Abstract
The Dlx genes encode a family of transcription factors important to the development of the vertebrate forebrain. These genes have very similar expression domains during the development of the telencephalon in mouse and play a role in GABAergic interneuron differentiation. We have used triple fluorescent in situ hybridization to study the relative expression domains of the dlx and gad1 genes in the zebrafish telencephalon and diencephalon. We also generated transgenic zebrafish with regulatory elements from the zebrafish dlx1a/2a locus. The zebrafish dlx regulatory elements recapitulated dlx expression in the forebrain and mimicked the relationship between the expression of the dlx genes and gad1. Finally, we show that a putative enhancer located downstream of dlx2b can also activate reporter gene expression in a tissue specific manner similar to endogenous dlx2b expression. Our results indicate the dlx genes are regulated by an evolutionarily conserved genetic pathway and may play a role in GABAergic interneuron differentiation in the zebrafish forebrain.
Keywords: dlx, Forebrain, Transgenesis, Zebrafish, Regulatory elements, Fluorescent in situ hybridization
INTRODUCTION
The Dlx genes encode homeodomain containing transcription factors involved in the development of the mammalian forebrain (Anderson et al. 1997a; Anderson et al. 1997b; Marin et al. 2000; Pleasure et al. 2000). They have partially overlapping expression in the developing telencephalon of the mouse and follow a distinct spatial and temporal expression pattern: Dlx1 and Dlx2 are expressed in the ventricular zone and subventricular zone; Dlx5 is expressed in the subventricular zone and Dlx6 is expressed laterally in the mantle zone (Liu et al. 1997; Anderson et al. 1999; Eisenstat et al. 1999; Yun et al. 2003). Dlx gene expression highly overlaps with that of glutamic acid decarboxylases (Gads), the enzymes responsible for the synthesis of γ-amino butyric acid (GABA). Furthermore, DLX proteins can induce Gad gene expression in vitro and in vivo (Anderson et al. 1999; Stühmer et al. 2002; Yun et al. 2003). Dlx1-/-/Dlx2-/- mutant mice show a loss of GABAergic interneuron differentiation in the ventral telencephalon, further implicating the Dlx genes in the differentiation of GABAergic interneurons (Anderson et al. 1997a).
Vertebrate Dlx genes are typically organized in convergently transcribed bigene pairs, separated by a short intergenic region usually less than 10kb (Zerucha et al. 2000; Sumiyama et al. 2002; Ghanem et al. 2003). Zebrafish have eight dlx genes of which six show similar bigene genomic arrangements to the mouse: dlx1a/dlx2a; dlx3b/4b; and dlx5a/6a clusters orthologous to Dlx1/2; Dlx3/4; Dlx5/6, respectively (Quint et al. 2000). Two additional genes, dlx2b and dlx4a, are thought to be duplicates of ancestral Dlx2 and Dlx4, respectively, after the teleost specific genome duplication event (Amores et al. 1998). The expression domains of two Dlx genes comprising a given bigene pair are very similar, both in mice and zebrafish, most likely due to shared regulatory regions (Liu et al. 1997; Eisenstat et al. 1999; Ellies et al. 1997). Regulatory regions have been identified both upstream of the transcription start sites and within the intergenic domains of a given bigene pair (Zerucha et al. 2000; Ghanem et al. 2003; Ghanem et al. 2007). The zebrafish dlx genes share similar genomic arrangements to the mouse, including the presence of the regulatory elements I12a and I12b in the dlx1a/2a intergenic region, Upstream regulatory element 2 (URE2) upstream of dlx1a, I56i and I56ii between dlx5a and dlx6a (Zerucha et al. 2000; Ghanem et al. 2003). Five dlx genes (all but dlx3b, dlx4b and dlx4a) are expressed in the zebrafish forebrain with very similar expression domains in the telencephalon and diencephalon (Akimenko et al. 1994; Ellies et al. 1997). In particular zebrafish dlx1a and dlx2a have been proposed to mark cells closer to the ventricular wall than the dlx5a and dlx6a genes, reminiscent of the spatial-temporal succession of Dlx gene expression pattern in mouse (Zerucha et al. 2000). However, the precise spatial relationship between different dlx expression domains and gad1 gene expression has not been determined.
In this study, we describe in detail the extent to which zebrafish dlx and gad1 expression patterns overlap in the developing forebrain. The gad1 gene, orthologous to Gad67 in mammals, is used as a marker for GABAergic interneuron differentiation. The spatial-temporal expression pattern described in the mouse telencephalon appears to be similar in the zebrafish, with dlx1a and dlx2a being expressed in the ventricular zone and dlx5a, dlx6a and gad1 expressed in more lateral differentiated cells. We tested whether the conserved regions linked to the dlx1a/2a cluster of the zebrafish have regulatory functions consistent to previously reported data in the mouse forebrain, by designing reporter constructs containing conserved sequences from the dlx1a/2a locus. We also utilized a previously described transgenic line, Tg(dlx5a/6aIG:GFP), showing these regulatory regions are sufficient to mimic the endogenous nested expression patterns of the dlx genes in the zebrafish forebrain (Zerucha et al. 2000). We further explored the regulation of the dlx2b gene, paralogous to the dlx2a gene in the zebrafish. A small domain containing two putative dlx binding sites had been identified downstream of dlx2b (Ghanem et al. 2003), denoted dlx2b downstream regulatory element (dlx2bDRE). The forebrain expression of dlx2b is at least partially recapitulated by reporter gene expression when under the regulation of dlx2bDRE.
