Abstract
Methods of comprehensive microarray-based aneuploidy screening in single cells are rapidly emerging. Whole-genome amplification (WGA) remains a critical component for these methods to be successful. A number of commercially available WGA kits have been independently utilized in previous single-cell microarray studies. However, direct comparison of their performance on single cells has not been conducted. The present study demonstrates that among previously published methods, a single-cell GenomePlex WGA protocol provides the best combination of speed and accuracy for single nucleotide polymorphism microarray-based copy number (CN) analysis when compared with a REPLI-g- or GenomiPhi-based protocol. Alternatively, for applications that do not have constraints on turnaround time and that are directed at accurate genotyping rather than CN assignments, a REPLI-g-based protocol may provide the best solution.
Keywords: whole-genome amplification, SNP microarray, copy number, single-cell genotyping, aneuploidy screening
Introduction
Many groups have developed whole-genome microarray-based methods to assess chromosome copy number (CN) in order to diagnose aneuploidy in human embryos from a single cell (Handyside et al., 2009; Vanneste et al., 2009; Gutierrez-Mateo et al., 2010; Johnson et al., 2010a; Treff et al., 2010a). These developments are in large part due to the failure of fluorescence in situ hybridization (FISH)-based methods to result in the expected clinical benefit of aneuploidy screening for the treatment of infertility (reviewed in Fritz, 2008). Genome-wide approaches are certainly more comprehensive than FISH (24 compared with ≤12 chromosomes, respectively) and some microarray-based methods have shown significantly improved consistency (Treff et al., 2010a,b) and predictive value for aneuploidy diagnosis (Scott et al., 2008; Northrop et al., 2010).
Some methods of 24 chromosome CN have also demonstrated accuracy of blinded predictions in single cells from a variety of cell lines with previously well-characterized karyotypic abnormalities (i.e. Treff et al., 2010a). Unfortunately, other studies have considered a method to be accurate by only establishing that two different methods of analysis indicate that an embryo is abnormal even if the results of the two tests indicate that the abnormalities involved completely different chromosomes (Gutierrez-Mateo et al., 2010). This may be inadequate to establish the accuracy of a test for single-cell 24 chromosome aneuploidy diagnosis. Even more troubling is the lack of any accurate calculations after analysis of single cells from cell lines with known abnormalities by technologies such as comparative genomic hybridization (CGH) or array-CGH. Some microarray-based studies have performed testing of single cells from cell lines (Vanneste et al., 2009; Johnson et al., 2010a). However, one study suggested that the method was accurate after evaluating only a small number of single cells (n= 7) from cell lines, was unable to obtain an interpretable result in 41% of blastomeres evaluated, and required analysis by two arrays; bacterial artificial chromosome (BAC) and single nucleotide polymorphism (SNP) (Vanneste et al., 2009). This may not represent sufficient validation, reliability or feasibility for routine clinical application. Although the second study involving cell lines (Johnson et al., 2010a) did evaluate a large sample size (n= 459), only a single type of abnormality (trisomy 21) was represented. This may also be inadequate to determine the accuracy of predicting aneuploidy for all 24 chromosomes.
Unfortunately, these and other preclinical validation considerations have gone overlooked during the development and implementation of many new technologies for 24 chromosome aneuploidy screening. Clinical studies have also been limited. For example, case–control and observational studies may not represent sufficient strength of evidence to determine the clinical validity of new technologies. This is particularly true in light of the experiences with FISH-based aneuploidy screening which was suggested to be clinically beneficial based on case–control studies (Gianaroli et al., 1997; Munne et al., 1999; Kahraman et al., 2000; Munne et al., 2003; Munne et al., 2006), despite failing to show a meaningful benefit in all randomized controlled clinical trials (reviewed in Fritz, 2008). Although similar case–control and observational clinical studies using comprehensive methods of aneuploidy screening have been reported (Wells et al., 2009; Munne et al., 2010; Rabinowitz et al., 2010; Schoolcraft et al., 2010a,b), randomized controlled trials have been limited. Indeed, class I strength of evidence for a significant improvement in clinical pregnancy and embryo implantation rates (i.e. Scott et al., 2010) should be made standard for any new aneuploidy screening technology prior to routine implementation. An equally important clinical trial involves a prospective blinded non-selection design (i.e. Scott et al., 2008) in which the negative predictive value of the test is determined. In other words, it is critical to know whether the test produces false-positive abnormal diagnoses in embryos that are otherwise capable of developing into chromosomally normal pregnancies. Such a study is important in confirming whether the test can be used to safely discard human embryos.
Whole-genome amplification
A critical step in every single-cell 24 chromosome aneuploidy screening method is whole-genome amplification (WGA). Single cells possess ∼6–7 pg of genomic DNA (gDNA) (Dolezel et al., 2003) and microarrays typically require nanogram amounts of DNA to proceed as recommended. This necessitates amplification of the genome by more than 1000-fold. Moreover, since these technologies have commonly been applied to quantitatively evaluate chromosomal CN, the WGA procedure must result in unbiased amplification to maintain relative quantities of DNA across the entire genome. Some methods of WGA and microarray-based molecular karyotyping rely upon the interpretation of qualitative genotypes rather than quantitative CN assignments (Handyside et al., 2009; Johnson et al., 2010a). In these situations and in applications where single gene disorders may be evaluated directly (Hellani, 2005; Burlet et al., 2006; Lledo et al., 2006; Panelli et al., 2006; Lledo et al., 2007; Ren et al., 2007; Renwick et al., 2007; Hellani et al., 2008a) or through microarray-based haplotype inheritance analyses (Handyside et al., 2009), genotyping fidelity is also a critical component of WGA.
