). Apart from the sigma factors and relatively small proteins (which are unlikely to display mechanistically complex enzymatic activities), the key known interactors of the E. coli RNA polymerase established through biochemical studies are NusA (7, 8) and RapA (6, 9). NusA – an essential RNA-binding protein in E. coli – has been described in the existing literature as a transcription elongation factor and a cofactor of antiterminators (10–13). The function of RapA (also known as HepA) – the first identified bacterial representative of the SWI/SNF protein superfamily – is not fully understood. Under most conditions (in liquid cultures), rapA deletion has little or no effect on cell growth (6, 14), protein expression (as judged by 2D electrophoresis; M.V. Sukhodolets, unpublished results), UV-sensitivity (14), or mutation rates (14). This apparent lack of a pronounced rapA effect in vivo points to functions other than general regulation of transcription, as proposed for RapA’s eukaryotic counterparts. However, rapA deletion results in a unique phenotype, rendering bacteria incapable of efficient growth on agar plates in high salt (15). In vitro, RapA promotes multi-round transcription in a salt concentration-specific fashion (15, 16). Salt-selectivity is therefore a shared feature of RapA effects observed in vivo and in vitro. Our recent work presented multiple, independent lines of evidence indicating that RapA binds and remodels RNA during transcription (17). This study also indicated that non-canonical DNA-RNA complexes such as DNA-RNA triplexes – the formation of which is enhanced in high salt (17) – could represent potential substrates for RapA, and we proposed that remodeling of non-canonical nucleic acid complexes could be the primary function of RapA and its homologs in vivo (17). A recently reported, 3.2Å-resolution structure of RapA was used to construct a putative structural model for a RapA-nucleic acid complex, suggesting that RapA is capable of simultaneous interactions with both double-stranded and single-stranded nucleic acids (18). This is consistent with the proposed role of RapA in the remodeling of non-canonical nucleic acid complexes.
In this work we constructed several new RapA mutations, focusing on the RapA domain harboring SWI/SNF homology motifs IV–VI (referred to herein as the SWI/SNF domain) and its interface with the domain containing the putative double-stranded nucleic acid template (dsT)-binding site (17, 18), and studied their effect on RapA’s ATP-hydrolytic and nucleic acid-remodeling activities. We demonstrate a significant effect of the mutations in this region on RapA’s ATP-hydrolytic and nucleic acid-remodeling activities. The effects of individual mutations on ATP hydrolysis loosely correlated with RapA’s nucleic acid-remodeling activity in in vitro transcription, indicating that the interaction between the SWI/SNF and dsT-binding domains may be important for the RapA-mediated remodeling of nonproductive transcription complexes.
In this study we have tested – for the first time in studies with RapA – the effects of RapA on the transcription of a model full-length E. coli mRNA containing intact 5’-and 3’-nontranslated sequences. The results presented herein are in accord with our existing model, in which RapA aids RNA polymerase in the displacement of nascent RNA from transcription complexes. We show that this enzymatic activity of RapA is unique and cannot be mimicked by other RNA-binding proteins with a demonstrated ability to promote transcriptional cycling, such as S1 (19). In this work we further developed our methods for fractionation and identification of in vitro transcription reaction intermediates. Using a non-denaturing (as well as a previously developed semi-denaturing) method of separation of the in vitro transcription reaction intermediates, we demonstrate that RapA promotes the formation of non-DNA-bound or aggregated RNA species during transcription. This work further refines our models for RapA function in vivo and – taken together with our previous study – establishes a new role in RNA management for a representative of the SWI/SNF protein superfamily. This finding not only increases our understanding of the fundamental process of transcription, but also brings a fresh perspective to studies demonstrating links between mutations in human SWI/SNF genes and cancer (20–24).
Materials and Methods
Enzymes
In this study, we have further improved the procedure for the isolation of native E. coli RNA polymerase and its accessory proteins. The modified procedure – in general reminiscent to that described in (25) – results in improved purity of the isolated proteins and complexes. The key changes, in comparison with the protocol described in (25), included (a) the utilization of two different FPLC instruments: a classic (now discontinued) FPLC system (Pegasus Scientific), set up in the cold room to carry out the initial, single-stranded DNA chromatography stage, and an ACTA FPLC system (GE Healthcare) set up and operated at room temperature via Unicorn software during the final chromatography stages (however, the Superloops were placed on ice during the Mono Q column loading stage), (b) the utilization of self-prepared (calf thymus) single-stranded DNA-Sepharose 4B instead of commercially available DNA-Agarose, to improve flow rates, (c) scaling down the procedure to 25–35 g of E. coli MG1655 cells per purification and using a Mono Q 5/5 column in order to achieve more uniform loading of the column, and (d) increasing the length (>2-fold) of the Superdex 200 column during the final, gel-exclusion chromatography stage. The schematic for the procedure is shown in Figure S1B; the detailed protocol will be described elsewhere. Recombinant, His-tagged wild-type and mutant RapA proteins were purified as described in Figures S1C and S2. Wild-type recombinant S1 and NusA were isolated as previously described (25).
PCR and DNA purification
DNA encompassing the slyD operon was amplified from MG1655 E. coli chromosomal DNA using MS696 (5’-ATTGTAAGCTTCCCGGGGAAACGCCACCGCCACATTATTGAGGCG) and MS697 (5’-CCGAAAGTGGATCCCGGGGCCCGGCCTGTCAGGCGCAGGATTCAATGGCG)D NA primers and the Expand High Fidelity PCR kit (Roche Diagnostics). Reactions were carried out as suggested by the manufacturer; the extension time was 2 min at 72°C. Because our previous work implied the possibility of non-canonical interactions between nucleic acids in transcription, we tested different methods for purification of PCR-generated DNA templates in order to rule out potential impurity and/or conformational heterogeneity in the isolated DNA; the amplified DNA used in key experiments was purified by FPLC, as described in Figure S1, using a custom-packed 0.3-ml Source Q column. Following the DNA amplification, the reaction mixture was diluted 4–5-fold with purified water (KD Medical, Molecular Biology Grade) and injected onto the column. The reaction products were eluted with a linear gradient of NaCl (0–1 M) in 20 ml of TGED buffer (0.01 M Tris-HCl, 5% glycerol, 0.1 mM EDTA, 0.1 mM dithiothreitol, pH 7.5), as shown in Figure S1. The amplified slyD DNA – confirmed by agarose gel electrophoresis (Figure S1A, insert) and DNA sequencing during the primer extension experiments (Figure 4A) – was precipitated with 7–8 volumes of ethanol; air-dried pellets were then dissolved in purified water to the concentrations indicated below.
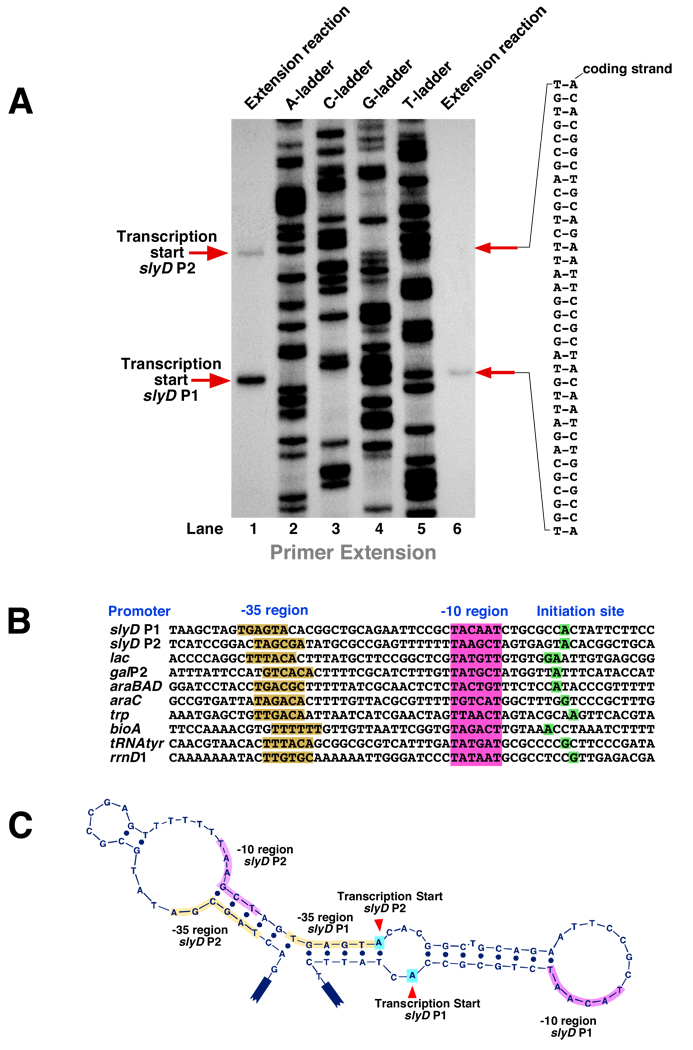
Figure 4. Mapping of the transcription initiation site(s) at the E. coli slyD operon. A.
Determination of the transcription start site(s) at the slyD operon. Primer extension reactions (lanes 1 and 6) resulting from extension of the DNA primer MS702 (see Materials and Methods) by M-MuLV Reverse Transcriptase from the slyD mRNA template(s) synthesized in vitro. Lanes 2–5 show the DNA sequencing ladder – the result of extension of the same primer by Taq DNA polymerase. B. Conserved elements (boxes shown in magenta and yellow) and the transcription start sites (green boxes) of the slyD P1 and P2 promoters. SlyD P1 and slyD P2 promoters are aligned against several well-studied E. coli promoters. C. The slyD operon’s conserved elements and the transcription start sites are shown on the coding DNA strand folded into the hypothetical lowest-energy secondary structure; the template strand is not shown. D. The nontranslated 5’-terminus of the slyD mRNA originating at the P2 promoter. The lowest-energy secondary structure is shown. The red arrowhead points to the 5’-terminus of the slyD mRNA originating at the P1 promoter. E. The effect of RapA on transcription at the slyD operon. In vitro transcription reactions were carried out as described in Materials and Methods. Medium-duration (15-min) reactions (lanes 1 and 2) and long-duration (60-min) reactions (lanes 3 and 4) were carried out in the presence or absence of excess (1 µM) wild-type RapA. Primer extension analysis of the RNAs synthesized in vitro was carried out as described in Materials and Methods. Note that RapA has little or no effect on P1-driven transcription, but has a positive (4–5-fold) stimulatory effect on transcription initiated at P2. The quantitated levels of reverse-transcribed RNAs are shown at the right.