RESULTS
Partially overlapping expression domains of dlx and gad1 genes in the zebrafish forebrain
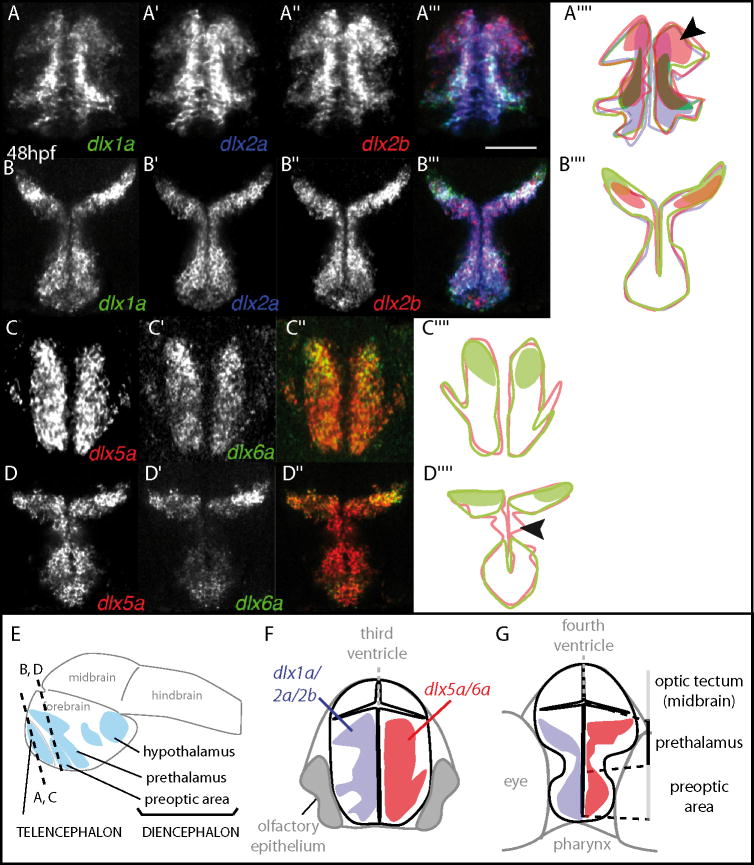
The zebrafish dlx genes from a bigene pair have been proposed to have highly overlapping expression domains within the forebrain (Zerucha et al. 2000). Currently there are no antibodies against specific zebrafish Dlx proteins except Dlx3b, therefore we relied on fluorescent RNA in situ hybridization to determine the extent to which the dlx genes expression domains overlap. We examined the dlx expression in the zebrafish forebrain from 24 hours post-fertilization (hpf) until 48 hpf. At 48 hpf, the forebrain is beginning to undergo secondary neurogenesis, and dlx and gad1 are expressed in the forebrain (Akimenko et al. 1994; Ellies et al. 1997; Martin et al. 1998). As observed by confocal imaging, the dlx1a, dlx2a, and the paralogous dlx2b genes are expressed in very similar domains in the subpallium of the zebrafish telencephalon at 48hpf (Fig. 1A-A’” and delimited by solid lines in Fig. 1A””). Within this domain, the three genes are expressed close to the ventricle, an area shown to be proliferative (Mueller and Wullimann, 2003). There may be quantitative differences within these domains as dlx2b appears to be more strongly expressed in the most dorsal domain of the subpallium (Fig. 1A” and colored zone in A””), while the dlx2a domain of expression is extended ventrally compared to the other two genes (see colored zone in Fig. 1A””). The three dlx genes also show virtually identical expression domains in the diencephalon, more specifically in the prethalamus and preoptic area (Fig. 1B-B’”), with an apparently higher level of expression for dlx1a and dlx2b in the dorsal part of the prethalamus compared to the rest of their expression domains (Fig. 1B””).
Figure 1. Similar forebrain expression of the two genes from a dlx cluster.
Single z transversal sections of triple fluorescent in situ hybridization shows dlx1a, dlx2a, and dlx2b expression domains at 48hpf in the telencephalon (A-A’”) and diencephalon (B-B’”), as well as dlx5a and dlx6a (C-C’” and D-D’”). Schematic summaries representing the overlaps in dlx expression are shown in A””, B””, C”” and D””: the solid lines define the boundaries of detectable expression; the zones of relatively higher expression are colored when applicable. The boundaries of expression domains are comparable for dlx1a, dlx2a and dlx2b but dlx2b specifically displays a more intense zone of expression in a dorsal-lateral domain (arrow in A””). Boundaries of expression domains for dlx5a and dlx6a are overall similar although dlx6a was hardly detected in the ventral domain of the prethalamus (arrowhead on D””). Levels of section are shown on the schematic in (E). Shared expression domains of clustered genes (and their paralogs) are mapped on a schematic representation of a brain section at the level of the telencephalon (F) and of the diencephalon (G): dlx1a/2a/2b in blue and dlx5a/6a in red. Scale bar: 100 μm.
The dlx5a and dlx6a genes display very similar expression domains in the subpallial telencephalon at 48hpf (Fig. 1C-C”) although dlx6a is highly expressed only in the dorsal-most part of the telencephalon. Both genes are expressed outside of the ventricular zone (Fig. 1F). In the diencephalon, transcripts of dlx5a/6a are localized throughout the prethalamus and preoptic area but dlx6a transcripts are strongly detected only in the dorsal-most part of the prethalamus (Fig. 1D). The expression limits of the dlx1a/2a/2b and dlx5a/6a genes appear to be, overall, very similar in the diencephalon (Fig. 1G). The highly overlapping domains of dlx forebrain expression are also observed at 24 and 36hpf (Suppl. 1 and Suppl. 2).
