Abstract
The LMO2 gene encodes a LIM-only protein and is a target of chromosomal translocations in human T-cell leukemia. Recently, two X-SCID patients treated by gene therapy to rescue T-cell lymphopoiesis developed T-cell leukemias with retroviral insertion into the LMO2 gene causing clonal T-cell proliferation. In view of the specificity of LMO2 in T-cell tumorigenesis, we investigated a possible role for Lmo2 in T-lymphopoiesis, using conditional knockout of mouse Lmo2 with loxP-flanked Lmo2 and Cre recombinase alleles driven by the promoters of the lymphoid-specific genes Rag1, CD19, and Lck. While efficient deletion of Lmo2 was observed, even in the earliest detectable lymphoid cell progenitors of the bone marrow, there was no disturbance of lymphopoiesis in either T- or B-cell lineages, and in contrast to Lmo2 transgenic mice, there were normal distributions of CD4− CD8− thymocytes. We conclude that there is no mandatory role for LMO2 in lymphoid development, implying that its specific role in T-cell tumorigenesis results from a reprogramming of gene expression after enforced expression in T-cell precursors.
Cancer formation is frequently the result of alteration of the normal cellular functions of proto-oncogenes (10). The T-cell oncogene LMO2 was first identified as a target of chromosomal translocations in human T-cell acute leukemias (2, 26). The gene encodes a LIM domain-only protein comprising two LIM domains (24) whose function is in protein interaction, binding to proteins such as TAL1/SCL and LDB1 in DNA-binding complexes that bind distinct bipartite target sites in normal hematopoietic (31) or T-cell tumor cells (7).
A remarkable feature of the LMO2 gene is its involvement exclusively in T-cell malignancies when abnormally expressed (24). There are three lines of evidence supporting this contention. First, chromosomal translocations activating LMO2 have only been observed in T-cell tumors. Indeed, a case of human T-cell acute leukemia has been described with both the Philadelphia chromosome (resulting in BCR-ABL fusion), normally the hallmark of myelogenous leukemia, and the LMO2-specific translocation t(11;14)(p13;q11) (6). Further, transgenic mice expressing Lmo2 in the lymphoid lineage or with overexpression in all tissues (16, 18, 22, 23) only developed T-cell malignancies, indicating that LMO2-mediated tumorigenesis is specific to the T-cell lineage. Finally, an unfortunate outcome of a recent gene therapy trial with retrovirally delivered interleukin-2 receptor γc subunit in X-linked severe combined immunodeficiency (X-SCID) patients (8) was the development of T-cell leukemia in two cases, following specific clonal expansion of T cells with retroviral insertion in the LMO2 gene (9, 9a). The specific outcome of T-cell leukemia in both these X-SCID patients, despite the use in the clinical protocol of the retroviral transduction of CD34-positive bone marrow progenitors, suggests that LMO2 may influence T-cell differentiation.
Additional evidence for a such role in T-cell lymphopoiesis comes from the finding that Lmo2 is normally expressed in immature CD4/CD8 double-negative thymocytes, before being downregulated as T cells mature (13). Moreover, transgenic mice with enforced Lmo2 expression show a differentiation block at the same CD4/CD8 double-negative thymocyte stage, preceding the appearance of clonal T-cell tumors (17, 18).
Gene targeting experiments have addressed the question of a putative role for Lmo2 in hematopoiesis (32, 35) but have been unable to answer whether Lmo2 plays a role in T-cell development. Null mutation of Lmo2 in mice causes embryonic lethality at around 10 days after inception, due to a failure of embryonic erythropoiesis (32). Thus, effects on lymphoid development could not be investigated. Similarly, the use of Lmo2 null embryonic stem cells to make chimeric mice showed that there was no embryonic stem cell-derived contribution to adult hematopoiesis in these animals (35), indicating that Lmo2 functions in the stem cells (i.e., the equivalent of the repopulating cells used in the X-SCID gene therapy) or their precursors. In both gene targeting strategies, any possible role for Lmo2 in lymphopoiesis could not be determined because the null cells fail to contribute to hematopoietic subcompartments.
An approach to assess a role for Lmo2 in lymphopoiesis is to use a conditional knockout strategy in which the Lmo2 gene is deleted in specific cells in the hematopoietic lineage. We used a deletional strategy based on flanking the Lmo2 gene with Cre recombinase recognition sites (loxP sites) and expressing Cre under the control of lymphoid-specific promoters (Rag1, CD19, or Lck). No changes in lymphoid development were found in the absence of the Lmo2 gene with this approach. Thus, despite its being a T-cell-specific oncogene, functional Lmo2 is not required for normal T-cell development.
MATERIALS AND METHODS
Conditional targeting of Lmo2.
Gene targeting was carried out with embryonic stem genomic DNA clones and homologous recombination after transfection into the CCB embryonic stem cell line as described (32, 35). The floxed Lmo2 allele in embryonic stem cells was made in two stages. In the first step, a targeting vector (pLmo2pLXP) was constructed which comprised the DNA extending from the 5′ end of exon 6 (which had an artificial XhoI site derived from a λ2001 genomic Lmo2 clone) to a PstI site ≈300 bp on the 5′ side of the most 3′ EcoRI site (Fig. 5A). pLmo2pLXP was assembled by cloning the XhoI-PstI fragment into the XhoI and XbaI sites of pBluescript after filling in the PstI and XbaI sites. A loxP-puromycin cassette (3) was cloned (BglII fragment with filled-in ends) into the unique XbaI site of the pBluescript subclone, after filling in the XbaI site. The MC1-tk fragment was added at the XhoI site to complete the pLmo2pLXP construction.
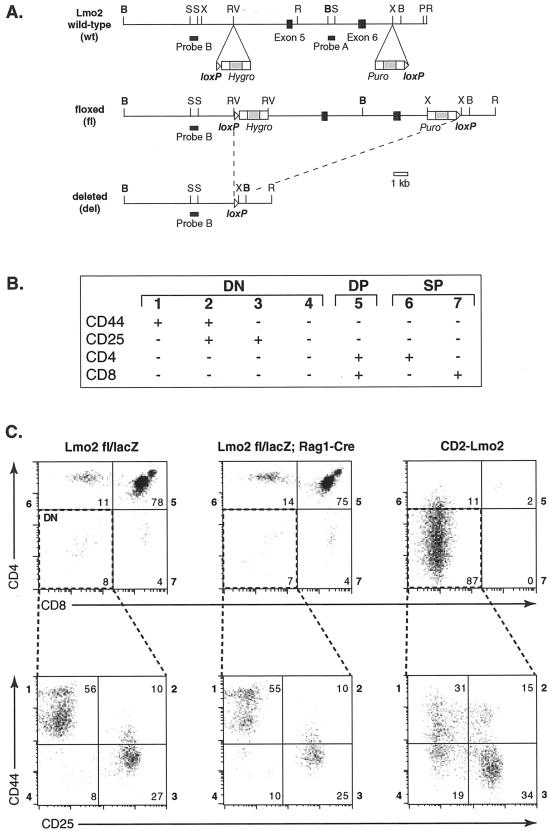
FIG. 5.
