Abstract
Neural crest cells migrate long distances and form divergent derivatives in vertebrate embryos. Despite previous efforts to identify genes upregulated in neural crest populations, transcription factors have proved to be elusive due to relatively low expression levels and often transient expression. We screened newly induced neural crest cells for early target genes with the aim of identifying transcriptional regulators and other developmentally important genes. This yielded numerous candidate regulators, including fourteen transcription factors, many of which were not previously associated with neural crest development. Quantitative real-time PCR confirmed upregulation of several transcription factors in newly induced neural crest populations in vitro. In a secondary screen by in situ hybridization, we verified the expression of >100 genes in the neural crest. We note that several of the transcription factors and other genes from the screen are expressed in other migratory cell populations and have been implicated in diverse forms of cancer.
INTRODUCTION
The neural crest is a cell population that forms at the border between the neural and non-neural ectoderm of vertebrate embryos. As the neural folds elevate, cells induced to become neural crest cells become incorporated into the dorsal neural tube, and subsequently undergo an epithelial to mesenchymal transition and depart from the future central nervous system. These multipotent precursors then migrate along prescribed and stereotypic pathways to defined and diverse sites in the periphery. Upon completion of migration, they differentiate into a wide variety of derivatives including melanocytes, peripheral neurons and glial, adrenomedullary cells, and much of the bone and cartilage of the face (Knecht and Bronner-Fraser, 2002; Le Douarin et al., 2007). As a population, the neural crest is of widespread interest because of its plasticity, mobility, and contribution to developmental disorders and cancers.
Previous work in a number of species, including mice, chick, frogs and zebrafish, has implicated a set of transcriptional regulators in conferring neural crest identity. This set of genes, termed neural crest specifier genes (Meulemans and Bronner-Fraser, 2004), include Sox9, Sox10, FoxD3, Snail2/Slug, c-Myc and AP2 (Spokony et al., 2002) (Honore et al., 2003) (Sasai et al., 2001) (Aoki et al., 2003) (Luo et al., 2003) (Bellmeyer et al., 2003). These transcription factors are expressed in premigratory and/or early migrating neural crest cells and thought to be involved in induction and survival of the premigratory population, their epithelial to mesenchymal transition from the neural tube, migratory ability, and cell lineage decisions (Anderson, 1994) (Cheung et al., 2005). However, it seems unlikely that these complicated processes would be controlled by the relatively few transcription factors identified to date in newly formed neural crest cells.
In a genomic analysis previously performed in our lab, a large number of genes were identified as upregulated in premigratory and migratory neural crest (Gammill and Bronner-Fraser, 2002). However, this yielded only two transcription factors that had not been previously associated with the neural crest. Because transcription factors are essential for understanding proximal portions of the gene cascades that regulate neural crest migration and differentiation, it is critical to identify the complete repertoire of genes in order to build testable gene regulatory networks (Davidson, 2006) (Ben-Tabou de-Leon and Davidson, 2007). Data from various organisms have led to the formulation of putative gene regulatory networks in different organisms (Mayor et al., 1995; Mayor and Aybar, 2001) (Meulemans and Bronner-Fraser, 2004) (Steventon et al., 2005). Further work in identifying the function of transcription factors and downstream genes will help refine and expand upon these networks. In turn, this work may give new insight into related gene circuits that may be operative in other migratory and metastatic cell types.
To identify putative neural crest transcription factors, we completed a subtractive screen for newly induced neural crest cells using a macroarrayed cDNA library from early chick embryos. This allowed identification of over 300 genes, many of which were not previously associated with neural crest development. As a secondary screen, we performed in situ hybridization to verify expression of 101 genes in premigratory and/or migrating neural crest cells. Quantitative PCR (QPCR) was used to confirm the level of upregulation of several transcription factors in newly induced neural crest cells. The results reveal many previously unknown transcriptional regulators in the neural crest and suggest interesting commonalities between neural crest cells, various metastatic cancers and other migratory cell types.
MATERIALS AND METHODS
Embryos and Explants
Fertile chicken eggs were incubated from 24 hours to three days to obtain embryos from HH stage 4 to HH stage 24. Embryos to be used for in situ hybridization were fixed overnight in 4% paraformaldehyde (PFA) at 4°C. They were then washed 4 × 5 minutes in PTW and dehydrated in a MeOH/PTW series at RT before being stored at −20°C in 100% MeOH for more than 3 days. Explants were dissected from embryos in Ca2+/Mg2+-free Tyrodes saline with .05% trypsin (Sigma). Conjugates were assembled and allowed to recover as previously described before embedding in collagen gels and culturing for 24 hours (Dickinson et al., 1995; Gammill and Bronner-Fraser, 2002).
RNA preparation and QPCR
RNA was prepared from tissues in collagen gels using Stratagene’s RNA isolation kit. Collagen gels were transferred to an Eppendorf microcentrifuge tube with forceps and triturated in lysis buffer.
Quantitative polymerase chain reaction (QPCR) was performed using the 96 well plate ABI 7000 QPCR machine in a TaqMan assay (Applied Biosciences) with Sybrgreen Itaq Supermix with ROX (BioRad), 300 nM of each primer, and 200–500 ng of cDNA in a 25 μL reaction volume. Gene specific primers were designed using the Primer Express program (Applied Biosystems) and synthesized by IDT.
