SUMMARY
Eukaryotic chromosome maintenance requires telomeric repeat synthesis by telomerase. It remains uncertain how telomerase domains interact to organize the active RNP and how this architecture establishes the specificity of the catalytic cycle. We combine human telomerase reconstitutions in vivo, affinity purifications, and discriminating activity assays to uncover a network of protein-protein and protein-RNA domain interactions. Notably, we find that complete single-repeat synthesis requires only a telomerase reverse transcriptase (TERT) core. Single-repeat synthesis does not require the TERT N-terminal (TEN) domain, but RNA-dependent positioning of the TEN domain captures substrate and allows repeat synthesis processivity. A TEN domain physically separate from the TERT core can capture even a minimal template-paired DNA substrate, with substrate association enhanced by the presence of a 5′ single-stranded extension. Our results provide insights into active enzyme architecture, explain biological variations of the catalytic cycle, and predict altered activities for TERT proteins of some eukaryotes.
Keywords: Telomerase ribonucleoprotein, reverse transcriptase activity, telomeric repeat synthesis, nucleic acid interaction, single-stranded DNA
INTRODUCTION
Most eukaryotic chromosomes terminate with a tract of telomeric repeats. These repeats associate with proteins to form chromatin structures that protect authentic telomeres from inappropriate recombination or repair (O’Sullivan and Karlseder, 2010). The erosion of telomeric repeats with each round of genome replication poses a challenge for long-term genome stability and cellular renewal. Almost universally, compensating de novo repeat synthesis is required for organismal viability. Acting as a specialized reverse transcriptase (RT), telomerase copies a template in the integral telomerase RNA subunit (TER) to elongate chromosome 3′ ends (Blackburn and Collins, 2010). The active site of TERT shares consensus motifs with retroelement RTs, but TERT also has telomerase-specific domains required for enzyme activity (Wyatt et al., 2010). Although TERT and TER can reconstitute an active RNP when combined in a heterologous cell lysate, endogenously functional telomerase holoenzymes are much larger multisubunit complexes assembled along a biogenesis pathway requiring cascades of assisting chaperones (Collins, 2008). For example, human telomerase holoenzyme biogenesis must load a monomer of human telomerase RNA (hTR) with two complete sets of H/ACA RNP proteins before the RNP can recruit a monomer of TERT (Errington et al., 2008; Egan and Collins, 2010). Lacking the extensive cohort of chaperones and cofactors that provide assembly specificity in vivo, in vitro reconstitution of human TERT and hTR typically yields heterogeneous complexes with varying enzyme properties (Mizuno et al., 2007).
Telomerase differs from retroviral RTs in its synthesis of a single-stranded rather than double-stranded product and its regeneration of the single-stranded RNA template (Blackburn and Collins, 2010). This more thermodynamically challenging activity obliges a catalytic cycle with complex single-stranded nucleic acid handling (Collins, 2009). The initial placement of template in the active site depends on positioning cues given by template-flanking regions of TER, as well as by the extent of template base-pairing with a substrate 3′ end (Wang et al., 1998; Miller and Collins, 2002). In the elongation phase, template transit through the active site is limited by 5′-flanking RNA secondary structure and TERT interaction (Tzfati et al., 2000; Lai et al., 2002; Chen and Greider, 2003). Coordinated with template handling, single-stranded regions of substrate or product DNA must also be threaded by the enzyme. As characterized predominantly for the Tetrahymena and human telomerase holoenzymes, a single-stranded DNA region 5′ of the template hybrid can reduce primer Km and/or promote repeat addition processivity (RAP) by increasing product retention through cycles of hybrid dissociation and template translocation (Collins, 1999). Human and Tetrahymena minimal TERT+TER RNPs do support some RAP, but telomerase holoenzyme and holoenzyme-interacting proteins other than TERT provide the highest-affinity sites of single-stranded DNA interaction (Wang et al., 2007; Min and Collins, 2009).
Telomerase-specific features of enzyme mechanism originate from gains of function by TER and TERT. TER roles in the catalytic cycle have been studied most extensively for Tetrahymena and human telomerases, exploiting the comprehensive secondary structure models derived by phylogenetic comparison (Chen and Greider, 2004) and activity assays optimized by use of minimized trans-complementing RNA domains in vitro (Collins, 2009). At least in the Tetrahymena and human systems, a TERT RNA binding domain (TRBD) adjacent to the RT domain is necessary and sufficient for high-affinity TERT-TER interaction (Lai et al., 2001). Also in both systems a lower-affinity TER interaction with the TERT far N-terminal/essential N-terminal (TEN) domain has been demonstrated in vitro (Moriarty et al., 2004; O’Connor et al., 2005), although its functional significance remains unknown. The TEN domain has been ascribed a role in sequence-specific recognition of single-stranded DNA based on mutagenesis-induced changes in primer use and weakly specific DNA crosslinking (Wyatt et al., 2010). The only direct evidence for DNA contact to the TEN domain in a catalytically active RNP derives from primer crosslinking prior to elongation-dependent radiolabeling, which identified a tryptophan side chain near the Tetrahymena TEN/TRBD boundary as a contact site for single-stranded DNA (Romi et al., 2007). However, side-chain substitution at this position has no functional impact on primer binding, alignment, or elongation (Romi et al., 2007; Jacobs et al., 2007). Thus, beyond base-pairing to the template, the principles of DNA recognition and positioning that are fundamental to the telomerase catalytic cycle remain undefined.
The low efficiency of active telomerase reconstitution in vitro is a major barrier to elucidating TERT domain functions and overall telomerase RNP architecture. The inability to produce a high yield of active recombinant enzyme has also hindered the development of a reliable screening platform for therapeutically useful small-molecule telomerase inhibitors or activators. High-resolution structures have been obtained only for isolated motifs of Tetrahymena or human TER, an inactive truncation of the Tetrahymena TRBD, a region within the Tetrahymena TEN domain, and a full-length insect protein with a TRBD, RT domain, and CTE (Jacobs et al., 2006; Rouda and Skordalakes, 2007; Gillis et al., 2008; Mitchell et al., 2010; Sekaran et al., 2010; Zhang et al., 2010). Here we address the architecture of human telomerase domain interactions using cellular RNP assembly to ensure physiological specificity and using trans-complementation assays to validate functional domain boundaries. Our studies reveal a complex domain interaction architecture that traps template hybrid and single-stranded product within the RNP. Our findings suggest a molecular rationale for known and inferred strategies of telomerase regulation and predict the evolutionary diversification of telomerase catalytic activity.