There are a variety of commercially available reagents to perform single-cell WGA that have aided in widespread utilization (Table I). For example, some groups have used a multiple displacement amplification (MDA) approach using QIAgen's ‘REPLI-g’ technology (Handyside et al., 2004, 2009; Sher et al., 2007, 2009) or GE Healthcare's ‘GenomiPhi’ technology (Le Caignec et al., 2006; Hellani et al., 2008b; Vanneste et al., 2009). MDA involves the use of a bacteriophage (Φ29) DNA polymerase that employs rolling circle amplification during incubation at a single temperature (isothermal) (Dean et al., 2002). Other groups have employed PCR-based amplification strategies using Sigma's ‘GenomePlex’ technology (Fiegler et al., 2007; Gutierrez-Mateo et al., 2010; Treff et al., 2010a). PCR-based WGA involves the use of a DNA polymerase from the thermophilic bacterium Thermus aquaticus and repeated cycling between temperatures appropriate to sequentially denature and elongate the DNA (Saiki et al., 1988). Interestingly, comparison studies of commercially available MDA and PCR-based WGA methods have only evaluated the performance on input DNA quantities that exceed those found in a single cell (Lovmar et al., 2003; Barker et al., 2004; Park et al., 2005). The present study performs the first direct comparison of commercially available single-cell WGA methodologies for amplification reliability, fidelity and accuracy by SNP microarray analysis.
Table I.
Comparison of notable 24 chromosome aneuploidy screening technologies.
| Characteristic | CGH |
SNP array |
Array-CGH |
SNP array + array-CGH | ||||
|---|---|---|---|---|---|---|---|---|
| Wells et al. (1999) | Sher et al. (2007) | Johnson et al. (2010a) | Treff et al. (2010a) | Handyside et al. (2009) | Gutierrez-Mateo et al. (2010) | Hellani et al. (2008b) | Vanneste et al. (2009) | |
| WGA method | PCR (custom DOP-PCR) | MDA (REPLI-g) | MDA (undisclosed) | PCR (GenomePlex) | MDA (REPLI-g) | PCR (GenomePlex/Sureplex) | MDA (GenomiPhi) | MDA (GenomiPhi) |
| Array method | NA | NA | 370K SNP | 250K SNP | 370K SNP | 2K CGH (BAC) | 44K oligonucleotide | 4K CGH (BAC) and 250K SNP |
| 2-day turnaround time | — | — | + | Treff et al. (2009a) | — | + | — | — |
| Cell line studya | — | — | + | + | — | — | — | + |
| Consistency studyb | Wells and Delhanty (2000) | — | + | + | — | — | — | + |
| FISH comparison study | Fragouli et al. (2008) | Keskintepe et al. (2007) | — | Treff et al. (2010b), Northrop et al. (2010) | — | + | + | — |
| Single gene disorder detectionc | — | — | Rabinowitz et al. (2009) | Treff et al. (2009b) | Handyside et al. (2010) | — | — | — |
| Chromosome translocation detection | — | — | Johnson et al. (2010b) | Treff et al. (2010c) | — | Escudero et al. (2010) | — | — |
| Observational or case–control study | Schoolcraft et al. (2010a) | +, Sher et al. (2009) | Rabinowitz et al. (2010) | Schoolcraft et al. (2010b) | — | Munne et al. (2010) | — | — |
| Non-selection study | — | — | — | Scott et al. (2008) | — | — | — | — |
| Randomized controlled study | — | — | — | Scott et al. (2010)d | — | — | — | — |
| Deliveries reported | Wells et al. (2009) | Sher et al. (2009) | — | Treff et al. (2009a) | — | + | — | — |
A ‘+’ symbol refers to the reference cited in the header of each respective column.
aAnalysis of accuracy on single cells with known karyotypes.
bAnalysis of multiple blastomeres from within the same embryos.
cDemonstrated ability to evaluate a monogenic disorder from the same biopsy.
dThis study included demonstrating equivalence of a real-time PCR protocol (Treff et al., 2009c) to the SNP microarray protocol (Treff et al., 2010a) prior to using it in a randomized controlled trial.
Materials and Methods
Experimental design
This study was designed to evaluate three commercially available methods of WGA on single cells. The evaluation was conducted using an SNP microarray platform with gDNA extracted from a large amount of cells serving as a benchmark for genotyping and CN accuracy on single cells from the same cell line.
Single-cell isolation
Four human fibroblast cell lines were obtained from the Coriell Cell Repository (Camden, NJ, USA). The karyotype of each cell line was different in the CN of the X chromosome and included a 46,XY cell line (GM00323) representing a chromosome X CN of 1, a 46,XX cell line (GM00321) representing a chromosome X CN of 2, a 47,XXX cell line (GM04626) representing a chromosome X CN of 3 and a 49,XXXXY cell line (GM00326) representing a chromosome X CN of 4. Cells were cultured in Eagle's minimum essential medium with 15% fetal bovine serum, 2× non-essential amino acid and 1% penicillin–streptomycin–glutamine (Invitrogen Corp., Carlsbad, CA, USA) at 37°C and 5% CO2. Single cells were isolated following treatment with trypsin/EDTA (Invitrogen) to detach the adherent fibroblast cultures as recommended. Single cells were then picked up in 1 μl of media using a 100 μm stripper tip (Midatlantic Diagnostics, Mount Laurel, NJ, USA) under a dissecting microscope and placed in the bottom of a 0.2 ml PCR tube (Ambion Inc., Austin, TX, USA) holding WGA method-specific solutions as described below. Thirty single cells were picked up from each cell line; 10 single cells for each WGA method. One microlitre of media was removed to serve as negative controls for each WGA method. gDNA was also extracted from each cell line immediately after single cells were obtained using the DNeasy Blood and Tissue Kit (Qiagen Inc., Valencia, CA, USA) as described by the manufacturer.