Conditional deletion of the Lmo2 gene has no effect on T-cell development. A floxed Lmo2 allele was made in embryonic stem cells with homologous recombination to add loxP sites flanking exons 5 and 6 of Lmo2 (exons encoding most of the Lmo2 protein). Mice were made with the floxed Lmo2 allele (Lmo2fl), and compound genotype individuals were generated carrying Lmo2fl on one chromosome and a null Lmo2allele on the other (Lmo2lacZ) and with a Rag1-Cre allele. Thymus cells were isolated from these mice at 8 to 16 weeks, and the surface marker profiles were examined by flow cytometry and compared with Lmo2fl;Lmo2lacZ littermates and with CD2-Lmo2 transgenic mice. (A) Restriction map of the floxed Lmo2 locus. A partial restriction map of the genomic region of Lmo2 is depicted in the top line, indicating the positions of Lmo2 exons 5 and 6 and those of two probes used to detect targeting events in the embryonic stem cells and subsequently in genomic DNA of mouse tissues. A loxP-hygromycin resistance marker cassette (3) was introduced at the EcoRV site in intron 4, and a loxP-puromycin resistance marker cassette (3) was introduced at the XbaI site 3′ of the last coding exon 6. The organization of the floxed Lmo2 allele is shown in the middle line. After Cre-mediated deletion through the loxP sites, a deleted allele is made, from which the two selectable markers are deleted. Restriction sites are B, BamHI; P, PstI; R, EcoRI; RV, EcoRV; S, SacI; and X, XbaI. (B) A representation of the T-cell development stages. DN, DP, and SP represent the CD4/CD8 status of the different stages, where DN = CD4 and CD8 negative. DP = CD4 and CD8 positive, SP6 = CD4 positive (CD4+ CD8−), and SP7 = CD8 positive (CD4− CD8+). (C) Thymocytes were prepared from mice of the indicated genotypes and labeled with fluorescent antibodies specific to the indicated T-cell markers. In the top panels, the distribution of CD4 and CD8 markers is shown. The quadrants representing T-cell stages 5 to 7 are indicated by the side of each quadrant and percent for each T-cell subpopulation within the quadrant. The double-negative T cells are in the bottom left quadrant in each case. For the lower panels, the double CD4- and CD8-negative cells were analyzed for CD44 and CD25 marker expression, to subdivide them into DN1 to DN4. For this analysis, the CD4+ CD8+ were first depleted with antibody-coated Dynabeads, resulting in CD4− CD8− double-negative thymocytes with purity of 98%, 96%, and 98% for Lmo2fl/lacZ, Lmo2fl/lacZ;Rag1-Cre, and CD2-Lmo2 mice, respectively. The double-negative T-cell stages are shown in bold alongside the FACS quadrants to which they are ascribed.
Embryonic stem cells were transfected with pLmo2pLXP, and puromycin-resistant, ganciclovir-sensitive clones were selected. Gene targeted events were identified with genomic DNA digested with XbaI and filter hybridized by standard methods as described with the Lmo2 genomic probe A (32). The targeting event was verified with a 300-bp PstI-EcoRI fragment (probe D) outside the 3′ side of the targeting vector with SphI digests of genomic DNA. This identified targeted embryonic stem clones with a loxP site downstream of Lmo2 exon 6. A verified, targeted embryonic stem clone (KO3PU/D3) was transfected in a second step to add a loxP site upstream of exon 5 to create the floxed Lmo2 allele.
A targeting vector (pKO73hyg) was constructed with the XbaI-EcoRI fragment spanning exon 5 of Lmo2 (see Fig. 5A) from a λ2001 clone isolated from an embryonic stem cell DNA-derived genomic library (32). This fragment was cloned into the XbaI and EcoRV sites of pBluescript (after filling in the EcoRV site of the genomic fragment) and a loxP-hygromycin cassette (3) was cloned into the unique EcoRV genomic site. MC1-tk (29) was cloned into the XbaI site to give the pKO73hyg targeting clone.
The embryonic stem clone KO3PU/D3 was transfected with pKO73hyg, and hygromycin-resistant, ganciclovir-sensitive clones were selected. Homologous recombination targeted cells were identified with genomic DNA digested with SacI, amplified by PCR, and including an nuclear localisation signal, an SV40 polyadenylation signal and MC1-neo-polyA (29), was cloned into the BamHI site to yield an in-frame fusion of the N-terminal three amino acids of Rag1 with Cre (Met-Ala-Gly-Cre).
Lck-Cre was made by PCR amplifying the whole Cre cDNA sequence with primers including terminal NcoI restrictions sites; the PCR product was cleaved with NcoI, the ends filled in and cloned into the blunt-ended BamHI site of the Lck transgenic expression clone (33). The transgenic fragment was excised with NotI and injected into pronuclei of fertilised eggs and transgenic strains were developed. One line in which detectable Cre protein was found was selected for further use.
The CD19-Cre targeting construct (a gift from Jurgen Roes and Klaus Rajewsky) has been described before (25). The plasmid was transfected into CCB embryonic stem cells, and targeted clones were identified by hybridization of BamHI digested DNA with a 0.9-=kb BamHI-PstI probe located about 2 kb 5′ of CD19 exon 1. Targeted embryonic stem clones were injected into C57BL6 blastocysts and chimeric mice bred to give germ line transmission of the CD19-Cre targeted allele.
Filter hybridization.
Genomic DNA was prepared from mouse tissues and 10 μg amounts were digested to completion with BamHI, separated on 0.8% agarose gels and transferred to nylon membranes after denaturation (19, 28). The membranes were hybridized with the Lmo2 genomic probes A or B (32) with random labeling (4) with [32P]dCTP.
Genomic PCR.
For genomic PCR of hematopoietic cells, 1000 cells were sorted with a MoFlo cell sorter (Dako Cytomation), directly into PCR tubes and analyzed with the following primers specific for Lmo2 genomic sequences; Primers for detection of deleted Lmo2 A5, 5′-GATGAATTCGTGTCTCTTTCTCCTGCAACTA-3′ A3, 5′-GATGGATCCAGGACGGTGGTTTGAGAACTGTTG-3′ Primers for detection of Lmo2 exon 5 B5, 5′-GTGGATGAGGTGCTGCAGATA-3′ B3, 5′-TCCGTCCCAGCTTGTAGTAGAG-3′.
PCR conditions used were 95°C for 2 min, followed by 30 cycles at 95°C for 30 s, 63°C for 30 secs and 72°C for 1 min and a final incubation at 72°C for 10 min. For multiplex PCR, it was necessary to add primers A5 and A3 in 4x excess to B5 and B3 to obtain equivalent amplifications for the deleted and undeleted Lmo2 alleles, respectively. A probe for detecting the deleted PCR product was made by amplification of a fully internal, 106bp fragment from the product of B5 plus B3 with primers KO73P plus KO73BN. The sequences of these primers were KO73P, 5′-GGAGATCTGACTGCTTAAGGTGGC-3′, and KO73BN, 5′-AGTAGAGCAAACATTCCCTCTGAG-3′.
Western detection of proteins.
