Abstract
The replication fork barrier site (RFB) is an ∼100-bp DNA sequence located near the 3′ end of the rRNA genes in the yeast Saccharomyces cerevisiae. The gene FOB1 is required for this RFB activity. FOB1 is also necessary for recombination in the ribosomal DNA (rDNA), including increase and decrease of rDNA repeat copy number, production of extrachromosomal rDNA circles, and possibly homogenization of the repeats. Despite the central role that Foblp plays in both replication fork blocking and rDNA recombination, the molecular mechanism by which Fob1p mediates these activities has not been determined. Here, I show by using chromatin immunoprecipitation, gel shift, footprinting, and atomic force microscopy assays that Fob1p directly binds to the RFB. Fob1p binds to two separated sequences in the RFB. A predicted zinc finger motif in Fob1p was shown to be essential for the RFB binding, replication fork blocking, and rDNA recombination activities. The RFB seems to wrap around Fob1p, and this wrapping structure may be important for function in the rDNA repeats.
Sites that cause replication fork pausing have been identified in many genomes and are known as replication fork-blocking sites. The replication fork-blocking site was first identified in bacteria, both in plasmids and in the genome. Plasmid R6K and the genome of Escherichia coli each have a replication termination sequence, Ter (for a review, see reference 18). Ter is a conserved 22-bp sequence that inhibits the replication fork in a polar fashion (18). The Bacillus subtilis genome has a counterpart of Ter, called RTS, although there is no similarity between the sequences (5, 34). In E. coli, the tus protein binds specifically to the Ter sequence, and the Tus-Ter complex inhibits the action of DNA helicases to arrest replication (14, 15, 28, 32). Ter is also known as a recombination hot spot that stimulates both DNA double-strand breaks (37) and recombination (16, 17; for a review, see reference 41). Therefore, one possible role of Ter is thought to be maintenance of genome stability.
In eukaryotic cells, replication fork-blocking sites have been identified in the rRNA gene repeats (rDNA) from yeast to human cells (for a review, see reference 41). They are called replication fork barriers (RFB) and inhibit replication forks in the direction opposite to rDNA transcription. The RFB has been studied most intensively in the yeast Saccharomyces cerevisiae. S. cerevisiae carries about 150 copies of the rDNA repeats. The RFB is located near the 3′ end of the 35S rDNA in one of the nontranscribed spacer (NTS) regions (NTS1) (Fig. 1A). In the other NTS region (NTS2), there is an autonomously replicating sequence (ARS) which functions as a replication origin. In the S phase of the cell cycle, replication starts at the ARS bidirectionally and the rightward-moving replication forks are arrested at the RFB (Fig. 1). However, the other forks can go through the RFB site because of RFB polarity, like for Ter in E. coli (4, 29). Therefore, one of the biological roles of the RFB is thought to be as a barrier to prevent collision between replication and transcription machineries. Recently, we observed such a collision in an RFB-deficient mutant (44) (see Discussion). Ward et al. identified two closely spaced sequences, the RFB1 and RFB2 sites, in the RFB (47). These sites are responsible for the major and minor replication fork-blocking activities, respectively, that are seen in two-dimensional (2D) gel electrophoresis. We also identified a third site (here it is named RFB3) between the HindIII and EcoRI sites by using a strain whose RFB1 and RFB2 sites (HpaI-HindIII) are deleted (31) (for details, see Fig. 3 and 5). In this paper, I call these sites collectively the RFB site.
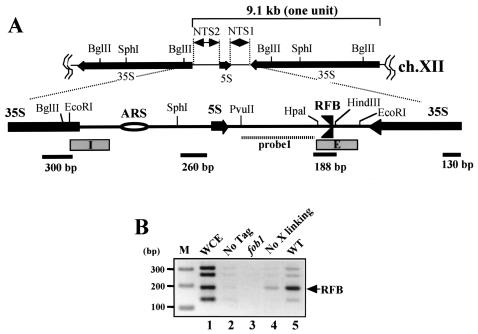
FIG. 1.
(A) Structure of rDNA repeats in S. cerevisiae. A single unit of rDNA consists of two transcribed genes (5S and 35S RNA genes) (the direction of transcription is indicated by arrows) and two nontranscribed regions (NTS1 and NTS2). The 35S rRNA gene is transcribed by PolI, while the 5S rRNA gene is transcribed by PolIII. The NTS and its surrounding regions are expanded. Two DNA elements related to DNA replication, the origin of replication (ARS) and the RFB, are located in NTS2 and NTS1, respectively. The RFB located near the end of the 35S rRNA gene allows progression of the replication fork in the direction of 35S rRNA transcription but not in the opposite direction (3, 35). Probe 1 (striped bar) is a probe used for Southern hybridization in 2D analysis. The lower bars show locations of fragments amplified by PCR for ChIP assay. The four fragments are located about every 1 kb. The lengths of the PCR products are shown below by bars. E and I (boxed) are elements of HOT1. (B) ChIP assay. The inverse image of PCR products resolved on ethidium bromide-stained 2.6% agarose gels is shown. Fob1-FLAGp was expressed in the fob1 strain (NOY408-1bf), and PCR was performed on chromatin fragments after the immunoprecipitation. Samples were prepared from the whole-cell extract (WCE) before the precipitation (lane 1), from NOY408-1b with Fob1p not tagged (lane 2), and from NOY408-1bf with Fob1-FLAGp not expressed (lane 3), with Fob1-FLAGp expressed but without prior cross-linking before precipitation (lane 4), and with Fob1-FLAGp expressed (lane 5). The position of the RFB fragment is indicated by an arrow. Lane M, 100-bp ladder marker (Invitrogen).
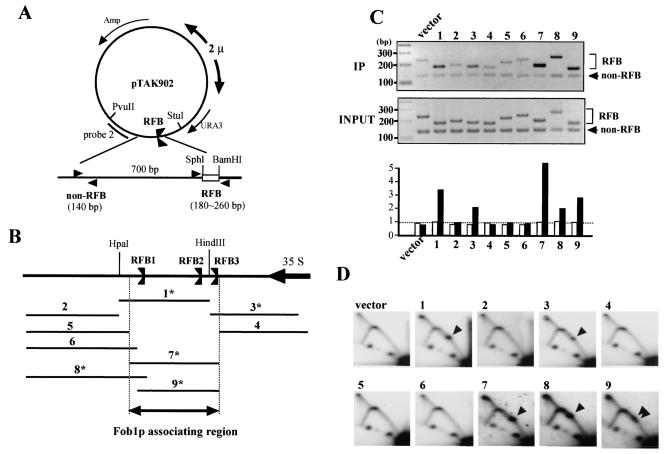
FIG. 3.