RESULTS
Delineation of Human TERT Domain Boundaries and Interdomain Interactions
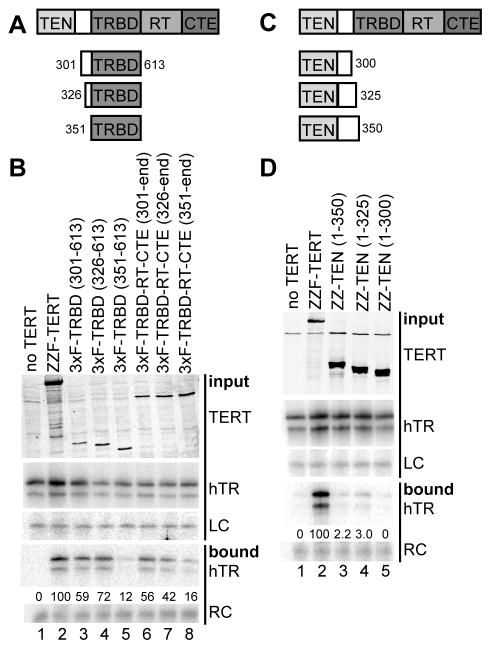
No individual domain or domain combination within human TERT has been produced recombinantly as a purified and functional polypeptide, leaving uncertain the boundaries between regions distinguished by primary sequence as TEN, TRBD, RT, and CTE. We introduced physical discontinuity as an approach to define TERT domain boundaries, analogous to the circular permutation and trans-complementation approaches used to define the functional architectures of Tetrahymena and human TERs. Non-overlapping combinations of TERT domains that together provided the full-length protein amino acid sequence were coexpressed in human VA-13 cells devoid of endogenous TERT and ectopically expressing hTR (see Figure 1A, lanes 5–12 for protein schematics). Cell extracts were then assayed for telomerase product synthesis using the sensitive PCR-amplified telomeric repeat amplification protocol (TRAP). Previous domain-multimerization studies of heterologously expressed human TERT suggest confusingly inconsistent TERT-TERT and TERT-hTR interaction requirements, likely due to the predominantly non-productive and non-physiological interactions permitted in recombinant expression systems such as rabbit reticulocyte lysate (RRL). However, with some variability in dependence on fragment endpoints, there is general support for productive trans-complementation of the TEN domain and TRBD-RT-CTE demonstrated using TERT fragments expressed both in RRL and by transfection of telomerase-negative human cells (Beattie et al., 2000; Beattie et al., 2001).
Figure 1. TERT Domain Interactions and Physically Separate Domain Function.
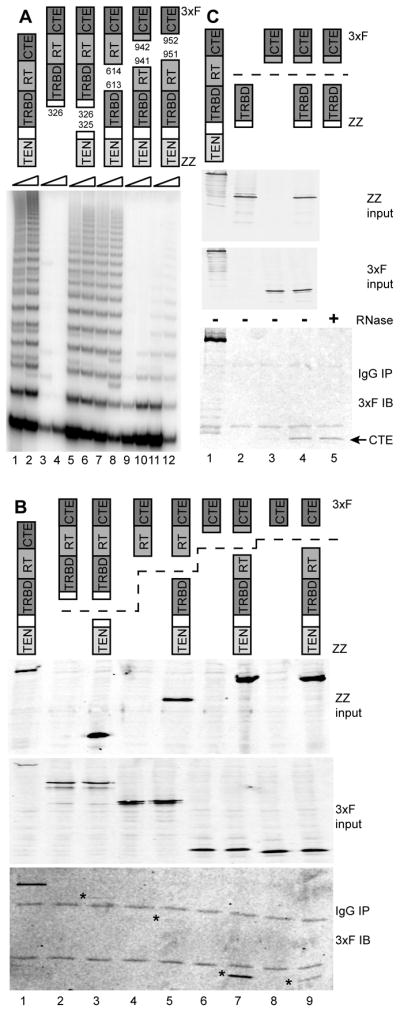
(A) Reconstitution of catalytic activity by TERT domain combinations. Amino acid positions of domain breakpoints are indicated. TRAP was performed using 0.3 or 1.0 μg total protein.
(B,C) Physical association of TERT domains. ZZ-tagged proteins were purified (IP) using IgG agarose, eluted, and examined for copurification of 3xF-tagged proteins by immunoblot (IB). In (B), asterisks mark the expected mobility of the 3xF-tagged protein. In (C), RNase A incubation was performed after binding to IgG agarose and before elution. See also Figure S1.
We modeled human TRBD boundaries based on the functionality of purified recombinant Tetrahymena TRBD (O’Connor et al., 2005). Vertebrate TERTs include a mutationally insensitive linker region between the TRBD and TEN domain of ~150 amino acids (Armbruster et al., 2001), which complicates the choice of a breakpoint by comparison to Tetrahymena TERT alone. Optimization based in part on human TRBD RNA-binding activity (see below) allowed the reconstitution of telomerase catalytic activity from physically separate TEN domain and TRBD-RT-CTE polypeptides or the TEN-TRBD and RT-CTE combination (Figure 1A, lanes 5–8). To investigate the RT domain/CTE boundary, we first exploited the structure of Tribolium TERT (Gillis et al., 2008). The domain breakpoint modeled to be least disruptive at a structural level, between amino acids 941 and 942, reconstituted very low telomerase activity from the physically separate TEN-TRBD-RT and CTE (Figure 1A, lanes 9–10), although it did reconstitute optimal interdomain protein interaction (see below). By minor adjustment of this domain boundary into the originally presumed CTE, to between amino acids 951 and 952, the combination of physically separate TEN-TRBD-RT and CTE produced reliably detectable catalytic activity (Figure 1A, lanes 11–12). It seems likely that the folding of both the RT domain and CTE was improved by inclusion of the small amino acid segment partitioned between them by the alternative backbone breakpoints. As a control for these complementation assays, TRAP assays of cell extracts containing any individual domain-truncated TERT gave only PCR artifacts rather than an evenly spaced telomeric repeat product ladder (Figure 1A, lanes 3–4; additional data not shown).
