Abstract
Acetylation and phosphorylation of the amino-terminal tails of the core histones fluctuate on a global scale in concert with other major events in chromosome metabolism. A ubiquitin ligase, the anaphase-promoting complex (APC), controls events in chromosome metabolism such as sister chromatid cohesion and may regulate H3 phosphorylation by targeting Aurora A, one of several S10-directed H3 kinases in vertebrate cells, for destruction by the proteasome. Our analysis of apc10Δ and apc11ts loss-of-function mutants reveals that the APC controls the global level of H3 S10 phosphorylation in cycling yeast cells. Surprisingly, it also regulates dephosphorylation of H3 and global deacetylation of H2B, H3, and H4 during exit from the cell cycle into G0. Genetic, biochemical, and microarray analyses suggest that APC-dependent cell cycle control of H3 phosphorylation is exerted at the level of an Aurora H3 kinase, Ipl1p, while APC-dependent transcriptional induction of GLC7, an essential H3 phosphatase, contributes to sustained H3 dephosphorylation upon cell cycle withdrawal. Collectively, our results establish that core histone acetylation state and H3 phosphorylation are physiologically regulated by the APC and suggest a model in which global reconfiguration of H3 phosphorylation state involves APC-dependent control of both an H3 kinase and a conserved phosphatase.
The flexible amino-terminal tails of the core histones are subject to a variety of site-specific posttranslational modifications (reviewed in reference 34). Best characterized is acetylation, which occurs at conserved lysine residues in all core histones. Phosphorylation of H3 at S10 can be mechanistically linked to H3 acetylation and has also been well characterized (23, 32, 48, 49). It is possible to manipulate the extent of tail modification of the core histones on a global scale by artificial means. In budding yeast, for example, deletion of histone acetyltransferase (HAT) genes results in global deacetylation of target histones, and deletion of histone deacetylases (HDACs) results in the reverse (7, 12, 31, 62, 65, 75). The effects of HDAC deletion on global histone acetylation in yeast can be mimicked in mammalian cells by treatment with HDAC inhibitors (82). Furthermore, mutations that partially cripple an H3 S10 kinase (Ipl1p) or the Glc7p catalytic subunit of an H3-directed type 1 protein phosphatase (PP1) globally affect the accumulation of S10-phosphorylated H3 in yeast (32). Consistent with the general observation that the global state of histone modification is sensitive to artificial manipulation of histone-modifying enzymes, it has been found that core histone acetylation and phosphorylation are physiologically regulated on a global scale. Typically the global reconfiguration of histone modification state under physiological circumstances coincides with other major events in chromatin metabolism. For example, S10 phosphorylation of H3 coincides with chromosome condensation during the cell cycle in many species (23), as does deacetylation of H3 and H4 in mammals (41). In yeast global H3 acetylation increases in S phase when the genome is being replicated (39).
Considering the possibility that global reconfiguration of histone modification state might generally accompany global transitions in chromosome organization, we explored the regulation of histone acetylation and phosphorylation in another physiological context that involves genome-wide effects on chromosome structure and function, namely, exit of yeast cells from proliferative growth into G0 arrest. Nutrient deprivation triggers yeast cells to exit into G0 from G1 phase of the cell cycle (78, 79). Cells develop G0 phenotypes through the orderly execution of a series of molecular events analogous to a differentiation program in higher eukaryotes (28, 78, 79). These events include a phase during which the proliferation rate is greatly reduced (the postdiauxic phase), followed by cell cycle arrest with a 1n content of DNA. Large-scale transcriptional reprogramming occurs during entry into G0 (14), such that the content of protein-coding poly(A)+ RNA is reduced at least threefold in terminally differentiated G0 cells (79). In addition, the chromosomes of G0 cells adopt a folded conformation not observed in cycling cells (60, 61, 79). Using immunoblotting, we show that substantial reconfiguration of histone phosphorylation and acetylation state occurs during the G0 program of development.
What is the nature of the regulatory network that coordinates reconfiguration of histone modification state with other events in cell cycle exit? Presumably it includes upstream sensors of the environmental conditions that trigger entry into G0, intermediate signaling components, and effectors. Little that might inform analysis of the physiological control of global histone acetylation state in G0 can be gleaned from the literature. However, what is known about cell cycle control of an H3 S10 kinase in metazoans is germane to our investigation of the global control of histone modification state in yeast. This enzyme, Aurora A (Aurora2), physically interacts with the amino-terminal tail of H3 and efficiently phosphorylates S10 in free and nucleosomal H3 (13, 70). During the division cycle, Aurora A is degraded in G1 and subsequently resynthesized. Its timely destruction is controlled by the anaphase-promoting complex (APC), or cyclosome, a highly conserved, multisubunit protein ubiquitin ligase (E3) (reviewed in references 26, 58, and 84). In G1 the APC ubiquitinates Aurora A, with the ubiquitin tag serving as the signal for rapid degradation of the kinase by the proteasome (9, 10, 30, 47).
Although the physiological significance of H3 phosphorylation by Aurora A in metazoans is yet to be clarified, the findings described above may be relevant to cell cycle control of the sole Aurora kinase in yeast, Ipl1p. Ipl1p is responsible for global H3 S10 phosphorylation during the cell cycle (32). Because its bulk expression declines in G1 when the APC is active, others have suggested that Ipl1p expression might be controlled by the APC in yeast (5). Perhaps, then, an APC-dependent mechanism contributes to the observed repression of Ipl1p activity in G1 (6). In this regard, it is noteworthy that Ipl1p contains four RxxL motifs, which may serve as recognition motifs for ubiquitination by the APC (29). Collectively these data provisionally place the APC in a network controlling H3 phosphorylation during the cell cycle. Accordingly, we considered the possibility that this ubiquitin ligase might also regulate the histone modification state in response to nutrient signaling cues. Some additional information further encouraged this thinking. (i) The growth response of cycling yeast cells to glucose availability is controlled by the APC (33). For example, a temperature-sensitive mutation of an APC subunit involved in substrate recognition and/or enzymatic processivity (Apc10p/Doc1p) (8, 57) is suppressed by growth without glucose (33). Genetic evidence supports a model in which glucose-dependent signals inhibit APC functions related to the control of anaphase onset and mitotic exit (33). In this model the APC is well placed to be a regulator of the histone modification state during the G1-to-G0 transition, especially given that glucose deprivation is a trigger for cell cycle withdrawal. (ii) Our findings concerning the regulation of nucleosome assembly also relate to the idea that the APC might control the histone modification state during entry into G0. Nucleosomes are assembled from soluble histones and naked DNA by replication-coupled and replication-independent mechanisms (35). We previously discovered that mutation of the conserved APC5 subunit of the APC inhibits replication-independent chromatin assembly in a yeast extract (25). The reaction in this system is also sensitive to mutation of amino-terminal lysines in H4 that can be acetylated in vivo, and H3 and H4 from G2/M cells are better substrates for assembly in the extract than are H3 and H4 from S-phase cells (3, 46, 50). It follows that a defect in histone metabolism may underlie the assembly defect of extract from apc5 mutant cells.
These general indications that the APC could be in a pathway controlling histone modification state in yeast are supported here by direct experimental observations. We find that the APC controls the S10 phosphorylation state of H3 during the cell cycle and in the course of cell cycle withdrawal following nutrient limitation. Whereas the APC likely controls the H3 phosphorylation state during the cell cycle by effects on Ipl1p, an APC-dependent mechanism that controls G0 transcription of the GLC7 H3 phosphatase contributes significantly to reconfiguration of the H3 phosphorylation state in G0. Surprisingly, the APC also regulates global acetylation of H2B, H3, and H4 in response to nutrient signaling cues. Collectively, our results establish that distinct APC-dependent mechanisms regulate the global modification state of histones in yeast during active proliferation and during exit from the cell cycle into G0.
MATERIALS AND METHODS
Yeast strains, growth, and flow cytometry analysis.
Haploid derivatives of S288C were grown in yeast extract-peptone-2% dextrose (YPD) at 24°C unless otherwise indicated. BY4741 (wild type; MATa ura3ΔO his3Δ1 leu2ΔO met15ΔO) and its isogenic partners generated in the yeast gene disruption project (apc10Δ::G418, apc9Δ::G418, and cdc26Δ::G418) were from Research Genetics. YAP160 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 apc11::HIS3 leu2-Δ1::APC11;LEU2) and its isogenic apc11-13 derivative (apc11::HIS3 leu2-Δ1::apc11-13;LEU2) were from A. Page (42). Kelly Tatchell provided the glc7-127 mutant (KT1640 [MATa leu2 ura3-52 his3 glc7-127]) and its wild-type partner (KT1112 [MATa leu2 ura3-52 his3]) (30). APC10 was disrupted in KT1112 and the glc7-127 mutant by using the apc10Δ::G418 cassette obtained by PCR from strain apc10Δ (forward and reverse 400-bp primers are 5′-CATCTACCCACTTGCGCTTGATGAAATTTT and 5′-GTTAAGCTTCCAAAGTAAACAACCGAATCC). Disruption was confirmed by PCR analysis. Ipl1p tagged at its amino terminus with the hemagglutinin (HA) epitope (high-copy LEU2 plasmid pCC1128 from Clarence Chan [36, 37]) or glutathione S-transferase (GST) (high-copy leu2-d URA3 plasmid pGST-IPL1, expression controlled by the CUP1 promoter [51]) was transformed into strains described in Results. Expression of GST-Ipl1p was induced by growth in the presence of 0.5 mM CuSO4 and confirmed by anti-GST immunoblotting. Flow cytometry was performed as described by Epstein and Cross (16) with a FACScan instrument (Becton Dickinson).
Preparation and assay of plasmid supercoiling extracts (69).
Extracts were prepared from cells grown to stationary phase in 500 ml of YPD. Assembly reactions in 20-μl reaction mixtures were performed for 30 min at 30°C with relaxed 32P-labeled pBluescript template. Supercoiling was analyzed by agarose gel electrophoresis at 4°C in Tris-acetate-EDTA running buffer.