Identification of Fob1p association sites in the RFB. (A) Structure of pTAK902.1 to -9, which contain various subfragments of the RFB. RFB subfragments were inserted (between SphI and BamHI sites) near the 2μm replication origin, where they are predicted to inhibit the clockwise-moving replication fork. Positions of PCR primer sets used for the ChIP assay are shown by arrowheads (non-RFB and RFB). (B) Positions of the subfragments which were inserted into the SphI-BamHI site of pTAK902. The RFB1, RFB2, and RFB3 sites and the direction of 35S rDNA transcription are indicated. Numbers with asterisks indicate fragments that were associated with Fob1p (C) and inhibited the replication fork in the plasmid (D). (C) ChIP analysis by Fob1-FLAGp. Samples were prepared from NOY408-1bf strains carrying pTAK901 (Fob1-FLAGp expression vector) and pTAK902.1 to -9. In the top panel, DNA immunoprecipitated (IP) by the anti-FLAG antibody was used as the template, and in the middle panel, total DNA (whole-cell extract) was used as the template. Primer sets used for the ChIP assay are shown in panel A. Non-RFB and RFB primer sets amplified the vector sequence (lower bands) and inserted RFB subfragments (upper bands), respectively. Lane numbers above the panel correspond to the inserted fragments (panel B). vector, no RFB insertion in pTAK902. In bottom panel, the ratios of RFB to non-RFB are plotted. White and black bars indicate the values for input and IP, respectively. The values are relative to that of input vector. (D) Replication fork-blocking activities of various RFB subfragments on plasmid pTAK902. 2D analysis was performed as for Fig. 2C. DNA was prepared from the strains used for panel C, digested with PvuII and StuI, and subjected to 2D analysis followed by Southern hybridization with probe 2 (A). Numbers correspond to those of the subfragments in panel B. Spots indicated by arrowheads show accumulation of Y-shaped DNA molecules at the RFB site. vector, no RFB insertion in pTAK902.
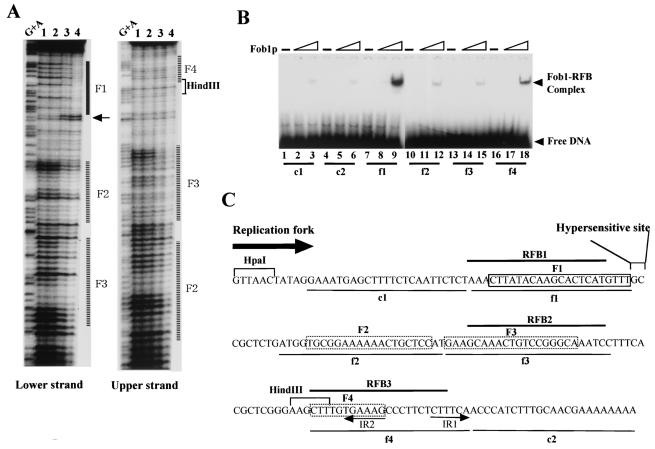
FIG. 5.
(A) DNase I footprinting assay with purified GST-Fob1p. A 32P-end-labeled RFB fragment (0.1 ng of fragment 7 in Fig. 3B) was mixed with purified GST-Fob1p and digested with DNase I. Samples were analyzed on an 8% (wt/vol) polyacrylamide sequencing gel. Amounts of GST-Fob1p were as follows: lane 1, 0 ng; lane 2, 2.5 ng; lane 3, 25 ng; lane 4, 250 ng. Lane G+A, Maxam-Gilbert sequencing reaction. (B) Detection of in vitro binding activity of GST-Fob1p to small subfragments of the RFB by gel shift assay. The gel shift assay was performed as for Fig. 4D. The positions of the 32P-end-labeled DNA fragments c1 to f4 are shown in panel C. Amounts of GST-Fob1p were as follows: lanes 1, 4, 7, 10, 13, and 16, 0 ng; lanes 2, 5, 8, 11, 14, and 17, 2.5 ng; lanes 3, 6, 9, 12, 15, and 18, 25 ng. (C) Summary of GST-Fob1p binding sites in the RFB. The DNA sequence around the RFB (∼180 bp) is shown. RFB1, RFB2, and RFB3 are indicated by solid lines above the sequence. c1 to f4, used for the gel shift assay in panel B, are shown below the sequence. Fob1p binding sequences, identified by the footprinting assay (A), are shown by boxes. The box (F1) indicates strong binding, and dotted boxes (F2 to F4) indicate weak binding. A DNase I-hypersensitive site next to F1 is indicated. The direction of the replication fork which is inhibited by the RFB is indicated by an arrow. IR1 and IR2 are inverted repeats.
Previous work showed that the gene FOB1 is required for the RFB activity (30). FOB1 was originally identified as a gene essential for HOT1 activity. HOT1 is a DNA element that stimulates genetic exchanges at nearby regions when inserted at a non-rDNA site (26). Two cis elements were subsequently identified as essential for HOT1 activity: the I element, which corresponds to the RNA polymerase I (PolI) promoter region, and the E element, which overlaps the enhancer for PolI transcription (Fig. 1A) (8) and the RFB. It was speculated that the RFB in HOT1 would be working as a recombination hot spot similar to Ter in E. coli; therefore, we first tried to isolate HOT1-deficient mutants and searched for RFB deficiency among them. One of the HOT1-defective mutants showed a blockless phenotype at the RFB (30). We named the mutated gene FOB1 (for fork blocking). (Interestingly, Ward et al. demonstrated that the fork-blocking event itself is not essential for the HOT1 activity [47]. Instead, Wai et al. found that FOB1 was required for the transcription of HOT1 by PolI [46], and the transcription seems to be necessary for the activation of HOT1 [19].)
In terms of rDNA recombination, the RFB site is thought to be a recombination hot spot that induces DNA double-strand breaks (31). FOB1 was shown to be essential for recombination in the rDNA (22, 23, 27, 36). FOB1-dependent recombination in the rDNA is known to be required for regulation of copy number, probably through unequal sister-chromatid recombination after a double-strand break at the RFB (27). Double-strand breaks also produce extrachromosomal rDNA circles, whose accumulation seems to be a cause of aging (42, 40). In fact, in a fob1 mutant, the life span is extended more than 50% compared to that of wild-type cells (6). As another aging gene, SIR2, whose gene product is a NAD-dependent histone deacetylase (20), is genetically downstream of FOB1 in the aging process, instability of the rDNA is probably an important component of this process (23).
We are interested in the molecular mechanisms of the RFB- and FOB1-dependent recombination and replication fork-blocking activity, and therefore in this study I aimed to resolve the enzymatic function of Fob1p. Perhaps the most likely function is RFB binding activity analogous to the action of Tus on the Ter sequence in E. coli. However, there are currently no reports showing that Fob1p binds to the RFB sequence. In this study, I demonstrate that Fob1p binds to the RFB and inhibits the replication fork in vivo and that this binding is characterized by an unusual wrapping activity.