We assayed for interdomain protein interactions using a tandem Protein A (ZZ) tag appended to the N-terminus of each TEN-domain-containing polypeptide and a triple FLAG tag (3xF) appended to the C-terminus of each CTE-containing polypeptide. Tags at either location do not inhibit human telomerase holoenzyme catalytic activity assayed in vitro, although C-terminal tagging can be deleterious for telomere maintenance in vivo (Counter et al., 1998). As a control, full-length TERT was N-terminally tagged with the ZZ and 3xF tag combination. Each 3xF-tagged TERT fragment was expressed in 293T cells alone or with the ZZ-tagged complementary TERT fragment, and then complexes with the ZZ-tagged TERT fragment were enriched from extract by immunopurification on IgG agarose. Immunoblot detection of the copurification of the 3xF-tagged TERT fragment revealed a particularly strong physical association of the isolated CTE with TEN-TRBD-RT (Figure 1B, lanes 7 and 9). Importantly, as a negative control, the CTE was not enriched from extracts lacking coexpressed ZZ-TEN-TRBD-RT (Figure 1B, lanes 6 and 8). Copurification of 3xF-tagged RT-CTE with ZZ-TEN-TRBD was also reproducibly detectable although less robust, while 3xF-tagged TRBD-RT-CTE copurification with ZZ-TEN was at the borderline of detection (Figure 1B, lanes 2–5; additional data not shown). These results were surprising in their inverse correlation to the rank order of catalytic activity reconstitution (Figure 1A).
The physical association results suggest a potentially direct CTE-TRBD interaction, given that any readily detectable copurification of protein domains is likely to occur without a requirement for the limiting amount of hTR. Consistent with a protein-protein interaction, CTE copurification with TEN-TRBD-RT was not influenced by the level of coexpressed hTR (data not shown). To further test the hTR-dependence of TRBD-CTE association, we coexpressed the protein domains, enriched ZZ-tagged TRBD from cell extract, and detected copurification of the 3xF-tagged CTE in the presence or absence of RNase A. RNase-insensitive interaction of the TRBD and CTE was indeed observed (Figure 1C, lanes 4–5). Although isolated domain interaction assays have the caveat of domain removal from endogenous full-length protein context, a human TERT CTE-TRBD interaction is consistent with the extensive interface of CTE-TRBD contact observed in the high-resolution structure of Tribolium TERT (Gillis et al., 2008). We also detected a reproducibly robust CTE-TEN domain interaction that unlike the CTE-TRBD interaction was sensitive to the location of the CTE epitope tag (Supplemental Figure S1). We note that intramolecular domain associations are favored by the physiological conditions of TERT expression (Errington et al., 2008), but TERT over-expression or conditions of in vitro reconstitution could encourage intermolecular domain complementation.
Multiple Protein-RNA Interactions Network the TER domains, TEN domain, and TRBD
To address how protein-RNA interactions contribute to creating the active RNP architecture, we characterized hTR interactions with TERT domains using in vivo reconstitution. Human TRBD domain boundaries were not addressed in the initial characterization of the TRBD (Lai et al., 2001) and would be uncertain by comparison to the Tetrahymena TRBD at the N-terminal edge contiguous with the vertebrate-enlarged TEN domain linker (see schematics in Figure 2A). We therefore compared hTR copurification with a series of TRBD or TRBD-RT-CTE polypeptides with different N-terminal boundaries. Full-length TERT or N-terminally 3xF-tagged TRBD or TRBD-RT-CTE proteins were coexpressed with hTR by transient transfection of 293T cells, followed by purification using FLAG antibody resin and detection of coenriched hTR and a recovery control added before RNA precipitation (Figure 2B). N-terminal TRBD truncation from position 326 to 351 substantially compromised but did not eliminate hTR interaction (Figure 2B, lanes 4–5 and 7–8). We then tested TEN domain polypeptides with the matching series of C-terminal breakpoints (Figure 2C), all of which accumulated as N-terminally ZZ-tagged proteins in cell extract (Figure 2D, top panel). Much weaker enrichment of hTR was detected, but the longer TEN domain polypeptides reproducibly copurified hTR above the background control (Figure 2D, compare lanes 3–4 to lane 1).
Figure 2. TERT Domain Interactions with hTR.
(A,C) Schematics of investigated domain boundaries.
(B,D) TRBD and TEN domain interaction with hTR. Input samples had comparable levels of tagged protein and hTR relative to the total RNA loading control (LC). FLAG-antibody bound and eluted samples had differential hTR recovery (quantified) relative to the precipitation recovery control (RC).
To investigate the sequence specificity of TEN domain association with hTR, and to establish that it is not an indirect result of TEN domain intermolecular association with the low level of endogenous full-length TERT, we examined the hTR region required for each TERT domain interaction. The affinity of full-length TERT or TRBD for hTR is primarily determined by interaction with hTR conserved region (CR) 4/5 (Figure 3A), in particular the P6.1 stem and a subset of flanking internal bulge residues between P5 and P6 (Lai et al., 2001; Robart and Collins, 2010). At least one lower-affinity interaction must occur between TERT and the hTR template/pseudoknot (t/PK) region, based on full-length TERT interaction with the hTR-U64 chimera replacing the entire hTR H/ACA domain with the U64 small nucleolar RNA (Mitchell and Collins, 2000). Here we compared TERT domain interactions with full-length hTR versus hTR internally deleted for the CR4/5 region (Figure 3A). We coexpressed each RNA with full-length TERT or N-terminally 3xF-tagged TEN, TRBD, or TRBD-RT-CTE by transient transfection of 293T cells. All of the domains and domain combinations accumulated comparably in cell extract (Figure 3B, top panel).
Figure 3. Multiple Specificities of TERT-hTR Interaction.

(A) Illustration of hTR secondary structure indicating paired stems (P) and relevant motifs. Shading indicates the deleted CR4/5 region.
(B) Distinct hTR domain specificity of TERT-hTR interactions. FLAG-antibody bound and eluted samples were analyzed as described for Figure 2.