Single-cell WGA
The GenomiPhi DNA amplification kit (GE Healthcare, Piscataway, NJ, USA) was used on single cells according to a previous publication (Le Caignec et al., 2006). One microlitre of culture media containing a single cell was loaded into 0.2 ml PCR tubes containing 2.5 μl alkaline lysis buffer [200 mM KOH and 50 mM DTT (Cui et al., 1989)]. The samples were stored at −80°C for at least 30 min and then incubated at 65°C for 10 min. Two and a half microlitres of neutralization buffer [0.9 M Tris–HCl, pH 8.3, 0.3 M KCl and 0.2 M HCl (Cui et al., 1989)] were then added to the sample to neutralize the lysis buffer. Nine microlitres of GenomiPhi sample buffer containing the random hexamer primers were added to the neutralized cell lysate, followed by 9 μl of GenomiPhi reaction buffer and 1 μl of GenomiPhi enzyme mix. The isothermal amplification was performed at 30°C for 3 h and the reaction was stopped upon incubation at 65°C for 10 min.
The REPLI-g Midi Kit (Qiagen) was used on single cells according to a previous publication (Handyside et al., 2004). Single cells in 1 μl of culture media were loaded into 0.2 ml PCR tubes containing 2.5 μl PBS buffer. Three and a half microlitres of buffer D2 were added followed by a 10 min incubation on ice and a 5 min incubation at 65°C. Three and a half microlitres of stop solution were added to stop the lysis reaction. A WGA master mix containing 10 μl nuclease free water, 29 μl reaction buffer and 1 μl DNA polymerase was added to the cell lysate followed by the isothermal amplification at 30°C for 16 h and inactivation at 65°C for 3 min.
The GenomPlex Single Cell Whole Genome Amplification Kit (WGA4; Sigma Aldrich, St Louis, MO, USA) was used on single cells as described in a previous publication (Fiegler et al., 2007). Single cells in 1 μl of culture media were loaded into 0.2 ml PCR tubes containing 7 μl of nuclease free water. One microlitre of alkaline lysis buffer was added followed by incubation at 65°C for 10 min to lyse the cell. One microlitre of neutralization buffer was added to neutralize the lysis buffer. WGA was performed following the manufacturer's instructions (Sigma Aldrich).
WGA DNA from each of the three methods described above was purified using the GenElute PCR Cleanup Kit (Sigma Aldrich) as described in the manufacturer's instructions.
Single-cell WGA reliability
The concentration of purified WGA DNA and gDNA was measured using a NanoDrop 8000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and DNA yield was calculated. One hundred nanograms of WGA DNA and gDNA were loaded to 2% E-Gel electrophoresis system (Invitrogen) and visualized with a Kodak Gel Logic 100 system (Kodak, Rochester, NY, USA). Successful WGA was defined as a single-cell sample that yielded more than the required input WGA DNA amount for SNP microarray-based analysis (250 ng). For each method, reliability was defined as the percentage of samples that met this definition.
Single-cell WGA genotyping fidelity
Three representative WGA DNA samples from each WGA method and each cell line were evaluated by SNP microarray analysis. Two hundred and fifty nanograms of WGA DNA or gDNA were processed with the GeneChip 250K NspI SNP microarray as instructed by the supplier (Affymetrix, Santa Clara, CA, USA). Genotypes of each SNP were obtained using the Dynamic Model Mapping Algorithm of the GeneChip Genotyping Analysis Software (GTYPE) 4.1 (Affymetrix). Genotyping coverage was defined as the percentage of SNPs which were successfully assigned a genotype. As such, the SNPs given a ‘no call’ assignment would contribute to reduced genotyping coverage. Genotyping accuracy was defined as the percentage of SNPs assigned a genotype that was equivalent to the genotype assigned to purified gDNA from the same cell line. Allele dropout (ADO) was defined as the number of SNPs that were assigned a homozygous genotype, despite being assigned a heterozygous genotype in the purified gDNA profiles from the same cell line.
Single-cell WGA CN accuracy
The same data used to evaluate genotyping accuracy above were also evaluated for CN accuracy by using the Copy Number Analysis Tool (CNAT) 4.0.1 (Affymetrix). The CN assignments of each sample were compared with those of the purified gDNA from the same cell line and to the known karyotype of each cell line as reported by the Coriell cell repository. Results were evaluated for accuracy at three levels of analysis; each individual SNP, each individual chromosome and each individual cell's 23 chromosome molecular karyotype. The overall CN assignment for a single chromosome was determined based on the SNP CN that represented the majority of the assignments within that chromosome (Treff et al., 2010a). Diagnostic accuracy was defined as the percentage of single cells given the correct whole chromosome specific gain, loss or euploid assignments.
Statistics and data repository
A Student's t-test was used to evaluate significance; α was set at 0.05. Variation was reported as ± 1 standard error of the mean (SEM). The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE24690 (http://www.ncbi.nlm.nih.gov/geo).
