Abstract
Background
Recently, new emergence of type I PRRSV has been reported in Korea by several research groups. Although specific subgroups of type I PRRSVs in Korea were observed in the previous phylogenetic analysis, there is a lack of information about the virulence of type I PRRSV recently isolated in Korea.
Methods
One type I PRRSV isolate (G2446, 3 times passaged in primarily cultured pulmonary macrophages) in Korea was experimentally infected in colostrum-deprived pigs. The pathological and serological evaluations were performed and compared to type II PRRSV strain (CP07-401-9, 5 times passaged in MARC-145 cell lines)-infected pigs, for 21 days post challenge (dpc).
Results
The pneumonia found in gross examination was more severe in type I PRRSV-infected pigs than type II PRRSV-infected pigs. Both groups showed bronchointerstitial pneumonia, mild multifocal perivascular lymphohistiocytic myocarditis and lymphadenopathy at 14 dpc. However, the unique histopathologic lesions were not found in the pigs experimentally infected with a Korean type I PRRSV isolate, when compared to previous data about classical pathology of PRRSV. The PRRS-specific antibodies were detected in the first week after challenge and viremia continued at least until 21 dpc in both groups.
Conclusion
The gross and histopathologic lesion in this study indicated that Korean type I PRRSV strain (G2446) caused classical PRRSV-specific lesions. Although this study evaluated one representative strain of Korean type I PRRSV, the results may provide information regarding the pathogenicity of type I PRRSV recently emerged in Korea.
Keywords: type I PRRSV, Korea, Experimental, infection, emerging
Background
Porcine reproductive and respiratory syndrome (PRRS) has spread worldwide and continues to be one of the most devastating diseases of swine throughout the world. PRRS is caused by a small, enveloped, positive strand RNA virus, PRRS virus (PRRSV), which belongs to the family Arteriviridae, genus Arterivirus [1]. Genetic and antigenic analyses have revealed two distinct PRRSV groups, the European (Type I) and the North American (Type II), with marked genetic and antigenic differences between the two genotypes as well as among viruses within each genotype [2-5].
PRRS has been experimentally induced with cell-culture-propagated virus in sows and pigs [6-8]. Also, it has been documented that PRRSV strains differ in virulence [9].
In the Republic of Korea, type II PRRSV infection was first described in 1993 [10]. Since then, there have been studies on the molecular characterization of type II PRRSV [11,12]. Recently, type I PRRSV infection occurred in Korean swine farms, and they showed unique characteristics in genetic analysis [13-16]. The type II PRRSV in Korea was suspected to be introduced from North America, and at least 4 different lineages of type II PRRSV were circulating in Korea [13]. In the nation-wide study, the Korean type I PRRSV (a term used to indicate type I PRRSVs in Korea) formed three distinct clusters from other type I PRRSV strains and cluster I was a predominant group [13,14]. Although the type I PRRSVs in Korea were included in panEuropean subgroup, they were further divided into three clusters (class I, II and III) in the phylogenetic analysis [4,14,15]. The class I was shown to be dominant strains in Korea. However, in spite of nation-wide phylogenetic analysis of the viruses, there is a lack of information about the virulence of type I PRRSV recently isolated in Korea. The aim of this study was to observe gross lesion, histopathological lesion and immunological properties in pigs after experimental infection of a type I PRRSV isolate especially belonged to 'class I', a dominant type I PRRSV in Korea.
Methods
Cells and viruses
In the case of type I PRRSV, tissue-culture-infective doses (TCID) were prepared as follows. A G2446 strain (Passage 3 in Pulmonary alveolar macrophages (PAM)) was prepared to viral concentration of 105 TCID50/ml using Dulbecco's modified Eagle's medium (DMEM) with 5% fetal bovine serum (FBS), penicillin (100 units/ml), streptomycin (100 μg/ml) and amphotericin B (0.25 μg/ml). In the case of type II PRRSV, a CP07-401-9 strain (Passage 5 in MARC-145 cells) was prepared to 105 TCID50/ml in the same media as described above for the Type I strain. The viruses were isolated from pigs in Korea and their sequence information was presented in the previous papers [13,15].