In vitro transcription
In vitro transcription reactions were carried out in either Buffer D (20 mM Tris-acetate, 10 mM magnesium acetate, 50 mM potassium acetate, 1 mM dithiothreitol, pH 7.9), Buffer C (50 mM Tris-HCl, 10 mM MgCl2, 100 mM NaCl, 1 mM dithiothreitol, pH 7.9), or Buffer B (50 mM Tris-HCl, 10 mM MgCl2, 200 mM NaCl, 1 mM dithiothreitol, pH 7.9); the transcription buffers are specified in Figure Legends. In our preliminary studies we found that the slyD mRNA was more efficiently synthesized in vitro in Buffer D than in the previously utilized Buffer B [17]). Since Buffer D is widely used as a standard buffer for a large number of bacterial restriction endonucleases and includes potassium and acetate (both key ions in E. coli), we switched to Buffer D as a more ‘physiological’ buffer for in vitro transcription with linear DNA templates containing the slyD operon. Typically, 14-µl in vitro transcription reactions included 1.4 µl 10x reaction buffer, 2–4 µl purified linear DNA template containing the slyD operon (20–90 µg/ml in purified water) or 1–2 µl supercoiled DNA template pCPGt3te (26) (0.3–0.4 µg/µl in purified water), 2 µl purified RNA polymerase holoenzyme (0.07–0.2 mg/ml in 1x TGED buffer, unless indicated otherwise in Figure Legends), 0.5–4 µl purified recombinant RapA (RapA concentrations are specified in Figure Legends), and purified water (KD Medical, Molecular Biology grade) to a volume of 12 µl. In vitro transcription was initiated by the addition of 2 µl of rNTP mix containing 1.4 mM each of ATP, GTP, CTP, UTP, and either [α-32P] ATP or [α-32P] UTP (MP Biomedicals). Reactions were incubated for 15 min at 37°C unless indicated otherwise. Transcription reactions were terminated by the addition of an equal volume (14 µl) of Stop solution (50% glycerol; 50 mM EDTA; Bromphenol Blue, 0.1%; pH 7.5), and 8-µl aliquots of the reactions were analyzed (without boiling, unless indicated otherwise) on either non-denaturing or semi-denaturing polyacrylamide gels. Semi-denaturing (0.4 mm-thick, 38 cm-long) gels were cast using the SequaGel kit (National Diagnostics); the exact polyacrylamide concentrations are specified in Figure Legends. Gibco BRL S2 sequencing gel apparatuses were used. Under semi-denaturing conditions (Figure 5), increasing gel thickness and reducing the sample volume led to a significant reduction in the yields of RapA-RNA and RNA polymerase-RapA-RNA complexes; we therefore believe that the complexes in question are not fully denatured by urea in the matrix of a 0.4 mm-thick gel during the course of these separations. Gels were typically run at 20–30 Watts until Bromphenol Blue reached the bottom of the gel, unless indicated otherwise. Non-denaturing (1 mm-thick, 32 cm-long) gels containing 1x TBE (KD Medical) were cast using ProtoGel polyacrylamide mix (National Diagnostics); Gibco BRL SA apparatuses were used. After electrophoresis, X-ray films (F-BX810; Phenix Research Products) were exposed to ‘wet’ gels covered with plastic wrap. With Kodak BioMax MS screens, typical exposure times were 8–40 hours at −80°C.
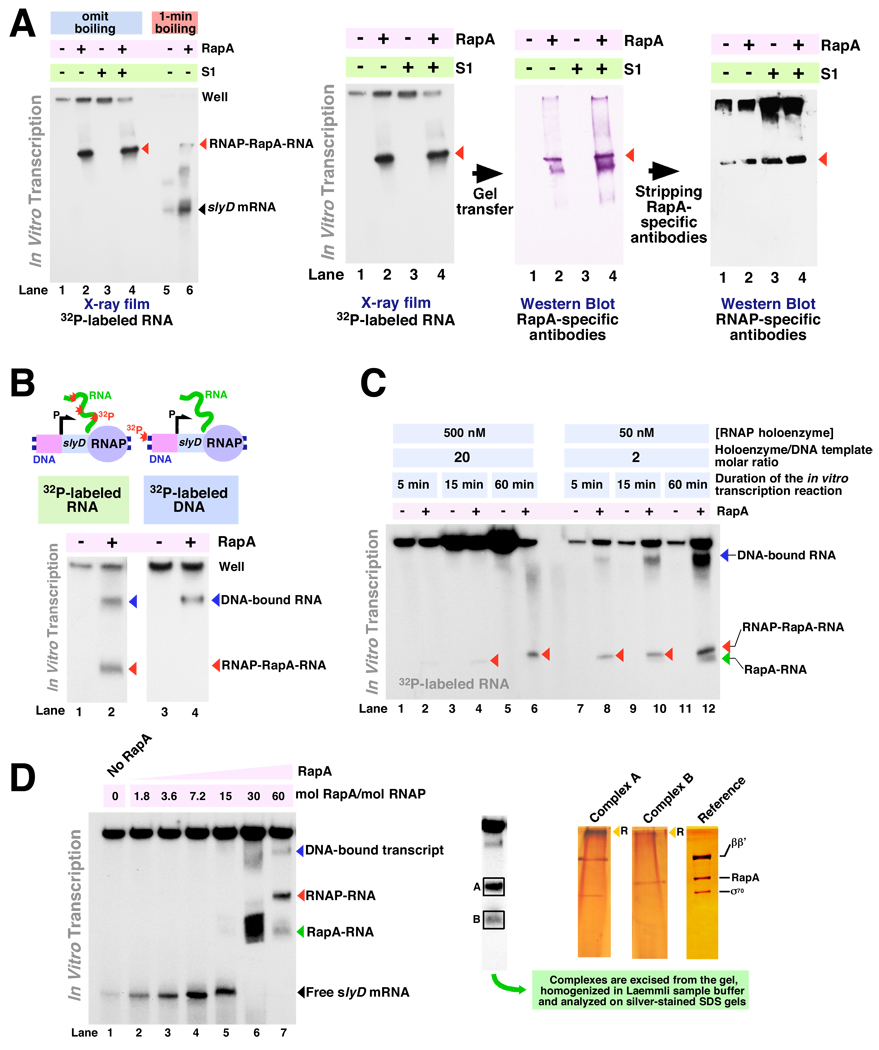
Figure 5. RapA contributes to the formation of free (non-DNA-bound or aggregated) RNA during in vitro transcription. I: Fractionation and identification of RapA-specific intermediates under semi-denaturing conditions. A.
Formation of RapA-specific complexes in in vitro transcription system with linear DNA templates encompassing the E. coli slyD operon. In vitro transcription reactions in the absence of effectors (lanes 1 and 5), in the presence of 1 µM of purified recombinant RapA (lanes 2 and 6), 1 µM purified recombinant S1 (lane 3), or 1 µM RapA plus 1 µM S1 (lane 4) were carried out as described in Materials and Methods, and the reaction products were separated on 6% SequaGel (National Diagnostics). Reaction conditions: Buffer D (see Materials and Methods); purified 965-nt linear DNA template containing the slyD operon (PCR MS696/697), 7 nM; RNA polymerase holoenzyme, 20 nM. Reactions not denatured by boiling are in lanes 1–4; reactions in lanes 5 and 6 are identical to those in lanes 1 and 2 except that the samples were subjected to a 1-min boiling step followed by placement on ice after the addition of Stop solution. RNA polymerase-RapA-transcript complexes are indicated with red arrowheads. Panels at the right: identification of the in vitro transcription reaction intermediates through a combination of uniform 32P RNA labeling and immunoassays. Following electrophoresis, X-ray films were exposed to ‘wet’ gels to visualize 32P-labeled RNA transcripts. Next, the contents of the gels were electroeluted onto Hybond-P membranes (Amersham Pharmacia Biotech) for immunoassays, first with RapA-specific and next with RNA polymerase-specific antibodies (sequentially, using the same membrane, after stripping the RapA-specific antibodies). B. Identification of DNA-bound complexes formed during in vitro transcription with linear DNA templates encompassing the slyD operon through selective 32P labeling of RNA or DNA. In vitro transcription reactions in the absence of RapA (lane 1) or in the presence of 1 µM of purified recombinant RapA (lane 2) were carried out as described in Materials and Methods. Reactions in lanes 3 and 4 were similar to those in lanes 1 and 2 except that 32P end-labeled DNA template and ‘cold’ rNTP mix was used; the experimental design is illustrated by the schematics above. The in vitro transcription reaction products were separated on 4.5% SequaGel (National Diagnostics). C. Kinetics of the formation of free (non-DNA-bound or aggregated) RNA polymerase-transcript complexes in the presence of RapA. In vitro transcription reactions were carried out as described in Materials and Methods in the absence of RapA (lanes 1, 3, 5, 7, 9, 11) or in the presence of 1 µM wild type recombinant RapA (lanes 2, 4, 6, 8, 10, 12), and the reaction products were resolved on 5% SequaGel (National Diagnostics). Reaction conditions: Buffer D; purified 965-nt linear DNA template containing the slyD operon (PCR MS696/697), 25 nM. The RNA polymerase holoenzyme concentrations are indicated in the figure. RNA polymerase-RapA-transcripts (red arrowhead), RapA-RNA adducts (green arrowhead), and DNA-bound transcripts (blue arrowhead) are also indicated. D. Titration with purified RapA of in vitro transcription reactions with the slyD operon. Reactions were carried out in Buffer D, as described in Materials and Methods, and the reaction products were resolved on 4.5% SequaGel (National Diagnostics). Linear DNA template (PCR 696/697), 40 nM; RNA polymerase holoenzyme, 50 nM. RNA polymerase-transcript (red arrowhead), RapA-RNA adduct (green arrowhead), and DNA-bound transcript (blue arrowhead) are indicated. Panels at the right: to confirm the identity of individual complexes, the bands containing 32P-labeled RNA transcript (highlighted by rectangle frames ‘A’ and ‘B’) were dissected from the gel and homogenized in ~200 µl of Laemmli sample buffer in Eppendorf tube-size disposable homogenizers. Following precipitation of the polyacrylamide slurry by centrifugation, the supernatants were analyzed on silver-stained SDS gels. The reference lane contained the 1:1 RNA polymerase holoenzyme-RapA complex. RapA, σ70, and the large RNA polymerase subunits are indicated; a silver-stained 7% SDS-polyacrylamide gel is shown. Yellow arrowheads (‘R’) indicate RNA, which was also visualized by silver staining.