Full-length hTR was robustly copurified with full-length TERT, TRBD, or TRBD-RT-CTE (Figure 3B, lanes 2, 4, and 5). As expected hTR was less efficiently copurified with the TEN domain. Nonetheless, hTR was coenriched with the TEN domain above the background control (Figure 3B, compare lanes 1 and 3). From extracts with coexpressed hTRΔCR4/5, a different profile of enrichment was observed. Full-length TERT copurified hTRΔCR4/5 and also copurified some endogenous full-length hTR (Figure 3B, lane 7). The TRBD and TRBD-RT-CTE proteins copurified a level of hTRΔCR4/5 substantially greater than the background control, but compared to full-length TERT they preferentially coenriched the endogenous full-length hTR (Figure 3B, lanes 9–10). This comparison reveals that deletion of the TEN domain reduced TERT interaction with the t/PK region. Inversely, the TEN domain alone did not preferentially enrich full-length hTR relative to hTRΔCR4/5 (Figure 3B, lane 8). This finding indicates that the specificity of TEN domain interaction with hTR derives from its association with the t/PK region, not CR4/5. Together these results imply at least three hTR-TERT interactions: high-affinity interaction of CR4/5 with the TRBD and lower-affinity interactions of the t/PK region with the TEN domain and TRBD. Attempts to define by mutagenesis the specific t/PK-region determinants of TEN domain or TRBD binding were inconclusive, due to reduced in vivo accumulation of stem-disrupted hTRΔCR4/5 variants, low copurification recovery of even the wild-type t/PK-region RNA with either protein domain alone, and the potential for indirect effects of stem disruptions on general t/PK folding.
TEN Domain Association with the TERT Core
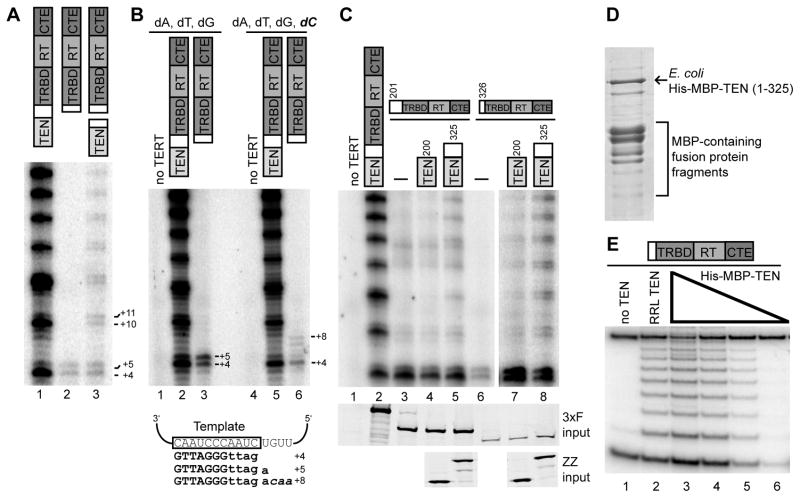
To investigate potential differences in RNP activity resulting from intramolecular versus intermolecular association of the TEN domain, we performed direct primer extension assays after FLAG antibody purification of N-terminally tagged full-length TERT, 3xF-tagged TRBD-RT-CTE, or 3xF-tagged TRBD-RT-CTE coexpressed with the trans-complementing ZZ-tagged TEN domain (see Figure 4A schematics). Purified RNPs were assayed for telomerase activity by radiolabeled nucleotide extension of the human telomeric-repeat primer substrate (T2AG3)3. To our surprise, the TRBD-RT-CTE RNP alone synthesized products representing primer elongation by complete single-repeat synthesis (Figure 4A, lane 2). This activity of this TRBD-RT-CTE TERT core was dependent on the coexpressed level of hTR, and the product ladder was offset appropriately dependent on the primer permutation of the telomeric repeat (Supplemental Figure S2A). Single-repeat synthesis would not be detected by a standard TRAP assay (Figure 1A, lanes 3–4), although it appears to have been detected by the TRAP conditions originally used in one laboratory (Beattie et al., 2000).
Figure 4. Activity of the TERT Core with or without or a Separate TEN Domain.
(A,B,C) Conversion of single-repeat synthesis to high RAP. Purified TERT or 3xF-TERT core (beginning at amino acid 326 unless indicated otherwise) with or without a coexpressed ZZ-tagged TEN domain (ending at amino acid 325 unless indicated otherwise) was assayed for primer extension. In (B), reactions in lanes 4–6 contained dCTP to allow incorporation of the additional nucleotides indicated in italic in the schematic below. In (C), lanes 7–8 are shown as a longer exposure of the same gel as adjacent lanes 1–6. See also Figure S2.
(D,E) Function of a bacterially expressed TEN domain. A Coomassie-stained gel of the partially purified protein is shown in (D). In (E), RRL-expressed 3xF-TRBD-RT-CTE was mixed with either RRL-expressed ZZ-TEN(1–325) as a positive control or a titration of partially purified His-MBP-TEN(1–325) prior to TRAP.
Notably, trans-complementation of the TERT core with the separately expressed TEN domain reconstituted processive repeat synthesis (Figure 4A, lane 3). The high RAP attained by the TERT core with a physically unlinked TEN domain suggests that cycles of dissociation of the template hybrid do not dissociate the TEN domain from the TERT core RNP. The overall level of processive repeat synthesis was lower with the trans-complementing versus intramolecular TEN domain, likely due at least in part to an incomplete stoichiometry of intermolecular TEN domain association. We observed that RAP reconstitution by TEN domain trans-complementation was sometimes stimulated by DNA primer addition prior to affinity purification. Therefore, unless noted otherwise (see below), trans-complementation assays were performed with DNA added to the extract and also to the activity assay. TERT core RNP single-repeat synthesis activity and TEN domain complementation to high RAP were observed independent of epitope tag position on the TERT core (Supplemental Figure S2B), with marginally higher activity recovered by purification of the N-terminally tagged TERT core used standardly in this work.
Close examination of the product profiles suggested that the minimal TERT core RNP has a reduced fidelity of template 5′ boundary definition. In standard primer extension assays, this altered specificity of template use is evident as an increased tendency for copying one nucleotide past the normal template 5′ boundary (note the relative strength of +5 versus +4 product, +11 versus +10 product, etc. in Figure 4A, lanes 2–3). Despite trans-complementing rescue of RAP, TEN domain expression did not rescue the altered template usage of the minimal TERT core RNP (Figure 4A). To demonstrate that the altered product profile resulted from template 5′ boundary bypass instead of enhanced product dissociation after the first nucleotide addition following template translocation, we performed activity assays that included dCTP. Inclusion of dCTP permits additional copying past the template 5′ boundary (see schematics in Figure 4B). The product profile of the full-length TERT RNP was insensitive to the presence of dCTP in the activity assay, but the +5 product of the TERT core RNP shifted to a series of longer products indicative of 5′ template boundary bypass (Figure 4B).