Experimental design
Two ml of 5 logTCID50/ml type I and type II PRRSV isolates, G2446 (GenBank ID: GU325647, p3, cluster I) and CP07-401-9 (GenBank ID: FJ972728, p5, vaccine-like), were inoculated into five colostrum-deprived pigs (3 weeks old) for each viral type via the intranasal route. Three pigs remained uninfected as a control group. Each group was maintained in a separate pen. After challenge, blood samples were collected at 4, 5, 6, 7, 8, 9, 10, 11, 12, 14 and 21 days post challenge (dpc). All animal experiments complied with the current laws of South Korea. Animal care and treatment were conducted in accordance with guidelines establishment by the Green Cross Veterinary Products Institutional Animal Care and Use Committee.
Serological tests
A commercial kit (HerdChek PRRS 2XR, IDEXX Inc., USA) was used, according to the manufacturer's instructions, to estimate the levels of PRRSV-specific antibodies. The IgG titer for PRRSV was indicated as S/P ratio which was the calculated value in the formula: (Sample O.D - Negative control O.D)/(Positive control O.D - Negative control O.D).
A commercial real time polymerase chain reaction kit (Ambion, USA) was used to detect viremia from sera during the study. To evaluate the serum neutralizing ability of both types of viruses, a Serum neutralizing (SN) test was performed based on the previous study [17]. In the case of a type I PRRSV isolate (G2446), PAM cells were used instead of MARC-145 cell lines.
Gross and histopathological tests
Pigs were euthanized to observe gross and histopathologic lesions at 14 and 21 dpc, respectively, as follows; three and two pigs from the challenged groups, and two and one pigs from the control group. The lung, heart, liver, spleen, kidney, ileum, and lymph nodes were collected and fixed with 4% formaldehyde solution. The RT-nested PCR was performed from the collected organs following previous paper [18]. The fixed organs were processed for histopathology, using hematoxylin and eosin staining.
Results and discussion
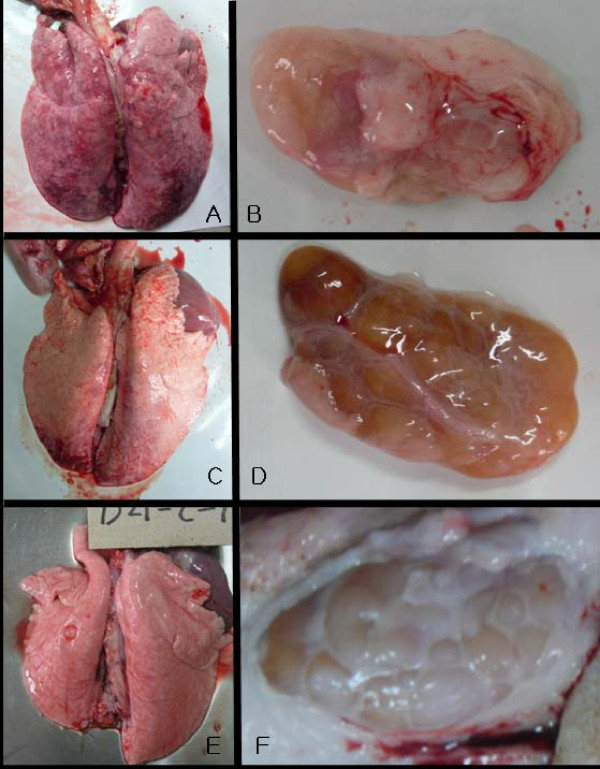
Gross lesions were enlarged tan lymph nodes and abnormal lungs in both challenged groups. In the lungs, the parenchyma was mottled or diffusely tan-red in the type I PRRSV-challenged pigs, while only mild regional mottling was observed in the type II PRRSV-challenged pigs (Figure 1).