During the exponential phase of the QPCR reaction a threshold and baseline was set according to the protocols of Applied Biosystems. The threshold cycle (CT) is defined as the cycle number at which the fluorescence generated by a reaction meets this threshold and is critical in determining the amount of the amplicon produced in the RT-QPCR reaction. The results for different samples were then interpolated on a line created by running standard curves for each primer set and then normalized against the results for a housekeeping gene, which is assumed to be a measure of the amount of cDNA used in each reaction. These calculations were performed according to the standard curve assay method which is detailed in the Applied Biosystems protocols. Each reaction plate was run with five point standard curves (three replicates each) for the gene of interest (GOI) and the chosen housekeeping gene, 18s. Each plate also held three non-template controls (in which no amplification was seen) for each set of primers. Standards are always run on the same plate as the unknown samples in order to ensure the validity of CT measurements.
QPCR primer sequences were as follows:
slug forward 5′-GGGCTGCTACCGTCTCCTCTA-3′
slug reverse 5′-GCTGACCCTTCCCAAAGATG-3′
gcnf forward 5′-TTCCCCCAAGATGTCATTGAC-3′
gcnf reverse 5′-TGATGGCTGCAATGCAGAAG-3′
elk-3 forward 5′-CAGCCCCACCTTTGGTTCTT-3′
elk-3 reverse 5′-TGAGGGCAATGGAACCGATA-3′
zf469 forward 5′-CCAGCTCCAAAGAAGCAACAA-3′
zf469 reverse 5′-TGCCGAGAAGAGTCCAAAGTG-3′
srebp2 forward 5′-GATCACAGGCTGGCTTCTCC-3′
srebp2 reverse 5′-GGCTTCCTGGCTCTGAATCA-3′
gtfIIe forward 5′-CCAAGTATGTCGTGCGTGGT-3′
gtfIIe reverse 5′-CCAGAGCCAAAGCGTGTTC-3′
In Situ Hybridization
Antisense, digoxygenin labeled RNA probes were prepared from clones categorized as being upregulated by the macroarray screen. The probes were hydrolyzed and in situs were performed as described (Xu, 1992). Embryos were visually examined at different angles to aid in scoring and photographed on a Zeiss Axioskop2 Plus. Selected embryos were then coarsely sectioned using a scalpel or finely sectioned using a cryostat (Bright) and mounted or immunostained.
Immunostaining
Wholemount in situs were post-fixed in 4% PFA (paraformaldehyde) + 0.02% glutaraldehyde and washed in PTW 4×15 minutes. They were nutated at 4°C overnight in 15% sucrose, kept for 8 hours at 37°C in 7.5% gelatin/15% sucrose and between 8 and 20 hours in 20% gelatin. The embryos were then embedded, frozen and cryosectioned at approximately 25 μM. Sections were briefly dried onto slides and degelatinized for 20 minutes at 42°C in PBS. They were then placed in PBS (phosphate buffered saline) + 0.1% Tween 20 + 5% goat serum block for hours at room temperature before an overnight 4°C incubation with 1:20 anti-HNK-1 antibody (American Type Culture Collection hybridoma). Sections were then washed several times in PBS + 0.1% Tween over several hours, blocked for 3 hours at RT with PTW (phosphate-buffered saline [PBS], pH 7.5, 0.1% Tween 20) and 5% goat serum, and incubated with 1:400 anti-IgM RRX goat anti mouse (Jackson ImmunoResearch) in PTW at 4°C overnight. Slides were then washed again in PTW several times, rinsed twice in dH20 and then coverslipped using PermaFluor (Immunotech).
RESULTS and DISCUSSION
In order to identify novel neural crest regulators, we performed a genome-level screen for genes upregulated in newly induced neural crest cells. Neural crest induction was recapitulated in vitro by culturing intermediate neural plate and non-neural ectoderm in apposition (Dickinson et al., 1995) (Selleck and Bronner-Fraser, 1996) (Gammill and Bronner-Fraser, 2002). cDNA enriched for neural crest genes was generated by subtracting intermediate neural plate and non-neural ectoderm cultured in isolation from tissue cultured as conjugates, and a macroarrayed early chick cDNA library screened as previously described (Gammill and Bronner-Fraser, 2002).
In this paper, we now describe the completion of the neural crest screen. We previously reported the early results from this screen and the identification of only three transcription factors, including snail-2 (slug) (Gammill and Bronner-Fraser, 2002). We and others went on to demonstrate the functional importance of candidate neural crest regulators from the screen, validating its relevance for neural crest development (Gammill et al., 2006) (Coles et al., 2006) (Gessert et al., 2007; Taneyhill et al., 2007) (Eroglu et al., 2006) (Suzuki et al., 2006) (the unknown from Gammill and Bronner-Fraser, 2002 was ultimately identified as cadherin-6b). In characterizing additional clones, we were particularly interested in identifying new transcription factors expressed in the neural crest. We reasoned that because transcription factors are generally the least abundant transcripts, they would be represented with the fewest clones in the arrayed library and would exhibit the weakest hybridization signals since they would have the lowest copy number in the hybridized cDNA. It seemed highly probable, therefore, that by picking all upregulated clones, not just the most obvious, that we should increase the number of transcription factors in our collection. As we previously performed only a sampling of upregulated clones (Gammill and Bronner-Fraser, 2002), here we comprehensively analyzed all genes with a two-fold or greater hybridization signal from the subtracted probe compared to the unsubtracted probe. It is important to appreciate that this differential hybridization signal is not a direct measure of gene expression levels, rather it reflects the efficiency of subtraction for that gene, which is determined by a variety of factors including differential expression levels, sequence polymorphism, and non-specific loss. Clones meeting this two-fold criterion were picked, sequenced, and compared to the Genbank non-redundant database and the GO ontology database. This resulted in the identification of 300 new candidate neural crest genes that fell into logical functional categories.