MATERIALS AND METHODS
Media, strains, and plasmids.
SD and SGal are synthetic media (24) containing 2% glucose and 2% galactose, respectively. Both SD and SGal were supplemented appropriately with amino acids and bases to satisfy nutritional requirements and also to retain unstable plasmids (24). YPGal medium is the same as yeast extract-peptone-dextrose (24) except that 2% glucose is replaced by 2% galactose.
The yeast strains and plasmids used are listed in Table 1. Disruption of FOB1 was described previously (30). Plasmid pTAK901 was constructed by inserting the PCR-amplified open reading frame (ORF) of FOB1 into the multicloning site of pESC-LEU (Stratagene). Point mutations were added to pTAK901 by site-directed mutagenesis (performed by Bio Dynamics Laboratory Inc., Tokyo, Japan) to construct pTAK991 to -995. pTAK900 was constructed by inserting the ORF of FOB1 into the BamHI-SalI site in pEG(KT), and glutathione S-transferase (GST) was added to the N terminus of Fob1p. Plasmids pTAK902.1 to -9 were constructed by inserting PCR products amplified from several regions around the RFB into the BamHI-SphI site of the YEp24 shuttle vector (New England Biolabs). pTAKc1 to -f4 were constructed by inserting annealed oligonucleotides into the BamHI-SphI site of YEp24. TAK899 was constructed by inserting a URA3 gene into the rDNA repeats.
TABLE 1.
Yeast strains and plasmids used
| Strain or plasmid | Genotype and comments | Reference or source |
|---|---|---|
| Strains | ||
| NOY408-1af | MATaade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 rpa135::LEU2 can1-100 fob1::HIS3 pNOY102 pNOY117 | 27 |
| NOY408-1b | MATaade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 | 39 |
| NOY408-1bf | Same as NOY408-1b except fob1::HIS3 | 27 |
| TAK899 | Same as NOY408-1bf except rDNA::URA3 | |
| YK9 | MATaura3-52 trp1-289 leu2-3-112 prb pep4::TRP1 | 33 |
| Plasmids | ||
| pNOY102 | Multicopy plasmid vector; URA3 GAL7-35S rDNA 2μm | 39 |
| pNOY117 | CEN6 ARSH4 TRP1 RPA135 | Nomura lab |
| pEG(KT) | Multicopy plasmid vector; URA3 Leu2-d GAL1-GST 2μm | 38 |
| pTAK900 | pEG(KT) carrying FOB1 | |
| pESC-LEU | Multicopy plasmid vector; LEU2 GAL1-FLAG 2μm | Stratagene |
| pTAK901 | pESC-LEU carrying FOB1 | |
| pTAK991 | Same as pTAK901 except the ORF of FOB1 has a point mutation (H159A) | |
| pTAK992 | Same as pTAK901 except the ORF of FOB1 has a point mutation (H164A) | |
| pTAK993 | Same as pTAK901 except the ORF of FOB1 has a point mutation (C193A) | |
| pTAK994 | Same as pTAK901 except the ORF of FOB1 has a point mutation (C196A) | |
| pTAK995 | Same as pTAK901 except the ORF of FOB1 has a point mutation (D291A) | |
| YEp24 | Multicopy plasmid vector; URA3 2μm pBR322 Ampr Tcr | New England Biolabs |
| pTAK902.1 to -9 | YEp24 carrying various fragments (1 to 9 in Fig. 3B) around the RFB | |
| pTAKc1 to -f4 | YEp24 carrying subfragments (24 bp, c1 to f4 in Fig. 5C) of the RFB | |
| Yeplac195 | Multicopy plasmid vector; URA3 2μm | 12 |
| Yep-FOB1 | Yeplac195 carrying FOB1 | 27 |
Marker loss assay.
TAK899 (∼105 cells) with various FOB1 plasmids (pTAK991 to -995) was grown to stationary phase in SGal lacking uracil at 30°C. The frequencies of Ura− recombinants were then determined by spotting aliquots of 10-fold serial dilutions of the cultures onto SD with and without 5-fluoro-orotic acid.
Purification of GST-Fob1p.
Purification of GST-Fob1p was performed as described previously (33). In short, a 2-liter YK9 culture with pTAK900 was grown to mid-logarithmic phase at 30°C in SD lacking uracil. The cells were transferred to YPGal and incubated for 4 h at 30°C. The cells were then harvested, destroyed with glass beads, and applied to a GST affinity column (Amersham Pharmacia Biotech.). The eluted fraction was further purified by gel filtration, and the GST-Fob1p fraction was collected.
DNA binding assay.
Gel shift assays were performed as follows. The RFB fragment (fragment 7 in Fig. 3B) and subfragments (70-bp DNA fragments, made by PCR with pTAKc1 to -f4 as the templates) which contain several parts (each 24 bp in length) of the RFB (see Fig. 5C) were used as the DNA target. These DNA fragments were end labeled with 32P and mixed with GST-Fob1p in 20 μl of phosphate buffer (20 mM sodium phosphate [pH 7.4], 1 mM EDTA, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 0.01% Triton X-100). The mixture was incubated at 30°C for 30 min. The DNA-Fob1p complex was visualized on 5% polyacrylamide gels at 4°C. Footprinting assays were performed with the SureTrack footprinting kit (Pharmacia) according to the manufacturer's instructions.
AFM imaging and analysis.
Atomic force microscopy (AFM) imaging was performed by the Research Institute of Biomolecule Metrology Co., Ltd., Tsukuba, Japan. Three micrograms of GST-Fob1p and 500 ng of the RFB fragment were mixed in 0.5 ml of phosphate buffer (20 mM sodium phosphate [pH 7.4], 1 mM EDTA, 2 M NaCl). The mixture was incubated at 25°C for 60 min and dialyzed in 20 ml of phosphate buffer (20 mM sodium phosphate [pH 7.4], 1 mM EDTA, 100 mM NaCl, 5 mM MgCl2) for 12 h. The mixture was then applied to a freshly cleaved mica surface, followed by successive washings with water and subsequent drying under nitrogen gas. Imaging was carried out with a Digital Instrument Nanoscope III with a type E scanner in the tapping mode.
Other methods.
The chromatin immunoprecipitation (ChIP) assay was based on methods described previously (45), except that Dynabead protein A was used (25). The RFB activity was analyzed by 2D gel electrophoresis as previously described (2). Determination of rDNA copy number was done as previously described (27).
RESULTS
Fob1p associates with the RFB in vivo.