Antibodies.
Fusions of residues 1 to 30 of yeast H3, 1 to 35 of yeast H2B, and 1 to 30 of yeast H4 with GST (plasmids from M. Grunstein [27]) were produced in Escherichia coli and used to immunize New Zealand White rabbits (3). Crude serum was used for H2B and H4 immunoblotting (H193 and H196, respectively). Anti-H3 immunoglobulin Gs (IgGs) (from serum H195) were affinity purified by using a fusion of residues 1 to 63 of yeast H3 to E. coli anthranilate synthase (63). The following antihistone antibodies were from Upstate Biotechnology: acetylated H3, acetylated H4, phospho-S10 H3, and full-length (bulk) H4; their product numbers are 06-599, 06-598, 06-570, and 07-108, respectively.
Immunoblotting.
Cells were grown to the desired optical density (OD) (the cell density was confirmed by hemocytometer counting), and the equivalent of 10 ml of cells at an OD at 600 nm (OD600) of 0.5 was pelleted. For routine immunoblotting, protein was precipitated with 25% trichloroacetic acid from cells lysed in 0.5% β-mercaptoethanol and 0.3 M NaOH, and the protein was then resuspended in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer. After incubation at 65°C for 10 min and vigorous vortexing, proteins were resolved in 15% SDS-polyacrylamide gels and electroblotted to nitrocellulose membranes. The membranes were then were incubated with primary antibody (in 3% bovine serum albumin-Tris-buffered saline-Tween 20),followed by detection with horseradish peroxidase-conjugated goat anti-rabbit (1/20,000) or anti-mouse (1/5,000) secondary antibody and enhanced chemiluminescence (AP Biotech). This analysis was also applied to cells arrested in G1 with 10 μg of α-factor per ml (supplemented at 90 min), in S phase with 0.1 M hydroxyurea, and in G2/M with 20 μg of nocodazlole per ml; arrest was confirmed by microscopic and FACScan analysis. The viability of apc10Δ cells is not affected by hydroxyurea or nocodazole treatment (V. Ramaswamy and M. C. Schultz, unpublished data). Primary antibody dilutions were as follows: H2B antiserum, 1/20,000; affinity-purified H3 IgG, 1/200; H4 antiserum, 1/20,000; K9/K14 diacetylated H3 IgG, 1/3,000; K5/K9/K12/K14 tetra-acetylated H4, 1:1,000; phospho-S10 H3 IgG, 1/2,000; and full-length H4 IgG, 1:1,000. Rabbit polyclonal anti-TATA binding protein (anti-TBP) antibody was used at a concentration of 1/5,000 in serum (20). Antiactin monoclonal antibody (C4) was obtained from Chemicon and used at a 1/1,000 dilution. Quantitation of immunoblots was performed by optical scanning with a Fluor-S Multiimager (Bio-Rad) and Quantity One version 4.2.3. software according to the manufacturer's instructions.
Microarray analysis and Northern blotting.
Biotin-labeled target cRNA was prepared in parallel from BY4741 and apc10Δ cells after 2 days of growth (between time points 6 and 7 in Fig. 1A) and hybridized to Affymetrix yeast S98 whole-genome oligonucleotide microarrays (all procedures were according to the manufacturer's instructions). Chips were processed and data were collected by using Affymetrix GeneChip system hardware running Microarray Suite 4.0 software. GeneSpring 4.1.5 was used for refined data analysis. Two independent experiments (r2 = 0.93; n = 1249) were performed for each comparison of the wild type versus the apc10Δ mutant; the fold change for each gene (http://fossil.biochem.ualberta.ca/SchultzLab/index.html) is the average of the two independent measurements. In comparisons described here, the fold change of expression for genes that normally respond to glucose depletion is the maximum recorded during the diauxic shift (twofold significance threshold) (14). For Northern blotting, total RNA was isolated at the indicated time points by hot-phenol extraction (20), resolved in a 0.8% formaldehyde-agarose gel, and then transferred to a nylon membrane and hybridized for 16 h at 65°C. The membrane was washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at room temperature for 10 min, followed by a 30-min wash in 0.1× SSC-0.1% SDS at 65°C. Fragments consisting of the open reading frames of genes were used as probes; they were prepared by PCR amplification from genomic DNA followed by random priming (Invitrogen). Probes were generated with the following primer pairs: 5′-TGTGGTGACATTCATGGG-3′ and 5′-CTTCCACAACTTGATGGG-3′ for GLC7, and 5′-GCTCAATCCAAGAGAGG-3′ and 5′-CCAAGGCGACGTAACATAG-3′ for ACT1. For quantitation, signals were captured by using a Typhoon phosphorimager (AP Biotech) and analyzed with ImageQuant 2.0 software. GLC7 expression levels were normalized to the ACT1 mRNA control.
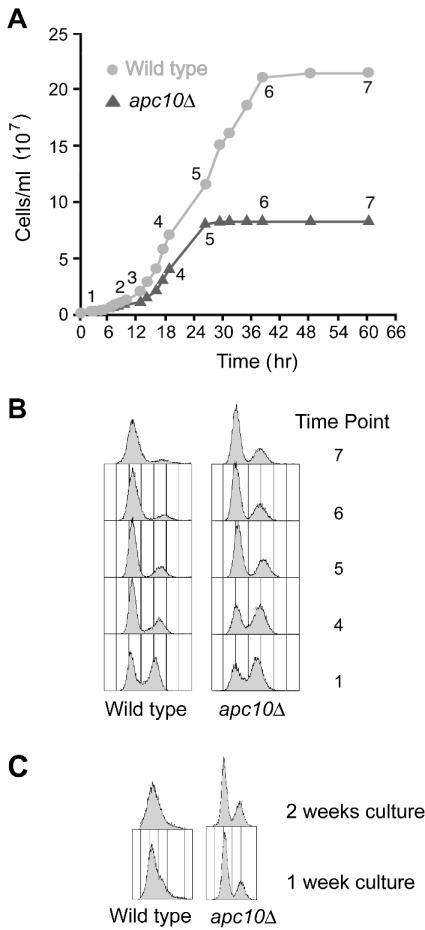
FIG. 1.
Growth properties of wild-type and apc10Δ cells in liquid culture. (A) Growth curves of wild-type and apc10Δ strains in rich medium. (B) Flow cytometry profiles of wild-type and apc10Δ cultures at the time points in panel A. (C) Flow cytometry profiles of wild-type and apc10Δ cultures after 1 and 2 weeks.
RESULTS
Changes in the amino-terminal modification state of histones are associated with execution of the G0 program.
The bulk expression and posttranslational modification of histones H2B, H3, and H4 were examined during culture of a standard laboratory strain, BY4741 (wild type), in rich liquid medium. Growth curves were obtained by direct counting of cells with a hemacytometer (Fig. 1A). We monitored viability by plating (not shown) and monitored cell cycle arrest by flow cytometry (Fig. 1B and C) to assess the suitability of strain BY4741 for studies of the response to nutrient withdrawal. We found that when proliferation has essentially stopped (60 h [time point 7]), ∼95% of cells in a BY4741 culture have a 1n content of DNA and the budding index is 8%; after 1 week in culture, all cells have a 1n content of DNA (Fig. 1C). At time point 7 and later (3 and 14 days), cells have the same viability as at the time of seeding. We conclude that BY4741 is a suitable strain for analysis of events in chromatin metabolism associated with normal execution of the G0 program of development.
In order to minimize histone proteolysis in this study, we trichloroacetic acid precipitated total protein from cells that had been instantaneously solubilized in NaOH immediately after recovery by centrifugation. Because the total protein content of cells declines with growth rate (Fig. 2A) (18), histone expression levels were determined on a per-cell basis (in all figures we compare cell equivalents of lysate, where the indicated 2× amount of lysate contains the protein from twice as many cells as 1× amount of lysate). We probed actin and TBP as loading controls; actin expression is uniform during the growth cycle, and, consistent with a previous report (76), TBP is repressed as cells enter G0 (Fig. 2A).
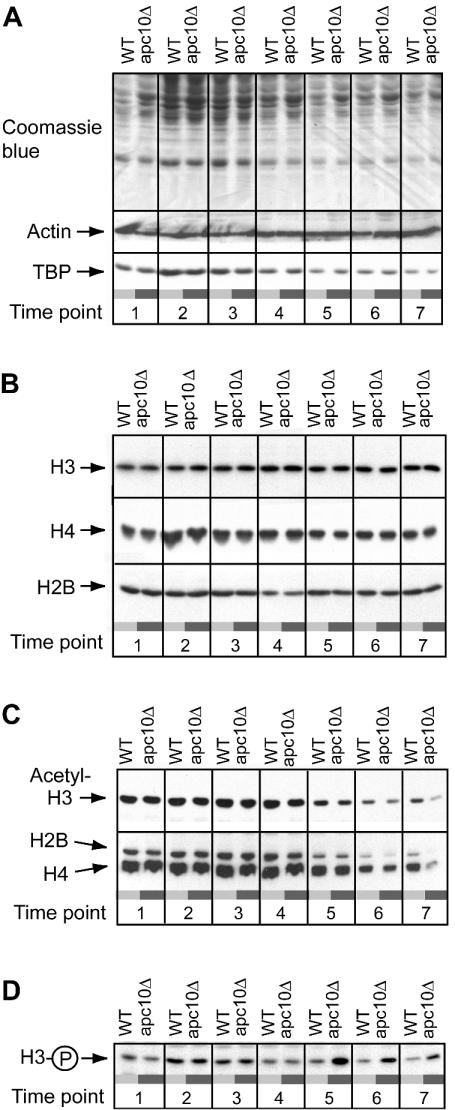
FIG. 2.