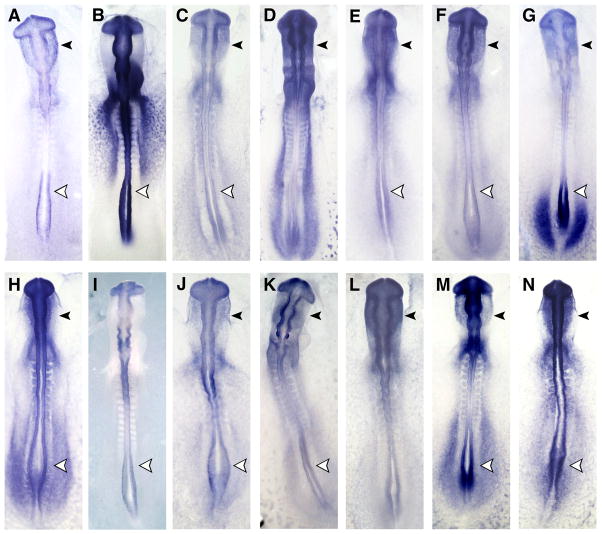
To independently confirm the specificity of our results, in situ hybridization analysis was performed at multiple stages of neural crest development on 112 of the identified genes from the most functionally interesting categories: chromatin, cytoskeletal, ECM, mitochondria, mitosis/cell cycle, nucleocytoplasmic export, protein production and degradation, receptors or downstream signaling molecules, rho pathway genes, RNA binding proteins, secreted signals or signal production, transcription coupled processes, transcription factors, transporters, and unknown genes (Figs. 1–6). This provided the additional advantage of revealing dynamic aspects of gene expression patterns over the course of neural crest development. In this way, we confirmed that 101 of 112 genes were strongly expressed in premigratory neural crest cells in the neural folds and/or in migrating neural crest relative to neighboring tissues. These clones were kept as verified candidate neural crest regulators (Table 1).
Figure 1.
Neural crest gene expression: In situ hybridization was performed to confirm the expression of genes seen to be upregulated in macroarray screen. Selected genes from different categories are shown in dorsal view with anterior at the top. (A) chromatin: safB1; (B) cytoskeletal: paranemin; (C) ECM: lamin B2; (D) mitochondria: hexokinase 2; (E) mitosis/cell cycle: TD-60; (F) Nucleocytoplamic export: importin 9; (G) protein production/degradation: makorin; (H) receptors/downstream signaling: MHC-B; (I) rho pathway: kinectin; (J) RNA binding proteins: HNRPM; (K) secreted signals/signal production: PEBP; (L) transport: OSBP2; (M) miscellaneous: PGK; (N) unknown: chEST872f20.
Figure 6.
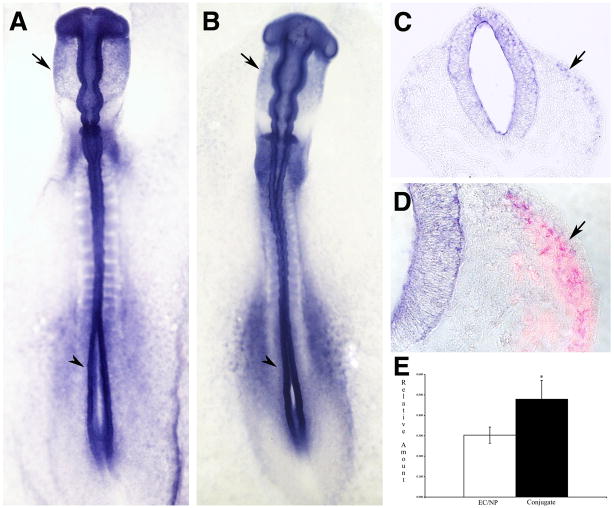
Neural crest expression of and QPCR results for SREBP2. (A-D) SREBP2 expression is seen specifically in premigratory (dorsal neural tube) and migratory neural crest. Shown are in situ hybridizations of 9 somite (A) and 18 somite (B-D) embryos in whole mount (A and B) and sections of whole mount stained embryos (C and D). Immunofluorescence for HNK-1 is in red (D). Arrowheads mark premigratatory neural crest, arrows mark migratory neural crest. (E) results of QPCR showing a significant difference between control and conjugated tissues (p = 0.030).
Table 1.
Summary of functional categories and genes identified.
Clones were organized into categories based upon functions assigned by sequence homology. Degree of homology is indicated by colored spots (
 = BLAST bit score > 200;
= BLAST bit score > 200;
 = bit score 80–200;
= bit score 80–200;
 = bit score 50–80;
= bit score 50–80;
 = bit score 40–50; ● = bit score < 40). In situ hybridization patterns in 4–16 somite embryos were scored for expression in neural folds (premigratory neural crest; pNC) and cranial migratory neural crest (mNC) under the assumption that the in vitro induced neural crest examined in the subtraction corresponds to a timepoint between these stages of neural crest development. General specificity of expression (SP) was also scored (−, expression in many tissues at varying levels; +/−, expression broad, but discrete patterns obvious; +, expression limited to only a few tissues; ++, expression highly specific).