One of the enzymatic functions of Fob1p was speculated to be RFB binding, analogous to the case for the E. coli Tus protein (15, 28). However, I was unable to detect such an activity by using in vitro assays such as gel shift and DNA footprinting assays with a yeast crude extract, even in a strain in which Fob1p was overproduced (T. Kobayashi et al., unpublished data). One of the reasons may be that Fob1p is localized in the nucleolus, especially near the rDNA region (6, 11), and therefore the amount of Fob1p in the soluble fraction may be too small to be detected. To determine if there is an association of Fob1p with the RFB site in vivo, I performed a ChIP assay with an antibody against a FLAG epitope which was fused to Fob1p. The Fob1-FLAGp was expressed by the galactose-inducible promoter in a fob1 mutant. For the assay, the DNA was cross-linked to the chromatin, precipitated with anti-FLAG antibodies, and detected by PCR with four primer sets. The positions of these PCR products are shown in Fig. 1A. The results of the PCRs are shown in Fig. 1B. The 188-bp fragment corresponding to the RFB region was amplified more than the other fragments (Fig. 1B, lane 5). In contrast, when total DNA (whole-cell extract) was used as the template, all four fragments were equally amplified (lane 1). In addition, in a strain whose Fob1 is not tagged (lane 2) or whose Fob1-FLAGp is not expressed (lane 3), there was little PCR amplification. These results indicate that Fob1p specifically associates with the RFB site in vivo. Surprisingly, weak association was observed in a control in which the cross-linking step was skipped (Fig. 1B, lane 4), suggesting that Fob1p is tightly associated with the RFB site.
A zinc finger motif is required for Fob1p association with the RFB, inhibition of the replication fork, and recombination in the rDNA.
Computer analysis (PHI-BLAST; National Center for Biotechnology Information) (7) predicted that Fob1p contains a zinc finger motif, which is known to bind to DNA. To examine whether this motif is essential for Fob1p association with the RFB, point mutations were made in the FOB1 ORF to disrupt the motif (C2H2) (Fig. 2A) and were expressed in a fob1 mutant. Expression of the mutated Fob1p was confirmed by Western blotting (data not shown). A ChIP assay similar to that described in the previous section was performed, and the results are shown in Fig. 2B. Fob1p association with the RFB was completely lost in the strains whose zinc finger motif is disrupted (Fig. 2B, lanes 3 to 6). In contrast, a control mutation in the integrase catalytic core-like structure (Fig. 2A), which was also predicted by computer analysis (PHI-BLAST; National Center for Biotechnology Information) (7), did not affect the association (Fig. 2B, lane 7).
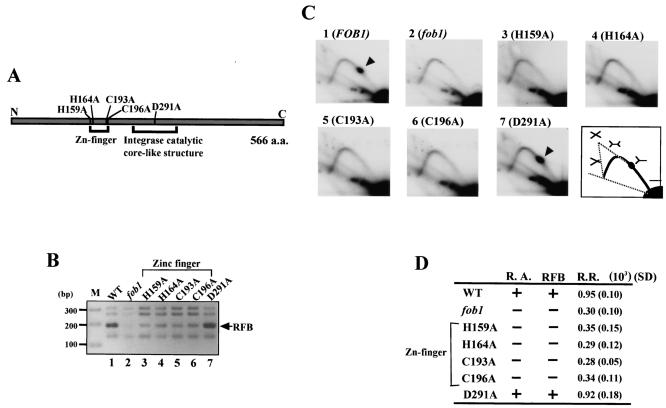
FIG. 2.
Effects of various FOB1 mutations. (A) Structure of Fob1p. The gray bar shows the 566-amino-acid (a.a.) Fob1p. Regions corresponding to the predicted zinc finger motif and integrase catalytic core-like structure are shown below the bar. Positions of mutated amino acids are indicated above the bar, and the mutations are labeled according to convention. The N and C termini are indicated. (B) ChIP assay with the mutated Fob1p constructs. Various mutated Fob1-FLAGp were expressed in the fob1 strain (NOY408-1bf), and the associated DNA was precipitated with an anti-FLAG monoclonal antibody (Stratagene) after cross-linking. Lane 1, Fob1-FLAGp was expressed; lane 2, Fob1-FLAGp was not expressed; lane 3 to 7, each mutated Fob1-FLAGp was expressed, as indicated. The position of the RFB fragment is indicated by an arrow. (C) Replication fork-blocking activity at the RFB site in the mutated FOB1strains was analyzed by 2D analysis. DNA was prepared from the strains used for panel B, digested with BglII and SphI, and subjected to 2D analysis followed by Southern hybridization with probe 1 (Fig. 1A). Spots indicated by arrowheads show accumulation of Y-shaped DNA molecules at the RFB site. A diagram of the migration pattern is shown in the lower right panel, with the structures of the replication intermediate molecules shown above the pattern. (D) Summary of the effects of the mutations in FOB1. R.A., RFB, and R.R., RFB-associating activity, replication fork-blocking activity, and recombination rate, respectively. + and −, wild-type level and fob1 level, respectively. The recombination rates were measured by determining the frequency of loss of a URA3 marker inserted in the rDNA repeats. The values are the averages from three independent experiments. Standard deviations (SD) are shown in parentheses.
Next, I examined the effect of these mutations on RFB activity by using 2D gel electrophoresis (2). The results are shown in Fig. 2C. When wild-type Fob1p was expressed, a spot which corresponds to Y-shaped replication intermediates stopped at the RFB was observed (panel 1). In contrast, when Fob1p was not expressed, this spot was not seen (panel 2), as the protein is essential for the RFB activity (30). When the zinc finger mutations of Fob1p were expressed, this spot could not be detected either (panels 3 to 6), although the control mutation did not affect RFB activity (panel 7). This suggests that the zinc finger is essential for the fork-blocking activity of Fob1p.
I also tested the effects of the mutations on rDNA recombination. A URA3 marker was integrated into the rDNA repeats, and the rate of loss was detected on a fluoro-orotic acid plate. The results are shown in Fig. 2D. Mutations in the zinc finger motif reduced the recombination rate to the level found in fob1 mutants. In contrast, the control mutation did not affect the rate (D291A). Taken together, these results suggest that the zinc finger motif of Fob1p is required for the association with the RFB and that this association is essential for both the replication fork-blocking and recombination activities at the RFB.
Fob1p associates with a broad region in the RFB in vivo to inhibit DNA replication.