Results and Discussion
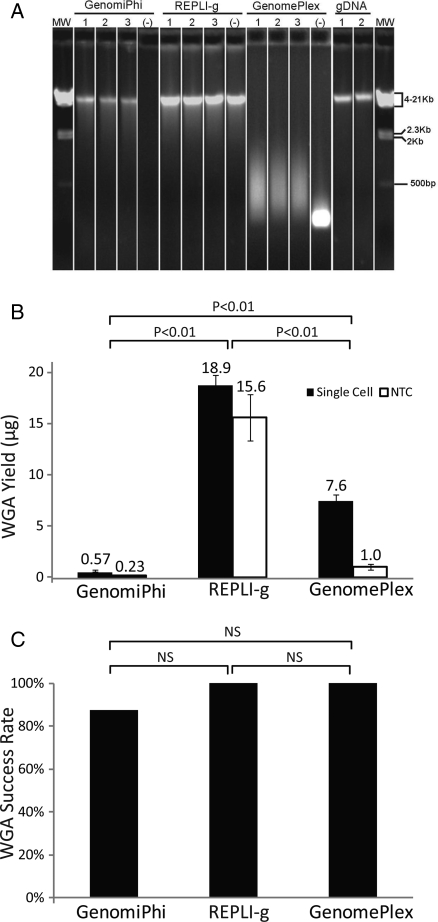
Both GenomiPhi and REPLI-g methods produced WGA DNA that was equivalent in molecular weight to that of the gDNA (Fig. 1A). However, a similar-sized DNA smear was detected from the no template controls amplified with REPLI-g. As a result, gel electrophoresis of REPLI-g WGA DNA alone was insufficient to determine whether amplification was successful. GenomPlex WGA DNA product size ranged from 100 to 1000 bp (Fig. 1A). The average WGA DNA yield from the GenomiPhi protocol was 0.57 ± 0.1 μg and significantly less (P< 0.01) than the 7.63 ± 0.4 μg from GenomePlex or the 18.93 ± 0.8 μg from REPLI-g (Fig. 1B). Similar quantities of DNA were detected from the no template controls amplified using the REPLI-g protocol (15.62 ± 2.3 μg). As a result, DNA quantification by UV spectroscopy of REPLI-g WGA DNA was also insufficient to determine whether specific amplification was successful. This is consistent with previous studies which have found non-specific primer-directed DNA amplification with no template control MDA reactions (Lage et al., 2003; Brukner et al., 2005). Eighty-eight per cent (35/40) of the single cells successfully amplified with the GenomiPhi method by yielding >250 ng of WGA DNA. REPLI-g and GenomePlex methods yielded >250 ng WGA DNA from 100% of the single cells (Fig. 1C). No significant difference in reliability of obtaining sufficient quantities of DNA for microarray analysis was observed between the three methods.
Figure 1.
Reliability of single-cell WGA. (A) Gel electrophoresis of purified reaction products of three representative samples and one no template control (-) from each of three single-cell WGA methods (GenomiPhi, REPLI-g and GenomePlex). Representative purified gDNA and molecular weight markers (MW) are included for size references. (B) The mean yield of amplification (±SEM) of 40 single cells (black bars) or 4 no template controls (NTC; white bars) from each of three single-cell WGA methods. (C) The rates of successful amplification of 40 single cells from each of three single-cell WGA methods.
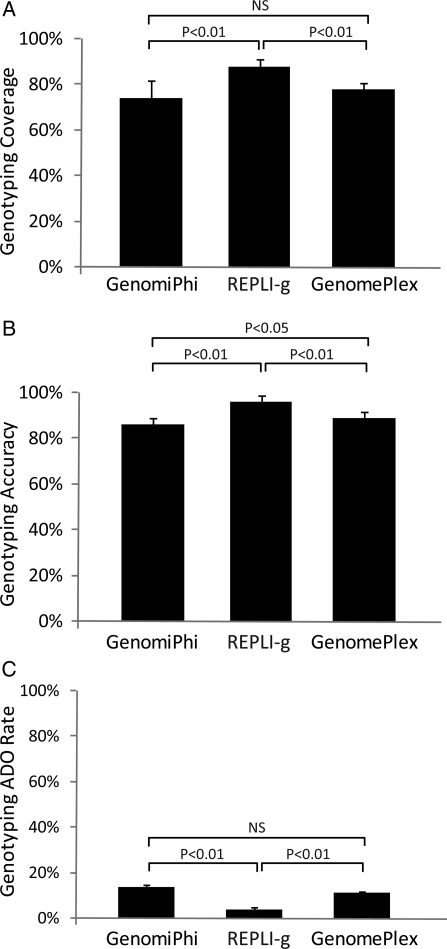
Single-cell WGA DNA provided an average of 74% genotyping coverage with the GenomiPhi protocol and 78% with GenomePlex, which were both significantly lower than the 88% obtained with REPLI-g (Fig. 2A). Single-cell WGA DNA genotypes provided an average of 86% accuracy with the GenomiPhi protocol, which was significantly less than the 89% accuracy obtained with GenomePlex (Fig. 2B). Both the GenomiPhi and GenomePlex protocols’ genotyping accuracy was significantly lower than the 96% obtained with REPLI-g (Fig. 2B). There was an average ADO rate of 14% using GenomiPhi and 11% using GenomePlex, both of which were significantly higher than the 4% obtained using REPLI-g (Fig. 2C). These results are applicable to performance of methods that require accurate genotyping and qualitative analysis of aneuploidy, such as those described by Johnson et al. (2010a) and Handyside et al. (2010), or in situations where one might consider using WGA DNA to genotype-specific genes of interest (i.e. for single gene disorder screening).
Figure 2.
Genotype fidelity of single-cell WGA. (A) The percentage of SNPs evaluated that were successfully assigned a genotype (genotyping coverage) for each of three single-cell WGA methods. (B) The percentage of SNPs assigned a genotype identical to the purified gDNA assignments (genotyping accuracy) for each of three single-cell WGA methods. (C) The percentage of SNPs assigned a homozygous genotype in the single cells but also assigned a heterozygous genotype in the purified gDNA samples (genotyping ADO rate) for each of three single-cell WGA methods.