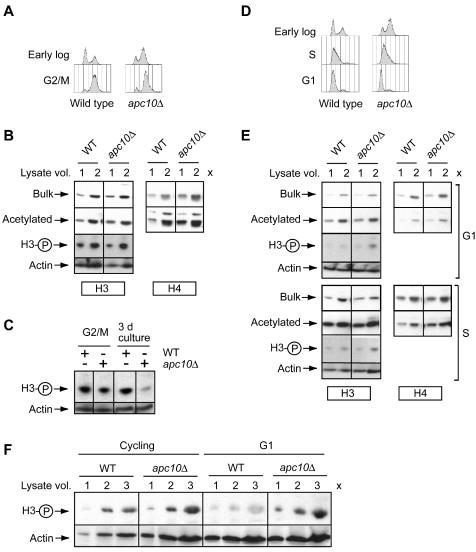
Developmental regulation of histone modification state in wild-type and apc10Δ cells. Proteins were resolved by SDS-polyacrylamide gel electrophoresis and detected by direct staining of the gel or by immunoblotting. (A) Total protein content (Coomassie blue staining) and expression of actin and TBP in wild-type (WT) and apc10Δ cells. (B) Immunoblotting analysis of bulk H2B, H3, and H4 expression. (C) Acetylation state of H2B, H3, and H4 as determined by immunoblotting. (D) S10 phosphorylation state of H3 as determined by immunoblotting. All time points are as in Fig. 1A.
Bulk histone expression was assessed by probing blots with antibodies raised against fusions of the amino-terminal tails of H2B, H3, and H4 with GST (Fig. 2B) and an antibody that recognizes both unmodified and acetylated calf thymus H4. As shown in Fig. 2B, each histone is resolved as a single band in all samples. No slower-migrating bands, which could be indicative of ubiquitination, were detected in our analysis of H2B (not shown). The bulk expression of H2B and H3 is essentially uniform throughout the growth cycle. H4 expression increases during the first 8 h of culture and then declines to a level at 18 h (time point 4) that is maintained until 95% of cells are arrested with a 1n DNA content (60 h [time point 7]).
Acetylated histones were detected in the same samples used to monitor bulk histone expression. Blots were probed with separate antibodies that recognize the K9/K14 diacetylated isoform of H3 and K5/K8/K12/K16 tetra-acetylated H4 (and to a lesser extent acetylated H2B) (see Materials and Methods). Whereas the bulk expression level of H2B, H3, and H4 does not change as cells with a 1n DNA content accumulate in a culture, amino-terminal lysine residues in H2B, H3, and H4 are progressively deacetylated (Fig. 2C). This deacetylation starts after 18 h and continues until the end point of the experiment. We obtained an estimate of the fold change in H3 and H4 acetylation levels by quantitation of immunoblots of serially diluted samples from cells at early and late time points. The results were normalized to the recovery of unmodified histone. This analysis revealed that H3 and H4 acetylation levels decline approximately 2.3- and 1.6-fold, respectively, in the course of the transition from log-phase growth (time point 3) to cessation of activate proliferation (time point 7).
Knowing that G0 is entered from G1 (74), when H3 is dephosphorylated (32), led us to suspect that H3 might shift towards the dephosphorylated state as a population of cells enters G0. This possibility was examined in the experiment shown in Fig. 2D. We indeed observed progressive dephosphorylation of H3 starting after 15 h of culture (time point 3), slightly before histone deacetylation is initiated. Quantitation revealed that H3 S10 phosphorylation is 1.9-fold lower at time point 7 (60 h) than at time point 4 (18 h) in the growth cycle. Therefore, execution of the G0 program is accompanied at its onset by global changes in the chromosome modification state due to decreased H2B, H3, and H4 acetylation and H3 S10 phosphorylation. These findings suggest that the balance of activity between opposing histone-modifying enzymes is dynamic during the growth cycle. This analysis, however, does not provide any insight into the nature of the regulatory system that must couple cell cycle and nutrient signaling cues to biochemical events that directly underlie the reconfiguration of the histone modification state.
Developmentally programmed reconfiguration of histone modification state is perturbed in APC mutants.
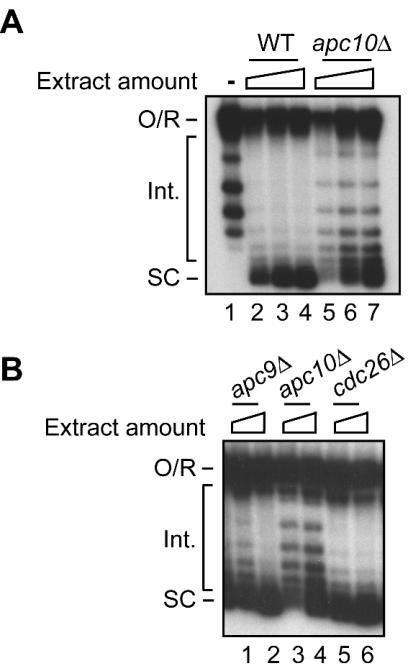
A critical component of the regulatory system that establishes the histone phenotype of early G0 cells was identified in experiments that extended previous studies of nucleosome assembly. These experiments exploited a crude yeast extract in which nucleosome deposition, as measured by plasmid supercoiling, is sensitive to the acetylation state of endogenous H4 (50, 67). Based on our previous work suggesting that mutation of APC5 is associated with defective supercoiling in vitro (25), we assayed the supercoiling capacity of stationary-phase extracts from mutants of three other well-characterized APC subunits (26). While relatively unaffected by deletion of either APC9 or CDC26, supercoiling activity was perturbed in extracts from a mutant lacking Apc10p/Doc1p, a processivity and substrate recognition subunit of the APC (Fig. 3). The supercoiling results suggest, among other possibilities, that defective chromatin assembly in vitro could be due to changes in the histone composition of extracts that stem from altered histone metabolism in vivo. While the link between chromatin assembly and histone modification has been not investigated further, related studies (see below) have uncovered a regulatory system in which the APC modulates the state of histone modification in vivo.
FIG. 3.
Plasmid supercoiling activity of extracts from APC mutants. (A) Wild-type strain (WT) compared to apc10Δ mutant. Twenty-five, 50, and 100 μg of extract protein was assayed for each strain. (B) Comparison of apc10Δ to apc9Δ and cdc26Δ APC mutants. Twenty-five and 50 μg of extract protein was assayed for each strain. The migration of open circular or relaxed DNA (O/R), intermediate topoisomers (Int.), and highly supercoiled species (SC) is indicated. Lane 1 in panel A is the input relaxed plasmid DNA.
The analysis of APC10 was extended by testing whether its deletion impairs the developmental regulation of histone modification. Figure 2 compares the states of histone modification in apc10Δ and wild-type cells during the growth cycle. Deletion of APC10 does not affect the bulk expression of H2B, H3, or H4 at any time in the growth cycle (Fig. 2B). However at the point when the proliferation rate of wild-type cells sharply declines (36 h [time point 6]), H2B, H3, and H4 are more comprehensively deacetylated in apc10Δ than wild-type cells (Fig. 2C). During further culture, this difference becomes more pronounced, so that at 60 h H3 and H4 are, respectively, 2.4- and 5-fold hypoacetylated in mutant compared to wild-type cells. Phosphorylation of H3 at S10 is also misregulated in apc10Δ cells (Fig. 2D). Surprisingly H3 becomes hyperphosphorylated at the apparent point of entry into stationary phase (2.4-fold over the wild-type level at time point 6) and then is slowly dephosphorylated as cells further accumulate in G0. While dephosphorylation is progressive, at the end point of this experiment H3 is still hyperphosphorylated at S10 in the mutant compared to the wild type (the wild-type H3 phosphorylation level is 1.8-fold lower than that in apc10Δ cells).
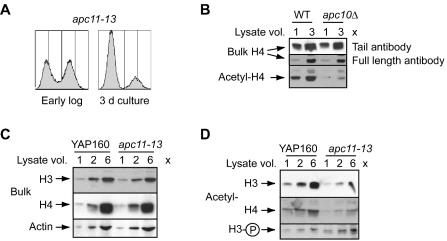
To confirm that aberrant histone acetylation and phosphorylation in apc10Δ cells reflects a defect in APC function, the regulation of these H3 and H4 modifications was examined in a strain with a conditional mutation of APC11, encoding the highly conserved RING-H2 subunit of the APC's ubiquitin ligase core (22, 26, 45, 58). Wild-type and apc11-13 (45) cells were grown to early stationary phase at the permissive temperature (24°C) (Fig. 4A), a condition that partially inactivates the APC in cycling apc11-13 cells but does not affect viability (45). Figure 4C and D show the immunoblotting results for H3 and H4 in apc11-13 cells, and for comparison, Fig. 4B shows the results for H4 in apc10Δ cells (in this instance, bulk H4 was also detected by an antibody that recognizes all forms of calf thymus H4). Clearly, the apc10Δ and apc11-13 mutations confer similar histone acetylation phenotypes; bulk H3 and H4 expression is not perturbed by the apc11-13 mutation (Fig. 4C), and both histones are aberrantly deacetylated in apc11-13 cells (Fig. 4D). Prolonged exposure of H4 immunoblots also reveals hypoacetylation of H2B in apc11-13 cells (not shown). Figure 4D further shows that S10 phosphorylation of H3 is elevated in apc11-13 cells in G0. Because histone acetylation and phosphorylation are perturbed when the catalytic function of the APC is compromised, it is likely that APC-dependent regulation of the chromosome covalent modification state involves ubiquitination of a target protein(s).
FIG. 4.
Mutation of a subunit of the catalytic core of the APC disrupts regulation of histone modification state. (A) Flow cytometry profile of apc11-13 cells during active proliferation (early log phase) and at the point of entry into G0 (3-day culture). (B) Expression of bulk and tetra-acetylated H4 in 3-day cultures of the indicated strains. Two different antibodies were used to detect bulk H4 (see text). WT, wild type. (C) Expression of total H3 and H4 in wild-type and apc11-13 cells cultured for 3 days. Actin was probed as the loading control. (D) Expression of acetylated H3 and H4 and S10-phosphorylated H3 in wild-type and apc11-13 cells cultured for 3 days.