Primer extension experiments
20-µl primer extension reactions included 2 µl 10x M-MuLV Reverse Transcriptase Reaction Buffer, 6000–10000 cpm (32P) end-labeled DNA primer MS702 (5’-CGCGCCAACAGCGACATCAAATTTGTCGCC), 2 µl dNTP mix (containing 1 mM each of dATP, dGTP, dCTP, and dTTP), 2 µl M-MuLV Reverse Transcriptase (New England Biolabs, 200,000 U/ml), 4 µl purified RNA from in vitro transcription reactions carried out as described above, and purified water to a final volume of 20 µl. Reactions were incubated for 30 min at 37°C and terminated and analyzed as described above for in vitro transcription reactions. Labeling and gel-purification of the DNA primer MS702 was performed as previously described for end-labeled RNA probes (17). Dideoxy DNA sequencing was carried out as described elsewhere.
Mutagenesis
The QuikChange™ site-directed mutagenesis kit (Stratagene) was used to construct the described RapA mutations. Primers LZ1 (5’-CAGCATCAGTGGCTGGTAGAAATGCTGGCCGCTTTCAACCTGCGCTTTGCGCTATTTGAT) and LZ2 (antiparallel to LZ1) were used to construct the RapA R221A/R222A double mutation. Primers BY3 (5’-GAGCAGCGTATTGGTCGTCTGGATGCTATCGGCGCGGCGCACGATATTCAGATCCATGTG) and BY4 (antiparallel to BY3) were used to construct the RapA R599A/Q602A mutation, and primers BY5 (5’-GCTATTAAAGTCTCCGGCATTATGGGCGCAGCTGCAAGTGCGGAAGATCGTGCTCGCGATATGCTC) and BY6 (antiparallel to BY5) were used to construct the RapA R457A/K458A mutation. Following amplification with mutagenic primers and DpnI digestion of the template DNA (pQE32-RapA [17]), aliquots of the reaction mixtures were used to transform XL10-Gold Ultracompetent Cells (Stratagene). Plasmid DNAs from individual clones were purified and analyzed. The constructs were confirmed by DNA sequencing. The expression of full-length RapA in individual clones was also verified by SDS-PAGE-Western Blot using RapA-specific antibodies (14). The purified plasmids carrying the constructed mutation(s) were then re-transformed into M15/pRep4 E. coli competent cells (Qiagen), and the His-tagged RapA protein was overproduced and purified as described above.
Western blotting was carried out as follows. Following electrophoresis, proteins were transferred onto either Immobilon-P (Millipore) or Hybond-P (Amersham Pharmacia Biotech) membranes (with similar results) in Mini-PROTEAN 3 Trans-Blot modules (Bio-Rad), typically, at 40–60 V overnight. Membranes were blocked with 5% Blotting Grade Blocker (Bio-Rad; #170–6404) in PBST (Calbiochem; #524653) for 30–60 min and incubated with primary antibodies (typically, a rabbit serum at a dilution of 1/1000 to 1/5000 in 1x PBST containing 1% Blotting Grade Blocker) for 1–5 hours at room temperature on an orbital shaker. Following three 5-min washes with 1x PBST, membranes were incubated with secondary antibodies (a donkey anti-rabbit Ig–horseradish peroxidase conjugate [Amersham Pharmacia Biotech], typically at a dilution of 1/2000 to 1/16000 in 1x PBST containing 1% Blotting Grade Blocker) for 30–60 min. Following three 5-min washes with 1x PBST, and 2–3 brief washes with water, the membranes were incubated for 30 sec with 2–4 ml of SuperSignal West Pico Chemiluminescent Substrate (Pierce; #34080), briefly drained, and placed in plastic wrap; X-ray films (Phenix Research Products; F-BX810) were then exposed to the luminescent membranes.
ATPase activity assays
ATPase activity assays were performed by measuring the amount of [α-32P]ADP released from [α-32P]ATP. Either Buffer D or Buffer C was used as a reaction buffer, with similar results; reaction buffers are specified in Figure Legends. Ten-µl reaction mixtures contained 1 µl of the 10x reaction buffer, 0.3–0.5 µg purified RapA, 50–1600 nM cold ATP, and 500–3000 cpm of [α-32P]ATP (MP Biomedicals; #32007U). Following incubation at 37°C (typically for 60 min, unless indicated otherwise in Figure Legends), 1.5–3 µl from each reaction was spotted on a PEI-cellulose plate (Analtech; #206016), and chromatography was carried out in 1 M LiCl, 1 M formic acid. Following chromatography, the plates were covered with plastic wrap, and X-ray films (Phenix Research Products; F-BX810) were exposed to the TLC plates.
E. coli strains
The previously constructed MG1655rapA− E. coli strain (14) was used in this work.
Results
Functional significance of RapA for bacterial growth
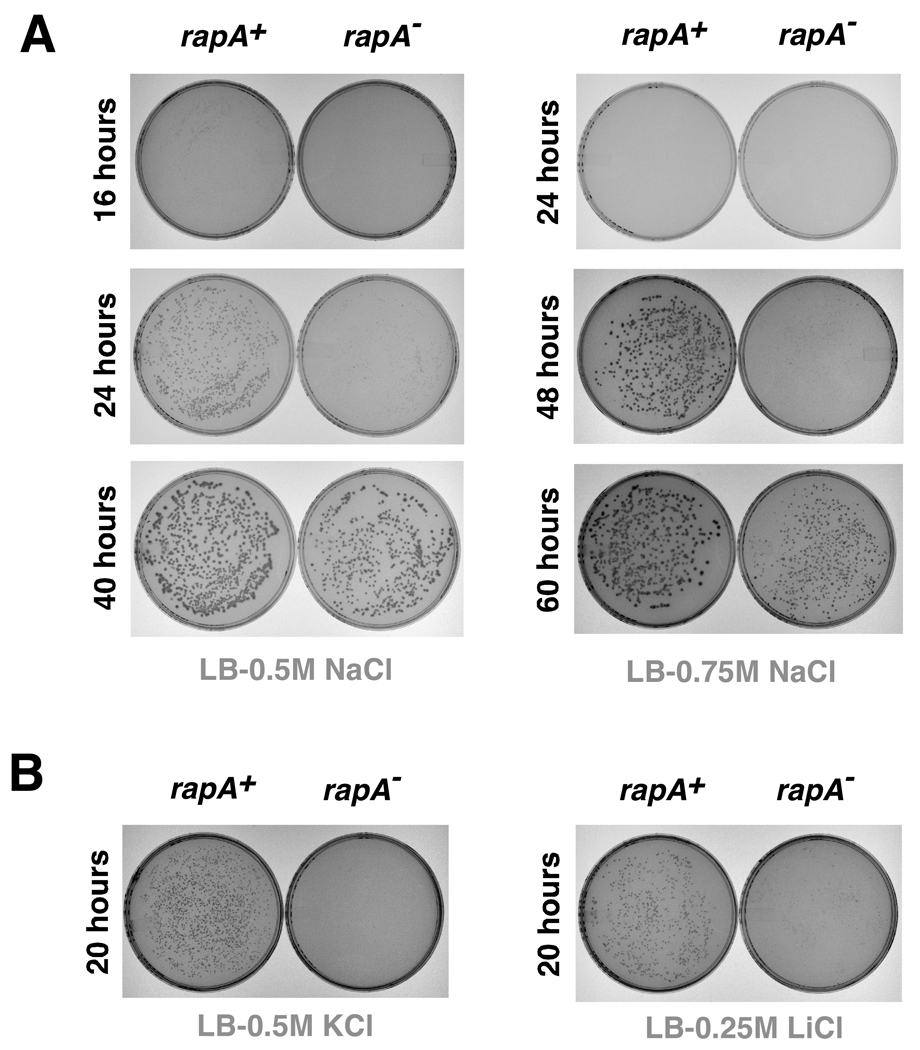
Although the rapA gene is not essential in E. coli (14), rapA deletion results in a unique phenotype manifested as the inability of E. coli to grow efficiently on LB agar containing 1M NaCl (15). We recently explained this phenotype in light of our newly rendered model for RapA function in the remodeling of noncanonical DNA-RNA complexes in transcription (17), the formation of which is enhanced in high salt (17). To study this phenotype further, we (a) analyzed the kinetics of growth of E. coli MG1655rapA+ and MG1655rapA− strains on LB-agar plates in the presence of NaCl and other salts, and (b) compared growth efficiencies of the two strains on agar plates with those in liquid media. Kinetic experiments indicated a pronounced effect of rapA on cell growth at ≥0.5 M NaCl. Prolonged incubations of the plates (20–40 hours) containing 0.5–0.75 M NaCl resulted in significant differences in colony size between the two strains (Figure 1A). The effects were more pronounced on LB-agar plates than on SOC-agar or Superbroth-agar plates (data not shown). The effect of salts other than NaCl on growth of the two strains in plated cultures is illustrated in Figure 1B. Interestingly, the growth defect associated with the rapA deletion mutation was not evident in the liquid media – under otherwise similar conditions – but was seen only in plated cultures. As an alternative means of increasing the intracellular ionic strength in liquid cultures, we monitored the kinetics of growth of the rapA+ and rapA−strains in the presence of polyethylene glycol. However, these experiments consistently showed little or no difference in growth rates between the two strains, even at relatively high polyethylene glycol concentrations (data not shown).
Figure 1. In vivo effects of the rapA deletion mutation. A.
Kinetics of the E. coli MG1655rapA+ and MG1655rapA− cells’ growth on LB-agar plates containing elevated concentrations of sodium chloride. B. Effects of KCl and LiCl on growth of the two strains on LB-agar.