Single cell suspensions were lysed by boiling in Laemmli sample buffer (0.25 M Tris-HCl, pH 6.8, 5% SDS, 10% glycerol, 100 mM dithiothreitol, 0.05% bromophenol blue) and protein equivalent to 106 cells was fractionated by 4 to 20% Tris-glycine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride filter. The filter was washed for 1 h in TBS-T (Tris-buffered saline with 0.1% Tween-20) with 4% powdered skimmed milk at room temperature before 1 h of incubation with a 1:1,000 dilution of anti-Cre (Novagen). The filter was washed six times with TBS-T, incubated with a 1:5000 dilution of horseradish peroxidase conjugated anti-rabbit antibody (Amersham) and washed six times with TBS-T. The filter was subjected to enhanced chemiluminescence analysis (Amersham) according to the manufacturer's recommendations.
Flow cytometry.
Single cell suspensions were prepared from the indicated tissues and preblocked with purified anti-CD16 (Fc-block); 106 cells were placed in FACS tubes and stained with phycoerythrin (PE)-conjugated anti-CD4, fluorine isothiocyanate (FITC)-conjugated anti-CD8, PE-anti-T-cell receptor β subunit, PE-anti-CD2, PE-anti-CD3, PE-anti-Thy 1.2, PE-anti-B220, FITC-anti-IgM, biotinylated anti-IgG1, PE-anti-Ter119, PE-anti-Mac1, PE-anti-Sca1, PE-anti-CD19 and FITC-anti-CD34 antibodies. CD4-CD8 double-negative thymocytes were prepared as described below and incubated with PE-anti-CD44, FITC-anti-CD25, and FITC-anti-T-cell receptor γ subunit antibodies.
Bone marrow B-cell development was monitored according to the protocol of Hardy et al. (11, 12); 107 cells were immunostained with allophytocyanin (APC)-conjugated anti-B220 and biotinylated anti-CD43 antibodies, combined with FITC-anti-CD24 and PE-anti-Ly51/BP-1 for stages A to C or FITC-anti-IgM and PE-anti-IgD for stages D to F. Biotinylated antibodies were detected by subsequent staining with a streptavidin-cytochrome conjugate. All antibodies were from BD Pharmingen. Cells were analyzed with a FACScan flow cytometer (BD Immunocytometry Systems) with dual color excitation when detecting APC-conjugated antibodies, or sorted with a MoFlo cell sorter (Dako Cytomation).
Cell selection with Dynabeads.
To perform cell enrichments or depletions, 5 × 107 cells were suspended in 1 ml of PBS containing 1% fetal bovine serum, incubated with 25 μl (107) each of CD4-, CD8-, or B220-linked Dynabeads (Dynal Biotech), or combinations, for 20 min at 4°C. Target cells were isolated on a magnetic particle concentrator, and the procedure repeated on the supernatant. In the case of positive selections, and bound cells were washed three times in the above medium before use.
RESULTS
Conditional gene deletion in lymphopoiesis.
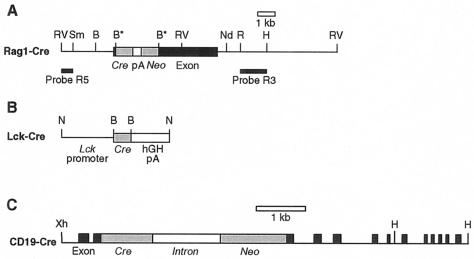
The Cre-loxP recombination system is an ideal tool for tissue specific gene deletion. Homologous recombination is used to add loxP sites flanking the gene of interest in mouse embryonic stem cells and later mice are generated which contain the floxed gene and express the Cre recombinase in specific cells. To facilitate lymphoid-specific deletion we used the promoters of three lymphoid specific genes, viz., Rag1, which is expressed early in lymphopoiesis and during development of both T and B-cell lineages, Lck which is expressed specifically in T cells and the B-cell gene CD19. We generated alleles of Rag1 and Lck controlling Cre expression, as well as utilizing a previously described CD19-Cre allele (25) (Fig. 1 A to C). In the case of the Rag1-Cre and CD19-Cre (25), homologous recombination was used to knock-in the Cre gene into the respective genes in embryonic stem cells, followed by generation of mice carrying the allele in their germ line (Fig. 1). An Lck promoter fragment (33) was used in the case of Lck-Cre to generate transgenic mice (see Materials and Methods).
FIG. 1.
Generation of cell-specific Cre expressing mouse lines. (A) Rag1-Cre was made by gene targeting knock-in of Cre into the Rag1 exon. A Rag1 targeting vector was constructed with a SmaI-NdeI fragment, encompassing the Rag1 coding exon, A BamHI site was created in exon 1 of Rag1 by mutation (B*), and the Cre cDNA was cloned in-frame with the Rag1 coding sequence. The simian virus 40 (SV40) polyadenylation site was cloned 3′ of Cre, and the neomycin resistance cassette MC1-neo-pA (Neo) was cloned downstream of this. This construct was used to target an endogenous Rag1 allele by homologous recombination to make embryonic stem cells with one targeted Rag1 allele. Mice were made from these embryonic stem cells with germ line transmission of the Rag1-Cre allele. These mice showed no variation in hematopoiesis, and homozygous Rag1-Cre mice failed to develop lymphocytes, as previously found for Rag1 knockout mice (21). (B) Generation of the Lck-Cre transgene. A Cre cDNA fragment was cloned into the unique BamHI site of a Lck promoter cassette (33).,(C) CD19-Cre comprises a knock-in of Cre into exon 2 of the mouse CD19 gene (25). Restriction sites are B, BamHI; H, HindIII; Nd, NdeI; N, NotI; R, EcoRI; RV, EcoRV; Sm, SmaI; S, SacI; X, XbaI; and Xh, XhoI.
The specificity of the three Cre lines was established with the Lmo2fl locus as a reporter in mice. A conditional null Lmo2 allele was made in embryonic stem cells with homologous recombination to add loxP sites within introns flanking the bulk of the coding region (exons 5 and 6, encoding codon 15 to the end of Lmo2) (Fig. 5A). Transient expression of Cre recombinase in these floxed embryonic stem cells (designated Lmo2fl) caused deletion in the Lmo2 coding region, as judged by a PCR based assay (data not shown). Mice were generated with the floxed Lmo2 allele and were bred to homozygosity, and maintained as such, with no detectable phenotypic alteration. These mice were used as a source of conditional null Lmo2 and were crossed with mice carrying a constitutive null Lmo2 allele, in which the lacZ gene had been knocked into Lmo2 exon 5 (34) (Lmo2lacZ), to give mice designated Lmo2fl/lacZ. In this line of mice, the Lmo2fl/lacZ genotype produces nophenotypic change (i.e., these mice are heterozygous Lmo2 null) but cells become homozygous Lmo2 null when Cre-mediated deletion of the Lmo2fl allele occurs. Thus, when Cre expression is restricted to any particular cell type, such as hematopoietic subsets, consequences of Lmo2 null mutation for those specific cells (and their descendants) can be monitored.