Figure 1.
The gross lesions of lungs and superficial inguinal lymph nodes in the challenged pigs at 14 dpc. A, lung of G2446 (type I)-challenged group; B, superficial inguinal lymph node of G2446(type I)-challenged group; C, lung of CP07-401-9 (type II)-challenged group; D, superficial inguinal lymph node of CP07-401-9 (type II)-challenged group; E, lung of control group at 21 dpc; F, superficial inguinal lymph node of control group at 21 dpc
Histopathological lesions were only found in lung, lymph node and heart tissue (Table 1). The lungs of both challenged groups shared the same pathological observations as follows;
Table 1.
Histopathological findings in the challenged pigs
| DPC* | Type | Pathological findings† | ||
|---|---|---|---|---|
| Lung | Inguinal Lymph Nodes | Heart | ||
| 14 | I | Moderate focal/multifocal bronchointerstitial pneumonia (3/3) | Lymphadenopathy (3/3) | Mild multifocal perivascular lymphohistiocytic myocarditis (1/3) |
| II | Moderate diffuse/multifocal bronchointerstitial pneumonia (2/3) | Lymphadenopathy (3/3) | Mild multifocal perivascular lymphohistiocytic myocarditis. (3/3) | |
| Control | No lesions (2/2) | No lesions (2/2) | No lesions (2/2) | |
| 21 | I | Moderate diffuse interstitial pneumonia (2/2), moderate peribronchiolar and perivascular lymphohistiocytic cuffing (2/2) | Lymphadenopathy (2/2) | Mild multifocal perivascular lymphohistiocytic myocarditis (1/2) |
| II | Mild to moderate diffuse interstitial pneumonia (2/2), severe peribronchiolar and perivascular lymphohistiocytic cuffing (1/2) | Lymphadenopathy (2/2) | Very mild multifocal perivascular lymphohistiocytic myocarditis (1/2) | |
| Control | No lesions (1/1) | No lesions (1/1) | No lesions (1/1) | |
* days post challenge
† No lesions were observed in liver, spleen, kidney and ileum.
(1) bronchointerstitial pneumonia, which was characterized by lymphohistiocytic inflammation and an accumulation of necrotic cells, likely originated from alveolar macrophages, in alveolar spaces, and (2) Type 2 pneumocyte hyperplasia in the alveolar septa. Mild multifocal perivascular lymphohistiocytic myocarditis was sometimes observed in both challenged groups as well. In the control group, there were no pathologic changes. The results of RT-nested PCR showed PRRSV-positive in all collected organs in both challenged groups during the study (data not shown).
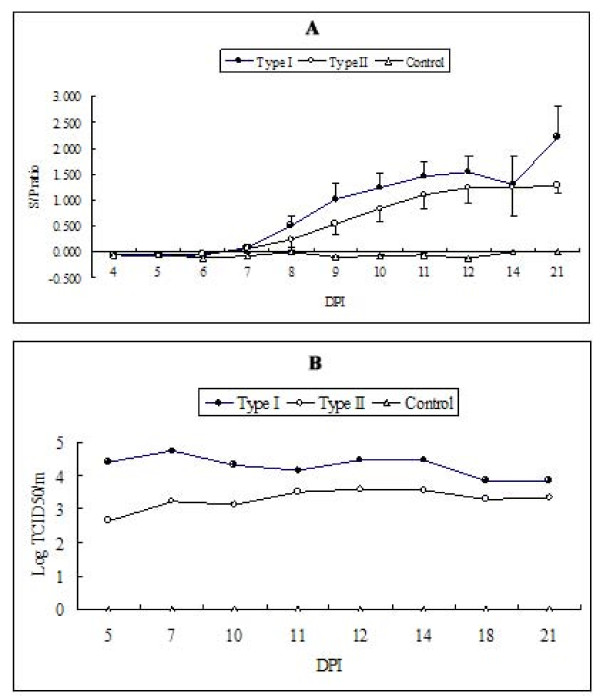
PRRSV-specific antibodies were first detected at 8 and 9 dpc in type I and type II PRRSV-challenged groups, respectively (Figure 2). Although it was not significant by Student's T test, mean s/p ratios remained higher in the type I PRRSV-challenged group than in the type II challenged group during the study. Even in the presence of PRRSV-specific IgG, no detectable level of SN titer was observed in either challenge group, and viremia was also maintained throughout the study.