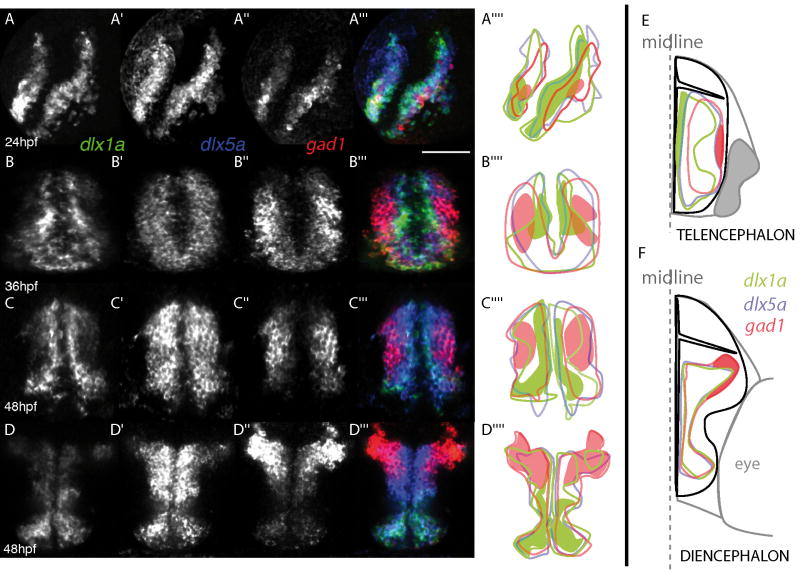
Consistent with previous reports, we find that dlx2a is transcribed in the ventricular zone of the zebrafish forebrain, where zash1a (ascl1a), gad1, and GABA expression can also be detected (Wullimann and Mueller, 2002; Mueller et al. 2006; Mueller et al. 2008). Based on the results presented in Fig. 1, it appears that expression of a gene in a dlx bigene cluster is representative of the second gene in the cluster. To further compare the expression domains of zebrafish dlx genes in the forebrain with areas of GABAergic interneuron differentiation, we studied the relative expression domains of one dlx gene from each bigene pair, dlx1a and dlx5a, alongside the specification marker gad1. The dlx1a, dlx5a, and gad1 genes are expressed in very comparable domains within the telencephalon and diencephalon at 24hpf (Fig. 2A). Beginning at 36hpf and continuing at least until 48hpf, overt differences are observed between dlx1a, dlx5a, and gad1 expression patterns within the telencephalon (Fig. 2B and C and see schematic on Fig. 2E). The medial limit of dlx1a expression was found at the level of the ventricle, while the medial limit of expression for both dlx5a and gad1 was found more lateral. As for the lateral limits of dlx5a and gad1 expression domains, these are found outside of the dlx1a expression domains. In addition, relatively stronger expression of dlx1a is detected close to the ventricle while stronger expression of gad1 is located in the lateral part of the telencephalon (see Fig. 2C””, and Fig. 2E). In the diencephalon, the dlx1a, dlx5a, and gad1 expression domains are very comparable in the preoptic area and prethalamus (Fig. 2D), although the limit of gad1 expression is found more dorsal than that of the dlx genes. The levels of transcripts are not homogenous within the dlx1a and gad1 domains, with dlx1a weakly expressed and gad1 highly expressed in the dorsal-most domain of the prethalamus and vice versa in the preoptic area (Fig. 2D””).
Figure 2. Expression domains of dlx genes with respect to gad1 expression.
Single z sections of triple fluorescent in situ hybridization for dlx1a, dlx5a, and gad1 in the telencephalon at 24hpf (A-A”, para-sagital section, anterior is on the left), 36hpf (B-B”, transverse section in the telencephalon, dorsal is to the top), and 48hpf (C-C”, transversal section in the telencephalon; D-D”, transversal section in the diencephalon; dorsal is to the top) with a colored merge of all three channels on A’”, B’”, C’” and D’”. For each section level, a schematic representation is given to localize the boundaries of each expression domain (solid line) and a domain of higher level of expression is represented as a colored surface. Panel E gives a schematic representation of the partially nested expression domains in the telencephalon (left half) and diencephalon (right half) at 48hpf, based on the data shown in panels C-C’” and D-D’”: the proximal (ventricular) domain where only dlx1a is detected is shown in green and the lateral domain where gad1 only is detected is colored red. Scale bar: 100 μm.
The activity of zebrafish dlx enhancers is similar to their mouse counterparts
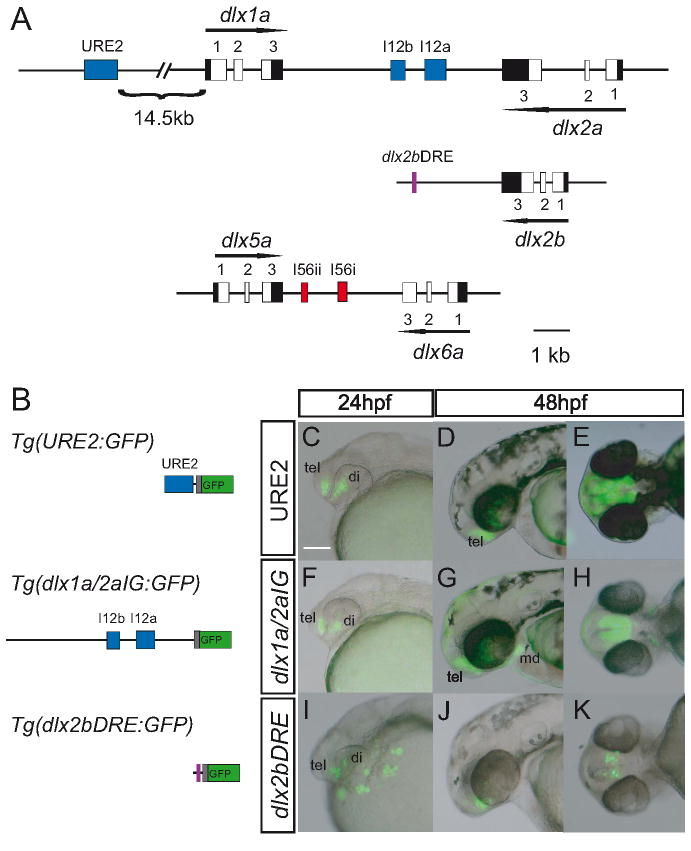
The I12a and I12b enhancers, located in the mouse Dlx1/2 intergenic region, have regulatory functions when tested in transgenic mice, with only I12b displaying forebrain activity (Zerucha et al. 2000; Ghanem et al. 2003; Ghanem et al. 2007). The mouse URE2 enhancer is located upstream of Dlx1 and drives expression in a number of tissues, one of which is the developing forebrain (Ghanem et al. 2007). To test if orthologous sequences from the zebrafish genome have regulatory function in the zebrafish, we designed two reporter constructs containing these genomic sequences (Fig. 3A, B). We selected 6kb of the dlx1a/2a intergenic region, including I12a and I12b, and 900bp encompassing URE2. These sequences were cloned upstream of a β-globin minimal promoter associated with the coding sequences of GFP to produce the Tg(dlx1a/2aIG:GFP) and the Tg(URE2dlx1a/2a:GFP) reporter transgenes, respectively.