We next investigated the role of the vertebrate-expanded linker region separating the TEN domain and TRBD. Removing the linker region from the TEN domain did not prevent trans-complementation of high RAP but did reduce its efficiency (Figure 4C, lanes 7–8; Supplemental Figure S2C). This remained true even if the linker region was fused to the TERT core (Figure 4C, lanes 3–5). Addition of the linker region to the TERT core improved the fidelity of 5′ template boundary definition (Figure 4C, compare lanes 3–5 and 6–8), as to some extent did moving the epitope tag on the TERT core to its C-terminus (Supplemental Figure S2B), but none of the TERT core RNPs had the precision of repeat synthesis demonstrated by full-length TERT. We conclude that the linker is not strictly necessary for either the single-repeat synthesis activity of the TERT core RNP or for high RAP by trans-complementation of the TEN domain. However, TEN domain folding and/or its productive interaction with the TERT core RNP are improved when the TEN domain polypeptide includes the linker region.
The isolated Tetrahymena TEN domain expressed in E. coli is soluble and when purified retains functionality for TER binding (O’Connor et al., 2005). We therefore tested the ability of the bacterially expressed human TEN domain to fold autonomously into a soluble form functional for trans-complementation of RAP. Although previous studies have failed to detect activity-stimulatory function for the bacterially expressed human TERT TEN domain (Sealey et al., 2010), TEN(1–325) expressed in fusion to a six-histidine and maltose binding protein (MBP) combination tag could be partially purified (Figure 4D) and added to RRL-expressed 3xF-tagged TERT core to reconstitute dose-dependent processive repeat synthesis activity detectable by TRAP (Figure 4E). Overall we conclude that the TEN domain is a structurally and functionally peripheral module that serves to increase the precision and processivity of repeat synthesis.
TER Motif Roles Within and Beyond the TERT Core RNP
One obvious role of non-template TER motifs is to provide binding affinity for TERT, as characterized above and in previous work. Less obvious yet equally critical roles are played by TER motifs that do not contribute binding affinity, some of which can still function to position active RNP domains relative to each other. Several evolutionarily conserved non-template regions of TER have mechanistically uncertain roles in the catalytic cycle. Our ability to detect catalytic activity independent of the TEN domain and to monitor productive positioning of a trans-complementing TEN domain through the gain of RAP provided an opportunity to discriminate TER motif functions that are independent of or dependent on the TEN domain.
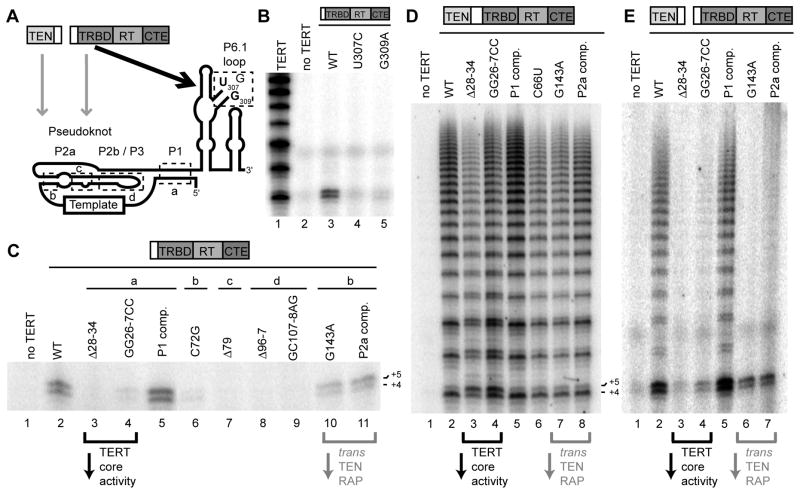
To investigate TERT core RNP activity dependence on conserved hTR motifs, we first reconstituted TERT core RNPs with hTR variants harboring sequence substitutions in the P6.1 loop nucleotides U307 and G309 (Figure 5A) that are strictly conserved in all sequenced vertebrate TERs (Podlevsky et al., 2008). P6.1 loop nucleotide substitutions do not affect hTR assembly with TERT in vitro or in vivo but nonetheless severely compromise the catalytic activity of full-length TERT RNPs (Chen et al., 2002; Robart and Collins, 2010). The U307C and G309A substitutions greatly reduced the single-repeat synthesis activity of the TERT core RNP (Figure 5B, lanes 3–5). We conclude that P6.1 loop residues have a critical role in template use that is independent of coordination with the TEN domain.
Figure 5. Distinct hTR Sequence Requirements for TERT Core, TERT Core and TEN Domain, or Full-length TERT RNP Activity.
(A) Schematic of TERT and hTR domain interactions and the hTR P6.1 loop nucleotides (in bold) and t/PK-region paired stem regions (in dashed boxes) investigated in this work.
(B,C,D,E) RNA sequence requirements for activity. RNPs were assayed for primer extension after purification on FLAG antibody resin. See also Figure S3.
We next investigated the catalytic activity of TERT core RNPs reconstituted with disease-linked hTR variants in the P1, P2, and P3 stems of the t/PK region (Figure 5A; substituted stem regions are boxed and labeled a–d). Disease-linked t/PK-region substitutions compromise the catalytic activity of full-length TERT RNPs without substantial impact on hTR assembly with TERT in vivo (Robart and Collins, 2010). When reconstituted as TERT core RNPs (Figure 5C), most of the disease-linked t/PK substitutions imposed an extent of activity inhibition parallel to that detected in full-length TERT context (Robart and Collins, 2010). One exception is the much more severe defect imposed by the P1 disruption Δ28–34 in the TERT core RNP compared to the full-length TERT RNP (compare Figures 5C and 5D, lanes 2–3). An independent P1 disruption also much more severely compromised the activity of the TERT core RNP than the full-length TERT RNP, and this inhibition was rescued by compensatory restoration of P1 base-pairing (Figures 5C and 5D, lanes 4–5). As noted previously (Chen and Greider, 2003; Robart and Collins, 2010), P1 disruption in the full-length TERT RNP allows template 5′ boundary bypass, evident in the first-repeat ratio of accurate +4 to extended +5 product that is perpetuated in each cycle of repeat synthesis (Figure 5D). We suggest that P1 integrity is important for the low-affinity TRBD contact with the t/PK region, which could contribute to template positioning in a manner particularly critical in TERT core RNP context (see Discussion). Loss of this interaction may be partially compensated in the full-length TERT enzyme by t/PK positioning through contact with the TEN domain.