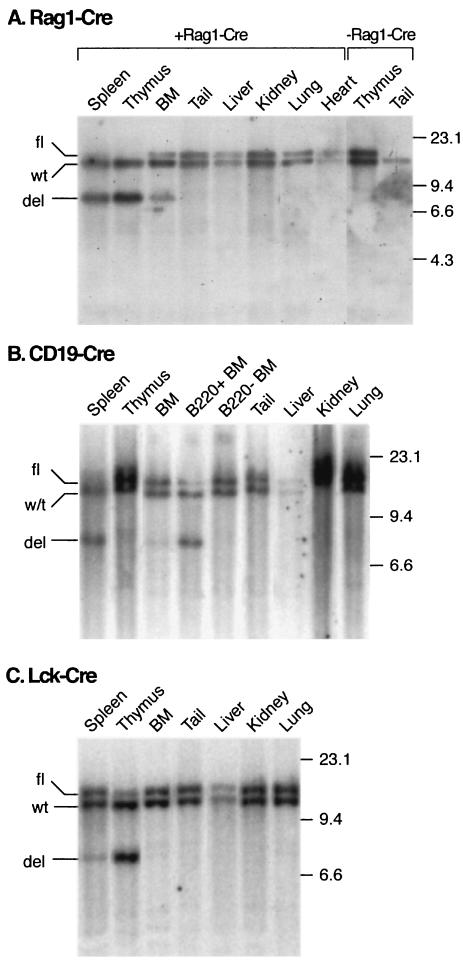
Lmo2fl;Cre mice were generated for each lymphoid Cre expressing line and initially whole tissue DNA genomic filter hybridization was used to compare Lmo2 gene deletion in hematopoietic tissues (i.e., spleen, thymus, or bone marrow). This showed that Lmo2fl deletion was specifically confined to the target lymphopoietic organs (Fig. 2). The presence of the Rag1-Cre allele caused essentially complete loss of the Lmo2fl allele in the spleen and thymus giving rise to a deleted allele (termed Lmo2del) but only partial deletion in bone marrow, implying that the Rag1-Cre allele causes complete deletion of Lmo2fl in developing lymphoid cells (Fig. 2A). The CD19-Cre allele caused significant deletion in spleen, but very little in thymus and bone marrow, consistent with activity in B cells (25). Indeed, purified B220+ B cells from bone marrow showed ≈80% deletion of the floxed Lmo2 allele (Fig. 2B). Conversely, the Lck-Cre allele caused ≈90% deletion in thymus but little deletion in bone marrow, implying that Cre function is restricted to T cells.
FIG. 2.
Detection of Lmo2 deletion in mouse tissues. Mice which were heterozygous for the floxed Lmo2 allele (Lmo2fl/wt mice) and which carried one of the three Cre alleles were euthanized, and genomic DNA was prepared from the indicated tissues. The DNA was digested with BamHI, fractionated on agarose gels, immobilized on nylon and hybridized with Lmo2 probe B (32) (Fig. 5A). Sizes of hybridizing bands were estimated by comigration of λ DNA cleaved with HindIII (shown in kilobases). The hybridizing bands are indicated corresponding to the wild-type Lmo2 allele (wt), the floxed Lmo2 allele (fl), and the deleted allele (del). (A) Lmo2fl/wt;Rag1-Cre mice. DNA from various tissues of Lmo2fl/wt;Rag1-Cre and Lmo2fl/wt mice were examined. DNA from tail biopsy material serves as a control for detection of the wild type (wt) allele and the targeted (i.e., floxed) allele. Rearrangements (deletions, del) were restricted to hematopoietic organs spleen, bone marrow (BM), and thymus. (B) Lmo2fl/wt;CD19-Cre mice. DNAs from various tissues of Lmo2fl/wt;CD19-Crewere analyzed together with DNA prepared from purified B220+ cells selected from bone marrow with anti-B220 Dynabeads (B220+ bone marrow). B220− bone marrow corresponds to DNA prepared from B220-depleted cells. (C) Lmo2fl/wt;Lck-Cre mice. DNAs from various tissues of Lmo2fl/wt;Lck-Cre mice were analyzed as shown. Clear evidence of rearrangements (deletions, del) is restricted to thymus and spleen DNA.
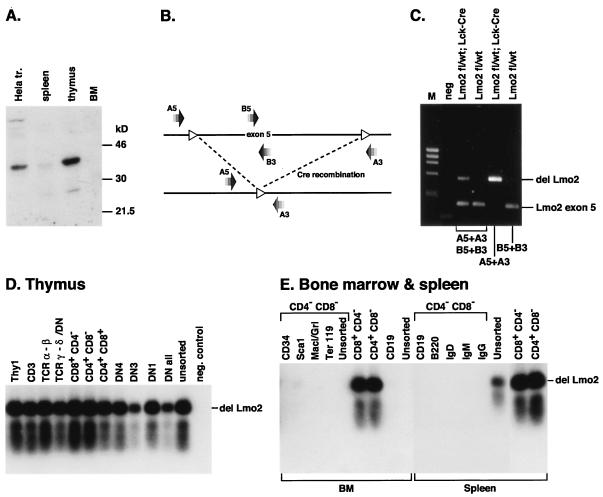
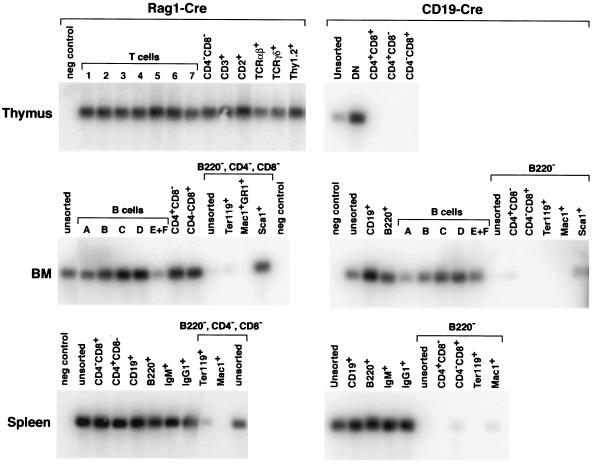
More detailed analyses of the cell specificity of the Cre expression was carried out with PCR based approaches (Fig. 3 and 4). A multiplex genomic PCR assay was developed to allow detection of the undeleted and deleted forms of Lmo2 by designing primers which either amplify an Lmo2 exon 5 fragment (from undeleted Lmo2) or the fused intronic sequences after deletion of the floxed Lmo2 allele (Fig. 3, B and C). Specific subsets of cells were selected based on surface marker expression, with a fluorescence activated cell sorter, from thymus (Fig. 3, D) or bone marrow and spleen (Fig. 3, E) of Lmo2fl;Lck-Cre mice and subjected to the PCR analysis. This showed Lmo2fl gene deletion in T cells of all maturities but no detectable deletion in B cells (B220, CD19, Ig+), myeloid cells (Mac1/Gr1+), erythroid (Ter119+) or progenitor cells (CD34+ or Sca1−), corroborating the T-cell specify of the Lck-Cre allele.
FIG. 3.