Figure 3. Regulatory elements of dlx1a/2a and dlx2b drive reporter gene expression in the zebrafish forebrain and branchial arches.
Genomic organization of the dlx1a/dlx2a and of the dlx2b loci (A). Exons are represented in white and UTRs in black. Intergenic regulatory elements are shown as blue (URE2 and I12b) and red boxes (I56i and I56ii). The dlx2bDRE is shown as a purple box. Schematic representation of the transgene constructs (containing the β-globin minimal promoter (grey) linked to GFP (green)) used to generate transgenic zebrafish (B). The Tg(dlx1a/2aIG:GFP) construct contains a 6kb of the intergenic fragment from the dlx1a/dlx2a intergenic region, the Tg(URE2dlx1a/2a:GFP) contains a 900 bp fragment encompassing the URE2 enhancer and the Tg(dlx2bDRE:GFP) contains a 200 base pair fragment downstream of dlx2b. Activity of the transgene constructs in the forebrain at 24hpf and 48hpf (C-E) Tg(URE2dlx1a/2a:GFP) transgene (F-H) Tg(dlx1a/2aIG:GFP) transgene (I-K) Tg(dlx2bDRE:GFP). tel, telencephalon; di, diencephalon; md, mandibular portion of branchial arches. Scale bar: 100 μm.
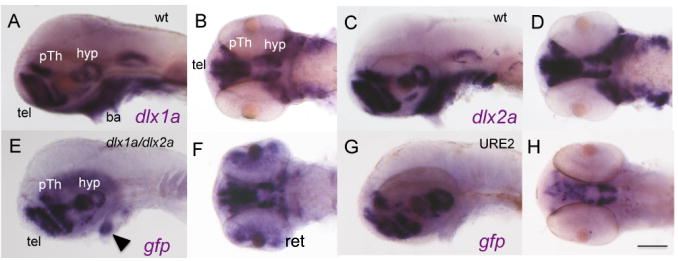
Embryos harboring the Tg(URE2dlx1a/2a:GFP) transgene show GFP expression in the telencephalon and diencephalon starting at approximately 24hpf (Fig. 3C). GFP expression can be detected in the telencephalon and diencephalon from 24hpf until at least 72hpf (Fig. 3D and E). Expression of GFP in the Tg(URE2dlx1a/2a:GFP) line can also be detected in the mesenchyme of the visceral arches at 96hpf (data not shown). The Tg(dlx1a/2aIG:GFP) embryos show GFP expression in domains highly similar to those observed in Tg(URE2dlx1a/2a:GFP) embryos with fluorescence in the telencephalon and diencephalon starting at 24 hpf (Fig. 3F). Transgene expression can also be detected in the jaw region starting at approximately 48hpf (Fig. 3G). The overall forebrain GFP expression of Tg(dlx1a/2aIG:GFP) and Tg(URE2dlx1a/2a:GFP) lines closely mimics the endogenous forebrain expression of the dlx1a and dlx2a genes (Fig. 4). However, the transgenes are only active in small domains within the branchial arches and not at all in the olfactory or otic placodes, other areas of known dlx1a/2a expression (Akimenko et al. 1994; Ellies et al. 1997). The Tg(dlx1a/2aIG:GFP) transgene also targets GFP expression in the retina at 48hpf, an area where expression of dlx1a or dlx2a has not been previously reported (Fig. 4F).
Figure 4. The dlx regulatory elements recapitulate endogenous dlx expression in the forebrain of the zebrafish at 48hpf.
Whole mount in situ with probes against dlx1a (A,B), dlx2a (C,D) and gfp (E, F) Tg(dlx1a/2aIG:GFP) is expressed in the telencephalon, ventral thalamus, and hypothalamus. There is also expression in the pharyngeal arches (arrowhead), which recapitulates a small portion of the endogenous dlx arch expression domain. G,H) URE2 activity closely resembles the expression of Tg(dlx1a/2aIG:GFP) in the telencephalon, ventral thalamus, and hypothalamus and mimics dlx expression in these tissues. tel, telencephalon; pTh, prethalamus; hyp, hypothalamus; di, diencephalon; md, mandible; ba, branchial arches. Scale bar: 100 μm.