Aberrant reconfiguration of histone modification state in APC mutants is not an indirect consequence of lethal metabolic disruption.
Three cellular phenotypes are associated with the failure of apc10Δ cells to properly reconfigure the modification state of the histones. These phenotypes, however, do not account for abnormal histone metabolism during the growth cycle. The failure of apc10Δ cultures to reach the same stationary-phase density as wild-type cultures (Fig. 1A) could account for abnormal histone metabolism in apc10Δ cells if the latter is generally associated with early cessation of proliferation in liquid culture. This possibility is ruled out by two observations: (i) other mutants (ubc1Δ and spt20Δ mutants) that stop proliferating at a low density in liquid culture have the wild-type pattern of H4 acetylation (not shown), and (ii) apc11-13 cells have clear defects in histone metabolism (Fig. 4) but proliferate to the same density as wild-type cells (not shown). Although wild-type and apc10Δ strains are equally viable after 4 days in culture (see Fig. 6B), long-term survival is severely compromised in the mutant (not shown). This observation raises the possibility that aberrant histone metabolism is caused by events that lead to inviability in G0. We exclude this possibility, however, because apc11-13 cells have the same histone phenotypes as apc10Δ cells but do not lose viability upon long-term culture (not shown).
FIG. 6.
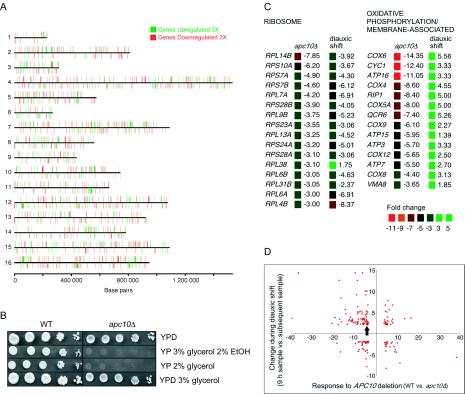
Misregulation of transcription in apc10Δ cells. (A) Physical locations of all repressed and induced genes in apc10Δ cells. (B) Serial dilutions of wild-type (WT) and apc10Δ cells on plates containing different carbon sources. Cells were cultured for 4 days in YPD prior to spotting. EtOH, ethanol. (C) Change in expression levels of genes in two keyword categories (http://pir.georgetown.edu/pirwww/search/searchdb.html) in response to deletion of APC10 in G0 (apc10Δ) and to nutrient depletion (diauxic shift) (14). (D) Global comparison of the fold change in expression levels of genes that respond both to deletion of APC10 in G0 (apc10Δ) and to nutrient depletion. GLC7 is indicated by the arrow.
As noted in the introduction, normal progression into G0 involves cell cycle exit to a state in which the nucleus has a 1n content of DNA. Thus, 2n cells account for only ∼5% of the total wild-type population after 2.5 days in culture (Fig. 1A, time point 7), and the budding index is 8%; after 1 week, 2n cells have essentially disappeared (Fig. 1B). In apc10Δ cultures, on the other hand, ∼20% of cells have a 2n content of DNA after 2.5 days (Figs. 1A and B), and the budding index is 18%. Furthermore, 2n apc10Δ cells in a growth-arrested culture are never recruited into the 1n population (Fig. 1C). These observations raise the possibility that APC mutants are grossly defective for execution of the G0 program, in which case a complex interplay of indirect effects and not an abnormal response to nutrient signaling cues could account for their histone phenotypes. We note, however, that in several important respects the growth cycle of apc10Δ cultures resembles that of wild-type cultures. Cultures of both strains shift from containing about the same proportion of 1n and 2n cells during active proliferation to having a predominance of 1n cells when proliferation has ceased (Fig. 1B), and total protein and TBP content decline with almost identical kinetics in wild-type and apc10Δ cultures (Fig. 2A). We conclude that the majority of cells in a growth-arrested apc10Δ culture have successfully executed at least some critical aspects of the normal developmental response to nutrient limitation. Although the 2n cells that persist in apc10Δ cultures may be in a G2/M-like state, the experiments outlined below reveal that the histone phenotypes of apc10Δ cells are not due to G2/M arrest.
Cell cycle regulation of histone modification state in apc10Δ cells: disruption of H3 S10 phosphorylation but not H3 and H4 acetylation.
The flow cytometry analysis of stationary-phase apc10Δ cultures reveals the presence of a subpopulation of 2n DNA cells. This result raises the possibility that when APC mutants are in G2/M, histone metabolism is disrupted and it is this disruption that accounts for the histone modification profile of a G0 population of apc10Δ cells. To explore this possibility, we examined H3 and H4 acetylation and H3 phosphorylation states in wild-type and apc10Δ cells uniformly arrested in G2/M by using nocodazole. As shown in Fig. 5A, G2/M cells are equally represented in wild-type and mutant cultures treated with nocodazole. Furthermore, there is no evidence of H3 and H4 deacetylation or H3 hyperphosphorylation in apc10Δ cells (Fig. 5B), in contrast to the situation in nutrient-limited cultures (Fig. 2 and 5C). We conclude that apc10Δ cells that cease proliferation in response to nutrient limitation do not inherit a pattern of H3 and H4 hypoacetylation or H3 S10 hyperphosphorylation that was established during G2/M of the cell cycle.
FIG. 5.
Cell cycle control of histone modification state by the APC. (A) Flow cytometry profiles of wild-type and apc10Δ cells arrested in G2/M. The profiles of early-log-phase cultures are shown for comparison. (B) Levels of acetylated H3 and H4 and S10-phosphorylated H3 in wild-type (WT) and apc10Δ cells arrested in G2/M. The loading controls are bulk H3, bulk H4, and actin. (C) Direct comparison of expression of S10-phosphorylated H3 in wild-type and apc10Δ cells in G2/M and early G0. The loading control is actin. (D) Flow cytometry profiles of wild-type and apc10Δ cells arrested G1 and S. The profiles of early-log-phase cultures are shown for comparison. (E) Levels of acetylated H3 and H4 and S10-phosphorylated H3 in wild-type and apc10Δ cells arrested in G1 and S. Loading controls are as in panel B. (F) Expression of S10-phosphorylated H3 in cycling (early-log-phase) and G1-arrested cells. Strains are as indicated.
We extended this analysis to characterization of the modification state of the histones in G1- and S-phase-arrested cells obtained by treatment with α-factor and hydroxyurea, respectively. By FACScan analysis, apc10Δ and wild-type cells respond identically to these treatments, with α-factor treatment yielding mostly G1 cells and hydroxyurea treatment yielding a population in which most cells have initiated DNA replication (Fig. 5D). Figure 5E shows that H3 and H4 acetylation does not differ between wild-type and mutant cells in either G1 or S phase. In contrast, H3 is significantly hyperphosphorylated in apc10Δ cells arrested in G1 (Fig. 5E and F). This result suggests that Apc10p is required during the mitotic cycle for normal dephosphorylation of H3 in G1. In summary, although one-fifth of stationary-phase apc10Δ cells are in a G2/M-like state, there is no defect in histone acetylation or phosphorylation during G2/M when apc10Δ cells are actively proliferating. On the other hand, APC10 is required for S10 dephosphorylation of H3 during G1 in the mitotic cell cycle and during exit from the cell cycle into G0.
mRNA expression is globally disrupted in apc10Δ cells under conditions of nutrient limitation.
The extent to which the histone phenotype of apc10Δ cells in 2-day cultures (between time points 6 and 7) is associated with aberrant gene expression was tested by using microarray technology. Fifteen percent (944) of the ∼6,200 known and predicted protein-coding genes are misregulated in mutant versus wild-type cells (≥3-fold significance threshold). This effect is similar in scale to that observed when wild-type cells enter the diauxic phase of the growth cycle (17% of genes affected) (14). It is not as profound as the effect of depleting H3 and H4 (25% of present genes affected) (80) and is in contrast to the modest response to deleting the GCN5 HAT, which impinges on only 4% of genes (43). From a global perspective, the consequence of APC10 deletion for gene expression is not specific to previously described chromosomal domains; APC10-responsive genes are distributed throughout the chromosomes (Fig. 6A). Neither is the effect on expression limited to induction or repression; of the protein-coding genes, 547 are repressed and 397 are induced. Also, because the net outcome in apc10Δ cells is repression of <3% of all genes (140) at a stage when bulk acetylation is reduced by at least 50%, the acetylation phenotype is unlikely to simply represent the sum of chromatin-remodeling events at individual genes that are misregulated in the mutant.
The pattern of transcriptional misregulation in apc10Δ cells is complex. The effects are not restricted to genes that normally change in activity during entry into G0 (the diauxic shift) (14). In fact, most genes in the apc10Δ gene set are not diauxic shift genes (66%; 620 of 924), and the fraction of diauxic shift genes in the mutant set (34%; 324 of 944) is similar to the fraction in the genome as a whole (∼28%; 1,740 of 6,200). The misregulated genes also do not substantially populate defined functional categories of gene product; at the threefold significance threshold, only one functional category (from PIR keywords categories in GeneSpring 4.1.5 [http://pir.georgetown.edu/pirwww/search/searchdb.html]) is represented in the apc10Δ gene set (oxidative phosphorylation/membrane associated; P = 0.024). Nonetheless, some phenotypes related to nutrient utilization are possibly a direct consequence of transcriptional misregulation in the apc10Δ mutant. For example, two enzymes required for ethanol and glycerol utilization (83) are strongly downregulated in apc10Δ cells (ADH2, 54-fold repression; ACS1, 8.7-fold repression), which lose viability when grown on 2% glycerol or 3% glycerol plus 2% ethanol (Fig. 6B).