To study in detail the Fob1p-associated region in the RFB, nine fragments of the RFB and the surrounding region were cloned into the SphI-BamHI site of the YEp24 shuttle vector (Fig. 3A and B). As a plasmid-cloned RFB was shown to inhibit the replication fork (4, 29), association of Fob1p should be observed on active RFB fragments in the plasmid. The plasmids were transformed into the yeast strain NOY408-1bf, which has Fob1-FLAGp (pTAK901), and a ChIP assay was performed as outlined above. Two PCR primer sets for the detection of the precipitated fragments were designed (Fig. 3A). One amplifies a vector region as an internal control for the ChIP assay, and the other amplifies the subcloned RFB fragments. The results of the ChIP assay are shown in Fig. 3C (upper panel). There are two bands in the ethidium bromide-stained gel in each lane. The lower band is the vector control fragment, and the upper bands correspond to the subcloned RFB fragments. The intensities of the bands were measured, ratios of the upper to the lower bands were calculated, and the values were plotted on a graph (Fig. 3C, lower panel). The intensities of fragments 1, 3, 7, 8, and 9 were higher than those of other fragments. In contrast, when total DNA (whole-cell extract) was used as a template for the PCR, the ratios were similar (Fig. 3C, middle and lower panels). The results of the ChIP assay are summarized in Fig. 3B, where fragments which were associated with Fob1p are indicated. As shown in Fig. 3B, Fob1p associates with a broad region of DNA corresponding to the RFB site.
Next, I investigated the DNA replication fork-blocking activity of the subcloned RFB fragments by 2D analysis. The results are shown in Fig. 3D. Fragments 1, 3, 7, 8, and 9 showed replication fork-blocking activity, and these fragments correspond precisely to the Fob1p-associated fragments identified in Fig. 3C. Interestingly, two spots are observed within fragment 9, although only one spot was observed for fragment 7. This could be because part of RFB1 was deleted in fragment 9 and thus RFB1 activity was reduced, allowing the RFB2 and RFB3 activities to be detected. This suggests that RFB1 is the strongest inhibitor of the replication fork and that most replication forks arrest at RFB1. RFB2 and RFB3 are adjacent, and therefore their spots are expected to be fused. These results strengthen the conclusion that Fob1p association with the RFB site is required for the replication-blocking activity.
Fob1p specifically binds to the RFB in vitro.
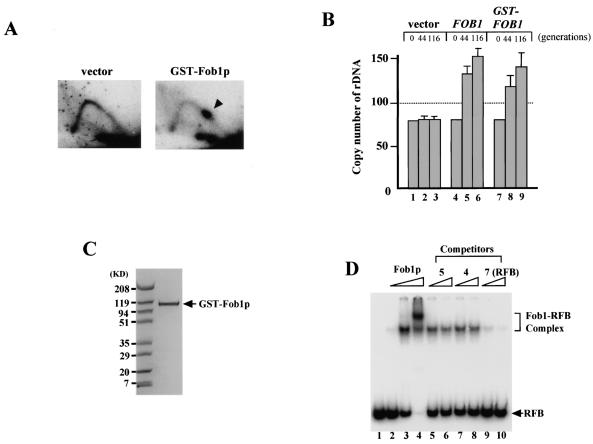
To examine whether Fob1p associates with the RFB itself or whether it requires other factors, I purified Fob1p for in vitro analysis. Fob1p with a GST tag was overexpressed in yeast, and GST-Fob1p was isolated with GST affinity column. A few contaminating bands were visible with Coomassie blue staining, and therefore the solution was treated by gel filtration to remove these bands. Figure 4C shows the purified GST-Fob1p separated by acrylamide gel electrophoresis and stained with Coomassie blue. As the fusion gene, GST-FOB1, could complement RFB and rDNA copy number expansion deficiencies in a fob1 mutant in vivo (Fig. 4A and B), I used the fusion protein for further in vitro analysis.
FIG. 4.
Binding activity of purified GST-Fob1p. (A) 2D analysis to detect replication fork-blocking activity of GST-Fob1p in vivo. DNA samples were prepared from NOY408-1bf carrying pEG(KT) (vector) or pTAK900 (GST-Fob1p). An arrowhead shows the accumulation of Y-shaped molecules, indicative of replication fork-blocking activity. (B) rDNA amplification activity of GST-Fob1p in vivo. NOY408-1af was transformed with pEG(KT) (vector), Yep-FOB1 (FOB1) or pTAK900 (GST-FOB1). At 44 and 116 generations after the introduction, DNA was isolated and rDNA copy number was determined. Generation 0 corresponds to DNA that was isolated before transformation. (C) Analysis of purified GST-Fob1p by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. GST-Fob1p was purified by using a GST-affinity column and gel filtration from a crude extract of YK9 with pTAK900. The fusion protein was applied to a 10 to 20% polyacrylamide gel and stained with Bio-Safe Coomassie blue (Bio-Rad). (D) Detection of in vitro binding activity of GST-Fob1p to the RFB fragment by gel shift assay. End-labeled RFB fragments (0.16 ng) (Fig. 3B, fragment 7) were mixed with 0 ng (lane 1) 0.25 ng (lane 2), 2.5 ng (lane 3), and 25 ng (lane 4) of GST-Fob1p, and the mixture was applied to a native 5% polyacrylamide gel. Lanes 5 to 10 show competition assays to detect the binding specificity of the RFB fragment. Here, 0.16 ng of end-labeled RFB fragments, 2.5 ng of GST-Fob1p, and one of the three kinds of cold competitor fragments (fragment 5, 4, or 7 [Fig. 3B]) were used in the assay. Fragments 5 and 4 are RFB flanking sequences, and fragment 7 is the RFB itself. Competitors were used at 1.6 ng (lanes 5, 7, and 9) or 3.2 ng (lanes 6, 8, and 10).
GST-Fob1p was used in a gel shift assay to see whether specific binding between Fob1p and the RFB site occurs. I used fragment 7 as the target RFB fragment because it showed the strongest association with Fob1p in vivo by ChIP analysis (Fig. 3C). GST-Fob1p was mixed with the 32P-end-labeled RFB fragment, and the resulting complex was detected by native acrylamide gel electrophoresis. The results are shown in Fig. 4D. Shifted bands corresponding to the RFB-Fob1p complex were identified (Fig. 4D, lanes 2 to 4). Next, to examine the specificity of the binding activity, non-RFB fragments (regions flanking the RFB; fragments 4 and 5 in Fig. 3B) were added as competitors. As shown in Fig. 4D (lanes 5 to 8), a 20-fold excess of competitor DNA did not reduce the amount of complex detected, even though unlabeled RFB resulted in complete competition (lanes 9 and 10). Therefore, Fob1p directly and specifically binds to the RFB.
Fob1p binds to two separate sequences in the RFB and interacts with several other sites.