dlx enhancer activity closely mimics endogenous dlx expression in the telencephalon
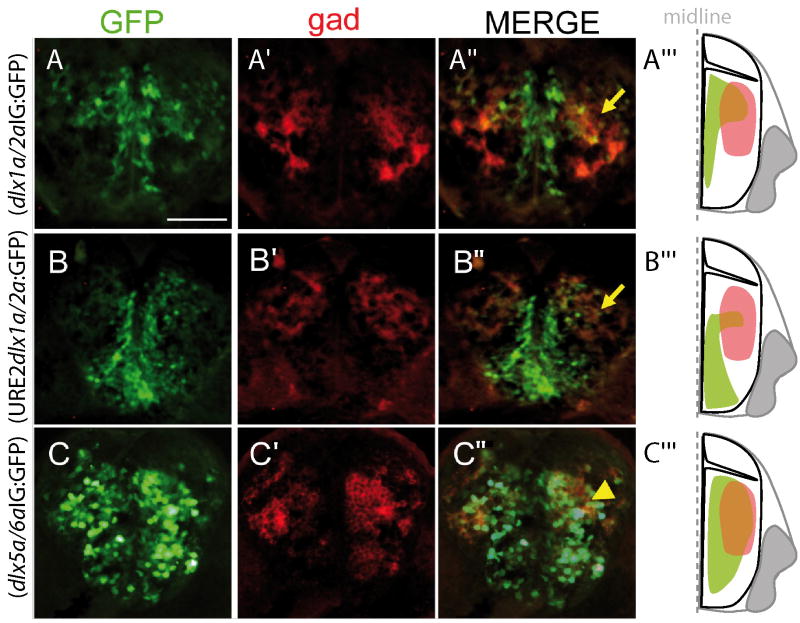
To determine if the dlx regulatory elements are active in cells that express Gad proteins, we co-labeled transverse sections of the zebrafish forebrain with antibodies against GFP and Gad proteins (Fig. 5). At 48hpf, Tg(dlx1a/2aIG:GFP) reporter expression is located close to the ventricle and dorsally in the telencephalon but Gad proteins are localized more laterally (Fig. 5A). Very few cells are co-expressing Gad and GFP and those rare cells are located in the dorsolateral domain of the telencephalon (Fig. 5A). Expression of the Tg(URE2dlx1a/2a:GFP) reporter transgene is detected close to the ventricle and ventrally in the subpallium of the telencephalon, with only very few cells co-expressing GFP and Gad, again located in a dorsolateral domain (Fig. 5B). To assess the activity of dlx5a/6a regulatory elements in the developing zebrafish forebrain we utilized the Tg(dlx5a/6aIG:GFP) transgenic line containing the two enhancers, I56i and I56ii, that recapitulate endogenous dlx5a/6a expression (Zerucha et al. 2000; Mione et al. 2008). The Tg(dlx5a/6aIG:GFP) embryos show reporter expression in the telencephalon and diencephalon of zebrafish embryos and most Gad positive cells co-express GFP (Fig. 5C, data not shown) (Mione et al. 2008). These data suggest that the enhancers associated with the dlx1a/2a bigene cluster drive expression in the ventricular zone of the subpallium, while the enhancers associated with the dlx5a/6a bigene cluster drive expression in more lateral cells expressing Gad proteins. The expression domains described for GFP in transgenic lines are therefore consistent with the spatially restricted expression of the dlx genes (Fig. 6).
Figure 5. Immunolocalization of GFP and Gad proteins on transverse cryosections of the telencephalon at 48hpf.
Expression of GFP (A, B, C) and Gad proteins (A’, B’, C’) is detected in the three transgenic lines, a merged image is presented in panels A”, B” and C” to show overlap (yellow arrows). An interpretative schematic representation of a hemi-section is presented in each case in panels A’”, B’” and C’”. Scale bars: 100 μm.
Figure 6.
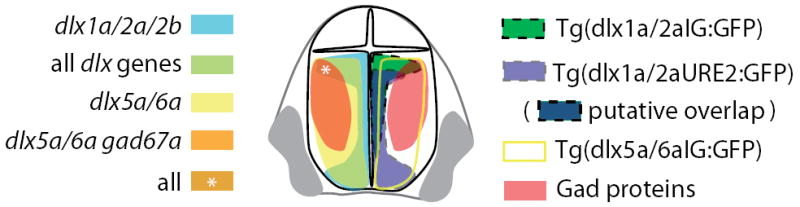
Schematic representation of the overlaps in dlx and gad expression (left) and of the activity of dlx regulatory elements (right) in the zebrafish forebrain.
Co-expression of paralogous genes and insights into the regulation of zebrafish dlx2b expression
The I12b and URE2 enhancers appear to be sufficient to recapitulate expression of dlx1a/dlx2a in the forebrain. The paralogous gene to dlx2a, dlx2b, is also expressed in similar domains of the forebrain (Fig. 1). These highly overlapping domains of expression for dlx1a, dlx2a, and dlx2b suggest the existence of a regulatory sequence at the dlx2b locus that would be similar, in sequence and/or in function, to those described for the dlx1a/2a locus. Phylogenetic footprinting of the dlx2b locus identified a 60bp sequence located in the 3’ region of the gene and showing similarity to other dlx enhancers, including two putative dlx transcription factor-binding sites (Ghanem et al. 2003). We amplified a 200 bp region downstream of dlx2b, including these 60bp, that we named dlx2b Downstream Regulatory Element (dlx2bDRE), and placed it in a β-globin:GFP transgene which was tested in primary transgenic zebrafish embryos (Fig. 3B). This transgene targets GFP expression in the telencephalon and diencephalon of 84% (77/92) of primary injected zebrafish at 24hpf (Fig. 3I). This expression persists until at least 48hpf in the telencephalon and diencephalon and closely resembles that observed in the Tg(URE2dlx1a/2a:GFP) and Tg(dlx1a/2aIG:GFP) transgenic lines (Fig. 3J and K).
DISCUSSION
Conserved activity of dlx regulatory elements between zebrafish and mouse
Several conserved regulatory elements have been identified that are sufficient to mimic many aspects of endogenous Dlx expression in the mouse (Zerucha et al. 2000; Ghanem et al. 2003; Ghanem et al. 2007). Similarly, the distinct expression domains of the dlx genes in the forebrain seem to be completely recapitulated by orthologous zebrafish enhancers. Therefore, our results show that zebrafish and mouse regulatory elements not only show strong sequence conservation, but also are sufficient to drive similar expression in homologous tissues (forebrain and pharyngeal arches) indicating their activity has been conserved throughout vertebrate evolution. The expression domains of the Tg(URE2dlx1a/2a:GFP) and Tg(dlx1a/2aIG:GFP) transgenes are similar in the forebrain, although the former shows an extended expression domain in the ventral subpallium while the latter has an extended expression domain in the dorsal subpallium. Heterologous transgenic experiments have indicated that zebrafish dlx5a/6a regulatory sequences are functional in the mouse forebrain, mimicking the endogenous Dlx gene expression patterns (Zerucha et al. 2000; Stühmer et al. 2002). Consistent with these results, the URE2 element from distantly related vertebrates has conserved function in the forebrain of zebrafish and mice (R.B.M., M.D.T, and M.E., unpublished observations). Taken altogether, the high levels of conservation of the sequence and regulatory function strongly suggest that the genetic cascades upstream of the Dlx genes are conserved between distantly related vertebrates.