Alterations of ATP-hydrolytic and nucleic acid-remodeling activities of RapA by mutations in the interface between RapA’s SWI/SNF and dsT-binding domains
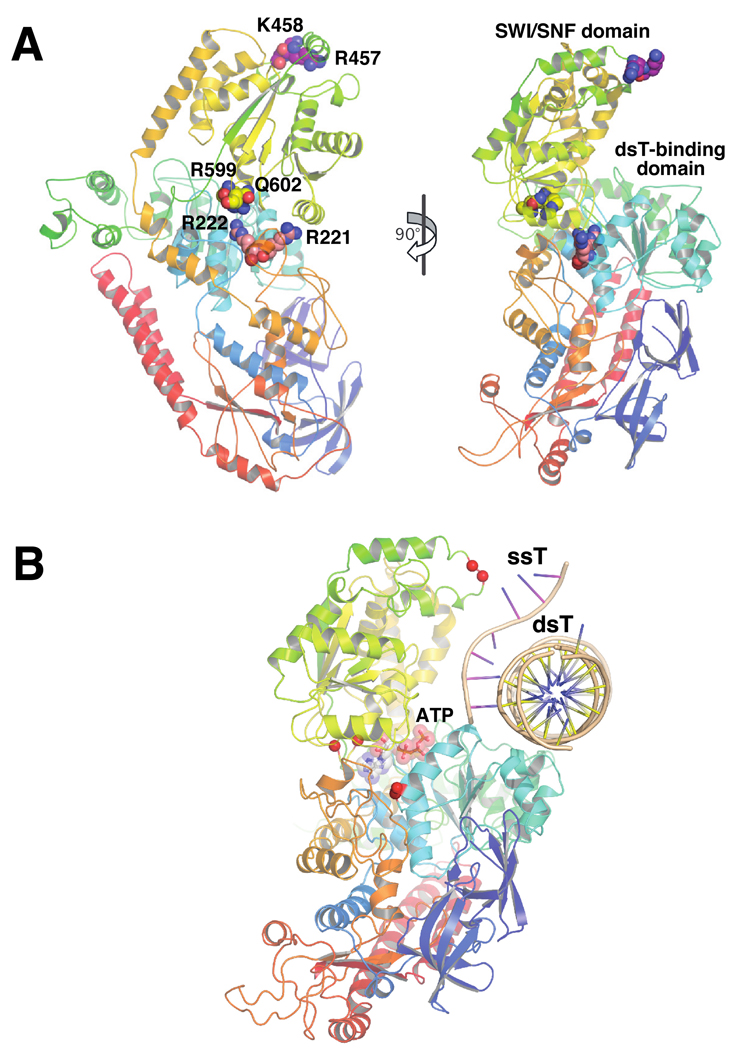
We sought to extend previously developed in vitro assays for analysis of the function of individual domains in RapA. Our primary focus was the RapA domain harboring SWI/SNF homology motifs IV–VI (referred to herein as the SWI/SNF domain), due to its obvious significance in defining the SWI/SNF protein superfamily. Studies with distant homologs of RapA have indicated that coordinated, ATP-driven re-orientation of the SWI/SNF domain with respect to the double-stranded nucleic acid-binding domain may enable translocation of SNF2 enzymes along nucleic acids (27, 28). We therefore sought to genetically alter the interface of the two domains in RapA (Figures 2A and 2B) in order to determine the effect of such modifications on RapA’s ATP-hydrolytic and nucleic acid-remodeling activities. To this aim, we constructed and purified two mutant RapA proteins (each carrying a double mutation): RapAR599A/Q602A (this conserved amino acid pair is located within the section of RapA’s SWI/SNF domain facing the dsT-binding domain [Figures 2A and 2B]), and RapAR221A/R222A (this moderately conserved arginine tandem is positioned in the section of RapA’s dsT-binding domain facing the SWI/SNF domain [Figures 2A and 2B]). Each double mutation – based on available structural data (18) – should result in destabilization of the interaction between RapA’s dsT-binding and SWI/SNF domains. The purification of both mutant proteins is described in Supplementary Data (Figures S2A and S2B). The second set of mutations targeted a potential single-stranded nucleic acid-binding site within RapA’s SWI/SNF domain. Recent work indicated that RapA’s SWI/SNF domain could be involved in binding single-stranded nucleic acids (18). The model of the RapA-nucleic acid complex introduced by Shaw et al. suggests that the loop within RapA’s SWI/SNF domain carrying a conserved tandem of positively charged amino acids (R457/K458 [Figures 2A and 2B]) may be involved in this interaction (18). We therefore constructed and purified a RapA mutant carrying alanine substitutions at these two amino acid positions (RapAR457A/K458A; Supplementary Data, Figure S2C).
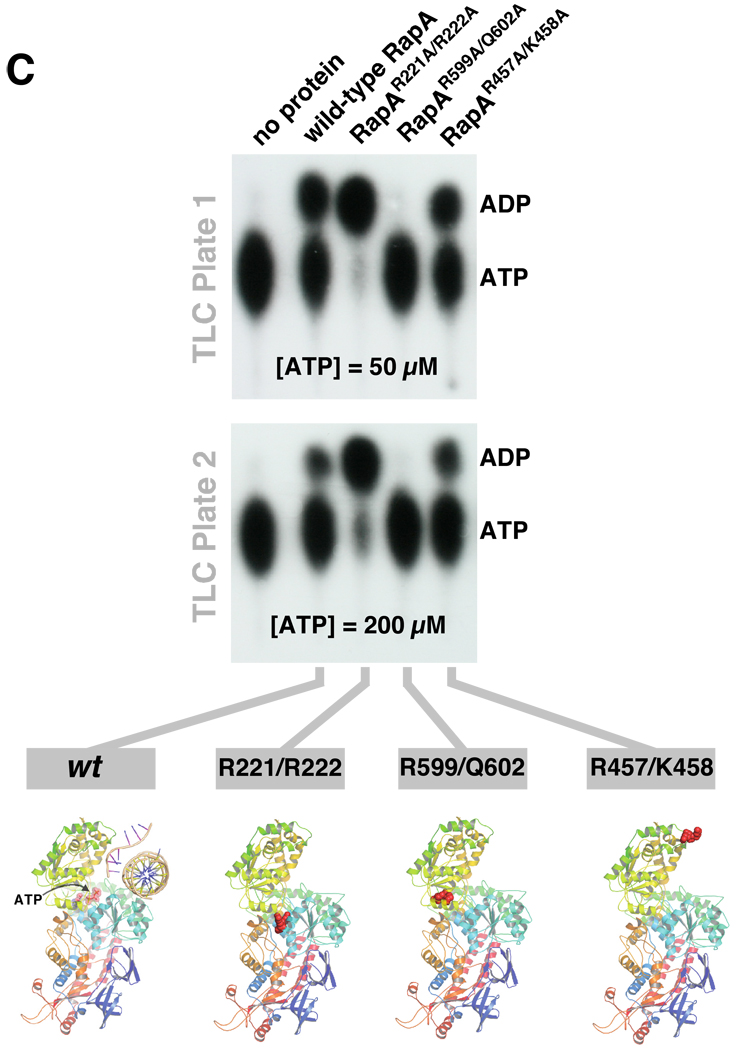
Figure 2. Effect of the mutations in RapA’s SWI/SNF domain and its interface with the putative double-stranded nucleic acid-binding domain on the ATP-hydrolytic activity of RapA.
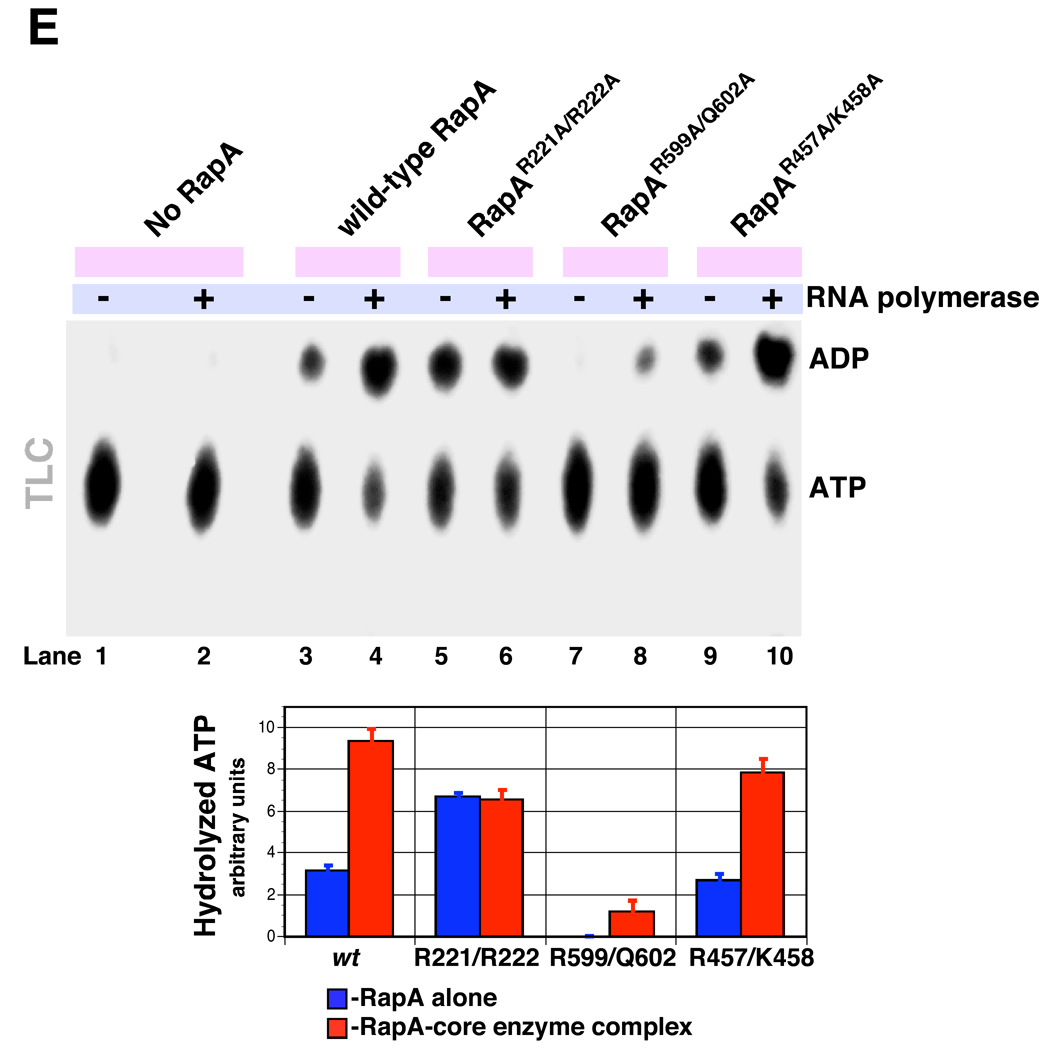
A and B Positions of the selected mutation sites in the RapA structure. For clarity, only the backbone of the RapA structure (18) is shown; the mutated amino acids are shown as spheres. Figure 2B also shows the hypothetical positions of a double-stranded nucleic acid template (dsT; a cross-sectional view is shown) and a single-stranded nucleic acid template (ssT). The dsT position (17, 18) is determined solely on the basis of modeling using the SsoRad54–DNA complex (47) as a template; no available data conclusively rule out RNA as a dsT. SsT binding by RapA’s SWI/SNF domain is also supported by the modeling data (18). The composite model shown in Figure 2B was constructed as described by Shaw et al. (18), essentially by mapping the ATP and nucleic acid-binding sites (both single- and double-stranded DNA) from available crystal structures of the ternary PcrA-ATP-DNA and binary SsoRad54-DNA complexes onto the E. coli RapA structure (PDB 3DMQ), followed by manual adjustment and energy minimization. C and D. Mutations in the interface of RapA’s putative dsT-binding and SWI/SNF domains significantly alter its ATP hydrolytic activity. ATPase assays were carried out in Buffer D, as described in Materials and Methods. Representative PEI-cellulose plates illustrating the ATP hydrolysis by the wild-type and recombinant mutant RapA proteins are shown in Figure 2C. Kinetic parameters (Km and Vmax) summarized in Figure 2D were obtained from the initial velocity of ATP hydrolysis vs. the ATP concentration plots (shown at the top) using Prism 5 Software (GraphPad). The parameters were calculated assuming classic Michaelis-Menten kinetics. ND: not detectable. E. Effect of the core RNA polymerase on ATP-hydrolytic activity of the wild-type and mutant RapA proteins. ATPase assays were carried out in Buffer D, as described in Materials and Methods. Reactions in the absence (lanes 1, 3, 5, 7, and 9) or in the presence of 2 mol of the purified core RNA polymerase per mol RapA (lanes 2, 4, 6, 8, 10) are shown. Quantitated results of the experiment are shown below; data represent the average of two independent sets of experiments.