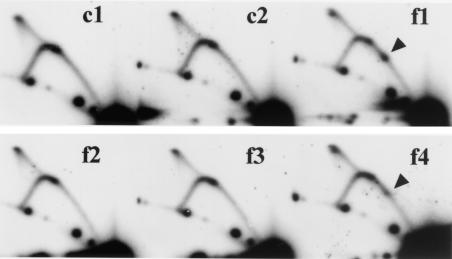
To identify precisely where in the RFB Fob1p binds, a DNase I footprinting assay was performed. GST-Fob1p was mixed with the RFB fragment (fragment 7 in Fig. 3B), and this complex was partially digested by DNase I, followed by denaturing gel electrophoresis. In the left panel of Fig. 5A, the 5′ end of the RFB fragment close to the HindIII site was labeled with 32P, and in the right panel, the other end was labeled. Therefore, protection of both strands by Fob1p against DNase I was determined. As shown in the left panel of Fig. 5A, apparent protection was detected in the F1 region, which corresponds to RFB1 (Fig. 5C). Beside F1 is a DNase I-hypersensitive site, which are often observed near protein binding sites. In regions F2 to F4, the protection is not so clear, but some degree of protection is observed. F3 and F4 are located in RFB2 and RFB3, respectively (Fig. 5C).
To investigate the binding activity of each region more precisely, I performed gel shift assays with small fragments (24 bp) from the RFB site, surrounded by nonyeast DNA. Fragments f1 to f4 and the controls (c1 and c2, which are regions flanking the RFB [Fig. 5C]) were cloned into the SphI-BamHI site of YEp24 (Fig. 3A), and PCR amplicons including the 24-bp sequences were amplified. The fragments were end labeled with 32P and used as substrates for the gel shift assay. The results are shown in Fig. 5B. GST-Fob1p binds to f1 and f4. However, binding affinities of the f2 and f3 fragments are at the same level as the control (c1 and c2). Fragment f1 showed about three times stronger binding affinity to Fob1p than f4 did. I conclude that GST-Fob1p independently binds to these two regions, which correspond to RFB1 and RFB3.
The small 24-bp fragments still have replication fork-blocking activity.
2D analysis was used to examine the replication fork-blocking activity of the small, 24-bp fragments as in Fig. 3D. The results are shown in Fig. 6. Although the intensities are not as strong as those in Fig. 3D, spots corresponding to the accumulation of Y-shaped replication intermediates arrested at the inserted fragments were detected in f1 and f4. Fragment f1 showed about three times stronger blocking activity than f4, corresponding to the results of the shift assay. These results further demonstrate that replication fork-blocking activity is associated with Fob1p binding activity and that binding to one region alone is sufficient for replication fork-blocking activity.
FIG. 6.
Replication fork-blocking activity of small RFB subfragments (24 bp). 2D analysis was performed as for Fig. 3D. DNA was prepared from NOY408-1bf with pTAK901 and pTAKc1 to -f4 (which carry the RFB fragments c1 to f4 [Fig. 5C]), digested with PvuII and StuI, and subjected to 2D analysis followed by Southern hybridization with probe 2 (Fig. 3A). Spots indicated by arrowheads show accumulation of Y-shaped DNA molecules at the RFB site.
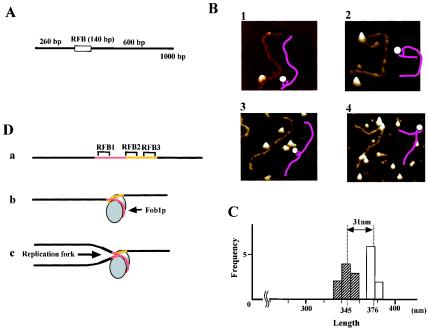
The RFB sequence wraps around Fob1p.
As Fob1p binds to both ends of the RFB sequence (f1 and f4 in Fig. 5B) and also interacts between these sites (f2 and f3 in Fig. 5A), I speculated that the RFB sequence may wrap around the protein. To test this, the Fob1p-RFB complex was observed by AFM. The substrate DNA was the RFB fragment (fragment 7, 140 bp) surrounded by nonyeast DNA fragments (Fig. 7A). The RFB site is located about one-third of the way along this fragment (the total length is 376 nm [1,000 bp]). Typical AFM images and the accompanying illustrations are shown in Fig. 7B. In the images, the DNA seems to be wrapping around Fob1p. Among the observed complexes, about 90% of the Fob1p molecules were located at the RFB site, and the average length of the DNA fragments was 31 nm (∼90 bp) shorter than that of the original (Fig. 7C). This length corresponds to the distance between RFB1 and RFB3. This further suggests that in these complexes, the DNA wraps around Fob1p (Fig. 7D, model b). Therefore, the RFB sequence may wrap around Fob1p in vivo, and this structure might be important for function (see Discussion).
FIG. 7.
Observation of the Fob1-RFB complex by AFM. (A) Structures of RFB fragments used in AFM analysis. The box shows the RFB fragment (fragment 7 from Fig. 3B), and bars indicate non-rDNA sequence. (B) AFM images of the Fob1p-RFB complex in a quick surface plot format. The illustrations drawn as pink lines (DNA) and white circles (Fob1p) are interpretations of the AFM images. (C) Frequency plot (histogram) of the length of the RFB fragment measured from AFM images. White and hatched boxes indicate the numbers of free DNA molecules and Fob1p-RFB complexes, respectively. The mean fragment lengths are indicated below the graph by dotted lines. (D) Models of the Fob1p-RFB complex. a, the RFB fragment without Fob1p. The RFB1 region is shown in red, and RFB2 and RFB3 are shown in yellow. b, the wrapping model. The RFB sequence wraps around Fob1p. Fob1p may be acting as a dimer in this structure. See text for details. c, a model for how the replication fork is inhibited by the RFB wrapping around Fob1p. The fork is inhibited mainly at RFB1.
DISCUSSION
In this study I demonstrated by using ChIP and gel shift assays that Fob1p binds to the RFB site. Furthermore, I demonstrated that Fob1p binds to two sites in the RFB and that the activity is dependent on a putative zinc finger motif. Finally, the RFB site seems to have an unusual wrapping action around Fob1p, reminiscent of a nucleosome structure. These findings explain the blockless phenotype in a fob1 mutant and therefore implicate Fob1p as the central player in replication fork blocking by binding directly to the DNA and then mediating the blocking of the replication fork. The wrapping structure also explains why the RFB requires a long DNA sequence (∼100 bp). I speculate that this wrapping structure is important for stable binding to a silenced locus by behaving like a nucleosome.
Defossez et al., using immunofluorescence microscopy, first reported the nucleolar localization of Fob1p (6). More recently, Gadal et al., using microscopy, also observed that Fob1p was located at the nucleolar-nucleoplasmic interface with the rDNA (11). DNA binding activity of Fob1p was also suggested by the discovery of a zinc finger motif, which often contributes to DNA binding activity (7). Here I show that this motif is responsible for the association (Fig. 2B). Dlakic predicted the 3D structure of Fob1p and pointed out the similarity of a domain to the integrase catalytic core-like structure of retrotransposons (Fig. 2A) (7). I mutated the conserved asparatic acid of this core structure to alanine (D291A) (Fig. 2A) to see the effect on the function of Fob1p. As shown in Fig. 2D, the mutation was not important, at least for DNA binding, replication fork-blocking, and recombination activities in the rDNA. Therefore, the predicted integrase catalytic core-like structure may have another, unknown function.