Spatial-temporal expression of dlx and gad genes is conserved between mouse and zebrafish
The Dlx genes have been shown to play an important role in the differentiation and migration of GABAergic interneurons in mice (Anderson et al. 1997a; Anderson et al. 1997b; Stühmer et al. 2002a; Stühmer et al. 2002b). The Dlx1/2 genes are expressed in immature neurons and Dlx5/6 in more mature neurons, eventually differentiating into Gad positive interneurons (Liu et al. 1997; Eisenstat et al.1999; Stühmer et al. 2002b; Yun et al. 2003). Previous studies of gene expression and cell proliferation revealed similarities in the topological relationships of mouse and zebrafish telencephalon (For reviews see: Wullimann and Mueller, 2004; Mueller and Wullimann, 2009; Wullimann, 2009). Thus, the ventricular (proximal) zone is a site of proliferation and expresses dlx2a (Mueller et al. 2002; Mueller et al. 2008). The expression domain of dlx5a (and by extension, dlx6a) is shifted laterally compared to dlx1a (dlx2a), and overlaps partially with the more lateral of gad1 expression domain. These relative expression patterns can be indicative of a temporal sequence of gene expression (dlx1a/2a →dlx5a/6a→ gad1) in cells moving from the proliferative zone to more lateral regions of the telencephalon and may be related to the various stages of GABAergic neuron differentiation. This apparent movement of cells from the ventricular zone to more lateral domains may be due to tangential and/or radial migration, as both modes of migration have been previously identified in the zebrafish telencephalon (Mueller et al. 2006; Mueller et al. 2008; Mione et al. 2008). As the domains of enhancer activity are consistent with gene expression data (Fig. 6), transgenic zebrafish lines may be useful to isolate neural progenitors from the forebrain at different stages of GABAergic interneuron differentiation.
Conserved genetic mechanisms lead to the highly overlapping expression domains of dlx1a/2a and dlx2b
The two Dlx genes of a bigene pair have highly overlapping expression domains that have been suggested to be due to shared regulatory regions (Ellies et al. 1997). In this study, we report the very similar expression domains of dlx1a, dlx2a and dlx2b in the forebrain starting at 24hpf and continuing until at least 48hpf. We explored the possibility that dlx2b expression was controlled by sequences resembling dlx1a/2a enhancers, such as URE2 or I12b. Sequence comparisons of the forebrain enhancers (I12b, I56i, and I56ii) to the dlx2b locus had identified a 60bp sequence containing two putative dlx binding sites located downstream of dlx2b (Ghanem et al. 2003). Given the position of this element, it could be paralogous to the I12b sequence located between dlx1a and dlx2a, but would have accumulated mutations since the whole-genome duplication event specific to teleosts. Here we show that this dlx2bDRE region is sufficient to drive reporter gene expression in the forebrain, with patterns similar to those observed with the dlx1a/2a transgenes, suggesting that similar regulatory mechanisms may be involved in dlx2b and dlx1a/2a expression. The dlx2bDRE forebrain activity may also be attributable to the presence of the two dlx binding sites within a conserved 60bp region. A region from the I56i enhancer containing two Dlx binding sites was sufficient to drive forebrain expression of a reporter gene in transgenic mice (Zerucha et al. 2000; Ghanem et al. 2003). The conservation of the dlx binding sites is suggestive of potential cross-regulation of dlx2b expression by dlx1a and/or dlx2a, as the mouse Dlx2 protein has been shown to bind directly to DNA sequences organized in a similar context (Zerucha et al. 2000; Zhou et al. 2004; Potter et al. 2009). Cross-regulation between dlx1a/dlx2a and dlx2b via the dlx2bDRE would explain the strong overlap in expression. Overall, the highly overlapping expression of the dlx1a/2a bigene pair with the unlinked dlx2b is suggestive of similar gene regulation mechanisms at the two loci.
In conclusion, we have described the spatial overlap between dlx bigene pairs and gad1 expression in the embryonic zebrafish forebrain. The dlx1a/2a genes are expressed close to the ventricle, dlx5a/6a are expressed throughout the subpallium, and gad1 is expressed in the lateral most regions consistent with the hypothesis that dlx genes play a role in GABAergic interneuron differentiation in the zebrafish forebrain, similarly to what is known in the mouse. The dlx regulatory elements have conserved function in the zebrafish and mouse forebrain, and mimic the spatial relationship between dlx and gad1 within the zebrafish telencephalon. These results support the idea that genetic pathways controlling dlx gene expression and GABAergic interneuron development, in the forebrain of mammals and teleosts, may be conserved over approximately 430 million years of evolution.
EXPERIMENTAL PROCEDURES
Zebrafish Husbandry
Wild type zebrafish were raised according to standard procedures (Westerfield, 2000). Embryos were staged as hours post-fertilization (hpf) according to specific criteria outlined by Kimmel et al. 1995. All experiments were carried out in accordance with animal care guidelines provided by the Canadian Council on Animal Care and the University of Ottawa animal care committees approved all protocols.
dlx transgene constructs
The dlx1a/2aIG:GFP transgene construct contains a six kilobase (kb) HindIII-HindIII region from the dlx1a/2a intergenic (IG) region and comprises two potential enhancers I12a and I12b (Ellies et al. 1997; Ghanem et al. 2003). This region was cloned into a modified SP72 vector (Promega, Madison, WI) containing a human β-globin minimal promoter immediately upstream of the green fluorescent protein (GFP) coding sequence. This β-globin-GFP cassette does not, on its own, produce any GFP expression in zebrafish. The URE2dlx1a/2a:GFP transgene construct contains 900 bp of a region 5’ to the dlx1a transcriptional start site (Ghanem et al. 2008). This region was amplified by PCR using the following oligonucleotides: 5’ GCAAAGCACAGAATTATTCT 3’ and 5’ CTTTTAGGGTTTTTGTTCGGA 3’. This 900 bp fragment was cloned into a second modified SP72 vector containing the β-globin:GFP cassette, and Tol2 transposase recognition sites.