Although the transcription phenotype of apc10Δ cells may represent a complex integration of direct effects of APC10 deletion with indirect growth effects, it is informative to consider the results in terms of current ideas about the relationship between the histone modification state and transcriptional regulation. For example, some changes in gene expression in apc10Δ cells (Fig. 6C) are consistent with a dependence of repression on histone deacetylation (38). The behavior of many nuclear ribosomal protein genes, which are normally downregulated during the diauxic shift by a mechanism involving HAT and HDAC complexes (42, 64), is a typical example; 15 of 16 such genes in the apc10Δ gene set are hyperrepressed in the mutant. Conversely, the dampened induction of many G0-activated genes (for example the 14 oxidative phosphorylation/membrane-associated genes in Fig. 6C) suggests interference with acetylation events that are normally required for induction during the diauxic shift. Transcription phenotypes in apc10Δ cells, however, do not universally reflect the expected association of deacetylation with repression (Fig. 6D). Of the 166 genes that are induced in apc10Δ cells and are responsive to nutrient limitation (14), 116 are normally repressed during the diauxic shift. Therefore, even in the context of global deacetylation, some genes in apc10Δ cells are not appropriately repressed. Fifty genes that are overexpressed in apc10Δ cells are normally induced during the diauxic shift. Such G0-activated genes therefore are hyperinduced in the APC10 null background.
Glc7p phosphatase is a potential effector in the APC-dependent pathway controlling H3 phosphorylation in G0.
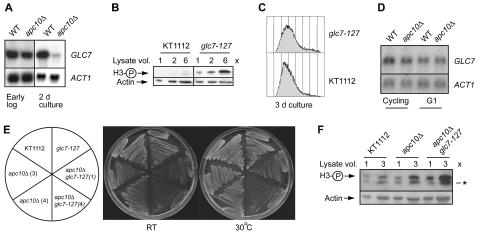
The microarray experiment revealed that in 2-day cultures, mRNA expression of only one histone-modifying enzyme is sensitive to APC10 deletion. That gene is GLC7, encoding the H3 S10 phosphatase of yeast. GLC7 mRNA is 4.2-fold downregulated in apc10Δ cells in late postdiauxic phase and early G0 (Fig. 6D), whereas IPL1 and SNF1, encoding the H3 S10 kinases with which it interacts genetically (32, 48, 67), are unaffected. We used Northern blotting to confirm and extend this observation. As shown in Fig. 7A, GLC7 expression is essentially identical in wild-type and apc10Δ cells in early log phase (lanes 1 and 2). On the other hand, consistent with the microarray result, GLC7 is strongly repressed (sevenfold) in early-stationary-phase apc10Δ cells (Fig. 7A, lanes 3 and 4).
FIG. 7.
Interactions of the APC with GLC7. (A) Expression of GLC7 mRNA in wild-type (WT) and apc10Δ cells during active proliferation (early log phase) and after 3 days in culture (cells entering G0). Five-microgram samples of total RNA were analyzed; ACT1 served as the loading control. (B) S10 phosphorylation state of H3 in glc7-127 cells entering G0. KT1112 is the wild-type strain. (C) Flow cytometry profile of wild-type (KT1112) and glc7-127 cells after 3 days in culture at the permissive temperature. (D) Expression of GLC7 mRNA in cycling (early-log-phase) and G1-arrested populations of wild-type and apc10Δ cells. ACT1 mRNA was probed as the loading control in 5-μg samples of total RNA. (E) Growth of wild-type (KT1112), glc7-127, apc10Δ, and apc10Δ glc7-127 cells after 3 days on YPD at room temperature (RT) and 30°C. Two different clones of apc10Δ and apc10Δ glc7-127 are shown. (F) Expression of S10-phosphorylated H3 in populations of wild-type (KT1112), apc10Δ, and apc10Δ glc7-127 strains after 3 days in culture. The loading control is actin. *, cross-reacting band observed with some lots of the phospho-H3 antibody.
These results support a model in which normal stationary-phase expression of GLC7 is dampened when APC10 is deleted. Consequently, the H3 S10 kinase/phosphatase expression ratio is tipped in favor of the kinases and, therefore, hyperphosphorylation of H3 (in the simplest case because Glc7p downregulation causes H3 phosphatase activity to decline). This model predicts that the S10 phosphorylation state of H3 in G0 is sensitive to Glc7p expression. In order to test this prediction, we examined H3 S10 phosphorylation in a previously described GLC7 mutant and its isogenic wild-type partner. The glc7-127 mutation evidently compromises the ability of the phosphatase to act on H3 in proliferating cells, even at the permissive temperature. Consequently phospho-H3 accumulates in proliferating glc7-127 cells (32). As shown in Fig. 7B, H3 phosphorylation is also substantially elevated in glc7-127 cells in early G0 (there was no effect on histone acetylation [V. Ramaswamy and M. C. Schultz, unpublished data]). To rule out the possibility that H3 is hyperphosphorylated in glc7-127 cultures in G0 because they include more G1-like (1n) cells than wild-type cultures (Fig. 5C to E), the DNA content of cells in 3-day cultures (the same cells used for immunoblotting) was analyzed by flow cytometry. Figure 7C reveals that 1n cells are not more abundant in glc7-127 cultures than in wild-type cultures. We conclude that compromising Glc7p-dependent phosphatase activity can contribute to increased H3 S10 phosphorylation in G0.
The results in Fig. 6C and 7A and the evidence that GLC7 transcription is normally induced in nutrient-deprived cells (14, 17) (see Discussion) suggest that transcriptional regulation of GLC7 contributes to developmental control of the H3 S10 phosphorylation state during execution of the G0 program. The effect of APC10 deletion on H3 S10 phosphorylation in G1 (Fig. 5E and F) raises the possibility that transcriptional regulation of GLC7 by the APC also contributes to the control of H3 phosphorylation during the cell cycle. This hypothesis was explored by measuring GLC7 expression in wild-type and apc10Δ cells arrested in G1 (Fig. 7D). When Northern blotting results are quantitated after normalization to actin expression, it is clear that there is no difference in GLC7 expression between G1-arrested wild-type and apc10Δ cells. Therefore, the mechanism that accounts for H3 S10 hyperphosphorylation in apc10Δ cells likely involves an APC-dependent effect on GLC7 transcription in G0 but not in G1.
GLC7 and APC10 are expected to interact genetically if they are in the same pathway that controls H3 phosphorylation in G0. To test this possibility, we created apc10Δ and apc10Δ glc7-127 mutants isogenic to the strains used in the experiments in Fig. 7B and C. Growth of these four strains was compared in plating assays, of which representative examples are shown in Fig. 7E (two independent isolates each of apc10Δ and apc10Δ glc7-127 mutants were used). Consistent with analysis in the BY4741 genetic background (Fig. 1A), APC10 disruption confers a slow growth phenotype (compare the wild type to the two apc10Δ strains). Combination of the apc10Δ and glc7-127 mutations has a synthetic effect, in which the double mutant grows more slowly than either the apc10Δ or the glc7-127 single mutant. This genetic interaction supports the hypothesis that GLC7 and APC10 both act in a pathway controlling the phosphorylation state of H3. Consistent with this interpretation, H3 hyperphosphorylation in G0 is accentuated in the glc7-127 apc10Δ double mutant compared to its wild-type and single mutant partners (Fig. 7F).
Ipl1p kinase is a potential effector in the APC-dependent pathway controlling H3 S10 phosphorylation during the cell cycle.
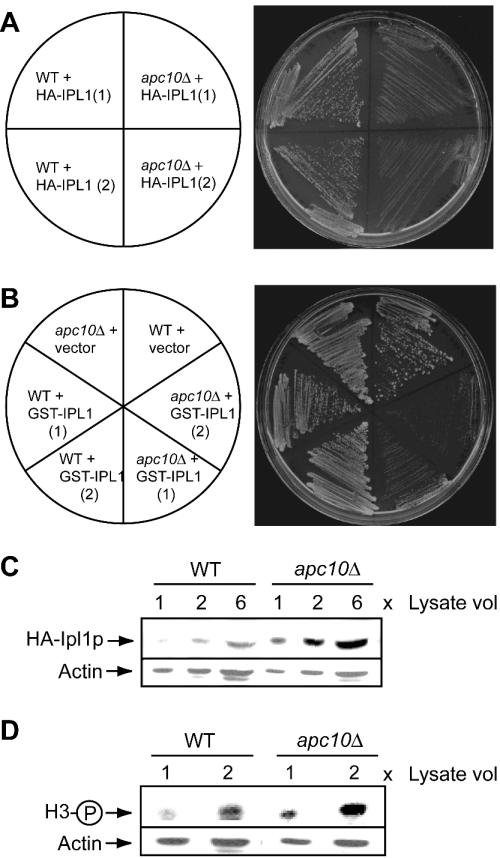
Results in the literature implicate Ipl1p in the control of H3 phosphorylation state in cycling cells, possibly as a component of a pathway involving the APC (see the introduction). Our genetic and biochemical analyses indeed support the existence of an APC-dependent mechanism for regulation of Ipl1p, as outlined below. First, IPL1 interacts genetically with APC10. High-copy-number vectors expressing Ipl1p tagged at the amino terminus with the HA epitope or with GST were used to transform wild-type and apc10Δ cells. As for Fig. 7E, cell growth was monitored by plating two independent isolates of each genotype, and a vector control was included in the experiments with GST-Ipl1p. Figure 8A and B show that overexpression of HA- or GST-tagged Ipl1p reproducibly impairs the growth of apc10Δ but not wild-type cells. Second, expression of Ipl1p is misregulated in apc10Δ cells. Immunoblotting was used to monitor the steady-state level of HA-Ipl1p in wild-type and apc10Δ strains in Fig. 8A. In cells arrested in G1 by treatment with α-factor, deletion of apc10Δ is associated with substantial accumulation of HA-Ipl1 (Fig. 8C). Accumulation of previously identified APC substrates is a well-established phenotype of some APC mutants (58). We conclude that expression of Ipl1p is limited in G1 by a mechanism requiring the APC. Because G0 is entered from the G1 phase of the cell cycle, in APC mutants it is possible that elevated G1 expression of Ipl1p persists in G0 and contributes to S10 hyperphosphorylation of H3 in the context of nutrient limitation. This hypothesis is supported by the observed modest but reproducible elevation of H3 S10 phosphorylation which accompanies overexpression of HA-Iplp in G1-arrested apc10Δ cells (Fig. 8D).