For all of the hTR variants that supported single-repeat synthesis by the TERT core, we tested whether TEN domain trans-complementation would confer RAP. Although P1 disruption reduced activity level overall, it did not inhibit conversion of single-repeat synthesis to high RAP (Figure 5E, lanes 3–4). In contrast, although the P2a G143A substitution did not prevent single-repeat synthesis by the TERT core RNP (Figure 5C, lane 10), it severely inhibited TEN domain trans-complementation of RAP (Figure 5E, lane 6). Combining the G143A substitution with the compensatory C66U substitution to restore P2a base-pairing did not completely rescue the trans-complementation defect (Figure 5E, lane 7). In full-length TERT RNP context, neither the G143A nor C66U substitution nor the compensatory combination was strongly inhibitory (Figure 5D, lanes 6–8), indicating that physical linkage of the TEN domain and TRBD partially overcomes the defect imposed by P2a substitution in the TERT core RNP. These results suggest that changes of P2a structure affect the RAP-stimulatory positioning of the TEN domain in a manner sensitized by TEN domain trans-complementation. This could be due to loss of a direct hTR-TEN domain interaction or more likely an indirect, dominant-negative effect of altered t/PK folding, as the P2a region is not conserved among vertebrate TERs (Podlevsky et al., 2008). We conclude that TEN domain recruitment and function depend on a multiplicity of factors including t/PK-region structure, hTR-stimulated TEN domain association with the TERT core (Supplemental Figure S3), and potential protein domain interactions (Supplemental Figure S1).
The TEN Domain Captures Substrate and Product
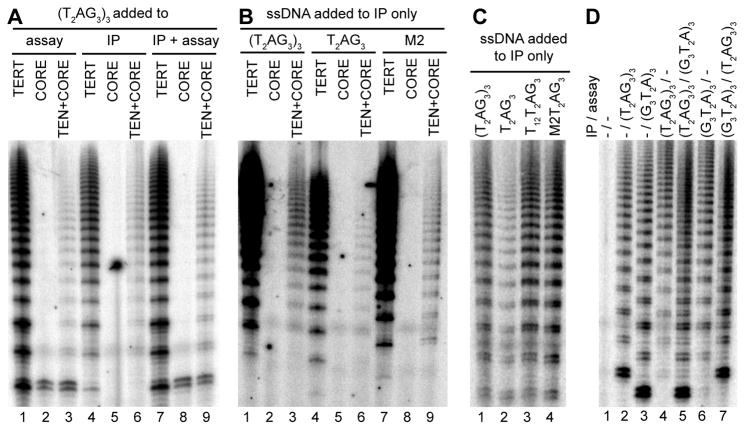
An excess of single-stranded DNA was included in the activity assay reactions above to discriminate multiple-repeat products as a consequence of RAP rather than re-initiation on a previously elongated product. To investigate the stability of telomerase RNP association with substrate DNA, we performed assays with DNA added to the extract prior to RNP purification without additional DNA added to the activity assay. The high-RAP activity of the full-length TERT RNP or the TERT core RNP with separately expressed TEN domain could be detected by elongation of a DNA substrate bound in extract, while the TERT core RNP alone could not capture substrate in a manner stable to purification (Figure 6A, lanes 4–6). As a control, DNA addition to the activity assay allowed repeat synthesis by all enzymes including the TERT core RNP (Figure 6A, lanes 1–3 and 7–9).
Figure 6. Substrate Capture by the TEN Domain.
DNA oligonucleotides were added to the RNP purification and/or activity assay as indicated.
To address whether stable substrate capture requires TEN domain recognition of single-stranded DNA, we tested whether various substrates added to cell extract would be retained during purification and subsequently elongated in the activity assay reaction. The TERT core RNP lacking the TEN domain was not able to capture any substrate in a manner stable to affinity purification, whereas the full-length TERT RNP and the TERT core RNP with trans-complementing TEN domain could both productively capture not only the three-repeat primer (T2AG3)3 but also the single-repeat primer T2AG3 or the non-telomeric TRAP primer M2 with a template-complementary GTT 3′ end (Figure 6B). These findings suggest that any DNA that can form a template hybrid can be captured in a TEN-domain-dependent manner. All captured substrates were elongated with a high-RAP product profile (with product intensity differences affecting the extent of the visible product ladder), which would be logical given that first-repeat synthesis endows all products with a single-stranded telomeric-repeat extension.
To assess whether the length and/or sequence of single-stranded extension from the template hybrid affects the efficiency of substrate capture, we compared the capture of substrates with the same 3′ single telomeric repeat but different 5′ regions. Any sequence added 5′ of the single-repeat T2AG3 increased the efficiency of substrate capture (Figure 6C). Increasing the DNA concentration added to cell extract before RNP purification did not provide the stimulation gained by increasing primer length (data not shown). As an additional control, we challenged RNPs purified in the presence of (T2AG3)3 or (G3T2A)3 with the alternate primer added to the activity assay. Challenge primers could be elongated but did not prevent elongation of the originally captured substrate (Figure 6D), consistent with purification of a mixture of substrate-captured and substrate-free RNP. Based on the overall analysis of substrate capture and elongation RAP, we suggest that the template hybrid and single-stranded product become trapped within the network of protein-protein and protein-RNA domain interactions that literally internalize a substrate-engaged active site within the active RNP (Figure 7).
Figure 7. Model for Active RNP Architecture.
Substrate capture and product trapping may result from TEN-domain-dependent internalization of the active site within the RNP. For clarity only the paired stems and some single-stranded connectors of the hTR t/PK region are depicted, without incorporation of helical twist.
DISCUSSION
Functional Architecture of the Active Human Telomerase RNP
We initiated these studies to probe interdomain communication during telomerase RNP assembly and over the catalytic cycle. Using physical and functional approaches, we uncovered a network of protein-protein and protein-RNA interactions that organize TERT core RNP domains and position the TEN domain for substrate capture and product retention. These studies yielded the observation that the human TERT TEN domain is dispensable for complete single-repeat synthesis. N-terminally truncated Saccharomyces cerevisiae TERTs can also retain nucleotide addition activity, particularly with substrates that should form a relatively stable template hybrid (Lue, 2005). Evolutionary divergence in the stability of template-product hybrid and the strength of duplex binding in the active site may mask a conserved functionality of the TERT core in an RT-like synthesis reaction. Importantly, the single-repeat synthesis activity of the human TERT core RNP could be converted to a full-length TERT RNP product profile by trans-complementation of the TEN domain. TEN domain association with the TERT core RNP remained stable to both affinity purification and to reiterative dissolution of the template hybrid during processive repeat synthesis. The linker between the TEN domain and TRBD was not critical for trans-complementation of high RAP, but it greatly stimulated the reconstitution of high-RAP activity when included as part of the TEN domain polypeptide.