Generating and visualization of transgenic zebrafish
Transgenic dlx1a/2aIG:GFP zebrafish lines were generated as described in Amsterdam et al. 1995. The dlx1a/2aIG:GFP construct was injected at 200ng/μl in standard DNA microinjection buffer (0.2 mM KCl, 0.1% phenol red). The URE2dlx1a/2a:GFP construct was co-injected at 50 ng/μl with 50 ng/μl of tol2 transposase mRNA following standard procedures as described by (Fisher et al. 2006). Approximately 200 zebrafish larvae expressing GFP were obtained for each construct and retained as founders. They were intercrossed and at least 100 embryos for each pair were screened for GFP fluorescence at 24-48 hpf. At least two independent transgenic lines were generated for dlx1a/2aIG:GFP and for URE2dlx1a/2a:GFP. In each case, the two transgenic lines obtained for a given construct yielded identical GFP expression patterns. GFP-expressing F1 embryos were raised to adulthood and intercrossed. Screening for GFP positive embryos was done using a Nikon NBZ 1500 dissecting microscope and imaging was done with a Nikon DXM 1200C digital camera.
In situ hybridization
Whole mount mRNA in situ hybridization was done as described in Thisse and Thisse (1998). The antisense mRNA probes were labeled with digoxygenin-11-UTP (Roche, 11277073910) and synthesized from cDNA clones: dlx1a (Ellies et al. 1997), dlx2a (Akimenko et al. 1994), dlx2b (Ellies et al. 1997), dlx5a (Akimenko et al. 1994), dlx6a (Ellies et al. 1997), gad1 (Mueller et al. 2008). The GFP probe was synthesized as complementary to the cDNA fragment that was amplified with the oligonucleotides 5’ AAGGGCGAGGAGCTGTTCAC 3’ and 5’ GAACTCCAGCAGGACCATGT 3’ and cloned into the pDrive vector (Qiagen, Valencia, CA).
Fluorescent RNA in situ hybridization was carried out with a protocol modified from those described previously (Jowett and Yan, 1996; Welten et al., 2006). DNP labeled probes were revealed with tyr-Cy5, digoxigenin labeled probes were revealed using tyr-Cy3, fluorescein labeled probes was revealed with tyr-fluorescein (available from Perkin-Elmer). Our full tissue labeling protocols can be found online: http://wiki.zfin.org/display/prot/Triple+Fluorescent+In+Situ
Imaging of fluorescent in situ hybridizations
Embryos were placed on glass slides and positioned under coverslips for confocal imaging. Confocal z-stacks were obtained by using a Zeiss LSM5 PASCAL (Carl Zeiss, Germany) and excitation lasers were at 488 (Fluorescein), 543 nm (Cy3), 633 nm (Cy5). Channels were acquired sequentially to avoid cross talk between the different filters. Images were processed using LSM Image Manager and Volocity LE software (Improvision).
Immunohistochemisty on zebrafish forebrain sections
Zebrafish embryos were collected and fixed with 4% PFA overnight, washed in PBS, and placed in PBS containing 30% sucrose to equilibrate. Embryos were then embedded in Shandon cryomatrix (Thermo Scientific, 2860051) and frozen at -20 °C. Cryosections were done on a Leica CM1850 (Leica Microsystems, Weltzar, Germany) at a thickness of 10 μm and stored at -20 °C until use. Sections were washed 3X in PBS 0.1% Triton X (PBST) for 10 minutes and blocked for 2 hours in 2% BSA in PBST. The sections were incubated with the primary antibody (see below) overnight in a humid chamber at 4 °C. After removal of the primary antibody with 3 X PBST washes, the appropriate secondary antibody was incubated on the slides for 2 hours at room temperature. After the final washes, the slides were mounted using Aquatex mounting media (VWR, 65036-62). Signals were visualized on a Nikon Eclipse E3600 stereomicroscope for both fluorescent stains. The following primary antibodies were used in this study: Rabbit anti-GFP (1:1000, Invitrogen, A-11122); Mouse anti-glutamic acid decarboxylase (BioMol International, GC3108). The following secondary antibodies were used in this study: Goat anti-rabbit Alexa Fluor488 (1:300, Invitrogen, A11008); Goat anti-mouse Alexa Fluor594 (1:300, Invitrogen, A11005).
Supplementary Material
tel, telencephalon; po, preoptic area; pTh, prethalamus. Scale bar: 100 μm.
B’”) Expression of dlx5a and dlx6a share overlapping domains with gad1 expression on the lateral boundary. C”) gad1 expression is found throught the ventral thalamus at 36hpf. tel, telencephalon; po, preoptic area; pTh, prethalamus. Scale bar: 100 μm.
Acknowledgments
We would like to thank Charles Kimmel for providing the reagents used for fluorescent in situ hybridizations. We would also like to thank Marie-Andrée Akimenko for critical reading of the manuscript. This work was supported by CIHR grant MOP14460 to ME and NIH grant HD22486 to Charles Kimmel, University of Oregon.