Mutations in the interface of RapA’s dsT-binding and SWI/SNF domains altered ATP hydrolysis by purified RapA, as expected. The RapA R599A/Q602A mutation abolished ATPase activity in purified mutant RapA (Figures 2C and 2D; the solubility and yields of RapAR599A/Q602A were comparable to those seen with wild-type RapA [Figures S2B and S2D]). In contrast, the RapA R221A/R222A mutation resulted in an increase in ATPase activity in the purified protein (Figures 2C and 2D); the effect was primarily due to an increase in kcat (Figure 2D). Because this is the first case to date in which a RapA mutation resulted in an increase in RapA’s specific ATPase activity, we carried out multiple, side-by-side purifications of wild-type RapA and RapAR221A/R222A; during the course of these trials we consistently observed higher specific ATPase activities with purified RapAR221A/R222A (a representative side-by-side comparison of specific ATPase activities in proteins obtained in an independent set of purification procedures is shown in [Supplementary Data Figure S3]). The RapA R457A/K458A double mutation had little or no effect on ATP binding (Km, ATP) and hydrolysis (Figure 2D). This result is expected and is consistent with the predicted position of these amino acids in the section of RapA’s SWI/SNF domain opposite from the putative ATP-binding site (18).
Next, we monitored ATP hydrolysis by wild-type and mutant RapA proteins in the presence or absence of ssRNA, a (ds)RNA-DNA hybrid, and dsDNA. There was no apparent effect of these nucleic acids on the rates of RapA-mediated ATP hydrolysis (Supplementary Data, Figure S4). This result is in accord with previously reported data indicating little or no modulation of RapA’s ATPase activity by nucleic acids (6) (in contrast to the Rad54 homolog from Sulfolobus solfataricus [SsoRad54], the ATPase activity of which is stimulated by nucleic acids [29]). Because it was reported that RNA polymerase exerts a stimulatory effect on kcat of the RapA ATPase (6) we also monitored the ATP hydrolysis by wild type and mutant RapA proteins in the presence or absence of RNA polymerase. We found that RapAR221A/R222A (the specific ATPase activity of which was comparable to that of wild-type RapA in the presence of the polymerase) was insensitive to the stimulatory effect of the polymerase (Figure 2E, compare lanes 3 and 4 to lanes 5 and 6). ATP hydrolysis was detectable in reactions containing RapAR599A/Q602A in the presence of the polymerase – an apparent partial recovery of the ATP-hydrolytic function in the RapAR599A/Q602A-core enzyme complex (Figure 2E, compare lanes 3 and 4 to lanes 7 and 8). The RapAR457A/K458A double mutant mimicked the behavior of wild-type RapA (Figure 2E, compare lanes 3 and 4 to lanes 9 and 10).
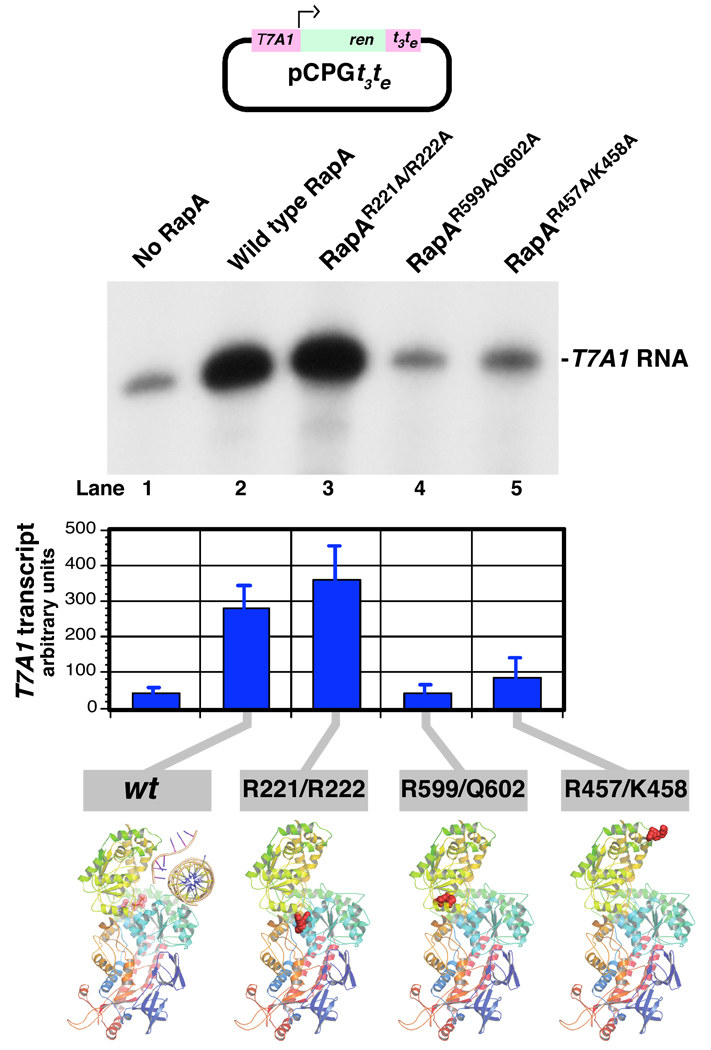
Next, we compared the transcription-stimulatory activities of the constructed RapA mutants with that of wild-type RapA. In this set of experiments we used an extensively characterized supercoiled DNA template (pCPGt3te) containing the T7A1 promoter (26; also see refs. 15–17). In the case of RapAR221A/R222A and RapAR599A/Q602A, the effects of the mutations on the transcription-stimulatory activity correlated with those on ATP-hydrolytic activity (i.e., there was little or no transcription-stimulatory effect in the presence of excess RapAR599A/Q602A and there was a transcription-stimulatory effect comparable to or exceeding that of wild-type RapA in the case of RapAR221A/R222A)(Figure 3). RapAR457A/K458A was largely ineffective in promoting transcriptional cycling under these conditions (Figure 3, compare lanes 2 and 5), even though its ATP-hydrolytic activity was comparable to that of wild-type RapA (Figures 2C and 2D).
Figure 3. Effect of the mutations in RapA’s SWI/SNF domain and its interface with the putative double-stranded nucleic acid-binding domain on RapA’s transcription-stimulatory activity.
In vitro transcription reactions with the supercoiled DNA template pCPGt3te (26) containing the T7A1 promoter were carried out in Buffer C for 60 min in the absence (lane 1) or in the presence of 0.36 µM recombinant wild-type (lane 2) or mutant RapA proteins (lanes 3–5) as described in Materials and Methods. Supercoiled DNA template, 0.032 µg/µl; purified RNA polymerase holoenzyme, 0.023 µg/µl. Following the addition of Stop solution, the reactions were denatured by boiling and the 32P-labeled RNA transcripts were fractionated on 8% SequaGel (National Diagnostics). Quantitated results of the experiment are shown at the bottom; data represent the average of two independent sets of experiments.
Selection and characterization of a model system for in vitro transcription of a complete E. coli gene
To date, the effects of RapA on the synthesis of relatively long and less structured (i.e. more physiologically relevant) RNAs have not been thoroughly studied. This apparent shortcoming of our previously used model systems prompted us, in the present work, to focus on the effects of RapA on the transcription of a complete E. coli model gene possessing intact 5’ and 3’ nontranslated sequences. Although a number of model systems for in vitro transcription of E. coli genes have been described, they did not fully meet our selection criteria, which included requirements for (a) a robust promoter resulting in a relatively high copy number of the translated protein product, (b) a transcribed gene constituting an independent transcription-translation unit (to avoid polar effects in genetic studies), (c) the exhibition of a relatively benign enzymatic activity by the translated protein, and (d) a size of ≤1kb (to facilitate the analysis of DNA-bound transcription complexes on polyacrylamide gels). After some preliminary work, we selected the E. coli slyD gene, which encodes a 21.2-kDa protein bearing the same name (referring to the observation that mutations in the slyD gene result in sensitivity to lysis by bacteriophage [30]). SlyD is an abundant protein chaperone with demonstrated peptidyl-prolyl cis-trans isomerase (rotamase) activity (31–33) and is a frequent contaminant during the metal affinity resin purification of His-tagged proteins (34–36). Indeed, we identified SlyD by mass spectrometry during the purification of recombinant RapA (Supplementary Data, Figure S1C, bottom panel). During the course of our preliminary work, we (a) amplified and purified DNA containing the slyD gene and its promoter and confirmed its ability to support a promoter-specific RNA polymerase transcriptional activity (see below, Figure 5 and Figure 6), and (b) mapped the slyD promoter(s) and determined the exact transcription sites for the RNA polymerase holoenzyme by the primer extension method (Figure 4). Primer extension indicated two distinct transcription start sites – both, conventionally, A’s (Figure 4A; the nontranslated 5’-termini of the slyD mRNAs are shown in Figure 4D). The tandem slyD promoters (denoted P1 and P2) share homology with known E. coli promoters (Figure 4B). Primer extension experiments indicated little or no non-promoter-specific initiation during in vitro transcription (Supplementary Data, Figure S5). We also (c) determined the approximate position of the slyD gene’s transcription terminator (loosely mapped to the tandem hairpins located immediately downstream from the slyD gene) (Supplementary Data, Figure S6); the efficiency of termination in vitro at this intrinsic slyD terminator was consistently high (>80%; Supplementary Data, Figure S6). Taken together, these results are in accord with the existing E. coli databases, which indicate that the slyD gene is transcribed and translated independently. In summary, the PCR-generated DNA templates encompassing the slyD operon met most of our criteria, so we proceeded to the experiments addressing the effects of RapA.