Figure 2.
PRRSV-specific serum IgG (A) and viremic titers (B). Error bars in type I and II-challenged groups indicate standard errors. For the quantitation of viremia, a commercial real time RT-PCR kit was used and virus titer was calculated using a standard curve generated from serially diluted type I or II virus isolates. Two serum samples from each dpc collection were stored until 21 dpc. At this point, they were pooled and quantitative RT-PCR was performed. Since no viral genome was detected in the control group, viral titer in control group sera was indicated as '0'.
In this study, the virulence of newly emerging type I PRRSV in Korea was evaluated in young pigs, using one type I PRRSV isolate. Our main aim was to evaluate the virulence of newly emergent type I PRRSV under experimental conditions. Although only one isolate of type I PRRSV was used in this study, the data may provide fundamental information for further comparative studies. Since the previous study had reported the observation of PRRSV-specific histopathological lesions from 3 to 21 dpc, we tested the challenged pigs until 21 dpc [19]. Previous pathogenicity study of type II PRRSV isolated in Korea showed that alveolar proteinaceous and karyorrhectic debris were interspersed with macrophages as well as mild type-II pneumocyte hyperplasia and hypertrophy [19]. The comparative study using two type II PRRSV and one type I PRRSV also revealed that both types of PRRSVs could induce lymphohistiocytic myocarditis, lymphadenopathy and encephalitis as well as lung lesions [9].
In agreement with the previous data, pathologic changes after experimental challenge with a Korean isolate of type I PRRSV (class I) were observed in the lungs, lymph nodes and heart, until 14 and 21 dpc. The pathologic changes by the Korean isolate of type I PRRSV in this study were also type 2 pneumocyte hyperplasia, necrotic debris in alveolar septa, perivascular lymphohistiocytic myocarditis and lymphadenopathy, which were all similar to previous reports. These data indicated that a type I PRRSV isolate, in this study, showed classical lesions of PRRSV infection and sufficiently induced acute disease in young pigs (3 weeks old). Pigs at this age are known to be in a vulnerable stage for PRRSV infection [20]. Although unique lesions of the Korean isolate of type I PRRSV was not found in this study, the severity of gross lesions in lungs was higher than that of the type II PRRSV strain (VR-2332-like). Therefore, type I PRRSV (G2446, class I) in this study may also present a risk for co-infections with other viruses and bacteria in the pigs during the nursery to growing period.
The seroconversion was also well-defined by ELISA around 8 dpc. Although mean S/P ratio was higher in type I PRRSV challenged pigs, it was not significant by student's T test, which led incapable of differentiating type I and II PRRSV infection in the field using IDEXX ELISA. Further consideration will be needed for type-specific serological methods to differentiate type I and II PRRSV infection in Korea. In this study, no SN titer was observed until 21 dpc in both group. Since SN titer for type I PRRSV was usually detected at around 35 days post inoculation, the lack of detection of SN antibodies in this study is probably due to the short period of time of experiment (21 days) [21]. Furthermore, the prolonged viremia of both types I and II PRRSV observed in this study raised concerns about their long-term transmission among pigs. Therefore, type-specific PRRSV surveillance and control policy are important to minimize the type I PRRSV transmission in Korea.