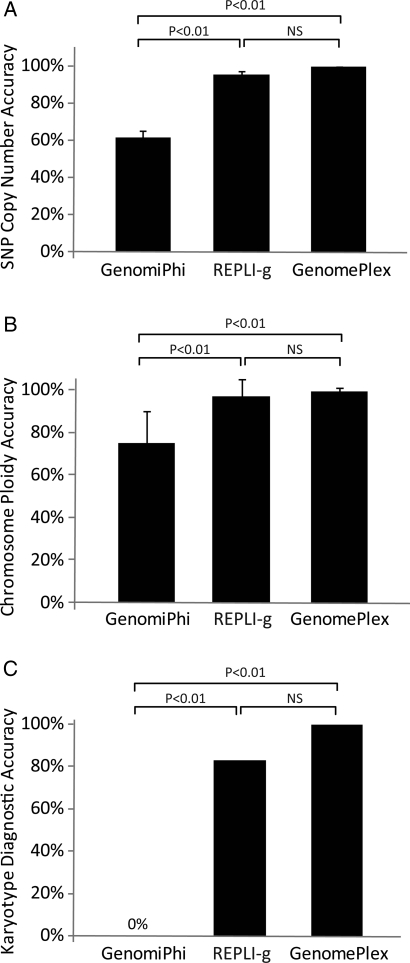
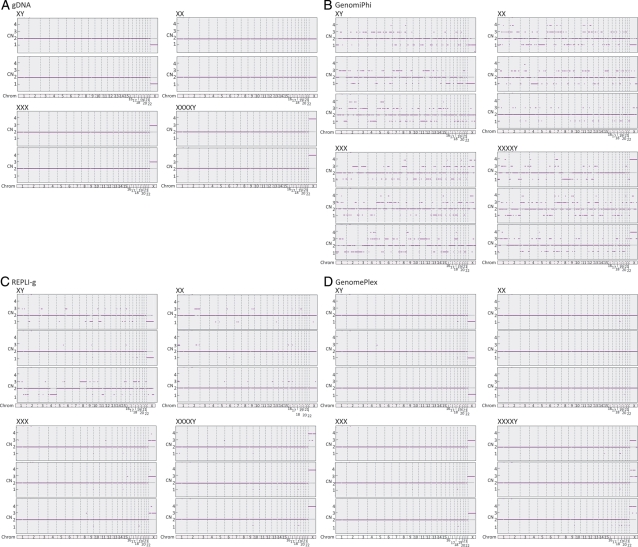
Similarity of single-cell CN assignments to assignments made on gDNA and as expected from the conventional karyotype data for each cell line were evaluated at three levels. For individual SNPs, 62% similarity was obtained using the GenomiPhi protocol, which was significantly less than the 95% similarity obtained using REPLI-g or the 99% similarity obtained using GenomePlex (Fig. 3A). For individual chromosomes, 75% similarity was obtained using the GenomiPhi protocol, which was significantly less than the 97% similarity obtained using REPLI-g or the 99% similarity obtained using GenomePlex (Fig. 3B). For single-cell molecular karyotyping diagnosis, 0% accuracy was obtained using the GenomiPhi protocol, which was significantly less than the 83% similarity obtained using REPLI-g or the 100% similarity obtained using GenomePlex (Fig. 3C). A comprehensive view of the gDNA and single-cell CN assignments is also displayed in Fig. 4 and reflects the levels of accuracy reported above. These results are of particular importance to the performance of methods that require accurate quantitative analysis of CN such as those reported by Le Caignec et al. (2006) and Vanneste et al. (2009), which used GenomiPhi technology, and Fiegler et al. (2007), Treff et al. (2010a) and Gutierrez-Mateo et al. (2010), which used GenomePlex technology.
Figure 3.
CN assignment accuracy of single-cell WGA. (A) The percentage of SNPs evaluated that were assigned the expected CN (SNP CN accuracy) for each of three single-cell WGA methods. (B) The percentage of chromosomes evaluated that were assigned the expected CN (chromosome ploidy accuracy) for each of three single-cell WGA methods. (C) The percentage of cells that were assigned the expected chromosome loss, gain or euploidy (karyotype diagnostic accuracy) for each of three single-cell WGA methods.
Figure 4.
SNP microarray-based CN graphs of (A) purified gDNA, or single cells amplified with (B) GenomiPhi, (C) REPLI-g or (D) GenomePlex protocols. Each panel includes analyses of each of four cell lines possessing one to four X chromosome copies.
The duration of amplification is also important when considering the application of single-cell WGA technology to clinical PGD. With WGA only one step is necessary to generate a diagnosis for the amplified sample, which also involves downstream microarray processing and analysis. For example, the most typical PGD application requires the completion of single-cell analysis within 24 h of initiating the procedure in order to avoid embryo cryopreservation. Therefore, although the REPLI-g protocol may be suitable for single-cell applications that do not have time constraints, the 16 h turnaround time may not allow for its routine use in PGD for aneuploidy screening. A more rapid turnaround time with isothermal MDA was represented in this study by the GenomiPhi protocol. Unfortunately, this shortened MDA protocol performed with the least reliability, fidelity and accuracy of all methods tested. In contrast, the GenomePlex protocol provided a more rapid turnaround time (4 h) which could be suitable for application to PGD and produced the highest CN assignment accuracy of all methods tested. Therefore, for applications requiring accurate and rapid CN analysis, such as PGD for aneuploidy screening, the GenomePlex protocol may be more appropriate than REPLI-g or GenomiPhi MDA-based protocols. However, for those applications requiring accurate genotyping analysis without time constraints, the REPLI-g protocol may be more appropriate than the GenomePlex or GenomiPhi protocols.
In summary, this study represents the first direct comparison of commercially available single-cell WGA method performance, a necessary step in all 24 chromosome aneuploidy screening technologies. Clinically relevant measurements of reliability, fidelity and accuracy were evaluated for each method. In general, a longer MDA protocol was better for genotyping accuracy than PCR, and PCR was better and faster than MDA for CN accuracy. Clinicians and laboratory directors should consider these and other critical pieces of evidence (presented in Table I and reviewed in Scott and Treff, 2010) when evaluating new technologies that intend to predict the chromosomal status and reproductive potential of human embryos.
Authors’ roles
N.R.T. and R.T.S. designed the study, N.R.T., J.S. and L.E.N. wrote the manuscript, and J.S., X.T. and L.E.N. performed the experiments and prepared the microarray data for publication.
Funding
Funding to pay the Open Access publication charges for this article was provided by Reproductive Medicine Associates of New Jersey.