= bit score 40–50; ● = bit score < 40). In situ hybridization patterns in 4–16 somite embryos were scored for expression in neural folds (premigratory neural crest; pNC) and cranial migratory neural crest (mNC) under the assumption that the in vitro induced neural crest examined in the subtraction corresponds to a timepoint between these stages of neural crest development. General specificity of expression (SP) was also scored (−, expression in many tissues at varying levels; +/−, expression broad, but discrete patterns obvious; +, expression limited to only a few tissues; ++, expression highly specific).
| A. Chromatin | pNC | mNC | SP | ||
|---|---|---|---|---|---|
|
|
H3, 3B | + | + | − | replacement histone; restricted to cells of meiotic prophase |
|
|
HP1BP74 | + | + | +/− | nucleosome assembly; DNA binding |
|
|
SAFB1 | + | + | +/− | may act in attaching base of chromatin loops to nuclear matrix |
| B. Cytoskeleton | |||||
|
|
Alpha E-catenin | + | + | +/− | major binding partner of beta-catenin |
|
|
CAP-23 | − | + | +/− | BASP1; regulates cell cortex actin dynamics |
|
|
MACF1 | + | + | +/− | may stabilize actin where microtubules and microfilaments meet |
|
|
NFM | + | + | +/− | neurofilament triplet M; intermediate filament family |
|
|
Nectin1 | + | +/− | +/− | part of adherens junctions; may interact with cadherin-catenin |
|
|
Nestin | + | + | +/− | NES; intermediate filament protein |
|
|
Merlin | − | + | +/− | gene responsible for neurofibromatosis-2 |
|
|
Nonerythroid α-spectrin | + | + | +/− | SPTAN1; filamentous cytoskeletal protein |
|
|
Paxillin | − | + | +/− | important for labile adhesions required for rapid cell migration |
|
|
Periplakin | − | + | +/− | thought to contact the intermediate filaments of keratinocytes |
|
|
PTK9L | + | + | +/− | protein contains an actin-binding site and an ATP-binding site |
|
|
TPM-γ | + | + | +/− | may alter actin filament organization, cell size, and shape |
| C. ECM | |||||
|
|
COL1A2 | + | − | +/− | fibrillar forming collagen; collagen α2 • • • • • • precursor |
|
|
COL4α5 | − | + | +/− | may be involved in Alport syndrome: deafness and renal problems |
|
|
COL4α6 | − | + | +/− | involved in basement membrane formation, Alport syndr variant |
|
|
LMNB2 | + | + | +/− | Lamin B2: intermediate filament protein |
| D. Mitochondria | |||||
|
|
BID | + | + | − | death agonist; member of BCL-2 family of cell death regulators |
|
|
Hexokinase 2 | − | + | +/− | catalyzes first committed step of glycolysis |
|
|
HADHB | + | + | − | part of multienzyme complex; mitochondrial fatty acid oxidation |
|
|
LDHA | + | − | +/− | promoter contains essential binding sites for HIF-1; |
|
|
Similar to BLCAP | + | + | +/− | seen in mitochondria; downregulated during bladder cancer |
|
|
Similar to NADH dehydrogenase | + | − | − | first enzyme in the mitochondrial electron transfer chain |
| E. Mitosis/cell cycle | |||||
|
|
CycB2 | + | + | +/− | essential for the control of the cell cycle at the G2/M transition |
|
|
CycB3 | + | + | +/− | nuclear sublocalization, belongs to cyclin family AB |
|
|
MCM2 | + | + | +/− | may play an important role in 2 crucial steps of the cell cycle |
|
|
MCM5 | + | + | +/− | forms complex with MCM2 |
|
|
Septin 9 | − | + | +/− | cell division control protein |
|
|
SEC14-like 1 | + | − | +/− | may be involved in E-cadherin-mediated adhesion and signaling |
|
|
TD-60 | + | + | +/− | RCC1 family; binds nucleotide-free form of Rac1 preferentially |
| F. Nucleocytoplasmic export | |||||
|
|
DEAD box polypeptide 23 | + | + | +/− | RNA helicase, nuclear pore protein, mRNA export |
|
|
Importin 9 | + | + | +/− | can mediate nuclear import of core histones into cell nuclei |
| G. Protein production/degradation | |||||
|
|
Cathespin B | − | + | − | intracellular degradation and turnover of proteins |
|
|
CBS | − | + | +/− | enzyme deficient in classical homocystinuria |
|
|
GARS-AIRS-GART | + | + | +/− | catalyzes 2nd, 3rd and 5th steps of de novo purine biosynthesis |
|
|
Glutathione S-transferase CL2 | + | + | − | conjugates reduced glutathione to hydrophobic electrophiles |
|
|
Makorin 1 | + | + | +/− | E3 ubiquitin ligase, promotes proteasomal degradation |
|
|
PSMD3 | − | + | − | proteasome 26S subunit, non-ATPase; subunit 3 |
|
|
Roxan | + | + | − | contains TPR, LD, and zf motifs, forms complex with eIF4G |
| H. Receptors/downstream signaling | |||||
|
|
Anamorsin (Ciapin) | + | + | +/− | critical in hematopoiesis, med. of cytokine-induced antiapoptosis |
|
|
Axl-related RTK (Rek) | + | − | +/− | Axl/Tyro3 family with a potential role in neural cell development |
|
|
MHC B complex protein | + | + | +/− | non-classical MHC protein, encodes G-protein like molecule |
|
|
FMIP | + | + | − | serum/glucocorticoid related kinase 2 |
|
|
GPR144 | + | − | +/− | G protein coupled receptor; integral part of membrane |
|
|
MHC Bcomplex protein | + | + | +/− | non-classical MHC protein, encodes G-protein like molecule |
|
|
Ptprf | + | + | − | Protein tyrosine phosphatase LAR: interacts with TRIO |
|
|
SPINT1 | + | − | + | member of Kunitz family of serine protease inhibitors |