Specificity of Cre recombinase expression in Lck-Cre mice. Transgenic Lck-Cre mice were bred with floxed Lmo2 mice, and deletion of the floxed allele was used as a reporter for Cre activity. Protein or DNA was isolated from the various sources indicated for Western detection analysis of Cre protein expression or genomic PCR analysis of Lmo2 deletion due to Cre activity. (A) Western blot analysis of Cre recombinase protein. Whole cell lysates from the indicated tissues of an Lck-Cre mouse or from HeLa cells transfected with a Cre expression construct (pGK-Cre, a gift from A. McKenzie) (30) (HeLa tr.), were prepared and subjected to Western detection analysis as described in Materials and Methods. BM, bone marrow. (B) Diagrammatic representation of Lmo2 PCR primer locations (see also Materials and Methods) with respect to the Lmo2 gene. Primers A5 plus A3 flank the loxP sites in the floxed Lmo2 allele and do not yield a PCR product from the intact floxed Lmo2 allele but do so when a deletion has occurred as the primers are brought into proximity of each other, yielding a 500-bp PCR fragment. Primers B5 and B3 are located in Lmo2 exon 5 and give rise to a 184-bp PCR fragment from intact Lmo2 (germ line or floxed). (C) Multiple PCR detection of Lmo2 and deleted Lmo2 alleles. Genomic DNA was prepared from the thymus of the indicated mice and used for PCR analysis with the indicated primers. PCR products were fractionated on 1.3% agarose gels and stained with ethidium bromide for photography. Lane M, φX174 DNA markers cut with HaeIII. (D and E) PCR analysis of genomic DNA from different subsets from thymus (D) and bone marrow or spleen (E) of an Lmo2fl/wt;Lck-Cre mouse. One thousand cells were sorted according to surface marker expression by FACS and directly used for multiplex PCR analysis, with primers A5, A3, B5, and B3. In some cases, to reduce potential background amplification from contaminating cells, cell populations were depleted with Dynabeads prior to sorting with specific antibody (CD4− CD8− bone marrow and spleen). DN represents CD4− CD8− double-negative thymocytes, and DN1, DN2, and DN3 are subsets of double-negative T cells defined by expression of CD44 and/or CD25 (see Fig. 5). PCR products were fractionated on agarose, transferred to nylon, and hybridized with the probe internal to the Lmo2del-specific PCR product of primers A5 and A3. As an internal control for amplification, all PCRs also gave rise to the Lmo2 exon 5 product as in C (data not shown).
FIG. 4.
Cell type specificity of Cre expression in Rag1-Cre and CD19-Cre mice. CD19-Cre or Rag1-Cre mice were bred with floxed Lmo2 mice to use the deletion of the floxed allele as a measure of Cre activity; 1,000 cells from thymus (top), bone marrow (BM, middle), or spleen (bottom) were obtained with the surface markers indicated and used directly for PCR analysis. The right-hand panels show data from Lmo2fl/wt;CD19-Cre mice, and the left-hand panels show data from the Lmo2fl/wt;Rag1-Cre mice. PCRs were performed with the same approach as in Fig. 3, and the products hybridized with the probe specific for the Lmo2del PCR product. B220-, CD4-, CD8-, or B220-depleted populations were used for FACS sorting from bone marrow or spleen where indicated. T-cell stages are labeled numerically, and B-cell stages are labeled alphabetically (12). As an internal control for amplification, all PCRs gave rise to the Lmo2 exon 2 PCR product as in Fig. 3C.
A similar analysis was performed on Lmo2fl/wt mice bearing either Rag1-Cre or CD19-Cre alleles (Fig. 4). In the case of Rag1-Cre, Lmo2del-derived PCR products were amplified from all stages of T cells (5) sorted from the thymus, from sorted spleen and bone marrow-derived mature T cells and from all B-cell stages (11) sorted from spleen and bone marrow (Fig. 4), whereas Ter119+ or Mac1/Gr1+ cells had little or no evidence of Cre activity. While some level of Lmo2 deletion was observed in Sca1+ cells, neither myeloid (Mac1+) nor erythroid (Ter119+) cells showed this deletion, hence this result presumably reflects the fact that Rag1 and Sca1 are coexpressed in the earliest detectable lymphoid progenitor cells (14). These data are consistent with the lymphoid specificity of the Rag1-Cre allele. The B-cell specificity of the CD19-Cre (25) was confirmed, as Lmo2del products were derived from all B-cell subsets of the bone marrow and spleen (Fig. 4). When these tissues were depleted of B cells, residual myeloid (Mac1/Gr1+), erythroid (Ter119+) and T cells (CD4 and/or CD8+) did not exhibit deletion of the floxed Lmo2.
Lmo2 has no mandatory role in lymphoid development.
The consequence of Lmo2 deletion in lymphoid cells was analyzed with Lmo2fl/lacZ mice crossed with one of the three Cre expressing strains. This strategy for Lmo2 gene deletion obviates any problem which might arise from inefficient Cre-mediated recombination since mice with one null Lmo2 allele (Lmo2lacZ) and one floxed Lmo2 allele (Lmo2fl) were used. Thus, all cells in which the floxed allele is deleted become homozygous for Lmo2 null mutation. Lmo2fl/lacZ;Cre mice were viable and exhibited no obvious hematopoietic defects, with normal bone marrow, spleen and thymus cell counts (data not shown). The possible importance of Lmo2 in T lymphopoiesis was examined by partitioning thymic T cells from Lmo2fl/lacZ;Rag1-Cre mice into various developmental stages with flow cytometry (illustrated in Fig. 5B, stages 1 to 7).
No significant alteration in T-cell stages 5, 6, or 7 was found in Lmo2fl/lacZ;Rag1-Cre mice compared with Lmo2fl/lacZ littermates (Fig. 5C). When the CD4− CD8− double-negative T cells were further analyzed for CD44 and CD25 expression, subfractionating the cells into DN1 to DN4 subsets, we could not detect any significant shift in the four T-cell populations (Fig. 5C, bottom). By contrast, age-matched CD2-Lmo2 transgenic mice, which have T-cell expression of Lmo2, showed an increase in CD4− CD8− double-negative thymocytes (17, 18), which display a predominant arrest at the stage DN3 of development as previously seen (7) (Fig. 5C). Thus, there is no detectable effect of Lmo2 gene deletion on T-cell development, while, paradoxically, enforced expression of Lmo2 in the T-cell lineage causes a profound developmental abnormality.
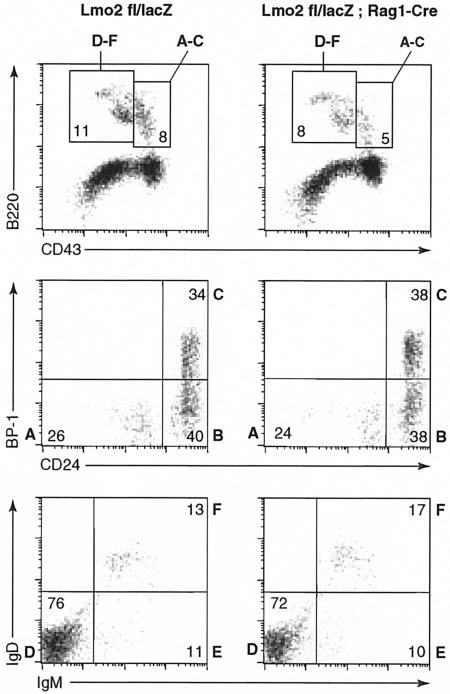
An analogous assessment of B-cell development was conducted on bone marrow-derived B cells (Fig. 6), as this is the site of B-cell lymphopoiesis in the adult mouse. When these were separated into B-cell subpopulations, by four-color immunocytometry with the established protocol (11), we found that the six B-cell stages of increasing maturity (Fig. 6A to F) were not significantly altered between Lmo2fl/lacZ;Rag1-Cre mice and Lmo2fl/lacZ littermates. As the Rag1 gene is expressed at early stages of both T and B lymphoid development, the lack of detectable effect on B lymphopoiesis argues that the presence of Lmo2 protein is not necessary for B lymphopoiesis to proceed to normally.