References
- Barker DL, Hansen MST, Faruqi AF, Giannola D, Irsula OR, Lasken RS, Latterich M, Makarov V, Oliphant A, Pinter JH, et al. Two methods of whole-genome amplification enable accurate genotyping across a 2320-SNP linkage panel. Genome Res. 2004;14:901–907. doi: 10.1101/gr.1949704. doi:10.1101/gr.1949704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukner I, Paquin B, Belouchi M, Labuda D, Krajinovic M. Self-priming arrest by modified random oligonucleotides facilitates the quality control of whole genome amplification. Anal Biochem. 2005;339:345–347. doi: 10.1016/j.ab.2005.01.005. doi:10.1016/j.ab.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Burlet P, Frydman N, Gigarel N, Kerbrat V, Tachdijian G, Feyereisen E, Bonnefont J, Frydman R, Munnich A, Steffann J. Multiple displacement amplification improves PGD for fragile X syndrome. Mol Hum Reprod. 2006;12:647–652. doi: 10.1093/molehr/gal069. doi:10.1093/molehr/gal069. [DOI] [PubMed] [Google Scholar]
- Cui XF, Li HH, Goradia TM, Lange K, Kasasian HH, Jr, Galas D, Arnheim N. Single-sperm typing: determination of genetic distance between the G gamma-globin and parathyroid hormone loci by using the polymerase chain reaction and allele-specific oligomers. Proc Natl Acad Sci USA. 1989;86:9389–9393. doi: 10.1073/pnas.86.23.9389. doi:10.1073/pnas.86.23.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean F, Hosono S, Fang L, Wu X, Faruqi A, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. doi:10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J, Bartos J, Voglmayr H, Greihuber J. Nuclear DNA content and genome size of trout and human. Cytometry A. 2003;51:127–128. doi: 10.1002/cyto.a.10013. doi:10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- Escudero T, Pere C, Fischer J, Prates R, Tormasi S, Santiago M. Preimplantation genetic diagnosis (PGD) for reciprocal translocations using array comparative genome hybridization (aCGH) Fertil Steril. 2010;94:S80–S81. [Google Scholar]
- Fiegler H, Geigl JB, Langer S, Rigler D, Porter K, Unger K, Carter NP, Speicher MR. High resolution array-CGH analysis of single cells. Nucleic Acids Res. 2007;35:1–10. doi: 10.1093/nar/gkl1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Lensi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod. 2008;23:2596–2608. doi: 10.1093/humrep/den287. doi:10.1093/humrep/den287. [DOI] [PubMed] [Google Scholar]
- Fritz MA. Perspectives on the efficacy and indications for preimplantation genetic screening: where are we now? Hum Reprod. 2008;23:2617–2621. doi: 10.1093/humrep/den400. doi:10.1093/humrep/den400. [DOI] [PubMed] [Google Scholar]
- Gianaroli L, Magli MC, Ferraretti A, Fiorentino A, Garrisi J, Munne S. Preimplantation genetic diagnosis increases the implantation rate in human in vitro fertilization by avoiding the transfer of chromosomally abnormal embryos. Fertil Steril. 1997;68:1128–1131. doi: 10.1016/s0015-0282(97)00412-3. doi:10.1016/S0015-0282(97)00412-3. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Mateo C, Colls P, Sanchez-Casas Padilla E, Escudero T, Prates R, Ketterson K, Wells D, Munne S. Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril. 2010 doi: 10.1016/j.fertnstert.2010.09.010. Oct. 23 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Handyside AH, Robinson MD, Simpson RJ, Omar MB, Shaw MA, Grudzinskas JG, Rutherford A. Isothermal whole genome amplification from single and small numbers of cells: a new era for preimplantation genetic diagnosis of inherited disease. MolHumReprod. 2004;10:767–772. doi: 10.1093/molehr/gah101. [DOI] [PubMed] [Google Scholar]
- Handyside AH, Harton GL, Mariani B, Thornhill AR, Affara NA, Shaw MA, Griffin DK. Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet. 2009 doi: 10.1136/jmg.2009.069971. [DOI] [PubMed] [Google Scholar]
- Handyside AH, Grifo J, Prates R, Tormasi S, Fisher JM, Munne S. Validation and first clinical application of karyomapping for preimplantation diagnosis (PGD) of Gaucher disease combined with 24 chromosome screening. Fertil Steril. 2010;94:S79–S80. doi:10.1016/j.fertnstert.2010.07.309. [Google Scholar]
- Hellani A. Clinical application of multiple displacement amplification in preimplantation genetic diagnosis. RBM Online. 2005;10:376–380. doi: 10.1016/s1472-6483(10)61799-3. [DOI] [PubMed] [Google Scholar]
- Hellani A, Sammour A, Johansson L, El-Sheikh A. Delivery of a normal baby after preimplantation genetic diagnosis for non-ketotic hyperglycinaemia. Reprod Biomed Online. 2008a;16:893–897. doi: 10.1016/s1472-6483(10)60158-7. doi:10.1016/S1472-6483(10)60158-7. [DOI] [PubMed] [Google Scholar]
- Hellani A, Abu-Amero K, Azouri J, El-Akoum S. Successful pregnancies after application of array-comparative genomic hybridization in PGS-aneuploidy screening. Reprod Biomed Online. 2008b;17:841–847. doi: 10.1016/s1472-6483(10)60413-0. doi:10.1016/S1472-6483(10)60413-0. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, Ross R, Alper M, Barrett B, Frederick J, et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24 h protocol. Hum Reprod. 2010a;25:1066–1075. doi: 10.1093/humrep/dep452. doi:10.1093/humrep/dep452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Hill M, Abae M, Frederick J, Swanson M, Rabinowitz M. First clinical application of DNA microarrays for translocations and inversions. Fertil Steril. 2010b;93:S13–S14. doi:10.1016/j.fertnstert.2010.01.095. [Google Scholar]