FIG. 8.
Interactions of the APC with IPL1. (A) Growth of wild-type (WT) and apc10Δ strains carrying the HA-IPL1 plasmid pCC1128. Growth was for 3 days at room temperature on medium lacking leucine. Two different transformants are shown. (B) Growth of wild-type and apc10Δ strains carrying the vector (pYEX) or a GST-IPL1 overexpression plasmid. Growth was for 3 days at room temperature on medium lacking uracil. Two different transformants are shown. (C) Anti-HA immunoblot analysis of G1-arrested wild-type and apc10Δ strains carrying the HA-IPL1 plasmid pCC1128. The loading control is actin. (D) Immunoblot analysis of the samples in panel C with antibody against S10-phosphorylated H3.
DISCUSSION
We show that the global reconfiguration of histone modification state in yeast is regulated by APC-dependent mechanisms during the cell cycle and during the response to nutrient limitation. Because mutation of a regulatory subunit of the APC (APC10 product) and mutation of a subunit of its catalytic core (APC11 product) confer similar defects in the control of histone modification state, it is likely that the APC functions as a ubiquitin ligase to modulate histone phosphorylation and acetylation. If this is true and APC-targeted proteins that control histone modification state are regulated by proteolysis, then crippling the proteasome should confer histone phenotypes similar to those observed for apc10Δ and apc11-13 cells. Analysis of H4 acetylation in a strain with a conditional mutation in the gene for an essential subunit of the 20S proteolytic core complex of the proteasome (PRE1) (4) bears out this prediction (V. Ramaswamy and M. C. Schultz, unpublished data). Published information about the physiological regulation of APC activity is consistent with the existence of an APC-dependent pathway controlling the histone modification state. (i) The APC is functional during G1 (26, 58, 84) when H3 is dephosphorylated by an APC-dependent mechanism. (ii) The APC in yeast is activated by glucose limitation (33), the best-characterized trigger of entry into G0. (iii) A highly active form of the APC can be isolated from G0 (terminally differentiated) mammalian cells (21, 58). Because G0 is normally entered from G1 (28, 78, 79), when the APC is active, the APC is evidently well positioned to modulate histone modification during cell cycle exit.
Global regulation of H3 phosphorylation state by mechanisms involving the APC.
H3 phosphorylation state fluctuates during the cell cycle in yeast by a mechanism that may involve regulation of the H3 S10 kinase Ipl1p (30). Ipl1p is downregulated in G1- compared to S-phase cells (5), Ipl1p-dependent kinase activity declines as cells enter G1 (6), and Ipl1p accumulates in G1-arrested apc10Δ cells (Fig. 8C). Collectively this evidence is consistent with the straightforward proposal, originally elaborated by Biggins et al. (5), that bulk expression of Ipl1p during the cell cycle is controlled by the APC. This regulatory mechanism contributes to cell cycle control of Aurora A-type H3 S10 kinases in higher organisms. Therefore, an APC-dependent pathway for cell cycle regulation of this family of kinases appears to be conserved in eukaryotes.
The information at hand indicates that APC-dependent regulation of H3 phosphatase in yeast contributes to physiological regulation of the H3 S10 phosphorylation state during the growth cycle. Specifically, because bulk Glc7p-dependent PP1 activity increases as cells enter quiescence (54) and is required for H3 dephosphorylation in G0, (Fig. 7), we propose that G0 induction of H3 S10 phosphatase activity is important for H3 dephosphorylation when nutrients become limiting (Fig. 2). How might the APC contribute to G0-induction of H3 phosphatase activity? An attractive possibility is based on the finding that G0 expression of GLC7 mRNA is repressed in apc10Δ cells (Fig. 6 and 7). Thus, we suggest that high-level expression of Glc7p in G0 is maintained, at least in part, by an APC-dependent mechanism that actively sustains GLC7 mRNA synthesis. High-level expression of Glc7p is then permissive for induction of Glc7p-dependent PP1 activity (54) and dephosphorylation of H3.
Consistent with our model that a nutrient-responsive pathway controls GLC7 mRNA expression in G0, GLC7 has many features in common with genes that are known to be upregulated when nutrients are limiting. Northern blotting and microarray experiments have shown that GLC7 is induced in G0 (14, 17), and its expression is sensitive to disruption of signaling by the target-of-rapamycin kinases, which is required for execution of the G0 program (24, 68). Furthermore the GLC7 promoter contains two optimally positioned copies of the stress response element that is commonly required for activation of G0-induced genes (44, 52). More generally speaking, the idea that an APC-dependent mechanism controls GLC7 transcription is consistent with recent evidence that the APC regulates expression of mammalian transcription factors SnoN (71, 77) and HOXC10 (19).
In the simplest model (described above), the APC contributes to maintenance of H3 dephosphorylation in quiescent yeast cells by controlling Glc7p-dependent H3 phosphatase activity. However, in other organisms phosphatases have been implicated in cell cycle control of H3 phosphorylation by virtue of their effects on H3 kinases. Most notably, fission yeast Aurora A and vertebrate Auroras A and B are inhibited by dephosphorylation, and the vertebrate Auroras are physically associated with phosphatases (53, 72). It follows that budding yeast Glc7p may regulate the H3 phosphorylation state in G0 by controlling H3 kinase activity. Accordingly, it will be interesting in the future to test whether Glc7p directly regulates Ipl1p in a pathway that includes the APC. Although cell cycle control of vertebrate and fission yeast Auroras by phosphatases has not been demonstrated, it may also be profitable, considering our results, to test whether any Aurora-directed phosphatase from these organisms is regulated by the APC.
The APC integrates cell cycle progression and cell cycle exit signals that control distinct effector mechanisms of H3 dephosphorylation.
Budding yeast cells enter G0 from G1 of the mitotic cell cycle (28, 78, 79). Therefore, cells enter G0 at a time in the cell cycle when the APC is active and an APC-dependent mechanism, most likely targeted degradation of Ipl1p, promotes H3 dephosphorylation. APC-dependent transcriptional induction of GLC7 subsequently contributes to sustained dephosphorylation of H3 in G0. This regulatory scheme has three interesting features. First, physiological regulation of H3 phosphorylation state is not restricted to APC-dependent mechanisms that impinge on the H3 kinase. Rather, both the H3 kinase and a phosphatase are regulated. Second, the regulatory mechanisms that contribute to global reconfiguration of H3 phosphorylation state differ according to the physiological circumstance. Fluctuation of kinase activity (6) in the context of constitutive phosphatase expression suffices to control H3 phosphorylation during the cell cycle. An additional mechanism, induction of the phosphatase, is engaged in response to nutrient limitation. While these mechanisms are distinct, they are tightly coupled in the context of G0 development: establishment of the hypophosphorylated state in G1 by regulation of the kinase is reinforced in G0 by regulation of the phosphatase. Third, the distinct mechanisms for controlling the H3 kinase/phosphatase ratio involve the same master regulator, the APC. Because it controls G1 dephosphorylation of H3, during every cell cycle the APC promotes a wave of chromosome modification that will be reinforced by APC-dependent induction of Glc7p phosphatase if nutrients become limiting. From a broad perspective, control of the histone modification state by the APC is reminiscent of its regulatory function during the active proliferation, when it controls sequential cell cycle transitions by distinct mechanisms (26, 58, 84).
Regulation of histone acetylation by the APC.
Rpd3p is the HDAC subunit of a conserved transcriptional repressor complex (2). In cycling yeast cells, Rpd3p participates in global histone deacetylation, perhaps by a mechanism involving binding of the Rpd3p complex to histones or histone-interacting proteins (42, 75). After this paper was first submitted, Sandmeier et al. (66) reported that H3 and H4 are not deacetylated when yeast rpd3Δ cells enter G0. Therefore, Rpd3p is also required for global deacetylation of H3 and H4 in cells that have ceased proliferation. Given that Rpd3p is involved in deacetylation of H2B (73), this enzyme may also contribute to the global deacetylation of H2B reported here (Fig. 2B). Relatively little is known about the regulation of Rpd3p. For example, it is not known if the intrinsic enzymatic activity of Rpd3p, its bulk expression, or its ability to globally bind to histones or histone-interacting proteins increases during cell cycle withdrawal. RPD3 mRNA is induced only modestly (if at all [14]) during the growth cycle, and its expression is not sensitive to deletion of APC10 (http://fossil.biochem.ualberta.ca/SchultzLab/index.html). Accordingly, global histone deacetylation in G0 must not involve APC-dependent transcriptional induction of RPD3 but rather must involve effects at the protein level (if indeed changes in Rpd3p expression or properties contribute to global reconfiguration of histone acetylation). If APC-dependent regulation of HAT activity plays a role in setting the level of global acetylation in G0, then multiple HATs must be affected, because neither Gcn5p nor Esa1p, whose deletion globally affects the histone modification state in cycling cells, can acetylate both H3 and H4 (62, 73). It is also possible that APC-dependent changes in the H3 S10 phosphorylation state globally influence H3 K9 acetylation (15, 48, 49), although preliminary results do not support this idea (V. Ramaswamy and M. C. Schultz, unpublished data).
A possible cause-effect relationship between chromatin assembly and APC-dependent reconfiguration of histone modification state.