FIG. 6.
Normal B-cell development occurs in the absence of Lmo2. Bone marrow cells were prepared from Lmo2fl/lacZ and Lmo2fl/lacZ;Rag1-Cre littermates, and B cells were separated into six stages of development by four-color immunocytometry with antibodies binding to B220, CD43, Ly51/BP-1, CD24 (HSA), BP-1, IgM, and IgD (11, 12). B-cell stages are shown in bold alongside the quadrants to which they are ascribed, and the cells in each subpopulation are indicated in each quadrant. The top panels show un-gated B220 and CD43 double fluorescence profiles with the location of the six B-cell stages (A to F, ranging from most immature to most mature IgM, IgD double positive). The separate stages are shown as gated subpopulations in middle (A to C) and lower panels (D to F).
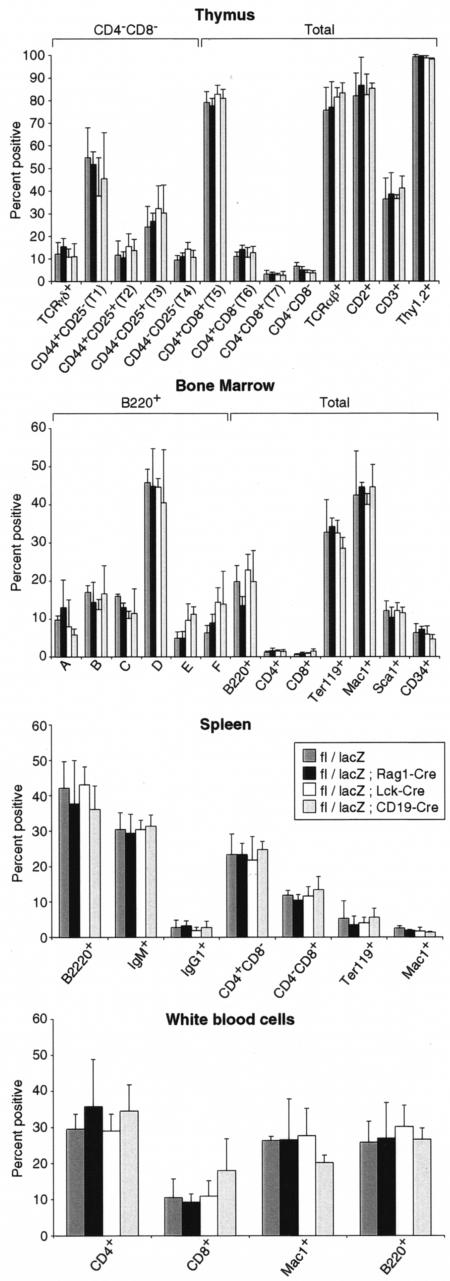
This analysis was taken further by comparing the proportions of hematopoietic cells with various surface marker phenotypes in mice with and without Lmo2 deletion. Lmo2fl/lacZ mice were produced containing either Rag1-Cre, CD19-Cre, or Lck-Cre alleles and various hematopoietic populations in thymus, spleen, bone marrow, or blood were compared with mice lacking Cre (Fig. 7). No significant alteration was observed in thymus, spleen, bone marrow, or white blood cell FACS profiles between the groups. These results argue against a compensatory molecule(s) present at specific stages of T- or B-cell development that could replace the functionality normally provided by Lmo2. The family of LIM-only proteins comprises four known members (24) whose sequence dissimilarities and expression patterns argue against their substituting for Lmo2.
FIG. 7.
Cumulative flow cytometric data of hematopoietic cells in mice lacking lymphoid Lmo2 expression. Hematopoietic cells from Lmo2fl/lacZ mice or Lmo2fl/lacZ mice carrying a Cre allele were prepared from thymus, bone marrow, spleen, or peripheral blood and analyzedby flow cytometry with antibodies against the indicated surface markers. The percentage of cells which were positive for each marker in each population are shown as the mean data from four mice in each group. Error bars denote one standard deviation of the mean.
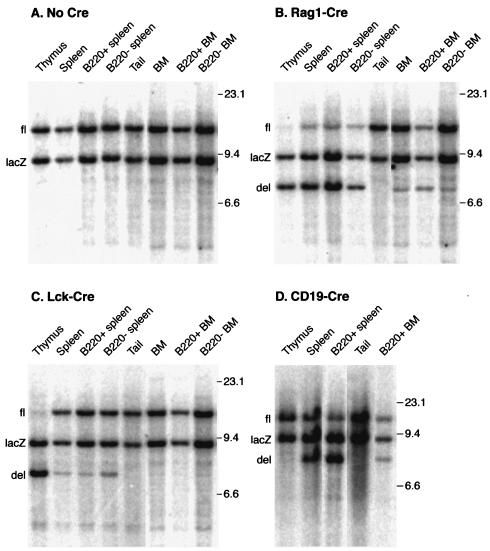
We assessed the extent of Lmo2 deletion in lymphoid cells of Lmo2fl/lacZ;Cre mice by Southern blotting (Fig. 8). Two hybridizing bands are found in Lmo2fl/lacZ DNA, corresponding to floxed (Fig. 8A, fl) and null (Fig. 8A, lacZ) Lmo2 alleles. Greater than 90% deletion was consistently found in thymus DNA of Lmo2fl/lacZ;Rag1-Cre and Lmo2fl/lacZ;Lck-Cre mice (Fig. 8B, C). The efficiency of Lmo2 deletion by the Rag1 controlled Cre in the B-cell lineage appears lower than in T cells, as the deletion in whole spleen DNA (predominantly B and T cells) of Lmo2fl/lacZ;Rag1-Cre mice was around 75%. Purified B220+ B cells from the spleen showed a similar level of Lmo2 deletion (Fig. 8B). Deletion in the similar B220+ B-cell population in Lmo2fl/lacZ;CD19-Cre mice was in excess of 90% (Fig. 8D). Coupled with the analysis of hematopoiesis (Fig. 5 to 7), the high proportions of Lmo2 homozygous null mutant cells supports the conclusion that both T and B lymphocytes can develop in the absence of Lmo2. Thus, while transgenic expression of Lmo2 has severe consequences for T-cell development, this protein is not mandatory for normal development of the T lymphoid system.
FIG. 8.