Figure 6. RapA contributes to the formation of free (non-DNA-bound or aggregated) RNA during in vitro transcription. II: Fractionation of transcription complexes under non-denaturing conditions. A.
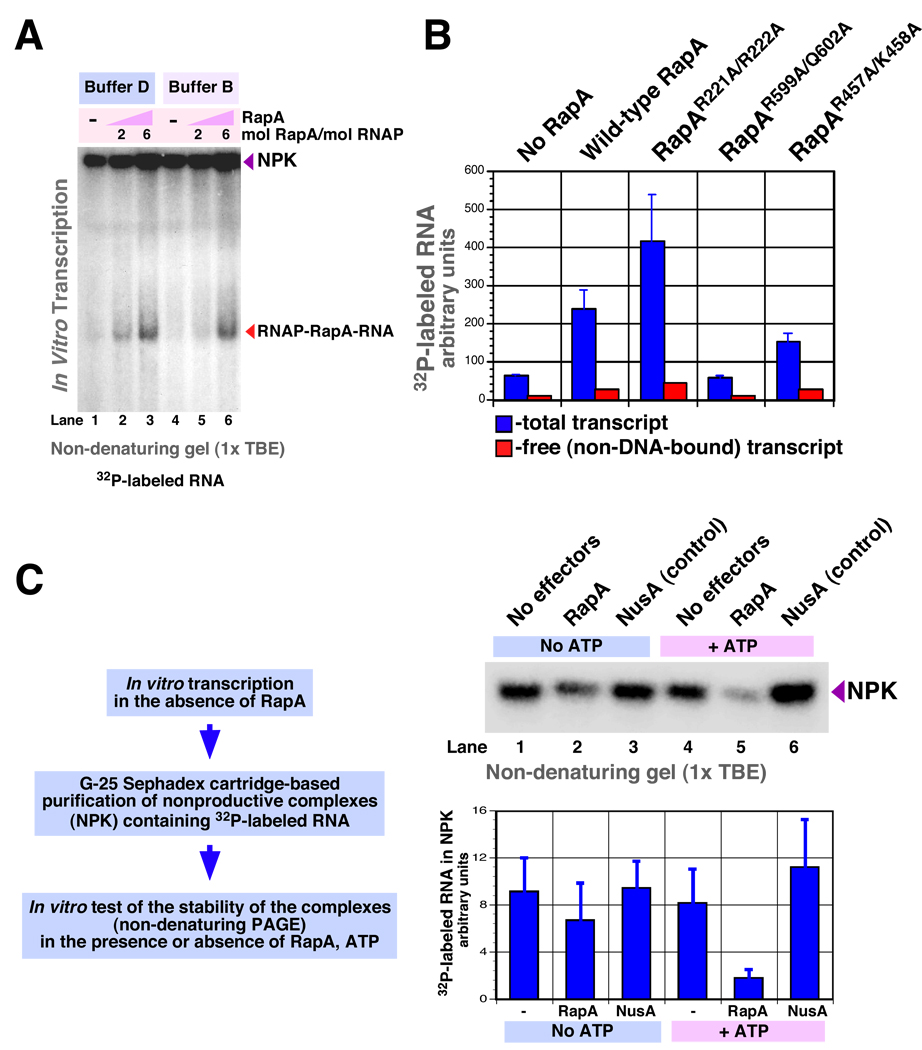
RapA promotes the formation of non-DNA-bound or aggregated RNA. In vitro transcription reactions were carried out for 30 min at 37°C. Reactions 1–3: Buffer D; reactions 4–6: Buffer B (see Materials and Methods). Other components: purified 965-nt linear DNA template containing the slyD operon (PCR MS696/697), 25 nM; RNA polymerase holoenzyme, 50 nM. The RapA/RNA polymerase holoenzyme molar ratios are indicated in the figure. Following the addition of Stop solution, the reaction products were fractionated on 5% ProtoGel (National Diagnostics) containing 1x TBE (KD Medical), using 0.5x TBE as a running buffer; denaturation of the reactions by boiling was omitted. B. Effect of the constructed mutations on RapA’s nucleic acid-remodeling activity. In vitro transcription reactions similar to those described in Figure 6A were carried out in the presence or absence of 8 mol of wild type or mutant RapA per mol RNA polymerase holoenzyme (50 nM). Reaction products were fractionated by non-denaturing PAGE on 8% ProtoGel, as described in Figure 6A. Graph shows the quantitated levels of total 32P-labeled RNA (blue columns) and non-DNA bound or aggregated RNA (red columns). Data represent the average of two independent sets of experiments. C. Effect of RapA on stability of purified nonproductive transcription complexes in the presence or absence of ATP. An in vitro transcription reaction similar to that described in Figure 6A, lane 1, was carried out (in the absence of RapA). Following a 30-min incubation, the reaction was diluted to 40 µl with ultrapure water (KD medical) and applied onto a MicroSpin™ G-25 column (Amersham Pharmacia Biotech) pre-equilibrated with ultrapure water. Following centrifugation for 2 min at 700 × g, approximately 40 µl of flow-through was recovered (typically containing 3000–8000 cpm 32P). To test the stability of the purified complexes, 10-µl reactions containing 1x Buffer C, 500–800 cpm transcription complexes purified as described above, 1 mM ATP (if present), and 0.2 µM purified RapA (if present) or NusA (if present, as a control) were incubated for 30 min at 37°C. Next, 4 µl of Stop solution was added to each reaction, and the amount of 32P-labeled RNA retained in the complexes was determined after the samples were fractionated by PAGE on 5% ProtoGel; electrophoresis was carried out as described in Figure 6A. The quantitated data (bottom panel) represent the average of two independent experiments.
Effect of RapA on transcription of the slyD gene in vitro and identification of the reaction intermediates
Semi-denaturing fractionation of in vitro transcription intermediates
In our previous work, semi-denaturing fractionation of in vitro transcription complexes was instrumental for the detection of RapA-transcript and RNA polymerase-RapA-transcript intermediates (17). We now used a similar approach to test what effect (if any) the addition of purified RapA would have on synthesis of the slyD mRNA. In the absence of RapA, RNA polymerase produced little or no detectable non-protein-bound or aggregated transcripts in the samples not denatured by boiling (Figure 5A, lanes 1 and 3). In contrast, reaction mixtures containing RapA consistently yielded non-DNA-bound or aggregated RNA (Figure 5). Next, we tested whether S1 – the RNA-binding ribosomal protein capable of stimulating transcriptional cycling (likely, via cooperative interaction with nascent RNA in a co-transcriptional manner [19]) – could mimic the observed activity of RapA. These experiments showed that excess S1 failed to promote an RNA release similar to that seen with RapA (resulting in the transcript’s appearance as non-DNA-bound or aggregated species in gel-fractionated reactions)(Figure 5A, compare lanes 2 and 3). On low-percent (<5%) polyacrylamide gels we were able to discriminate clearly between DNA-bound and non-DNA-bound transcripts through selective 32P RNA or DNA labeling (Figure 5B). The kinetics experiments demonstrated the RNA displacement at early (5-min) time-points (Figure 5C, lanes 7 and 8). An increase in the RNA polymerase/DNA template molar ratio, while boosting the overall yield of the transcript, promoted the formation of large complexes incorporating nascent RNA. Nevertheless, the activity of RapA was also apparent at late time points under these conditions (Figure 5C, lanes 3–6). Titration of the in vitro transcription reactions with purified RapA and identification of the RapA-specific reaction intermediates is shown in Figure 5D.
Non-denaturing fractionation of in vitro transcription intermediates
To rule out the possibility that the presence of urea accounted for the observed RNA displacement, we carried out PAGE-based fractionations of the in vitro transcription mixtures under non-denaturing conditions (in the absence of urea)(Figure 6A). The ability of RapA to displace 32P-labeled RNA from transcription complexes or aggregates was apparent on non-denaturing gels (Figure 6A); however, a loss of definition in the separation of individual complexes was apparent under non-denaturing conditions.
Next, we compared wild type RapA with mutant RapA proteins side-by-side in in vitro transcription reactions with DNA templates containing the slyD operon (Figure 6B). In general, the effects of wild-type and mutant RapA proteins on the levels of free (non-DNA-bound or aggregated) slyD mRNA observed under these conditions were comparable to those seen in in vitro transcription studies with supercoiled DNA template containing the T7A1 promoter (in which the reaction products were fractionated in the presence of urea [Figure 3]).
We carried out non-denaturing (G-25 Sephadex cartridge-based) purifications of non-productive transcription complexes containing 32P-labeled RNA transcript (Figure 6C, right panels) and tested their stability in the presence or absence of RapA and ATP. These experiments indicated an ATP-dependent reduction in the levels of DNA-bound or aggregated RNA in the presence of RapA (Figure 6C, graph).
Discussion
RapA, a prokaryotic representative of the SWI/SNF superfamily, was identifed through biochemical studies as a subunit of the E. coli RNA polymerase complex (6, 14). In vitro, excess RapA promotes multi-round transcription (15, 16). To investigate RapA’s nucleic acid substrate specificity and the mechanism of its transcription-stimulatory activity, our group analyzed nucleic acid-binding and remodeling activities of RNA polymerase-RapA complexes (17). In this work we sought (i) further advances in genetic analyses of RapA (specifically, to extend genetic approaches to understanding the explicit functions of individual RapA domains), (ii) further development of biochemical methods for trapping functionally significant transcriptional intermediates, and (iii) to introduce a model system for in vitro transcription of a complete E. coli gene for future biochemical and genetic studies.
Functional significance of RapA
To obtain additional insights into rapA function in vivo, we have further studied the E. coli (MG1655) rapA deletion strain. Importantly, we demonstrate that the pronounced slow-growth phenotype resulting from the rapA deletion mutation are seen only in plated (but not in liquid) cultures. We speculate that this may be due to the E. coli cell’s increased ability to utilize its ionic pumps in liquid media in order to lower the intracellular salt concentration (thus reducing the potentially negative impact of salt-induced interactions between nucleic acids). To date, this is the first study demonstrating that the distinct slow-growth phenotype associated with the rapA deletion mutation can be observed at physiological osmolarity (Figure 1B). Based on this evidence, we argue that the perception of rapA as a gene that contributes only marginally to E. coli physiology may not be fully correct.