Because some APC mutants have a chromatin assembly defect, it is possible that histone replacement (1) contributes to the global reconfiguration of the histone modification state in G0. For example, perhaps the APC activates a chromatin assembly pathway that preferentially deposits acetylated histones. When this pathway is inactivated under conditions of nutrient limitation or by mutation of the APC, other assembly pathways that preferentially use less modified histones could predominate. Against a background of degradation of free hyperacetylated histones, this mechanism would drive global replacement of hyper- with hypoacetylated histones. This idea is consistent with our evidence that chromatin assembly in vitro is responsive to the state of histone modification (3, 50) and that extracts from APC10 (this study) and APC5 (25) mutants have a defect in assembly activity. In order to critically test this possibility, it will be necessary to identify the assembly factors that are active in the crude yeast assembly system and to test their use of modified versus unmodified histones during active proliferation and in G0. It could be profitable to focus on Asf1p, a highly conserved chromatin assembly factor (35). asf1Δ has low viability in G0, and, as observed for apc10Δ and apc11-13 mutants, an abnormally high proportion of 2n cells accumulate in asf1Δ cultures as nutrients are depleted (81).
Work performed in the Harkness lab (Fig. 8C in reference 25) suggests that steady-state expression of amino-terminally unacetylated H3 increases when cycling cells harboring a mutant allele of the APC5 gene (apc5CA) are shifted to 37°C. In contrast, we have no conclusive (reproducible) evidence that perturbation of the APC interferes with bulk H3 expression or acetylation state in actively proliferating cells, and H3 phosphorylation is affected only in G1 of the cell cycle. Interestingly, cultures of the wild-type S288C strain used by Harkness et al. cease expanding at a substantially lower OD (OD600 of ∼5 [Fig. 1C in reference 25]) than typically observed for “wild-type” S288C derivatives and other laboratory strains grown at 30°C (OD600 of ∼10 [11, 18, 66, 76, 78]). Harkness et al. also showed that temperature has a profound effect on the growth cycle of wild-type cells. Specifically, the time at which cultures cease expanding is advanced by ∼32 h when cells are incubated at 37°C as opposed to room temperature (or 30°C), and cultures do not grow beyond an OD600 of ∼3.5. The latter result is surprising, because their medium can support proliferation to an OD600 of ∼5 (at room temperature or 30°C) and we find that S288C cells (strain BY4741) reach the same density at room temperature and 37°C (not shown). In our view these findings provide a basis for resolving the apparent discrepancy between the results presented here and by Harkness et al. (25). We propose that APC5 and apc5CA cells enter G0 at a relatively low density because the combination of the strain background and the conditions used by Harkness et al. promotes an abnormal response to nutrient limitation. Thus, the accumulation of unmodified H3 in apc5CA cells at low density may principally reflect a defect in the APC-dependent network that we find controls the histone modification state in response to nutrient signaling cues (rather than exclusive misregulation of cell cycle-dependent events).
Functional significance of global regulation of histone modification state.
Why is H3 phosphorylated during the cell cycle? Why in cycling cells is the global level of histone acetylation set to the observed level? Despite intense study in recent years, these questions await definitive answers (see, for example, references 23, 40, and 59). Accordingly, a comprehensive account of the functional significance of the changes in histone modification state that occur upon cell cycle exit in response to nutrient limitation is currently beyond our grasp. Nonetheless, at least in the case of the ribosomal protein genes, it seems highly likely that APC-dependent histone deacetylation contributes directly to transcriptional repression in G0. Thus, histone deacetylation by a mechanism involving the Rpd3p HDAC and Esa1p HAT complexes is a critical step in repression of ribosomal protein genes in response to nutrient limitation (42, 64), and we find that global deacetylation is associated with their hyperrepression in apc10Δ cells. On a more speculative note, an intriguing possibility regarding the global organization of chromosomes is worthy of consideration. There is evidence from sedimentation velocity analysis of chromosome complexes that the overall structure of chromosomes changes when cells enter G0 (60, 61). Perhaps this structural transformation, which admittedly is not well characterized at this time, involves genome-wide reconfiguration of the chemical modification state of the chromosomes by the APC-dependent mechanisms defined here. By extension, the observed level of global histone modification in cycling cells may help to establish a specific overall structure of chromosomes that is permissive for global events in chromosome metabolism that occur in cycling but not quiescent cells. Such events could include replication, sister chromatid cohesion and separation, and high overall transcriptional activity (in proliferating yeast cells, transcription occurs throughout the chromosomes and involves 76% of all genes [74]).
A considerable body of evidence suggests that global H3 phosphorylation in Tetrahymena and metazoans is important for chromosome condensation. The fact that similar kinases and phosphatases control the global H3 phosphorylation state in yeast and vertebrates heightens the expectation that H3 phosphorylation also regulates chromosome condensation in yeast. Mutation of H3 S10 to alanine, however, has no effect on chromosome dynamics in cycling yeast cells, even when another nearby site of phosphorylation in vivo (S28) is also changed to alanine (32). Importantly, this result does not exclude the possibility that global H3 phosphorylation regulates chromosome structure in G0. For example, the level of H3 phosphorylation set by APC-dependent events in G1 may be just below a threshold required to change the global structure of chromosomes. Persistent APC-dependent H3 dephosphorylation in the context of global histone deacetylation might then trigger global changes in chromosome structure when the G0 program is initiated. Accordingly, studies of chromosome metabolism in yeast that focus on histone modification during the G1-to-G0 transition may shed new light on the control of chromosome structure in higher organisms, including the possible role of misregulation of Aurora kinases in chromosome instability of some tumor types (55, 56).
Acknowledgments
V. Ramaswamy and J. S. Williams contributed equally to this work.
We are grateful to M. Grunstein and A. Carmen for expression plasmids and advice on the generation and use of antihistone antisera. A. Page, J. Leverson, T. Hunter, H. Steiner, M. Ellison, K. Tatchell, D. Wolf, C. Chan, and J. Stone kindly made available various yeast reagents. We thank T. Harkness, D. Hockman, A. Ghavidel, J. Yang, J. Cooper, and R. Moudgil for technical advice and assistance.
This work was supported by grants to M.C.S. from the Canadian Institutes for Health Research and the Alberta Heritage Foundation for Medical Research. V.R. and R.S. were supported by summer studentships from the AHFMR.
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 2.Alland, L., G. David, H. Shen-Li, J. Potes, R. Muhle, H. C. Lee, H. Hou, Jr., K. Chen, and R. A. DePinho. 2002. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol. Cell. Biol. 22:2743-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altheim, B. A., and M. C. Schultz. 1999. Histone modification governs the cell cycle regulation of a replication-independent chromatin assembly pathway in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:1345-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumeister, W., J. Walz, F. Zuhl, and E. Seemuller. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92:367-380. [DOI] [PubMed] [Google Scholar]
- 5.Biggins, S., F. F. Severin, N. Bhalla, I. Sassoon, A. A. Hyman, and A. W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buvelot, S., S. Y. Tatsutani, D. Vermaak, and S. Biggins. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmen, A. A., P. R. Griffin, J. R. Calaycay, S. E. Rundlett, Y. Suka, and M. Grunstein. 1999. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. USA 96:12356-12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, C. W., and D. O. Morgan. 2002. The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat. Cell. Biol. 4:880-887. [DOI] [PubMed] [Google Scholar]
- 9.Castro, A., Y. Arlot-Bonnemains, S. Vigneron, J. C. Labbe, C. Prigent, and T. Lorca. 2002. APC/Fizzy-Related targets Aurora-A kinase for proteolysis. EMBO Rep. 3:457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castro, A., S. Vigneron, C. Bernis, J. C. Labbe, C. Prigent, and T. Lorca. 2002. The D-box-activating domain (DAD) is a new proteolysis signal that stimulates the silent D-box sequence of Aurora-A. EMBO Rep. 3:1209-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choder, M. 1991. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 5:2315-2326. [DOI] [PubMed] [Google Scholar]
- 12.Clarke, A. S., J. E. Lowell, S. J. Jacobson, and L. Pillus. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19:2515-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crosio, C., G. M. Fimia, R. Loury, M. Kimura, Y. Okano, H. Zhou, S. Sen, C. D. Allis, and P. Sassone-Corsi. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 15.Edmondson, D. G., J. K. Davie, J. Zhou, B. Mirnikjoo, K. Tatchell, and S. Y. Dent. 2002. Site-specific loss of acetylation upon phosphorylation of histone H3. J. Biol. Chem. 277:29496-29502. [DOI] [PubMed] [Google Scholar]
- 16.Epstein, C. B., and F. R. Cross. 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6:1695-1706. [DOI] [PubMed] [Google Scholar]
- 17.Feng, Z. H., S. E. Wilson, Z. Y. Peng, K. K. Schlender, E. M. Reimann, and R. J. Trumbly. 1991. The yeast GLC7 gene required for glycogen accumulation encodes a type 1 protein phosphatase. J. Biol. Chem. 266:23796-23801. [PubMed] [Google Scholar]
- 18.Fuge, E. K., E. L. Braun, and M. Werner-Washburne. 1994. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J. Bacteriol. 176:5802-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabellini, D., I. N. Colaluca, H. C. Vodermaier, G. Biamonti, M. Giacca, A. Falaschi, S. Riva, and F. A. Peverali. 2003. Early mitotic degradation of the homeoprotein HOXC10 is potentially linked to cell cycle progression. EMBO J. 22:3715-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghavidel, A., and M. C. Schultz. 2001. TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell 106:575-584. [DOI] [PubMed] [Google Scholar]