We previously reported that there is a 69-bp minimum essential sequence required for inhibition of the replication fork with the plasmid-cloned RFB (29). However, here I demonstrate that a 24-bp sequence (f1 in Fig. 6, corresponding to RFB1) from the original 69-bp sequence is enough to arrest the fork. In addition, I found another replication fork-blocking activity outside the 69-bp sequence (f4 in Fig. 6, corresponding to RFB3). One reason for the increased sensitivity in detecting blocking activity in this system is that Fob1p is overexpressed by the galactose-inducible promoter. As there are ∼150 copies of the rDNA, each with an RFB site, with wild-type levels of Fob1p there might be not enough Fob1p to inhibit the replication forks on all of the RFB plasmids. Ward et al. identified two regions (RFB1 and RFB2) responsible for replication fork-blocking activity as two separated spots in a 2D analysis (47). RFB3 was identified by deletion analysis in a strain with only two copies of the rDNA (31). After deletion of HpaI-HindIII fragment which contains RFB1 and RFB2, weak replication fork-blocking activity (RFB3) still remains near the HindIII site. Ward et al. pointed out that in addition to the RFB2 sequence, an inverted repeat region (IR1 and IR2 in Fig. 5C) overlapping RFB3 is also required for the second spot in 2D analysis (47). Brewer et al. also noted the importance of the HindIII-EcoRI fragment (Fig. 1A), which includes RFB3, for the RFB activity (4). Here I show that a 24-bp sequence, corresponding to RFB3, is capable of binding to Fob1p and inhibiting the replication fork. However, I detected neither RFB activity nor Fob1p association of the RFB2 fragment, although RFB2 did interact with Fob1p in the footprinting assay (Fig. 5A). From these data, I speculate that the second spot observed in the 2D analysis (4, 47) (Fig. 3D) is the sum of RFB2 and RFB3 activities and that for the activity of RFB2, Fob1p association with RFB3 is necessary. This association makes it possible that RFB2 interacts with Fob1p to inhibit replication. The reason that the second spot disappeared with modification of the RFB2 sequence (47) may be that RFB3 by itself is not strong enough to be detected in their system.
We hypothesize that Fob1p acts as a barrier to avoid collision between the replication and transcription machineries. Recently, we detected such a collision in a fob1 derivative strain whose rDNA copy number is reduced to ∼20 (44). In this strain, most of the rDNA copies are expected to be transcribed (9), and the collision occurs frequently enough to be detected as a region of slow replication fork progression by 2D analysis. Such collision may impose a burden by inducing damage in the rDNA. Actually, the stability of the rDNA was reduced, and recombination frequently took place despite FOB1 deficiency in this strain. Recombination repair, which includes new DNA synthesis, may result in errors more frequently than normal DNA replication (43). Therefore, the RFB function may have evolved to prevent harmful mutations that result from collision (44).
In this study I demonstrated that Fob1p binds to the RFB. However, the molecular mechanism by which this complex inhibits the replication fork is still unknown. I speculate that one possible target of Fob1p may be DNA helicases, similar to the action of Tus in E. coli (1, 13, 32), although there is no homology between Fob1p and Tus. Ivessa et al. identified two DNA helicases (Rrm3p and Pif1p) which appear to function in rDNA replication (21). In an RRM3 mutant, replication stops at several sites in the rDNA. Moreover, the RFB activity doubles in the mutant. However, in a PIF1 mutant, the RFB activity decreases about threefold. Although the mechanism by which the helicases influence RFB activity is unknown, it is possible that their action is a target for Fob1p.
The binding action of Fob1p is unusual. AFM images appear to show the RFB sequence wrapped around the protein (Fig. 7D, model b). The distance between RFB1 and RFB3 (∼90 bp) suggests that Fob1p may act as a dimer in this wrapping structure, although it is difficult to know the circumference of Fob1p from its molecular weight. The binding activity of Fob1p was eliminated by disruption of the zinc finger motif, and therefore the motif seems to bind both RFB1 and RFB3 independently. This is consistent with Fob1p activity as a dimmer. The results in Fig. 3D and 6 suggest that RFB1 provides the strongest inhibition of the replication fork (Fig. 7D, model c). This is supported by its stronger binding activity to Fob1p in vitro (Fig. 5B). Therefore, the DNA binding site of Fob1p has different affinities for RFB1 and RFB2. Determination of the crystal structure of the protein will help reveal the precise structure of the Fob1p-RFB complex. It is noted that the GST tag fused to Fob1p is known to dimerize proteins. Therefore, there is a possibility that the tag induces dimerization, thus causing the RFB sequence to wrap around the GST-Fob1p dimer. However, the existence of adjacent RFB1 and RFB3 sites suggests that dimerization of Fob1p occurs in nature.
This wrapping structure itself is not essential for the RFB activity in the plasmid system, because small RFB fragments (24 bp of RFB1 or RFB3) inhibited the replication fork (Fig. 6). However, in the rDNA, the wrapping structure may be important for function. The RFB region is known to be silenced by Sir2p, where the chromatin structure becomes condensed (10). In this environment, the wrapping structure may be helpful for Fob1p to access the silenced chromatin structure and stay there. As shown in Fig. 1B, Fob1p association in the rDNA is detectable by ChIP assay without cross-linking. This indicates that the association is stably maintained during the high-salt washing process. Therefore, I speculate that by increasing the length of associating region, the wrapping nucleosome-like structure may allow Fob1p to bind the DNA and be stably maintained in the rDNA chromatin. Nucleosome mapping (48) around the RFB by using micrococcal nuclease indicated that nucleosomes are located around the RFB region, and, interestingly, the RFB site was also protected from the nuclease (unpublished data). This pattern of protection is consistent with a nucleosome-like wrapping structure of the Fob1p-RFB complex.
Acknowledgments
I thank M. Nomura (UCI, Irvine, Calif.) and T. Horiuchi (NIBB, Okazaki, Japan) for critical reading of the manuscript. I thank Y. Kamimura and H. Araki (National Institute of Genetics, Mishima, Japan) for technical advice on the ChIP assay, T. Yasuda and F. Hanaoka (RIKEN, Wako, Japan) for technical advice on the gel shift assay, and A. Ganley of our laboratory for preparation of the manuscript and discussion. Strain YK9 and pEG(KT) were kindly provided by Y. Kawasaki (Osaka University, Osaka, Japan).