References
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: Part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Force A, Yan Y-, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JLR. Interneuron migration from basal forebrain to neocortex: Dependence on dlx genes. Science. 1997a;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JLR. Mutations of the homeobox genes dlx-1 and dlx-2 disrupt the striatal subventricular zone and differentiation of late-born striatal neurons. Neuron. 1997b;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- Anderson S, Mione M, Yun K, Rubenstein JL. Differential origins of neocortical projection and local circuit neurons: Role of dlx genes in neocortical interneuronogenesis. Cereb Cortex. 1999;9:646–654. doi: 10.1093/cercor/9.6.646. [DOI] [PubMed] [Google Scholar]
- Eisenstat DD, Liu J-, Mione M, Zhong W, Yu G, Anderson SA, Ghattas I, Puelles L, Rubenstein JLR. DLX-1, DLX-2 and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Ellies DL, Stock DW, Hatch G, Giroux G, Weiss KM, Ekker M. Relationship between the genomic organization and the overlapping embryonic expression patterns of the zebrafish dlx genes. Genomics. 1997;45:580–590. doi: 10.1006/geno.1997.4978. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Ghanem N, Jarinova O, Amores A, Hatch G, Park BK, Rubenstein JLR, Ekker M. Regulatory roles of conserved intergenic domains in vertebrate dlx bigene clusters. Genome Res. 2003;13:533–543. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem N, Yu M, Long J, Hatch G, Rubenstein JL, Ekker M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J Neurosci. 2007;27:5012–5022. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett T, Yan YL. Double fluorescent in situ hybridization to zebrafish embryos. Trends Genet. 1996;12:387–389. doi: 10.1016/s0168-9525(96)90091-8. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of Embryonic Development of the Zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Liu JK, Ghattas I, Liu S, Chen S, Rubenstein JLR. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev Dyn. 1997;210:498–512. doi: 10.1002/(SICI)1097-0177(199712)210:4<498::AID-AJA12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SC, Heinrich G, Sandell JH. Sequence and Expression of Glutamic Acid Decarboxylase Isoforms in the Developing Zebrafish. J Comp Neurol. 1998;396:253–266. [PubMed] [Google Scholar]
- Mione M, Baldessari D, Deflorian G, Nappo G, Santoriello C. How neuronal migration contributes to the morphogenesis of the CNS: Insights from the zebrafish. Dev Neurosci. 2008;30:65–81. doi: 10.1159/000109853. [DOI] [PubMed] [Google Scholar]
- Mueller T, Wullimann MF. Anatomy of neurogenesis in the early zebrafish brain. Brain Res Dev Brain Res. 2003;140:137–155. doi: 10.1016/s0165-3806(02)00583-7. [DOI] [PubMed] [Google Scholar]
- Mueller T, Vernier P, Wullimann MF. A phylotypic stage in vertebrate brain development: GABA cell patterns in zebrafish compared with mouse. J Comp Neurol. 2006;494:620–634. doi: 10.1002/cne.20824. [DOI] [PubMed] [Google Scholar]
- Mueller T, Wullimann MF, Guo S. Early teleostean basal ganglia development visualized by zebrafish Dlx2a, Lhx6, Lhx7, Tbr2 (eomesa), and GAD67 gene expression. J Comp Neurol. 2008;507:1245–1257. doi: 10.1002/cne.21604. [DOI] [PubMed] [Google Scholar]
- Mueller T, Wullimann MF. An Evolutionary Interpretation of Teleostean Forebrain Anatomy. Brain Behav Evol. 2009;74:30–42. doi: 10.1159/000229011. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JLR. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Potter GB, Petryniak MA, Shevchenko E, McKinsey GL, Ekker M, Rubenstein JLR. Generation of Cre-Transgenic Mice using Dlx1/Dlx2 Enhancers and their Characterization in GABAergic Interneurons. Mol Cell Neurosci. 2009;40:167–186. doi: 10.1016/j.mcn.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint E, Zerucha T, Ekker M. Differential expression of orthologous dlx genes in zebrafish and mice: Implications for the evolution of the dlx homeobox gene family. J Exp Zool (Mol Dev Evol) 2000;288:235–241. doi: 10.1002/1097-010x(20001015)288:3<235::aid-jez4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Stühmer T, Anderson SA, Ekker M, Rubenstein JLR. Ectopic expression of the dlx genes induces glutamic acid decarboxylase and dlx expression. Development. 2002a;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- Stühmer T, Puelles L, Ekker M, Rubenstein JL. Expression from a dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb Cortex. 2002b;12:75–85. doi: 10.1093/cercor/12.1.75. [DOI] [PubMed] [Google Scholar]
- Sumiyama K, Irvine SQ, Stock DW, Weiss KM, Kawasaki K, Shimuzu N, Shashikant CS, Miller W, Ruddle FH. Genomic structure and functional control of the Dlx3-7 bigene cluster. Proc Natl Acad Sci USA. 2002;99:780–785. doi: 10.1073/pnas.012584999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High resolution whole-mount in situ hybridization. Zebrafish science monitor. 1998;5:8–9. [Google Scholar]
- Welten MC, de Haan SB, van den Boogert N, Noordermeer JN, Lamers GE, Spaink HP, Meijer AH, Verbeek FJ. ZebraFISH: Fluorescent in situ hybridization protocol and three-dimensional imaging of gene expression patterns. Zebrafish. 2006;3:465–476. doi: 10.1089/zeb.2006.3.465. [DOI] [PubMed] [Google Scholar]
- Westerfield M. A guide for the laboratory use of zebrafish (danio rerio) 4. Eugene, Oregon: University of Oregon Press; 2000. The zebrafish book. [Google Scholar]
- Wullimann MF, Mueller T. Expression of zash-1a in the postembryonic zebrafish brain allows comparison to mouse Mash1 domains. Brain Res Gene Expr Patterns. 2002;1:187–192. doi: 10.1016/s1567-133x(02)00016-9. [DOI] [PubMed] [Google Scholar]
- Wullimann MF, Mueller T. Teleostean and Mammalian Forebrains Contrasted: Evidence from Genes to Behavior. J Comp Neurol. 2004;475:143–162. doi: 10.1002/cne.20183. [DOI] [PubMed] [Google Scholar]
- Wullimann M. Secondary neurogenesis and telencephalic organization in zebrafish and mice: A brief review. Integrative Zoology. 2009;4:123–133. doi: 10.1111/j.1749-4877.2008.00140.x. [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the Notch Signaling by Mash1 and Dlx1/2 Regulates Sequential Specification and Differentiation of Progenitor Cell Types in the Subcortical Telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Zerucha T, Stühmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JLR, Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QP, Le TN, Qiu X, Spencer V, de Melo J, Du G, Plews M, Fonseca M, Sun JM, Davie JR, Eisenstat DD. Identification of a Direct Dlx Homeodomain Target in the Developing Mouse Forebrain and Retina by Optimization of Chromatin Immunoprecipitation. Nucleic Acids Res. 2004;32:884–892. doi: 10.1093/nar/gkh233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
tel, telencephalon; po, preoptic area; pTh, prethalamus. Scale bar: 100 μm.
B’”) Expression of dlx5a and dlx6a share overlapping domains with gad1 expression on the lateral boundary. C”) gad1 expression is found throught the ventral thalamus at 36hpf. tel, telencephalon; po, preoptic area; pTh, prethalamus. Scale bar: 100 μm.