- 21.Gieffers, C., B. H. Peters, E. R. Kramer, C. G. Dotti, and J. M. Peters. 1999. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc. Natl. Acad. Sci. USA 96:1317-11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gmachl, M., C. Gieffers, A. V. Podtelejnikov, M. Mann, and J. M. Peters. 2000. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 97:8973-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hans, F., and S. Dimitrov. 2001. Histone H3 phosphorylation and cell division. Oncogene 20:3021-3027. [DOI] [PubMed] [Google Scholar]
- 24.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harkness, T. A., G. F. Davies, V. Ramaswamy, and T. G. Arnason. 2002. The ubiquitin-dependent targeting pathway in Saccharomyces cerevisiae plays a critical role in multiple chromatin assembly regulatory steps. Genetics 162:615-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harper, J. W., J. L. Burton, and M. J. Solomon. 2002. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 16:2179-2206. [DOI] [PubMed] [Google Scholar]
- 27.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583-592. [DOI] [PubMed] [Google Scholar]
- 28.Herman, P. K. 2002. Stationary phase in yeast. Curr. Opin. Microbiol. 5:602-607. [DOI] [PubMed] [Google Scholar]
- 29.Hildebrandt, E. R., and M. A. Hoyt. 2001. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol. Biol. Cell 12:3402-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honda, K., H. Mihara, Y. Kato, A. Yamaguchi, H. Tanaka, H. Yasuda, K. Furukawa, and T. Urano. 2000. Degradation of human Aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene 19:2812-2819. [DOI] [PubMed] [Google Scholar]
- 31.Howe, L., D. Auston, P. Grant, S. John, R. G. Cook, J. L. Workman, and L. Pillus. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 33.Irniger, S., M. Bäumer, and G. H. Braus. 2000. Glucose and ras activity influence the ubiquitin ligases APC/C and SCF in Saccharomyces cerevisiae. Genetics 154:1509-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 35.Kadam, S., and B. M. Emerson. 2002. Mechanisms of chromatin assembly and transcription. Curr. Opin. Cell Biol. 14:262-268. [DOI] [PubMed] [Google Scholar]
- 36.Kang, J., I. M. Cheeseman, G. Kallstrom, S. Velmurugan, G. Barnes, and C. S. Chan. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155:763-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, J. H., J. S. Kang, and C. S. Chan. 1999. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145:1381-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knoepfler, P. S., and R. N. Eisenman. 1999. Sin meets NuRD and other tails of repression. Cell 99:447-450. [DOI] [PubMed] [Google Scholar]
- 39.Krebs, J. E., M. H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13:412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kristjuhan, A., J. Walker, N. Suka, M. Grunstein, D. Roberts, B. R. Cairns, and J. Q. Svejstrup. 2002. Transcriptional inhibition of genes with severe histone H3 hypoacetylation in the coding region. Mol. Cell 10:925-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruhlak, M. J., M. J. Hendzel, W. Fischle, N. R. Bertos, S. Hameed, X. J. Yang, E. Verdin, and D. P. Bazett-Jones. 2001. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J. Biol. Chem. 276:38307-38319. [DOI] [PubMed] [Google Scholar]
- 42.Kurdistani, S. K., D. Robyr, S. Tavazoie, and M. Grunstein. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 43.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 44.Lenssen, E., U. Oberholzer, J. Labarre, C. De Virgilio, and M. A. Collart. 2002. Saccharomyces cerevisiae Ccr4-Not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol. Microbiol. 43:1023-1037. [DOI] [PubMed] [Google Scholar]
- 45.Leverson, J. D., C. A. Joazeiro, A. M. Page, H. Huang, P. Hieter, and T. Hunter. 2000. The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol. Biol. Cell 11:2315-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling, X., T. A. Harkness, M. C. Schultz, G. Fisher-Adams, and M. Grunstein. 1996. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 15:686-699. [DOI] [PubMed] [Google Scholar]
- 47.Littlepage, L. E., and J. V. Ruderman. 2002. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 16:2274-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo, W.-S., L. Duggan, N. C. T. Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142-1146. [DOI] [PubMed] [Google Scholar]
- 49.Lo, W.-S., R. C. Trievel, J. R. Rojas, L. Duggan, J. Y. Hsu, C. D. Allis, R. Marmorstein, and S. L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine14. Mol. Cell 5:917-926. [DOI] [PubMed] [Google Scholar]
- 50.Ma, X.-J., J. Wu, B. A. Altheim, M. C. Schultz, and M. Grunstein. 1998. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl. Acad. Sci. USA 95:6693-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martzen, M. R., S. M. McCraith, S. L. Spinelli, F. M. Torres, S. Fields, E. J. Grayhack, and E. M. Phizicky. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286:1153-1155. [DOI] [PubMed] [Google Scholar]
- 52.Moskvina, E., C. Schuller, C. T. Maurer, W. H. Mager, and H. Ruis. 1998. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast 14:1041-1050. [DOI] [PubMed] [Google Scholar]
- 53.Murnion, M. E., R. R. Adams, D. M. Callister, C. D. Allis, W. C. Earnshaw, and J. R. Swedlow. 2001. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276:26656-26665. [DOI] [PubMed] [Google Scholar]
- 54.Nigavekar, S. S., Y. S. Tan, and J. F. Cannon. 2002. Glc8 is a glucose-repressible activator of Glc7 protein phosphatase-1. Arch. Biochem. Biophys. 404:71-79. [DOI] [PubMed] [Google Scholar]
- 55.Nigg, E. A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell. Biol. 2:21-32. [DOI] [PubMed] [Google Scholar]
- 56.Ota, T., S. Suto, H. Katayama, Z. B. Han, F. Suzuki, M. Maeda, M. Tanino, Y. Terada, Y., and M. Tatsuka. 2002. Increased mitotic phosphorylation of histone H3 attributable to AIM-1/Aurora-B overexpression contributes to chromosome number instability. Cancer Res. 62:5168-5177. [PubMed] [Google Scholar]
- 57.Passmore, L. A., E. A. McCormack, S. W. Au, A. Paul, K. R. Willison, J. W. Harper, and D. Barford. 2003. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 22:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peters, J.-M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9:931-943. [DOI] [PubMed] [Google Scholar]
- 59.Peterson, C. L. 2001. Chromatin: mysteries solved? Biochem. Cell. Biol. 79:219-225. [PubMed] [Google Scholar]
- 60.Piñon, R. 1979. Folded chromosomes in meiotic yeast cells: analysis of early meiotic events. J. Mol. Biol. 129:433-437. [DOI] [PubMed] [Google Scholar]
- 61.Piñon, R., and Y. Salts. 1977. Isolation of folded chromosomes from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 74:2850-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 63.Robinson, K. M., A. von Kieckebusch-Guck, and B. D. Lemire. 1991. Isolation and characterization of a Saccharomyces cerevisiae mutant disrupted for the succinate dehydrogenase flavoprotein subunit. J. Biol. Chem. 266:21347-21350. [PubMed] [Google Scholar]
- 64.Rohde, J. R., and M. E. Cardenas. 2003. The Tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rundlett, S. E., A. A. Carmen, R. Kobayashi, S. Bavykin, B. M. Turner, and M. Grunstein. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93:14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sandmeier, J. J., S. French, Y. Osheim, W. L. Cheung, C. M. Gallo, A. L. Beyer, and J. S. Smith. 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21:4959-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanz, P., G. R. Alms, T. A. J. Haystead, and M. Carlson. 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20:1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103:253-262. [DOI] [PubMed] [Google Scholar]
- 69.Schultz, M. C. 1999. Chromatin assembly in yeast cell-free extracts. Methods 17:161-172. [DOI] [PubMed] [Google Scholar]
- 70.Scrittori, L., F. Hans, D. Angelov, M. Charra, C. Prigent, and S. Dimitrov. 2001. pEg2 aurora-A kinase, histone H3 phosphorylation, and chromosome assembly in Xenopus egg extract. J. Biol. Chem. 276:30002-30010. [DOI] [PubMed] [Google Scholar]
- 71.Stroschein, S. L., S. Bonni, J. L. Wrana, and K. Luo. 2001. Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 15:2822-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sugiyama, K. Sugiura, T. Hara, K. Sugimoto, H. Shima, K. Honda, K. Furukawa, S. Yamashita, and T. Urano. 2002. Aurora-B associated protein phosphatases as negative regulators of kinase activation. Oncogene 21:3103-3111. [DOI] [PubMed] [Google Scholar]
- 73.Suka, N., Y. Suka, A. A. Carmen, J. Wu, and M. Grunstein. 1997. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8:473-479. [DOI] [PubMed] [Google Scholar]
- 74.Velculescu, V. E., L. Zhang, W. Zhou, J. Vogelstein, M. A. Basrai, D. E. Bassett, P. Hieter, B. Vogelstein, and K. W. Kinzler. 1997. Characterization of the yeast transcriptome. Cell 88:243-251. [DOI] [PubMed] [Google Scholar]
- 75.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 76.Walker, S. S., W.-C. Shen, J. C. Reese, L. M. Apone, and M. R. Green. 1997. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell 90:607-614. [DOI] [PubMed] [Google Scholar]
- 77.Wan, Y., X. Liu, and M. W. Kirschner. 2001. The anaphase-promoting complex mediates TGF-β signaling by targeting SnoN for destruction. Mol. Cell 8:1027-1039. [DOI] [PubMed] [Google Scholar]
- 78.Werner-Washburne, M., E. L. Braun, M. E. Crawford, and N. M. Peck. 1996. Stationary phase in Saccharomyces cerevisiae. Mol. Microbiol. 19:1159-1166. [DOI] [PubMed] [Google Scholar]
- 79.Werner-Washburne, M., E. L. Braun, G. C. Johnston, and R. A Singer. 1993. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57:383-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wyrick, J. J., F. C. Holstege, E. G. Jennings, H. C. Causton, D. Shore, M. Grunstein, E. S. Lander, and R. A. Young. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402:418-421. [DOI] [PubMed] [Google Scholar]
- 81.Yamaki, M., T. Umehara, T. Chimura, and M. Horikoshi. 2001. Cell death with predominant apoptotic features in Saccharomyces cerevisiae mediated by deletion of the histone chaperone ASF1/CIA1. Genes Cells 6:1043-1054. [DOI] [PubMed] [Google Scholar]
- 82.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990.Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 83.Young, E. T., N. Kacherovsky, and K. Van Riper. 2002. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J. Biol. Chem. 277:38095-38103. [DOI] [PubMed] [Google Scholar]
- 84.Zachariae, W., and K. Nasmyth. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13:2039-2058. [DOI] [PubMed] [Google Scholar]