This work was supported in part by grants-in-aid for scientific research 13141205, 13480234, and 14380332 from the Ministry of Education, Science and Culture, Japan; by a grant from the Ministry of Health and Welfare, Japan; and by a grant from the Human Frontier Science Organization, France.
REFERENCES
- 1.Bedrosian, C. L., and D. Bastia. 1991. Escherichia coli replication terminator protein impedes simian virus 40 (SV40) DNA replication fork movement and SV40 large tumor antigen helicase activity in vitro at a prokaryotic terminus sequence. Proc. Natl. Acad. Sci. USA 88:2618-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 3.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 4.Brewer, B. J., D. Lockshon, and W. L. Fangman. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71:267-276. [DOI] [PubMed] [Google Scholar]
- 5.Carrigan, C. M., J. A. Haarsma, M. T. Smith, and R. G. Wake. 1987. Sequence features of the replication terminus of the Bacillus subtilis chromosome. Nucleic Acids Res. 15:8501-8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Defossez, P. A., R. Prusty, M. Kaeberlein, S. J. Lin, P. Ferrigno, P. A. Silver, R. L. Keil, and L. Guarente. 1999. Elimination of replication fork block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3:447-455. [DOI] [PubMed] [Google Scholar]
- 7.Dlakic, M. 2002. A model of the replication fork blocking protein Fob1p based on the catalytic core domain of retroviral integrases. Protein Sci. 11:1274-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elion, E. A., and J. R. Warner. 1984. The major promoter element of rRNA transcription in yeast lies 2kb upstream. Cell 39:663-673. [DOI] [PubMed] [Google Scholar]
- 9.French, S. L., Y. N. Osheim, F. Cioci, M. Nomura, and A. L. Beyer. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritze, C. E., K. Verschueren, R. Strich, and R. E. Esposito. 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16:6495-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadal, O., S. Labarre, C. Boschiero, and P. Thuriaux. 2002. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 21:5498-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 13.Hidaka, M., T. Kobayashi, Y. Ishimi, M. Seki, T. Enomoto, M. Abdel-Monem, and T. Horiuchi. 1992. Termination complex in Escherichia coli inhibits SV40 DNA replication in vitro by impeding the action of T antigen helicase. J. Biol. Chem. 267:5361-5365. [PubMed] [Google Scholar]
- 14.Hidaka, M., T. Kobayashi, S. Takenaka, H. Takeya, and T. Horiuchi. 1989. Purification of a DNA replication terminus (ter) site-binding protein in Escherichia coli and identification of the structural gene. J. Biol. Chem. 264:21031-22107. [PubMed] [Google Scholar]
- 15.Hill, T. M., M. L. Tecklenburg, A. J. Pelletier, and P. L. Kuempel. 1989. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc. Natl. Acad. Sci. USA 86:1593-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horiuchi, T., and Y. Fujimura. 1995. Recombinational rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J. Bacteriol. 177:783-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horiuchi, T., Y. Fujimura, H. Nishitani, T. Kobayashi, and M. Hidaka. 1994. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J. Bacteriol. 176:4656-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horiuchi, T., M. Hidaka, and T. Kobayashi. 1991. Termination of chromosome duplication in bacteria, p. 25-46. In A. lshihama and H. Yoshikawa (ed.), Control of cell growth and division. Japan Scientific Society Press, Tokyo, Japan.
- 19.Huang, G. S., and R. L. Keil. 1995. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics 141:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai, S.-I., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 21.Ivessa, A. S., J. Q. Zhou, and V. A. Zakian. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100:479-489. [DOI] [PubMed] [Google Scholar]
- 22.Johzuka, K., and T. Horiuchi. 2002. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells 7:99-113. [DOI] [PubMed] [Google Scholar]
- 23.Kaeberlein, M., M. McVey, and L. Guarente. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics, p. 207-209. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Kamimura, Y., Y.-S. Tak, A. Sugino, and H. Araki. 2001. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20:2097-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keil, R. L., and G. S. Roeder. 1984. Cis-acting recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39:377-386. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, T., D. J. Heck, M. Nomura, and T. Horiuchi. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi, T., M. Hidaka, and T. Horiuchi. 1989. Evidence of a ter specific binding protein essential for the termination reaction of DNA replication in Escherichia coli. EMBO J. 8:2435-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi, T., M. Hidaka, M. Nishizawa, and T. Horiuchi. 1992. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol. Gen. Genet. 233:355-362. [DOI] [PubMed] [Google Scholar]
- 30.Kobayaski, T., and T. Horiuchi. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1:465-474. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi, T., M. Nomura, and T. Horiuchi. 2001. Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:136-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, E. H., A. Kornberg, M. Hidaka, T. Kobayashi, and T. Horiuchi. 1989. Escherichia coli replication termination protein impedes the action of helicases. Proc. Natl. Acad. Sci. USA 86:9104-9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei, M., Y. Kawasaki, and B. K. Tye. 1996. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae Mol. Cell. Biol. 16:5081-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, P. J., M. T. Smith, and R. G. Wake. 1989. A protein involved in termination of chromosome replication in Bacillus subtilis binds specifically to the terC site. J. Bacteriol. 171:3564-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linskens, M. H. K., and J. A. Huberman. 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4927-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merker, R. J., and H. L. Klein. 2002. hpr1Δ affects ribosomal DNA recombination and cell life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:421-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell, D. A., T. K. Marshall, and R. J. Deschenes. 1993. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9:715-722. [DOI] [PubMed] [Google Scholar]
- 39.Nogi, Y., R. Yano, and M. Nomura. 1991. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. USA 88:3962-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothstein, R., and S. Gangloff. 1999. The shuffling of a mortal coil. Nat. Genet. 22:4-6. [DOI] [PubMed] [Google Scholar]
- 41.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 42.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91:1033-1042. [DOI] [PubMed] [Google Scholar]
- 43.Strathern, J. N., B. K. Shafer, and C. B. McGill. 1995. DNA synthesis errors associated with double-strand-break repair. Genetics 140:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi, Y., T. Horiuchi, and T. Kobayashi. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17:1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka, T., D. Knapp, and K. Nasmyth. 1997. Loading of a Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90:649-660. [DOI] [PubMed] [Google Scholar]
- 46.Wai, H., K. Johzuka, L. Vu, K. Eliason, T. Kobayashi, T. Horiuchi, and M. Nomura. 2001. Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol. Cell. Biol. 21:5541-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward, T. R., M. L. Hoang, R. Prusty, C. K. Lau, R. L. Keil, W. L. Fangman, and B. J. Brewer. 2000. Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: shared sequences but independent activities. Mol. Cell. Biol. 20:4948-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, C. 1980. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286:854-860. [DOI] [PubMed] [Google Scholar]