| I. Rho pathway | |||||
|
|
KTN1 | + | − | + | mediator of RhoG and RhoA activity |
|
|
Trio | + | + | +/− | Interacts with PTPRF; displays two Rho-GEF domains |
| J. RNA binding proteins | |||||
|
|
Akap1 | + | +/− | +/− | bifunctional helical element: for ER and mitochondrion targeting |
|
|
ANKRD17 isoform a | + | − | +/− | Studies suggest that this protein is involved in liver development |
|
|
G3BP1 | + | + | +/− | element of the Ras signal transduction pathway |
|
|
EWS | + | + | +/− | mutated in Ewing sarcoma: cancer thought to have NCC origin |
|
|
HnrpA2/B1 | + | + | +/− | involved with pre-mrna processing |
|
|
Hnrp M | + | + | +/− | pre-mRNA-binding proteins in vivo |
|
|
Lark | + | + | +/− | has RT zinc finger; regulates protein production; RNA binding |
|
|
NONO | + | − | − | mammalian homolog of nonAdiss; nuclear RNA binding protein |
|
|
WBSCR20A | + | + | +/− | p120; Proliferation-associated nucleolar protein |
|
|
SH3DBP | + | + | +/− | binds Ras-GTPase-activating protein; associates with SH3 domain |
| K. Secreted signals/signal production | |||||
|
|
Collapsin-2 | + | − | + | Sema3D |
|
|
CFR | + | + | +/− | p70; ER and golgi localization |
|
|
PEBP | + | + | +/− | RAF kinase inhibitor protein; regulates Raf/MEK/ERK module |
|
|
Vav2 | + | + | +/− | oncogene; activates c-fos serum response element and CD69 |
| L. Transcription Coupled Processes | |||||
|
|
Rel assoc pp40 | + | + | +/− | functionally related to I kappa B (inhibitor of the NF-kappa B) |
| M. Transcription Factors | |||||
|
|
Ccar1 | + | + | +/− | DNA binding transcriptional regulator; conserved SAP domain |
|
|
CTCF | + | + | +/− | CCCTC-binding factor (zinc finger protein); represses c-myc |
|
|
Elk3 | − | + | + | ETS domain transcription factor family; TCF subfamily |
|
|
GTFIIE | + | + | +/− | part of a multiprotein complex near the transcription start site |
|
|
GCNF | + | + | +/− | essential transcription factor in vertebrate embryogenesis |
|
|
ILF2 | + | + | +/− | binds NFAT and stimulates its ability to enhance gene expression |
|
|
ILF3 | + | + | − | subunit of nuclear factor of activated T cells, dimerizes with ILF2 |
|
|
PHD | + | + | +/− | has PHD-finger domain; folds into interleaved type of Zn-finger |
|
|
SRF | − | + | + | Serum Response Factor: binds specific sequences as a homo-dimer |
|
|
SREBP2 | + | + | +/− | bHLH protein that stimulates transcription by binding a SRE |
|
|
Sim to TCF20 | + | + | +/− | stromylesin 1 PDGF-responsive element-binding protein |
|
|
WHSC1L1 | + | + | +/− | Wolf-Hirschhorn syndrome candidate 1-like 1; multiple ZF repeats |
|
|
Sim to Zinc finger protein 3 | + | + | − | binds and regulates the J and/or S elements in MHC II promoter |
|
|
ZFP469 | + | + | +/− | may be involved in txnl regulation; potential nuclear localization |
| N. Transporters | |||||
|
|
ATP1A1 | + | + | +/− | plays a very important role in nerve cell membranes |
|
|
HEPH, variant 2 | + | + | − | Cu-containing iron oxidase; iron export from intestinal enterocytes |
|
|
OSBP2 | + | + | +/− | may mediate oxysterol cytotoxicity in tissues |
|
|
RANBP3 | + | + | − | promote export complexes of the CRM1/NES/RanGTP type |
|
|
VWCD1 | + | − | + | Ca++-channel protein; important in excitation-contraction coupling |
| O. Miscellaneous | |||||
|
|
Adat1 | − | + | +/− | participate in the pre-mRNA editing of nuclear transcripts |
|
|
Adh5 | + | + | +/− | class III : glutathione-dependent formaldehyde dehydrogenase |
|
|
Aldehyde dehydrogenase 9a1 | + | +/− | − | catalyzes dehydrogenation of γ-aminobutyraldehyde to GABA |
|
|
sim to autoantigen NGP-1 | − | + | +/− | gtpase that associates with pre-60s ribosomal subunits in nucleolus |
|
|
GTBP/mutS | + | + | +/− | forms heterodimer with hMSH2; binds G/T mismatches in dsDNA |
|
|
Heparin binding protein RI-HB | + | − | − | RA-induced heparin-binding protein; essential for embryogenesis |
|
|
JIF-1 | + | + | − | interacts with presinillin1 to suppress function of c-Jun homodimers |
|
|
PGK | + | + | +/− | catalyses 1,3-diphosphoglycerate is to 3-phosphoglycerate |
|
|
RNH1 | + | − | +/− | blocks function of ribonucleases |
|
|
Taldo1 | + | + | − | key enzyme of the pentose phosphate pathway |
|
|
Tumor protein D52-like 2 | − | + | +/− | involved in cell proliferation, Ca2+-mediated signal transduction |
| P. Unknown/EST/nohomology | |||||
|
|
finished cDNA ChEST648l12 | + | + | +/− | similar to Homo sapiens K+ channel tetramerization protein |
|
|
finished cDNA ChEST872f20 | + | + | − | |
|
|
Gallus sim to hypoth LOC419534 | + | + | +/− | |
|
|
Gallus sim to FLJ00068 protein | + | − | + | |
|
|
Hypothetical protein MGC15523 | + | − | − |
Transcription factors
By completing the screen, we identified several additional transcription factors expressed in the neural crest. These include: Ccar1, CTCF, Elk-3, GCNF, GTFIIE, ILF2, ILF3, PhD, SRF, SREBP2, Similar to Tcf20/AR1/SPBP, WHSC1L1, ZFP3, and ZFP469. All of the genes were expressed in premigratory neural crest cells within the neural folds and/or migrating neural crest cells moving through the periphery.