- Kahraman S, Bahce M, Samli H, Imirzahoglu N, Yakism K, Cengiz G, Donmez E. Healthy births and ongoing pregnancies obtained by preimplantation genetic diagnosis in patients with advanced maternal age and recurrent implantation failure. Hum Reprod. 2000;15:2003–2007. doi: 10.1093/humrep/15.9.2003. doi:10.1093/humrep/15.9.2003. [DOI] [PubMed] [Google Scholar]
- Keskintepe L, Sher G, Keskintepe M. Reproductive oocyte/embryo genetic analysis: comparison between fluorescence in-situ hybridization and comparative genomic hybridization. Reprod Biomed Online. 2007;15:303–309. doi: 10.1016/s1472-6483(10)60343-4. doi:10.1016/S1472-6483(10)60343-4. [DOI] [PubMed] [Google Scholar]
- Lage JM, Leamon JH, Pejovic T, Hamann S, Lacey M, Dillon D, Segraves R, Vossbrinck B, González A, Pinkel D, et al. Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res. 2003;13:294–307. doi: 10.1101/gr.377203. doi:10.1101/gr.377203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Caignec C, Spits C, Sermon K, De Rycke M, Thienpont B, Debrock S, Staessen C, Moreau Y, Fryns JP, Van Steirteghem A, et al. Single-cell chromosomal imbalances detection by array CGH. Nucleic Acids Res. 2006;34:e68. doi: 10.1093/nar/gkl336. doi:10.1093/nar/gkl336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lledo B, Ten J, Galan F, Bernabeu R. Preimplantation genetic diagnosis of Marfan syndrome using multiple displacement amplification. Fertil Steril. 2006;86:949–955. doi: 10.1016/j.fertnstert.2006.03.036. doi:10.1016/j.fertnstert.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Lledo B, Bernabeu R, Ten J, Galan F, Cioffi L. Preimplantation genetic diagnosis of X-linked adrenoleukodystrophy with gender determination using multiple displacement amplification. Fertil Steril. 2007;88:1327–1333. doi: 10.1016/j.fertnstert.2007.01.034. doi:10.1016/j.fertnstert.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Lovmar L, Fredriksson M, Liljedahl U, Sigurdsson S, Syvanen AC. Quantitative evaluation by minisequencing and microarrays reveals accurate multiplexed SNP genotyping of whole genome amplified DNA. Nucleic Acids Res. 2003;31:129–139. doi: 10.1093/nar/gng129. doi:10.1093/nar/gng129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munne S, Magli MC, Cohen J, Morton NE, Sadowy S, Gianaroli L, Tucker M, Marquez C, Sable D, Ferraretti A, et al. Positive outcome after preimplantation diagnosis of aneuploidy in human embryos. Hum Reprod. 1999;14:2191–2199. doi: 10.1093/humrep/14.9.2191. doi:10.1093/humrep/14.9.2191. [DOI] [PubMed] [Google Scholar]
- Munne S, Sandalinas M, Escudero T, Velilla E, Walmsley R, Sadowy S, Cohen J, Sable D. Improved implantation after preimplantation genetic diagnosis of aneuploidy. Reprod Biomed Online. 2003;7:91–97. doi: 10.1016/s1472-6483(10)61735-x. doi:10.1016/S1472-6483(10)61735-X. [DOI] [PubMed] [Google Scholar]
- Munne S, Fisher JM, Warner A, Chen S, Zouves C, Cohen J, Group at RCP. Preimplantation genetic diagnosis significantly reduces pregnancy loss in infertile couples: a multicenter study. Fertil Steril. 2006;85:S178–S179. doi: 10.1016/j.fertnstert.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Munne S, Surrey ES, Grifo J, Marut E, Opsahl M, Taylor TH. Preimplantation genetic diagnosis using a-CGH significantly increases ongoing pregnancy rates per transfer. Fertil Steril. 2010;94:S81. doi:10.1016/j.fertnstert.2010.07.314. [Google Scholar]
- Northrop LE, Treff NR, Levy B, Scott RT., Jr SNP microarray-based 24 chromosome aneuploidy screening demonstrates that cleavage-stage FISH poorly predicts aneuploidy in embryos that develop to morphologically normal blastocysts. Mol Hum Reprod. 2010;16:590–600. doi: 10.1093/molehr/gaq037. doi:10.1093/molehr/gaq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panelli S, Damiani G, Espen L, Micheli G, Sgaramella V. Towards the analysis of the genomes of single cells: Further characterisation of the multiple displacement amplification. Gene. 2006;372:1–7. doi: 10.1016/j.gene.2006.01.032. doi:10.1016/j.gene.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Park JW, Beaty TH, Boyce P, Scott AF, McIntosh I. Comparing whole-genome amplification methods and sources of biological samples for single-nucleotide polymorphism genotyping. Clin Chem. 2005;5:1520–1523. doi: 10.1373/clinchem.2004.047076. doi:10.1373/clinchem.2004.047076. [DOI] [PubMed] [Google Scholar]
- Rabinowitz M, Behr D, Potter D, Ross R, Alper M, Banjevic M. Parental support for single gene PGD and simultaneous 24-chromosome screening reduces risks of allele misdiagnosis and transfer of aneuploid embryos. Fertil Steril. 2009;92:S202. doi:10.1016/j.fertnstert.2009.07.1448. [Google Scholar]
- Rabinowitz M, Beltsos A, Potter D, Bush M, Givens C, Smotrich D. Effects of advanced maternal age are abrogated in 122 patients undergoing transfer of embryos with euploid microarray screening results at cleavage stage. Ferti Steril. 2010;94:S80. doi:10.1016/j.fertnstert.2010.07.310. [Google Scholar]
- Ren Z, Zhou C, Xu Y, Deng J, Zeng H, Zeng Y. Mutation and haplotype analysis of Duchene muscular dystrophy by single cell multiple displacement amplification. Mol Hum Reprod. 2007;13:431–436. doi: 10.1093/molehr/gam020. doi:10.1093/molehr/gam020. [DOI] [PubMed] [Google Scholar]