Deletion of Lmo2 in mouse lymphoid cells expressing Cre recombinase. Genomic DNA was extracted from the indicated tissues of Lmo2fl/lacZ mice (No Cre) or from Lmo2fl/lacZ;Rag1-Cre (Rag1-Cre), Lmo2fl/lacZ;Lck-Cre (Lck-Cre), or Lmo2fl/lacZ;CD19-Cre (CD19-Cre) and digested with BamHI followed by filter hybridization analysis with Lmo2 probe B (Fig. 5A). In some cases of spleen and bone marrow, B220-expressing cells were purified with anti-B220 Dynabeads and DNA was prepared from these (B220+) and from the B220-depleted (B220−) populations. Genomic DNA bands are indicated, corresponding to Lmo2 floxed (fl), wild-type (wt), lacZ, and deleted (del) alleles. Cells in which deletion of the floxed Lmo2 occurs become homozygous null for Lmo2, as the second allele in these mice was a preexisting null allele (lacZ knock-in). Sizes (in kilobases) were estimated by coelectrophoresis of λ DNA cleaved with HindIII.
DISCUSSION
No obligatory requirement for Lmo2 in lymphopoiesis.
In this study we assessed the role of the acute T-cell leukemia oncogene Lmo2 in normal lymphoid development in mice, by creating specific null mutations of the gene in lymphopoiesis. This was achieved with three Cre expressing mouse lines in which Cre was synthesized from Rag1, Lck, or CD19 genes. We could find no evidence that loss of the Lmo2 gene had any effect on lymphoid development. This contrasts with previous data with Lmo2 null chimaeric mice in which no hematopoiesis was observed, implying a necessary role for Lmo2 in hematopoietic stem cells or in the generation of those cells from precursors (35) but none in lymphoid development. A later role in the myeloid-erythroid lineage has not yet been specifically examined.
The lack of an obligatory role for Lmo2 in T-cell development is paradoxical in view of the specific association of activated LMO2 with T-cell tumors and the normal expression of the gene in DN1 and DN2 T cells (13, 15). It is especially intriguing because enforced expression of Lmo2 in transgenic mice, under the control of the T-cell CD2 promoter, causes a block of T-cell differentiation at the DN3 stage (7), which is the stage at which Lmo2 is normally downregulated (13). This implies that the inability to shut off the function of Lmo2 at the DN2/DN3 differentiation boundary inhibits further passage through the T-cell differentiation pathway and in turn implies a normal role for Lmo2 at this T-cell differentiation stage. Our data show that there is no such discernible role for Lmo2.
LMO2 is a T-cell-specific oncogene.
LMO2 has only been implicated thus far in T-cell malignancies. Its discovery by association with recurrent chromosomal translocations in T-cell acute leukemias (24) was followed by its verification as a T-cell-specific oncogene in transgenic models where enforced Lmo2 expression was not limited to T cells (23). The recent development of leukemia by LMO2 activation via retroviral insertion in two X-SCID gene therapy patients (9a) has reinforced this T-cell tropism, as both patients developed T-cell acute leukemias (9).
A possible role for LMO2 in guiding differentiation into the T-cell lineage is indicated by these findings because the source of the cells for the retroviral transduction were CD34+ multipotential bone marrow cells (8). If there is no mandatory role for LMO2 in T-lymphoid development, it suggests that its specific role in T-cell tumorigenesis results from a reprogramming of gene expression after enforced expression in T-cell precursors. Examination of Lmo2-associated DNA binding complexes in transgenic mouse leukemic cells showed that Lmo2 forms complexes which differ in composition from those found in erythroid cells (7). Such complexes could be involved in the transcriptional regulation of target genes leading to a block in T-cell development and onset of leukemia. However, our current results imply that such complexes do not play a role in normal lymphoid development, implying either that they do not form during normal T lymphopoiesis, or that they form but are not functional.
It has been shown that in Lmo2-Tal1/Scl double transgenic mice have increased efficacy in T-cell leukemia development (17). In addition, another member of the family of LIM-only T-cell oncogenes, Lmo1, cooperates with Tal1/Scl in T-cell tumorigenesis and causes an inhibition of the E2A-HEB-mediated expression of the pre-T-cell receptor α-chain. This is thought to contribute to the arrested thymic development seen in Tal1/Scl transgenic mice (13). Moreover, mice lacking E2A develop leukemia of similar phenotype to CD2-Lmo2 transgenic mice (1). Hence, Lmo2-Tal1/Scl-induced leukemogenesis may be due to an abrogation of E2A protein function in T-cell development. By contrast, quantitative PCR analysis showed that regulation of pre-T-cell receptor α chain was normal in Lmo2fl/lacZ;Lck-Cre mice, implying that Lmo2 does not regulate this receptor subunit during normal T-cell development (data not shown).
LMO2 and retroviral insertion in X-SCID gene therapy.
The X-SCID gene therapy patients who developed leukemia as a result of retroviral activation of LMO2, had been infused with retrovirally transduced CD34+ progenitors to rescue the hematopoietic defect in these patients, with a retrovirus expressing the interleukin-2 receptor common γ chain which is absent in X-SCID (8). As a number of different chromosomal translocation genes have been identified which appear specific to T-cell acute leukemia (20), it is not clear why only LMO2 has been found in X-SCID leukemia cases.
One hypothesis is that there is cooperation between γ chain and LMO2 protein functions, perhaps with the former dictating cell division and the latter cell differentiation. In this situation, LMO2 expression is enforced by the presence of retrovirus (9, 9a), in an analogous way to the transgenic Lmo2 mouse models.
It is intriguing that the LMO2-associated chromosomal translocations in humans appear to be mediated by mistakes of the RAG recombinase (27). The Rag1 gene is expressed in the DN2/DN3 stages of T-cell differentiation and furthermore transgenic enforced Lmo2 expression results in accumulation of DN3 T cells. These combined observations suggest that the chromosomal translocations occur in humans by RAG-mediated errors in DN2/DN3 cells and the resultant enforced LMO2 expression leads to inhibition in T-cell development and onset of leukemia (24). By contrast, however, Lmo2 expression early in normal T-cell development is likely to be a remnant of expression in hematopoietic precursors, as it is not required for normal lymphoid maturation.
Acknowledgments
This work was supported by the Medical Research Council and M.P.M. was funded by the Leukemia Research Fund (United Kingdom).
We thank James Cruickshank, Lynn Grift, Gareth King, Theresa Langford, Angela Middleton, Claire Pearce, and Charlotte Rickett for animal husbandry, Annette Lenton for production of the art work, and N. Lobato for commenting on the manuscript. We thank J. Roes and K. Rajewsky for the CD19-Cre-tk plasmid, X. Gu and K. Rajewsky for the Cre plasmid, and R. Perlmutter for the Lck transgenic cassette.