RapA mutagenesis
RapA mutations described in this work targeted the interface of RapA’s putative double-stranded nucleic template (dsT)-binding and SWI/SNF domains and the cluster of positively charged amino acids within RapA’s SWI/SNF domain (R457/K458) – a tentative single-stranded nucleic acid-binding site (and a possible binding site for an RNA strand invading duplex DNA in our model for RapA function [17]) (Figure 2B; ssT). Mutations at the interface of RapA’s dsT-binding and SWI/SNF domains altered RapA’s ATPase activity (Figures 2C and 2D). These results are consistent with the predicted position of RapA’s ATP-binding site at the interface of the two aforementioned domains (18). The constructed mutations also altered the transcription-stimulatory activity of RapA and its ability to displace RNA from transcription complexes. In general, the effects of individual mutations on ATP hydrolysis correlated with RapA’s transcription-stimulatory activity, indicating that the interaction between the SWI/SNF and dsT-binding domains may be important for RapA-mediated remodeling of nonproductive transcription complexes. The significant negative effect of the R599A/Q602A double mutation (in the recently reported RapA structure [18], these two amino acids are in close proximity) on ATP hydrolysis and the concomitant loss of the protein’s transcription-stimulatory activity may be due to a general relaxation of the structure of the SWI/SNF domain (as a result of the loss of an internal stabilizing link) and its expansion into the ATP-binding cleft. Remarkably, the interaction with RNA polymerase enabled partial restoration of RapAR599A/Q602A’s ATP-hydrolytic function; this result may point to the possibility of re-orientation of RapA’s SWI/SNF domain with respect to the (putative) double-stranded nucleic acid-binding domain, as proposed for the SsoRad54 protein (27). The effect of the R221A/R222A mutation – which generated a protein with transcription-stimulatory activity exceeding that of wild-type RapA (albeit, insensitive to RNA polymerase regulation) – also seems to support the notion that the stimulation of RapA’s ATP-hydrolytic activity by RNA polymerase may involve re-orientation of RapA’s SWI/SNF domain with respect to the double-stranded nucleic acid-binding domain.
Effects of RapA on in vitro transcription of the slyD mRNA
In this work, we developed an in vitro transcription system of a complete model E. coli gene (slyD). We monitored the in vitro synthesis of slyD mRNA and fractionated in vitro transcription reaction intermediates (identified through a combination of selective 32P-labeling and immunoassays) by PAGE. Our data unambiguously demonstrate that RapA promotes the formation of non-DNA-bound or aggregated RNA species during in vitro transcription (Figure 5 and Figure 6). This result is in accord with the results of our previous study utilizing supercoiled DNA templates and an all-native-enzymes system (17). Under most conditions, RapA promoted the formation of (non-DNA-bound) RNA polymerase-transcript complexes. Previously reported RapA-transcript complexes were also detected during the course of this study (e. g. see Figures 5C and 5D). It seems unlikely that the formation of non-DNA-bound or aggregated RNA species in the presence of RapA was due to a general increase in the yield of RNA. (i) Generally, in this set of experiments (Figure 5 and Figure 6) we avoided extended incubations of the in vitro transcription mixtures in order to minimize potential contributions of RapA to transcriptional cycling. (ii) RapA promoted the formation of non-DNA-bound or aggregated transcript (effectively redistributing RNA in the system) under conditions in which there was clearly no increase in the transcript yield in the presence of RapA (for example, see Figure 5C, lanes 3–6). (iii) The experiments with the purified non-productive transcription complexes (Figure 6C) also indicate that RapA contributes to transcript release. Under most experimental conditions, little or no free (non-RNA polymerase-bound or aggregated) slyD mRNA was generated during in vitro transcription in the absence of RapA, perhaps pointing to mechanistic limitations in the in vitro synthesis of longer, less structured RNAs.
The function(s) of RapA
The homology of RapA to SWI/SNF proteins and more distant DEAD/H family helicases (37, 38) points to roles in nucleic acid remodeling. RapA’s status as an accessory RNA polymerase subunit (6, 14, 15, 17) indicates that this nucleic acid-remodeling activity is likely linked to transcription. In accord with this, RapA promotes the disruption of noncanonical DNA-RNA complexes (putative DNA-RNA triplexes) in an ATP-dependent manner (17). RNA is a likely substrate of RapA, based on multiple, independent lines of evidence summarized in our recent work (17). In that study, we proposed that RapA could track DNA along with the RNA polymerase core enzyme in order to disrupt non-canonical DNA-RNA complexes (17). The salt-selectivity of RapA’s in vivo and in vitro activities supports this model for RapA function.
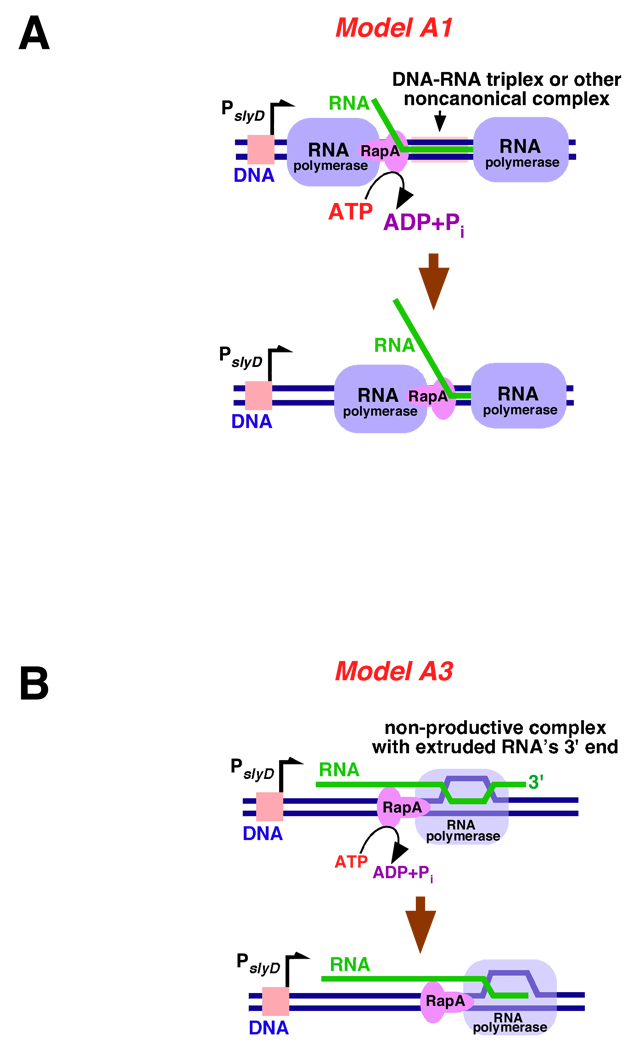
The results presented herein indicate that RapA contributes to the formation of free (non-DNA-bound or aggregated) RNA species during transcription. RNA polymerase without RapA failed to efficiently generate slyD mRNA from linear DNA templates containing the E. coli slyD operon. Our recent studies suggested that nonproductive interactions by nascent RNA could represent a key obstacle to continuous transcriptional cycling (19; also see 17). In the latter work we proposed that noncanonical complexes between dsDNA and transcript RNA (such as dsDNA-RNA triplexes) could form during transcription and impede the activity of ‘trailing’ RNA polymerase molecules. The formation of putative dsDNA-ssRNA triplexes is supported by a classic work by Roberts and Crothers (39), as well as several recent studies (17, 40, 41). RapA-mediated displacement of relatively long transcripts from DNA (as observed in this study under non-denaturing conditions [Figure 6]) argues that RapA may possess an independent nucleic acid-remodeling activity. The possibility that relatively long transcripts could be wrapped around DNA (in a less orderly fashion than that found in a conventional Hoogsteen triplex) as a result of the polymerase’s tracking of a helical, linear DNA molecule has been previously considered (42). RapA could disrupt these noncanonical DNA-RNA complexes by either riding with the polymerase while trawling the RNA (Figure 7A, Model A1 [the ‘minesweeper’ model]) or acting as a dsDNA-ssRNA topoisomerase (Model A2; schematic is not shown). Well-documented, nonproductive ternary complexes in which nascent RNA’s 3’-end is extruded (43–45) also could represent potential substrates for RapA. RapA could contribute to RNA release by either extracting the misaligned RNA or transporting the polymerase along DNA (thus acting as a translocase), both scenarios resulting in re-alignment of the core enzyme’s active site with the 3’-end of the RNA (Figure 7B, Model A3) and subsequent RNA release. Finally, the possibility of RapA disrupting complexes formed between transcripts synthesized on different DNA molecules (‘in trans’)(Models B; schematic is not shown) also cannot be entirely ruled out.
Figure 7. Models explaining possible mechanisms of the RapA-mediated redistribution of RNA during in vitro transcription.
See text for a detailed discussion of the proposed models. A. Model A1: RapA lifts RNA transcript from DNA’s major groove and/or unwraps RNA from DNA in a processive manner. B. Model A3: RapA dissociates nonproductive ternary (DNA-RNA polymerase-RNA) complexes with the extruded 3’-end of RNA by either extracting the misaligned RNA or transporting the polymerase along DNA; both activities could contribute to redistribution of RNA in the system and re-initiation of transcription.
Both the current study and our preceding work point to RNA as a substrate of RapA. It is therefore interesting to question whether the RNA-directed activities of RapA may overlap to some extent with those of Rho – an extensively studied RNA-binding protein with established roles in transcription termination (for review and a complete set of references see [46]) – particularly, in light of Rho’s known ATP-dependent helicase activity with respect to DNA-RNA hybrids and its demonstrated ability to enhance RNA release from transcription elongation complexes. Testing RapA and Rho side-by-side may shed new light on the functions of these two proteins and is likely to be a part of our future studies of E. coli transcription.
In summary, in this work we (a) reported in vivo data demonstrating functional significance of the rapA gene under physiological osmolarity, (b) introduced and characterized a model system for the in vitro synthesis of E. coli slyD mRNA and further developed techniques for non-denaturing fractionation of transcription intermediates, and (c) described several new mutations in RapA’s SWI/SNF domain and its interface with the (putative) double-stranded nucleic acid template-binding domain. The RapA R221A/R222A and R599A/Q602A mutations described in this study (both of which resulted in alterations in RapA’s ATPase activity with no apparent effect on protein folding or solubility) represent a valuable asset for future studies. Both mutant proteins, along with the ribosomal protein S1, could serve as controls for ATP-dependent and ATP-independent RapA activities. Finally, (d) in this work we further refined our models for RapA function in vivo. Our future studies will address the mechanistic aspects of RapA’s enzymatic activity in terms of its compatibility with one or more of the models proposed herein. We will also carry out genetic studies designed to identify potential rapA interactors in E. coli.