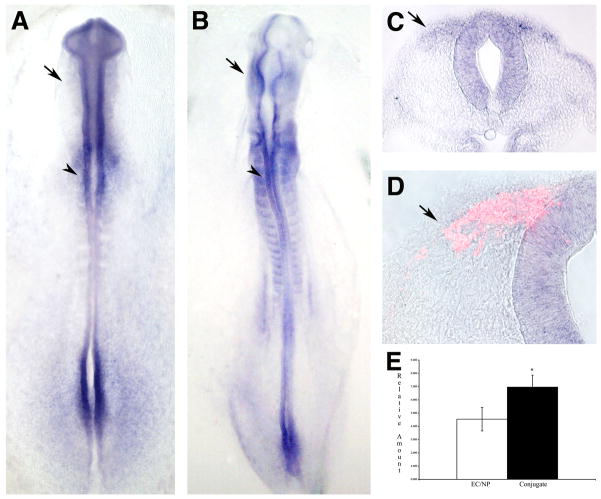
We verified the upregulation of a subset of transcription factors by quantitative polymerase chain reaction (QPCR). Intermediate neural plate and non-neural ectoderm were dissected and cultured as isolates or as conjugates in exactly the same manner as the subtracted probe was generated. We then quantitated the expression level of known (Snail2/Slug) and novel (identified in our screen) transcription factors in isolates and conjugates. The QPCR results showed that Snail2/Slug levels in the conjugated tissues consistently were upregulated by 5–7 fold (5.998 ± 0.642 s.d. in conjugates compared with 0.775 ± 0.059 s.d. in isolates, p=0.00015 by Student’s t test for our samples) (Fig. 2). Most of the transcription factors identified in our screen were upregulated approximately two-fold in the conjugates by QPCR (Figs. 2–6, Table 2). These levels were reproducible across replicates and experiments. This minimal induction likely reflects a variety of factors. Some genes may be expressed at low levels in isolates, and were upregulated relative to background levels in conjugates. Other genes may not have reached maximal expression levels at the endpoint of the assay, which represented premigratory crest (Dickinson et al., 1995), and so the difference by QPCR likely appeared less dramatic than by in situ (for example in the case of Elk-3, Fig. 3). Moreover, in all cases the conjugates contained neural and ectodermal cells in addition to newly induced neural crest cells, which will have diluted the neural crest signal. In sum, these results confirm that the transcription factors identified in the screen were upregulated in conjugates, and illustrate the advantages of the spatial and temporal information available when screening clones by in situ hybridization.
Figure 2.
Neural crest expression of and QPCR results for Snail 2 (slug). (A-D) Snail2 expression is seen specifically in premigratory (dorsal neural tube) and migratory neural crest. Shown are in situ hybridizations of 10 somite (A) and 13 somite (B-D) embryos in whole mount (A and B) and sections of whole mount stained embryos (C and D). Immunofluorescence for HNK-1 is in red (D). Arrowheads mark premigratatory neural crest, arrows mark migratory neural crest. Scale bars = X. (E) Results of conjugate QPCR showing a very highly significant difference between control and conjugate tissues (p = 0.00015).
Table 2.
QPCR Results.
| Slug | Elk-3 | GCNF | GTF2E | SREBP2 | |
|---|---|---|---|---|---|
| Isolates Avg | 0.775 | 2.086 | 0.303 | 1.826 | 4.539 |
| Isolates StDev | 0.059 | 0.416 | 0.040 | 0.413 | 0.886 |
| Conjugates Avg | 5.998 | 3.786 | 0.479 | 3.171 | 6.960 |
| Conjugates StdDev | 0.642 | 0.243 | 0.092 | 0.664 | 0.909 |
| P-value | 0.00015 | 0.005 | 0.0384 | 0.041 | 0.030 |
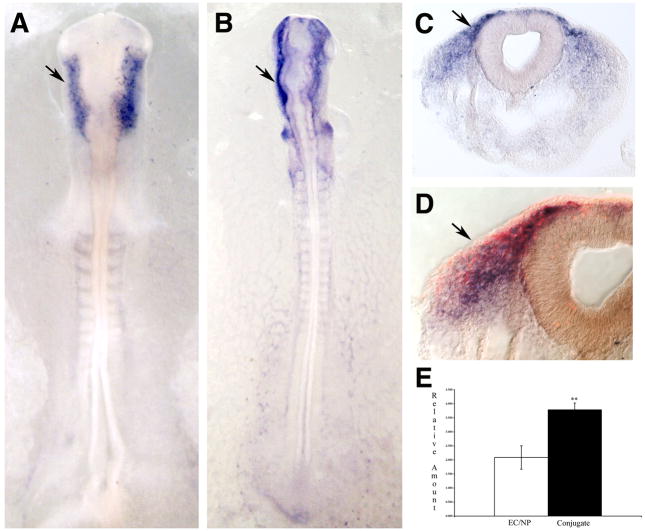
Figure 3.