REFERENCES
- 1.Bain, G., I. Engel, E. C. R. Maandag, H. P. J. Te Riele, J. R. Voland, L. L. Shart, J. Chun, B. Huey, D. Pinkel, and C. Murre. 1997. E2A deficiency leads to abnormalities in αβ T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 17:4782-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boehm, T., L. Foroni, Y. Kaneko, M. P. Perutz, and T. H. Rabbitts. 1991. The rhombotin family of cysteine-rich LIM-domain oncogenes: Distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc. Natl. Acad. Sci. USA 88:4367-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins, E. C., R. Pannell, E. M. Simpson, A. Forster, and T. H. Rabbitts. 2000. Interchromosomal recombination of Mll and. Af9 genes mediated by cre-loxP in mouse development. EMBO Rep. 1:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feinberg, A. P., and B. A. Vogelstein. 1983. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 5.Fischer, A., and B. Malissen. 1998. Natural and engineered disorders of lymphocyte development. Science 280:237-243. [DOI] [PubMed] [Google Scholar]
- 6.Fizzotti, M., E. Y. Chen, M. P. Link, A. J. Carroll, L. Foroni, T. H. Rabbitts, W. M. Crist, and S. S. Clark. 1994. Simultaneous expression of RBTN2 and BCR-ABL oncogenes in a human T-cell acute leukemia with a t(11;14)(p13;q11) and a late appearing Philadelphia chromosome. Leukaemia 8:1124-1130. [PubMed] [Google Scholar]
- 7.Grutz, G., K. Bucher, I. Lavenir, R. Larson, T. Larson, and T. H. Rabbitts. 1998. The oncogenic T-cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 17:4594-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hacein-Bey-Abina, S., F. Le Deist, F. Carlier, C. Bouneaud, C. Hue, J. De Villartay, A. Thrasher, N. Wulffraat, R. Sorensen, S. Dupuis-Girod, A. Fischer, E. Davies, W. Kuis, L. Leiva, and M. Cavazzana-Calvo. 2002. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 346:1185-1193. [DOI] [PubMed] [Google Scholar]
- 9.Hacein-Bey-Abina, S., C. von Kalle, M. Schmidt, F. Le Deist, N. Wulffraat, E. McIntyre, I. Radford, J. L. Villeval, C. C. Fraser, M. Cavazzana-Calvo, and A. Fischer. 2003. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 348:255-256. [DOI] [PubMed] [Google Scholar]
- 9a.Hacein-Bey-Abina, S., C. von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. MacIntyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LM02-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-67. [DOI] [PubMed] [Google Scholar]
- 11.Hardy, R. R., C. E. Carmack, S. A. Shinton, J. D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro-B and pre-pro-B-cell stages in normal mouse bone marrow. J. Exp Med. 173:1213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy, R. R., and K. Hayakawa. 2001. B-cell development pathways. Annu. Rev. Immunol. 19:595-621. [DOI] [PubMed] [Google Scholar]
- 13.Herblot, S., A.-M. Steff, P. Hugo, P. D. Aplan, and T. Hoang. 2000. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-Tα chain expression. Nat. Immunol. 1:138-144. [DOI] [PubMed] [Google Scholar]
- 14.Igarashi, H., S. C. Gregory, T. Yokota, N. Sakaguchi, and P. W. Kincade. 2002. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity 17:117-130. [DOI] [PubMed] [Google Scholar]
- 15.Kenny, D. A., L. W. Jurata, Y. Saga, and G. N. Gill. 1998. Identification and characterization of LMO4, an LMO gene with a novel pattern of expression during embryogenesis. Proc. Natl. Acad. Sci. USA 95:11257-11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larson, R., P. Fisch, T. Larson, I. Lavenir, T. Langford, G. King, and T. H. Rabbitts. 1994. T-cell tumours with disparate phenotypes develop with long latency in mice transgenic for rbtn2. Oncogene 9:3675-3681. [PubMed] [Google Scholar]
- 17.Larson, R. C., I. Lavenir, T. A. Larson, R. Baer, A. J. Warren, I. Wadman, K. Nottage, and T. H. Rabbitts. 1996. Protein dimerisation between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T-cell tumorigenesis in transgenic mice. EMBO J. 15:1021-1027. [PMC free article] [PubMed] [Google Scholar]
- 18.Larson, R. C., H. Osada, T. A. Larson, I. Lavenir, and T. H. Rabbitts. 1995. The oncogenic LIM protein Rbtn2 causes thymic developmental aberrations that precede malignancy in transgenic mice. Oncogene 11:853-862. [PubMed] [Google Scholar]
- 19.LeFranc, M.-P., A. Forster, R. Baer, M. A. Stinson, and T. H. Rabbitts. 1986. Diversity and rearrangement of the human T-cell rearranging γ genes: Nine germ-line variable genes belonging to two subgroups. Cell 45:237-246. [DOI] [PubMed] [Google Scholar]
- 20.Look, A. T. 1997. The metabolic basis of inherited disease, p. 109-141. McGraw-Hill, New York, N.Y.
- 21.Mombaerts, P., J. Iancomini, R. S. Johnson, K. Herrup, S. TRonegawa, and V. E. Papaloannou. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68:869-877. [DOI] [PubMed] [Google Scholar]
- 22.Neale, G. A., J. E. Rehg, and R. M. Goorha. 1997. Disruption of T-cell differentiation precedes T-cell tumor formation in LMO2 (rhombotin-2) transgenic mice. Leukaemia 11:289-290. [PubMed] [Google Scholar]
- 23.Neale, G. A., J. E. Rehg, and R. M. Goorha. 1995. Ectopic expression of rhombotin-2 causes selective expansion of the thymus and T-cell tumours in transgenic mice. Blood 86:3060-3071. [PubMed] [Google Scholar]
- 24.Rabbitts, T. H. 1998. LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev. 12:2651-2657. [DOI] [PubMed] [Google Scholar]
- 25.Rickert, R. C., K. Rajewsky, and J. Roes. 1995. Impairment of T-cell-dependent B-cell responses and B-1 cell development in CD19-deficient mice. Nature. 376:352-355. [DOI] [PubMed] [Google Scholar]
- 26.Royer-Pokora, B., U. Loos, and W.-D. Ludwig. 1991. TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11). Oncogene 6:1887-1893. [PubMed] [Google Scholar]
- 27.Sanchez-Garcia, I., Y. Kaneko, R. Gonzalez-Sarmiento, K. Campbell, L. White, T. Boehm, and T. H. Rabbitts. 1991. A study of chromosome 11p13 translocations involving T-cell receptorβ and T-cell receptorδ in human T-cell leukaemia. Oncogene 6:577-582. [PubMed] [Google Scholar]
- 28.Southern, E. M. 1975. Detection of Specific Sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 29.Thomas, K. R., and M. R. Capecchi. 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51:503-512. [DOI] [PubMed] [Google Scholar]
- 30.Torres, R. M., and R. Kuhn. 1997. Laboratory protocols for conditional gene targeting. Oxford University Press, Oxford, England.
- 31.Wadman, I. A., H. Osada, G. G. Grutz, A. D. Agulnick, H. Westphal, A. Forster, and T. H. Rabbitts. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren, A. J., W. H. Colledge, M. B. L. Carlton, M. J. Evans, A. J. H. Smith, and T. H. Rabbitts. 1994. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell 78:45-58. [DOI] [PubMed] [Google Scholar]
- 33.Wildin, R. S., H. U. Wang, K. A. Forbush, and R. M. Perlmutter. 1995. Functional dissection of the murine lck distal promoter. J. Immunol. 155:1286-1295. [PubMed] [Google Scholar]
- 34.Yamada, Y., R. Pannell, A. Forster, and T. H. Rabbitts. 2000. The oncogenic LIM-only transcription factor Lmo2 regulates angiogenesis but not vasculogenesis. Proc. Natl. Acad. Sci. USA 97:320-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada, Y., A. W. Warren, C. Dobson, A. Forster, R. Pannell, and T. H. Rabbitts. 1998. The T-cell leukaemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. USA 95:3890-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]