- Renwick P, Lewis C, Abbs S, Ogilivie C. Determination of the genetic status of cleavage-stage human embryos by microsatellite marker analysis following multiple displacement amplification. Prenat Diagn. 2007;27:206–215. doi: 10.1002/pd.1638. doi:10.1002/pd.1638. [DOI] [PubMed] [Google Scholar]
- Saiki R, Gelfand D, Stoffel S, Scharf S, Higuchi R, Horn G, Mullis K, Erlich H. Primer directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. doi:10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schoolcraft WB, Treff NR, Ferry K, Stevens JM, Katz-Jaffe MG, Scott RT. Clinical application of comprehensive chromosomal screening at the blastocyst stage. Fert Steril. 2010a;94:S23. doi: 10.1016/j.fertnstert.2009.10.015. doi:10.1016/j.fertnstert.2010.07.090. [DOI] [PubMed] [Google Scholar]
- Schoolcraft WB, Treff NR, Ferry K, Stevens JM, Katz-Jaffe MG, Scott RT. First clinical application of SNP microarray based 24 chromosome aneuploidy screening of human blastocysts. Fertil Steril. 2010b;4:S23–S24. doi:10.1016/j.fertnstert.2010.07.090. [Google Scholar]
- Scott RT, Jr, Treff NR. Assessing the reproductive competence of individual embryos: a proposal for the validation of new ‘-omics’ technologies. Fertil Steril. 2010;94:791–794. doi: 10.1016/j.fertnstert.2010.03.041. [DOI] [PubMed] [Google Scholar]
- Scott RT, Jr, Miller KA, Olivares R, Su J, Fratterelli J, Treff NR. Microarray based 24 chromosome preimplantation genetic diagnosis (mPGD) is highly predictive of the reproductive potential of human embryos: a prospective blinded non-selection trial. Fertil Steril. 2008;90:22. doi:10.1016/j.fertnstert.2008.07.438. [Google Scholar]
- Scott RT, Jr, Tao X, Taylor D, Ferry K, Treff N. A prospective randomized controlled trial demonstrating significantly increased clinical pregnancy rates following 24 chromosome aneuploidy screening: biopsy and analysis on day 5 with fresh transfer. Fertil Steril. 2010;94:S2. doi:10.1016/j.fertnstert.2010.07.007. [Google Scholar]
- Sher G, Keskintepe L, Keskintepe M, Ginsburg M, Maassarani G, Yakut T, Baltaci V, Kotze D, Unsal E. Oocyte karyotyping by comparative genomic hybridization provides a highly reliable method for selecting ‘competent’ embryos, markedly improving in vitro fertilization outcome: a multiphase study. Fertil Steril. 2007;87:1033–1040. doi: 10.1016/j.fertnstert.2006.08.108. doi:10.1016/j.fertnstert.2006.08.108. [DOI] [PubMed] [Google Scholar]
- Sher G, Keskintepe L, Keskintepe M, Maassarani G, Tortoriello D, Brody S. Genetic analysis of human embryos by metaphase comparative genomic hybridization (mCGH) improves efficiency of IVF by increasing embryo implantation rate and reducing multiple pregnancies and spontaneous miscarriages. Fertil Steril. 2009;92:1886–1894. doi: 10.1016/j.fertnstert.2008.11.029. doi:10.1016/j.fertnstert.2008.11.029. [DOI] [PubMed] [Google Scholar]
- Treff NR, Su J, Tao X, Miller K, Scott RT. First IVF babies born after rapid 24 chromosome embryo aneuploidy screening and fresh embryo transfer. Fertil Steril. 2009a;92:S49. [Google Scholar]
- Treff N, Tao X, Su J, Northrop LE, Kamani M, Bergh P, Miller K, Levy B, Scott R. SNP microarray based concurrent screening of 24 chromosome aneuploidy, unbalanced translocations, and single gene disorders in human embryos: first application of comprehensive triple factor PGD. Biol Reprod. 2009b;81:188. doi:10.1095/biolreprod.108.072629. [Google Scholar]
- Treff NR, Tao X, Lonczak A, Su J, Taylor D, Scott R. Four hour 24 chromosome aneuploidy screening using high throughput PCR SNP allele ratio analyses. Fertil Steril. 2009c;92:S49–S50. [Google Scholar]
- Treff NR, Su J, Tao X, Levy B, Scott RT., Jr Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil Steril. 2010a;94:2017–2021. doi: 10.1016/j.fertnstert.2010.01.052. doi:10.1016/j.fertnstert.2010.01.052. [DOI] [PubMed] [Google Scholar]
- Treff NR, Levy B, Su J, Northrop LE, Tao X, Scott RT., Jr SNP microarray-based 24 chromosome aneuploidy screening is significantly more consistent than FISH. Mol Hum Reprod. 2010b;16:583–589. doi: 10.1093/molehr/gaq039. doi:10.1093/molehr/gaq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treff N, Northrop LE, Kasabwala K, Su J, Levy B, Scott RT. SNP microarray based concurrent screening of 24 chromosome aneuploidy and unbalanced translocations in preimplantation human embryos. Fertil Steril. 2010c doi: 10.1016/j.fertnstert.2010.11.004. Nov. 29 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–583. doi: 10.1038/nm.1924. doi:10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod. 2000;6:1055–1062. doi: 10.1093/molehr/6.11.1055. doi:10.1093/molehr/6.11.1055. [DOI] [PubMed] [Google Scholar]
- Wells D, Sherlock JK, Handyside AH, Delhanty JD. Detailed chromosomal and molecular genetic analysis of single cells by whole genome amplification and comparative genomic hybridization. Nucleic Acids Res. 1999;27:1214–1218. doi: 10.1093/nar/27.4.1214. doi:10.1093/nar/27.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D, Fragouli E, Alfarawaty S, Munne S, Schoolcraft WB, Katz-Jaffe M. Highly significant improvement in embryo implantation and increased live birth rate achieved after comprehensive chromosomal screening: implications for single embryo transfer. Fertil Steril. 2009;94:S79. doi:10.1016/j.fertnstert.2009.07.305. [Google Scholar]