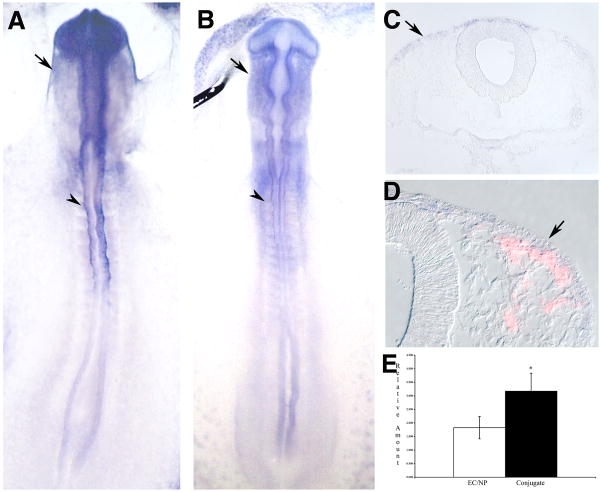
Neural crest expression of and QPCR results for Elk-3. (A-D) Elk-3 expression is seen specifically in premigratory (dorsal neural tube) and migratory neural crest. Shown are in situ hybridizations of 10 somite (A) and 16 somite (B-D) embryos in whole mount (A and B) and sections of whole mount stained embryos (C and D). Immunofluorescence for HNK-1 is in red (D). Arrowheads mark premigratatory neural crest, arrows mark migratory neural crest.(E) QPCR results showing a highly significant difference between control and conjugated tissues (p = 0.005).
Two of the transcription factors, Elk-3 (Fig. 3) and Serum Response Factor (SRF), were expressed in migrating neural crest cells but absent from the premigratory neural crest. Elk-3 (also known as Sap-2, ERP and Net) is a member of the ternary complex factor (TCF) subfamily of the Ets transcription factors. With the exception of migrating neural crest cells and some cranial mesenchyme, Elk-3 mRNA was not observed in other tissues at the developmental stages examined. Mice with a homozygous mutation resulting in the deletion of the DNA binding domain in Elk-3 have defects in both vasculature and wound healing (Ayadi et al., 2001). Elk-3 has further been shown to act as a repressor of PAI-1 to allow cellular migration (Buchwalter et al., 2005). The pattern of SRF is similar to that of Elk-3. SRF regulates the expression of cytoskeletal genes and is required for neuronal migration in the mouse forebrain (Alberti et al., 2005). When examined closely, SRF deficient murine ES cells display impaired migration, adhesion and cell spreading (Schratt et al., 2002), consistent with an important role in cell movement.
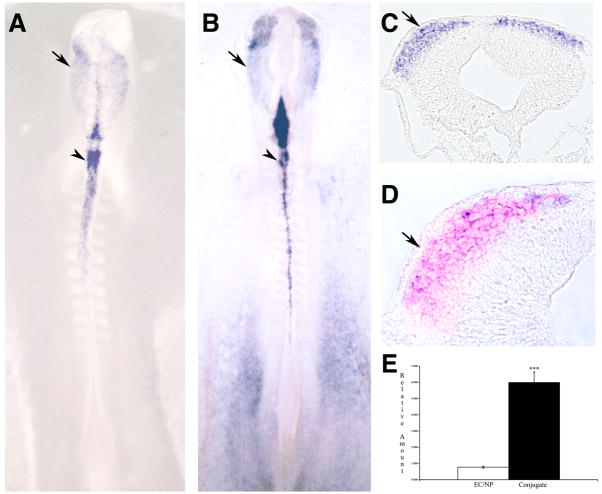
The majority of transcription factors identified in our screen were expressed in both premigratory and migratory neural crest cells. Several appear to be transcriptional activators (TCF20, GTFIIH and SREBP2), or of unknown function (ZFP3, ZFP469). TCF20 (AR1, SPBP) is a nuclear factor that can enhance the transcriptional activity of various transcription factors, such as c-Jun, Ets, Sp1, and Pax6, which suggests that TCF20 is a transcriptional activator. It probably acts as a coactivator for structurally and functionally disparate transcription factors binding to target sequences in promoters or enhancers (Rekdal et al., 2000). Germ cell nuclear factor (GCNF, NR6A1) (Fig. 4) is a nuclear orphan receptor that was first described in the mouse testis. GCNF is expressed in the neural plate and the neural crest of Xenopus midneurula embryos, with later expression in the central nervous system and branchial arches (Joos et al., 1996). When GCNF is depleted from the Xenopus embryo using a morpholino, there is improper formation of the neural tube as well as defective differentiation in the head (Barreto et al., 2003). GTFIIE (Fig. 5) is the alpha subunit (56kDa) of general transcription factor IIE is a part of a tetramer that is required to recruit TFIIH and in the opening of an RNA polymerase II promoter (Holstege et al., 1996). SREBP2 (Fig. 6) is a member of a family of basic-helix-loop-helix-leucine zipper (bHLH-ZIP) transcription factors that recognize sterol regulatory element 1 (SRE-1) (Hua et al., 1993). Little is known about PhD finger protein 12 except that it can bind DNA and is shown by GO gene ontology to have transcription factor activity. Similarly, ZFP3 and ZFP469 are zinc-finger transcription factors about which not much is known.