In addition to conserved TERT interdomain interactions, our results suggest evolutionary conservation of multiple TERT domain interactions with TER. With the same overall topology proposed here for human telomerase, the Tetrahymena TRBD binds to both the t/PK region and a stem-loop near the RNA 3′ end (Blackburn and Collins, 2010). Human TERT-hTR interaction is most sensitive to CR4/5 disruption while Tetrahymena TERT-TER interaction is most sensitive to disruption of the template-adjacent stem of the t/PK region (Mitchell and Collins, 2000; Lai et al., 2001). This evolutionary change in the relative affinity of individual TRBD-TER interactions should not have a major physiological consequence, because in vivo RNP biogenesis first assembles other proteins between the TER sites of TRBD interaction that can improve the synergy of subsequent TRBD-TER associations (Stone et al., 2007; Egan and Collins, 2010). In the active human RNP, TRBD interaction with P1 would account for why template copying does not reach the edge of P1: as in the Tetrahymena RNP, protein-RNA interaction acts as a steric barrier to template transit of the active site (Lai et al., 2002). In addition to conserved TRBD-TER interactions, our findings suggest a conserved TEN domain interaction with the t/PK region. Previous studies using RRL led to the conclusion that the human TERT TEN domain binds P1 (Moriarty et al., 2004; Moriarty et al, 2005), but results here suggest otherwise because P1 disruption reduced TERT core RNP activity rather than TEN domain trans-complementation of this activity to high RAP. Ultimately a combination of TEN domain protein-RNA, protein-protein, and protein-DNA interactions may be necessary to direct productive TEN domain positioning.
Implications for Altered Specificity of Enzyme Activity with Assembly, Regulation, and Evolution
Heterologously expressed TERTs have been reported to form a diverse collection of intermolecular associations that do not necessarily occur in the physiologically active RNP. Results above suggests a controlled order-of-addition strategy for improving the structural homogeneity of recombinant telomerase: TERT core expression under conditions that favor intramolecular folding, RNP assembly under conditions that promote interaction of both RNA domains with the same TERT core, and subsequent intermolecular association of the TEN domain. It seems highly likely that in vivo regulation controls the ability of telomerase to engage DNA substrates, potentially through control of TEN domain conformation. From this perspective, the vertebrate TERT expansion of the linker between the TEN domain and TRBD may be a signature of increased regulation. Prior to template hybrid formation, an open TEN domain conformation may be beneficial for substrate sampling. Conversion to the substrate-trapping TEN domain conformation could then be favored only after template hybrid formation with a chromosome terminus. In human cells or extracts, Hsp90 inhibition reduces RNP assembly and association with primer (Holt et al., 1999; Keppler et al., 2006). S. cerevisiae Hsp82 and coordinated factors also affect telomerase association with substrates (DeZwaan and Freeman, 2010). Previous observations can be unified by proposing Hsp90 control of the TEN domain linker: drug inhibition or depletion of Hsp90 could affect the conformational freedom of the TEN domain to allow TRBD access to assembly with hTR, substrate access to the template, and release of enzyme-product interaction. Reconstituted telomerase RNPs may be most sensitive to Hsp90 inhibition if they assemble with a cis-docked rather than trans-docked TEN domain, accounting for the differential Hsp90 inhibitor dependence of distinct populations of reconstituted active human telomerase (Mizuno et al., 2007).
Different cellular modes of telomerase action have been proposed including processive repeat synthesis, single-repeat synthesis, DNA repair (potentially through DNA synthesis), and chromosome end-capping (independent of DNA synthesis). Our results suggest that a telomerase holoenzyme could be constrained in TEN domain positioning to support only single-repeat synthesis. Also, because the human TERT core RNP lacking the TEN domain can copy a much longer than normal template region (almost twice the normal length in reactions with dCTP), TEN domain regulation may explain the different extents of template copying by S. cerevisiae telomerase at sites of chromosome healing versus established telomeres: a greater extent of the template is copied in the initial elongation of a broken chromosome than is copied for telomere maintenance (Kramer and Haber, 1993; Singer and Gottschling, 1994). Furthermore, it is plausible that some conformation of the TEN domain allows a template hybrid to form that cannot support elongation yet can confer chromosome end-capping activity (Chan and Blackburn, 2002). Regulatory factors could inhibit TEN domain docking on the TERT core by binding to the TEN domain or template-adjacent TER motifs. This antagonism is one of many mechanisms that could account for human telomere shortening upon overexpression of hnRNP C, which interacts with the hTR region between the template and P1 (Fu and Collins, 2007).
Finally, our findings predict altered specificities of activity for the domain-truncated TERT proteins of some eukaryotes (Blackburn and Collins, 2010). For example, insect TERTs have no N-terminal extension from their compact TRBD. We speculate that a TEN-less insect TERT could copy a long template without restriction from TEN domain trapping of template hybrid in the active site, potentially generating a duplex product like a retroelement RT. Evolutionary truncation of the TEN domain would not preclude a telomere maintenance function for insect TERTs, but our findings suggest that it would preclude the processive repeat synthesis typically necessary for TRAP detection of a product ladder.
EXPERIMENTAL PROCEDURES
Extract Preparation and Purifications
Whole-cell extracts were made by three cycles of freeze-thaw lysis in HLB buffer (20 mM HEPES at pH 8.0, 2 mM MgCl2, 10% glycerol, 0.2 mM EGTA, 0.1% NP-40, 1 mM DTT) followed by adjustment to 0.4 M NaCl and centrifugation to clear the extract. Affinity purifications were performed from extract adjusted to a final concentration of 150 mM NaCl. Rabbit IgG agarose or FLAG M2 antibody resin was added followed by end-over-end incubation at 4°C for 2 h. If single-stranded DNA was added to extract prior to RNP purification, a final concentration of 1 μM was used. Bound samples were washed three times with HLB containing 150 mM NaCl, 0.1% Triton X-100, and 0.2% CHAPS at room temperature. For domain interaction experiments, bound RNPs were eluted using protease (for ZZ-tagged proteins) or a triple-copy FLAG peptide as described (Egan and Collins, 2010). For bacterial expression, the TEN domain with N-terminal six-histidine and MBP tags was produced in BL21 (DE3) RP cells and partially purified by binding to and elution from Ni-NTA resin.