In conclusion, the Korean isolate of type I PRRSV used in this study could induce classical PRRSV-specific lesions and serological properties. Although only one strain of Korean type I PRRSV was evaluated in this study, the results could provide information about the virulence of recently emerging Korean type I PRRSV. However, the virulence of PRRSV infection could be differed among the same types of PRRSV strains [9]. So, further comparative studies based on this study are needed to be followed.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
HKK: Attending the experiments and writing a manuscript, CSL: Attending the experiments and sample collection, BKK: Organized the animal experiment in the facility, MJY: conducted animal care and sample collection, HJM: conducted animal care and sample collection, SJP: Attributed to statistical analysis, VGN: participated writing a manuscript for introduction, DSS: Idea development and designing all experiments, BKP: final correction of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Hye Kwon Kim, Email: khk1329@snu.ac.kr.
Chul Seung Lee, Email: lcs0610@hanmail.net.
Bo Kyu Kang, Email: suyun@gcvp.co.kr.
Min Ju Yeom, Email: paransearo@naver.com.
Hyoung Joon Moon, Email: mhj1219@naver.com.
Seong Jun Park, Email: juni1212@snu.ac.kr.
Van Giap Nguyen, Email: nvgiap@snu.ac.kr.
Dae Sub Song, Email: sds1@kribb.re.kr.
Bong Kyun Park, Email: parkx026@snu.ac.kr.
Acknowledgements
This study was supported by the Technology Development Program for Agriculture and Forestry from the Ministry of Agriculture and Forestry, Republic of Korea and by a grant (Code# 20070401034009) from the Biogreen 21 Program, Rural Development Administration, Korea. We also thank Dr. K. Jung in Ohio State University for interpreting histopathologic lesions.
References
- Cavanagh D. Nidovirales: A new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- Meng XJ, Paul PS, Halbur PG, Lum MA. Phylogenetic analyses of the putative M(ORF-6)-gene and N(ORF-7)-gene of porcine reproductive and respiratory syndrome virus (PRRSV) - implication for the existence of 2 genotypes of PRRSV in the USA and Europe. Arch Virol. 1995;140:745–755. doi: 10.1007/BF01309962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh MP, Elam MR, Kakach LT. Comparison of the structural protein-coding sequences of the VR-2332 and Lelystad virus-strains of the PRRS virus. Arch Virol. 1995;140:1451–1460. doi: 10.1007/BF01322671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadejek T, Oleksiewicz MB, Potapchuk D, Podgorska K. Porcine reproductive and respiratory syndrome virus strains of exceptional diversity in Eastern Europe support the definition of new genetic subtypes. J Gen Virol. 2006;87:1835–1841. doi: 10.1099/vir.0.81782-0. [DOI] [PubMed] [Google Scholar]
- Yoshii M, Kaku Y, Murakami Y, Shimizu M, Kato K, Ikeda H. Genetic variation and geographic distribution of porcine reproductive and respiratory syndrome virus in Japan. Arch Virol. 2005;150:2313–2324. doi: 10.1007/s00705-005-0549-2. [DOI] [PubMed] [Google Scholar]
- Christianson WT, Collins JE, Benfield DA, Harris L, Gorcyca DE, Chladek DW, Morrison RB, Joo HS. Experimental reproduction of swine infertility and respiratory syndrome in pregnant sows. Am J Vet Res. 1992;53:485–488. [PubMed] [Google Scholar]
- Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS, Gorcyca D, Chladek D. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North-America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest. 1992;4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparison of the pathogenicity of 2 US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 1995;32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- Halbur PG, Paul PS, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparative pathogenicity of nine US porcine reproductive and respiratory syndrome virus (PRRSV) isolates in a five-week-old cesarean-derived, colostrum-deprived pig model. J Vet Diagn Invest. 1996;8:11–20. doi: 10.1177/104063879600800103. [DOI] [PubMed] [Google Scholar]