Supplementary Material
Acknowledgements
We thank Timothy Gallaher at the University of Southern California School of Pharmacy for the mass spectrometry analyses of RapA and SlyD, Karen Sukhodolets, and anonymous reviewers for helpful comments and suggestions. This work was supported in part by Grant Number R15GM081803 from the National Institute of General Medical Sciences (to M.V.S.) (the content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health), a Welch Foundation grant (V-0004), and departmental funds.
Funding: this work was supported in part by Grant Number R15GM081803 from the National Institute of General Medical Sciences (to M.V.S.), a Welch Foundation grant (V-0004), and departmental funds
Abbreviations
- ATP
adenosine-5’-triphosphate
- CTP
cytidine-5’-triphosphate
- ds
double-stranded
- FPLC
fast protein liquid chromatography
- GTP
guanosine-5’-triphosphate
- RNAP
RNA polymerase
- PAGE
polyacrylamide gel electrophoresis
- PEI
polyethylenimine
- PBS
phosphate buffered saline
- PBST
PBS-Tween
- RNAP
RNA polymerase
- ss
single-stranded
- TBE
Tris-borate-EDTA
- TLC
thin layer chromatography
- UTP
uridine-5’-triphosphate
References
- 1.Burgess RR, Erickson B, Gentry D, Gribskov MM, Hager D, Lesley S, Strickland M, Thompson N. RNA polymerase and the Regulation of Transcription. New York: Elsevier Science Publishing Co., Inc.; 1987. pp. 3–15. [Google Scholar]
- 2.Gross CA, Lonetto M, Losick R. In: Transcriptional Regulation. Yamamoto K, McKnight C, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 129–176. [Google Scholar]
- 3.Maeda H, Fujita N, Ishihama A. Competition among seven Escherichia coli σ subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 2000;28:3497–3503. doi: 10.1093/nar/28.18.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess RR, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 5.Hager DA, Jin DJ, Burgess RR. Use of Mono Q high-resolution ion-exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- 6.Sukhodolets MV, Jin D. RapA, a novel RNA polymerase-associated protein, is a bacterial homolog of SWI2/SNF2. J. Biol. Chem. 1998;273:7018–7023. doi: 10.1074/jbc.273.12.7018. [DOI] [PubMed] [Google Scholar]
- 7.Greenblatt J, Li J. Interaction of the sigma factor and the nusA gene protein of E. coli with RNA polymerase in the initiation-termination cycle of transcription. Cell. 1981;24:421–428. doi: 10.1016/0092-8674(81)90332-9. [DOI] [PubMed] [Google Scholar]
- 8.Friedman DI, Gottesman M. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1983. pp. 21–51. [Google Scholar]
- 9.Muzzin O, Campbell EA, Xia L, Severinova E, Darst SA, Severinov K. Disruption of Escherichia coli hepA, an RNA polymerase-associated protein, causes UV-sensitivity. J. Biol. Chem. 1998;273:15157–15161. doi: 10.1074/jbc.273.24.15157. [DOI] [PubMed] [Google Scholar]
- 10.Horwitz RJ, Li J, Greenblatt J. An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell. 1987;51:631–641. doi: 10.1016/0092-8674(87)90132-2. [DOI] [PubMed] [Google Scholar]
- 11.Squires CL, Greenblatt J, Li J, Condon C, Squires CL. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl. Acad. Sci. USA. 1993;90:970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das A, Pal M, Mena JG, Whalen W, Wolska K, Crossley R, Rees W, von Hippel PH, Costantino N, Court D, et al. Components of multiprotein-RNA complex that controls transcription elongation in Escherichia coli phage lambda. Methods in Enzymology. 1996;274:374–402. doi: 10.1016/s0076-6879(96)74032-6. [DOI] [PubMed] [Google Scholar]
- 13.Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 2001;107:437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 14.Sukhodolets M, Jin D. Interaction between RNA polymerase and RapA, a bacterial homolog of SWI2/SNF2. J. Biol. Chem. 2000;275:22090–22097. doi: 10.1074/jbc.M000056200. [DOI] [PubMed] [Google Scholar]
- 15.Sukhodolets MV, Cabrera JE, Zhi H, Jin DJ. RapA, a bacterial homolog of SWI2/SNF2, stimulates RNA polymerase recycling in transcription. Genes Dev. 2001;15:3300–3341. doi: 10.1101/gad.936701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sukhodolets MV, Garges S, Jin DJ. Purification and activity assays of RapA, the RNA polymerase-associated homolog of the SWI/SNF superfamily. Methods Enzymol. 2003;370:283–290. doi: 10.1016/S0076-6879(03)70025-1. [DOI] [PubMed] [Google Scholar]
- 17.McKinley BA, Sukhodolets MV. Escherichia coli RNA polymerase-associated SWI/SNF protein RapA: evidence for RNA-directed binding and remodeling activity. Nucleic Acids Res. 2007;35:7044–7060. doi: 10.1093/nar/gkm747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw G, Gan, Jianhua, Zhou Y, Zhi H, Subburaman P, Zhang R, Joachimiak A, Jin D, Ji X. Structure of RapA, a SWI2/SNF2 protein that recycles RNA polymerase during transcription. Structure. 2008;16:1417–1427. doi: 10.1016/j.str.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sukhodolets MV, Garges S, Adhya S. Ribosomal protein S1 promotes transcriptional cycling. RNA. 2006;12:1505–1513. doi: 10.1261/rna.2321606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive pediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 21.Sevenet N, Lellouch-Tubiana A, Schofield D, Hoang-Xuan K, Gessler M, Birnbaum D, Jeanpierre C, Jouvet A, Delattre O. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum. Mol. Genet. 1999;8:2359–2368. doi: 10.1093/hmg/8.13.2359. [DOI] [PubMed] [Google Scholar]
- 22.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, Smith TW, Imbalzano AN, Jones SN. Disruption of INI1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts CMW, Leroux MM, Fleming MD, Orkin S. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene. Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- 25.Sukhodolets MV, Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- 26.Reynolds R, Bermudez-Cruz RM, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J. Mol. Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- 27.Lewis R, Durr H, Hopfner KP, Michaelis J. Conformational changes of a Swi2/Snf2 ATPase during its mechano-chemical cycle. Nucleic Acids Res. 2008;36:1881–1890. doi: 10.1093/nar/gkn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopfner KP, Michaelis J. Mechanisms of nucleic acid translocases: lessons from structural biology and single-molecule biophysics. Curr. Opin. Struct. Biol. 2007;17:87–95. doi: 10.1016/j.sbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Haseltine CA, Kowalczykowski SC. An archaeal Rad54 protein remodels DNA and stimulates DNA strand exchange by RadA. Nucleic Acids Res. 2009;37:2757–2770. doi: 10.1093/nar/gkp068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roof WD, Young R. jX174 lysis requires slyD, a host gene, which is related to the FKBP family of peptidyl-prolyl cis-trans isomerases. FEMS Microbiol. Rev. 1995;17:213–218. doi: 10.1111/j.1574-6976.1995.tb00204.x. [DOI] [PubMed] [Google Scholar]
- 31.Hottenrott S, Schumann T, Pluckthun A, Fischer G, Rahfeld JU. Escherichia coli SlyD is a metal ion-regulated peptidyl-prolyl cis/trans-isomerase. J. Biol. Chem. 1997;272:15697–15701. doi: 10.1074/jbc.272.25.15697. [DOI] [PubMed] [Google Scholar]
- 32.Scholz C, Eckert B, Hagn F, Schaarschmidt J, Balbach J, Schmid FX. SlyD proteins from different species exhibit high prolyl isomerase and chaperone activities. Biochemistry. 2006;45:20–33. doi: 10.1021/bi051922n. [DOI] [PubMed] [Google Scholar]
- 33.Leach MR, Zhang JW, Zamble DB. The role of complex formation between the Escherichia coli hydrogenase accessory factors HypB and SlyD. J. Biol. Chem. 2007;282:16177–16186. doi: 10.1074/jbc.M610834200. [DOI] [PubMed] [Google Scholar]
- 34.Mitterauer T, Nanoff C, Ahorn H, Freissmuth M, Hohenegger M. Metal-dependent nucleotide binding to the Escherichia coli rotamase SlyD. Biochem J. 1999;342:33–39. doi: 10.1042/0264-6021:3420033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee S, Shukla A, Guptasarma P. Single-step purification of a protein-folding catalyst, the SlyD peptidyl prolyl isomerase (PPI), from cytoplasmic extracts of Escherichia coli. Biotechnol Appl. Biochem. 2003;37:183–186. doi: 10.1042/ba20020044. [DOI] [PubMed] [Google Scholar]
- 36.Parsy CB, Chapman CJ, Barnes AC, Robertson JF, Murray A. Two-step method to isolate target recombinant protein from co-purified bacterial contaminant SlyD after immobilized metal affinity chromatography. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 2007;853:314–319. doi: 10.1016/j.jchromb.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 37.Bork P, Koonin EV. An expanding family of helicases within the DEAD/H superfamily. Nucleic Acids Res. 1993;21:751–752. doi: 10.1093/nar/21.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolsto AB, Bork P, Kvaloy K, Lindback T, Gronstadt A, Kristensen T, Sander C. Prokaryotic members of a new family of putative helicases with similarity to transcription activator SNF2. J. Mol. Biol. 1995;230:684–688. doi: 10.1006/jmbi.1993.1185. [DOI] [PubMed] [Google Scholar]
- 39.Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 40.Morvan F, Imbach JL, Rayner B. Comparative stability of eight triple helices formed by differently modified DNA or RNA pyrimidine strands and a DNA hairpin. Antisense Nucleic Acid Drug Dev. 1997;7:327–334. doi: 10.1089/oli.1.1997.7.327. [DOI] [PubMed] [Google Scholar]
- 41.Ivanov S, Alekseev Y, Bertrand JR, Malvy C, Gottikh MB. Formation of stable triplexes between purine RNA and pyrimidine oligodeoxyxylonucleotides. Nucleic Acids Res. 2003;31:4256–4263. doi: 10.1093/nar/gkg443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Masse E, Baaklini I. The problem of hypernegative supercoiling and R-loop formation in transcription. Front. Biosci. 2003;8:210–221. doi: 10.2741/970. [DOI] [PubMed] [Google Scholar]
- 43.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3’ end of the RNA intact and extruded. Proc. Natl. Acad. Sci. USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marr M, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Molecular Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 45.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 46.Banerjee S, Chalissery J, Bandey I, Sen R. Rho-dependent transcription termination: more questions than answers. J. Microbiol. 2006;44:11–22. [PMC free article] [PubMed] [Google Scholar]
- 47.Durr H, Korner C, Muller M, Hickman V, Hopfner K-P. X-Ray structure of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.