Activity Assays
TRAP assays were performed with samples diluted into HLB buffer (Errington et al., 2008). Primer extension activity assays were performed in a 20 μl reaction volume with RNPs immobilized on affinity resin washed into HLB buffer with no NaCl. Reaction buffer contained additional final concentrations of 50 mM Tris-acetate at pH 8.0, 4 mM MgCl2, 5 mM DTT, 250 μM dTTP, 250 μM dATP, 5 μM unlabeled dGTP, and 0.5 μL of 3,000 Ci/mmole α32P-dGTP. For activity assays investigating template boundary bypass, 250 μM dCTP was included in the reaction. If single-stranded DNA was added to the activity assay, a final concentration of 500 nM was used. Unless indicated otherwise the elongation primer was (T2AG3)3. M2 has the sequence AATCCGTCGAGCAGAGTT. Reactions were incubated at 30°C for 1 h. Products were extracted, precipitated, and resolved on an 11% (19:1), 0.6X TBE, 7 M urea gel; dried gels were imaged by Typhoon.
Supplemental Experimental Procedures describe expression constructs and subunit detection details.
Supplementary Material
Acknowledgments
We thank Kwan Chow, Barbara Eckert, Emily Egan, and Alec Sexton for plasmids, discussions, and comments on the manuscript. This research was funded by NIH HL079585.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armbruster BN, Banik SSR, Guo C, Smith AC, Counter CM. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol Cell Biol. 2001;22:7775–7786. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie TL, Zhou W, Robinson MO, Harrington L. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol Biol Cell. 2000;11:3329–3340. doi: 10.1091/mbc.11.10.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie TL, Zhou W, Robinson MO, Harrington L. Functional multimerization of the human telomerase reverse transcriptase. Mol Cell Biol. 2001;21:6151–6160. doi: 10.1128/MCB.21.18.6151-6160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Collins K. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a003558. 10.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Blackburn EH. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Greider CW. An emerging consensus for telomerase RNA structure. Proc Natl Acad Sci USA. 2004;101:14683–14684. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Opperman KK, Greider CW. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 2002;30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. Ciliate telomerase biochemistry. Annu Rev Biochem. 1999;68:187–218. doi: 10.1146/annurev.biochem.68.1.187. [DOI] [PubMed] [Google Scholar]
- Collins K. Physiological assembly and activity of human telomerase complexes. Mech Ageing Dev. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. Forms and functions of telomerase RNA. In: Walter NG, Woodson SA, Batey RT, editors. Non-Protein Coding RNAs. Berlin: Springer-Verlag; 2009. pp. 285–301. [Google Scholar]
- Counter CM, Hahn WC, Wei W, Caddle SD, Beijersbergen RL, Lansdorp PM, Sedivy JM, Weinberg RA. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc Natl Acad Sci USA. 1998;95:14723–14728. doi: 10.1073/pnas.95.25.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan DC, Freeman BC. HSP90 manages the ends. Trends Biochem Sci. 2010;35:384–391. doi: 10.1016/j.tibs.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol. 2010;30:2775–2786. doi: 10.1128/MCB.00151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington TM, Fu D, Wong JM, Collins K. Disease-associated human telomerase RNA variants show loss of function for telomere synthesis without dominant-negative interference. Mol Cell Biol. 2008;28:6510–6520. doi: 10.1128/MCB.00777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28:773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. Erratum. Nat Struct Mol Biol. 2007;14:984. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- Keppler BR, Grady AT, Jarstfer MB. The biochemical role of the heat shock protein 90 chaperone complex in establishing human telomerase activity. J Biol Chem. 2006;281:19840–19848. doi: 10.1074/jbc.M511067200. [DOI] [PubMed] [Google Scholar]
- Kramer KM, Haber JE. New telomeres in yeast are initiated with a highly selected subset of TG1–3 repeats. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF. A physical and functional constituent of telomerase anchor site. J Biol Chem. 2005;280:26586–26591. doi: 10.1074/jbc.M503028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. Proc Natl Acad Sci USA. 2002;99:6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase in vivo and in vitro. Mol Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Khurts S, Seki T, Hirota Y, Kaneko S, Murakami S. Human telomerase exists in two distinct active complexes in vivo. J Biochem. 2007;141:641–652. doi: 10.1093/jb/mvm071. [DOI] [PubMed] [Google Scholar]
- Moriarty TJ, Marie-Egyptienne DT, Autexier C. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol Cell Biol. 2004;24:3720–3733. doi: 10.1128/MCB.24.9.3720-3733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty TJ, Ward RJ, Taboski MA, Autexier C. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol Biol Cell. 2005;16:3152–3161. doi: 10.1091/mbc.E05-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. The telomerase database. Nucleic Acids Res. 2008;36:D339–343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Collins K. Investigation of human telomerase holoenzyme assembly, activity, and processivity using disease-linked subunit variants. J Biol Chem. 2010;285:4375–4386. doi: 10.1074/jbc.M109.088575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romi E, Baran N, Gantman M, Shmoish M, Min B, Collins K, Manor H. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc Natl Acad Sci USA. 2007;104:8791–8796. doi: 10.1073/pnas.0703157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: Implications for RNA recognition and binding. Structure. 2007;13:1403–1412. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Sealey DC, Zheng L, Taboski MA, Cruickshank J, Ikura M, Harrington LA. The N-terminus of hTERT contains a DNA-binding domain and is required for telomerase activity and cellular immortalization. Nucleic Acids Res. 2010;38:2019–2035. doi: 10.1093/nar/gkp1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekaran VG, Soares J, Jarstfer MB. Structures of telomerase subunits provide functional insights. Biochim Biophys Acta. 2010;1804:1190–1201. doi: 10.1016/j.bbapap.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Stone MS, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Wang H, Gilley D, Blackburn EH. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J. 1998;17:1152–1160. doi: 10.1093/emboj/17.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010;38:5609–5622. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. Inaugural Article: Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc Natl Acad Sci USA. 2010;107:18761–18768. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.