- Kweon CH, Kwon BJ, Lee HJ, Cho JJ, Hwang EK, Shin JH, Yoon YD, Kang YB, An SH, Kim YH, Huh W, Jun MH, Wensvoort G. Isolation of porcine reproductive and respiratory syndrome virus(PRRSV) in Korea. Kor J Vet Res. 1994;34:77–83. [Google Scholar]
- Cha SH, Choi EJ, Park JH, Yoon SR, Song JY, Kwon JH, Song HJ, Yoon KJ. Molecular characterization of recent Korean porcine reproductive and respiratory syndrome (PRRS) viruses and comparison to other Asian PRRS viruses. Vet Microbiol. 2006;117:248–257. doi: 10.1016/j.vetmic.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Yoon SH, Song JY, Lee CH, Choi EJ, Cho IS, Kim B. Genetic characterization of the Korean porcine reproductive and respiratory syndrome viruses based on the nucleocapsid protein gene (ORF7) sequences. Arch Virol. 2008;153:627–635. doi: 10.1007/s00705-007-0027-0. [DOI] [PubMed] [Google Scholar]
- Kim HK, Yang JS, Moon HJ, Park SJ, Luo Y, Lee CS, Song DS, Kang BK, Ann SK, Jun CH, Park BK. Genetic analysis of ORF5 of recent Korean porcine reproductive and respiratory syndrome viruses (PRRSVs) in viremic sera collected from MLV-vaccinating or non-vaccinating farms. J Vet Sci. 2009;10:121–130. doi: 10.4142/jvs.2009.10.2.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Roh IS, Choi EJ, Lee C, Lee CH, Lee KH, Lee KK, Song YK, Lee OS, Park CK. A molecular analysis of European porcine reproductive and respiratory syndrome virus isolated in South Korea. Vet Microbiol. 2010;143:394–400. doi: 10.1016/j.vetmic.2009.11.039. [DOI] [PubMed] [Google Scholar]
- Lee C, Kim H, Kang B, Yeom M, Han S, Moon H, Park S, Kim H, Song D, Park B. Prevalence and phylogenetic analysis of the isolated type I porcine reproductive and respiratory syndrome virus from 2007 to 2008 in Korea. Virus genes. 2010;40:225–230. doi: 10.1007/s11262-009-0433-3. [DOI] [PubMed] [Google Scholar]
- Nam E, Park CK, Kim SH, Joo YS, Yeo SG, Lee C. Complete genomic characterization of a European type 1 porcine reproductive and respiratory syndrome virus isolate in Korea. Arch Virol. 2009;154:629–638. doi: 10.1007/s00705-009-0347-3. [DOI] [PubMed] [Google Scholar]
- Kim WI, Lee DS, Johnson W, Roof M, Cha SH, Yoon KJ. Effect of genotypic and biotypic differences among PRRS viruses on the serologic assessment of pigs for virus infection. Vet Microbiol. 2007;123:1–14. doi: 10.1016/j.vetmic.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Kono Y, Kanno T, Shimizu M, Yamada S, Ohashi S, Nakamine M, Shirai J. Nested PCR for detection and typing of porcine reproductive and respiratory syndrome (PRRS) virus in pigs. J Vet Med Sci. 1996;58:941–946. doi: 10.1292/jvms.58.10_941. [DOI] [PubMed] [Google Scholar]
- Cheon DS, Chae C. Distribution of a Korean strain of porcine reproductive and respiratory syndrome virus in experimentally infected pigs, as demonstrated immunohistochemically and by In-situ hybridization. J Comp Pathol. 1999;120:79–88. doi: 10.1053/jcpa.1998.0257. [DOI] [PubMed] [Google Scholar]
- Klinge KL, Vaughn EM, Roof MB, Bautista EM, Murtaugh MP. Age-dependent resistance to Porcine reproductive and respiratory syndrome virus replication in swine. Virol J. 2009;6:177. doi: 10.1186/1743-422X-6-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarque GG, Nauwynck HJ, Van Reeth K, Pensaert MB. Effect of cellular changes and onset of humoral immunity on the replication of porcine reproductive and respiratory syndrome virus in the lungs of pigs. J Gen Virol. 2000;81:1327–1334. doi: 10.1099/0022-1317-81-5-1327. [DOI] [PubMed] [Google Scholar]