Figure 4.
Neural crest expression of and QPCR results for GCNF. (A-D) GCNF expression is seen specifically in premigratory (dorsal neural tube) and migratory neural crest. Shown are in situ hybridizations of 12 somite (A) and 15 somite (B-D) embryos in whole mount (A and B) and sections of whole mount stained embryos (C and D). Immunofluorescence for HNK-1 is in red (D). Arrowheads mark premigratatory neural crest, arrows mark migratory neural crest. (E) results of QPCR showing a significant difference between control and conjugated tissues (p = 0.0384).
Figure 5.
Neural crest expression of and QPCR results for GTF2E. (A-D) GTF2E expression is seen specifically in premigratory (dorsal neural tube) and migratory neural crest. Shown are in situ hybridizations of 10 somite (A) and 12 somite (B-D) embryos in whole mount (A and B) and sections of whole mount stained embryos (C and D). Immunofluorescence for HNK-1 is in red (D). Arrowheads mark premigratatory neural crest, arrows mark migratory neural crest. (E) results of QPCR showing a significant difference between control and conjugated tissues (p = 0.041).
Intriguingly, many of the transcription factors identified in our screen also are abnormally regulated in cancer cells. This is not surprising since many of the characteristics of a neural crest cell – motility, the ability to undergo an epithelial to mesenchymal transition, the ability to break through the basal lamina and form many derivatives – are also features of cancer cells. Genes found in both neural crest and associated with tumorigenesis include Ccar1, CTCF, ILF2 and WHSC1L1. For example, Cell Cycle and Apoptosis Regulatory Protein-1 (Ccar1 or Carp1) was first identified by a yeast two hybrid screen and found to interact with DED caspases (McDonald and El-Deiry, 2004). It mediates apoptosis signaling by CD437, a retinoid that causes cell cycle arrest and apoptosis in a number of cancer cells including HBC (human breast carcinoma) by utilizing an undefined retinoic acid receptor/retinoid X receptor-independent mechanism. Reduced levels of Ccar1 result in inhibition of apoptosis by CD437 or adriamycin, whereas increased expression of Ccar1 causes elevated levels of cyclin-dependent kinase inhibitor p21WAF1/CIP1 and apoptosis (Rishi et al., 2006). CTCF is an evolutionarily conserved zinc finger containing transcription factor and insulator binding protein that was originally identified and characterized as a transcriptional repressor of the c-Myc oncogene. It has been shown to regulate transcriptional activation as well as repression, and in addition is involved in cell-cycle regulation, apoptosis and cell differentiation (Pugacheva et al., 2006) (Ladomery and Dellaire, 2002). In Xenopus, CTCF was shown to have strong expression in the neural tube and developing brain as well as later expression in the branchial arches (Burke et al., 2002). Mutations of CTCF have been identified in Wilm’s tumors, as well as in breast and prostate cancers (Pugacheva et al., 2006) (Ladomery and Dellaire, 2002). ILF2 (NF45) associates with ILF3 (NF90) in the nucleus to regulate IL-2 gene transcription (Zhao et al., 2005) (Reichman et al., 2002). Both of these proteins are widely expressed in normal tissues and are seen at higher levels in the brain (Zhao et al., 2005) (Buaas et al., 1999). ILF2 is increased in lymphoma, leukemia, and hepatocellular carcinoma cell lines while ILF3 expression is increased in nasopharyngeal carcinomas, suggestive of a role in cell proliferation and the cell cycle (Zhao et al., 2005). WHSC1L1 (Wolf-Hirschhorn syndrome candidate 1 like 1) is another gene that is frequently rearranged in cancers, including breast cancer. WHSC1L1 is closely related to WHSC1, a disorder in which patients display a craniofacial defect known as ‘Greek helmet face’ (Bergemann et al., 2005). It had previously been suggested that this gene may be involved in embryonic development as well as cancers (Stec et al., 2001), and our identification of the gene in the neural crest is consistent with a possible role in craniofacial development.
Other genes implicated in metastasis
In addition to the transcription factors we identified, several genes in other categories that are likely to be downstream targets also have been associated with various types of cancer. Of these Lactate dehydrogenase A, Septin 9, and Cathepsin B were observed in premigratory but not in migrating neural crest cells. LDHA (lactate dehydrogenase A) was originally shown to be required for cell transformation by c-Myc (Hirota et al., 1990) (Lewis et al., 2000). Additionally, injection of cells co-expressing both LDHA and Rcl are able to form tumors in nude mice in a similar manner to c-Myc expressing cells, suggesting that these genes are downstream of c-Myc (although neither Rcl or LDHA overexpression alone induces tumorigenesis) (Lewis et al., 2000). Similarly, Septin 9 (Sept9, Sint1, Septin D1, MSF, MLL septin-like fusion) was originally described as a fusion partner gene of MLL in acute myeloid leukemia and maps to a region commonly affected in breast and ovarian cancers. The septin gene is a member of a family of nucleotide-binding proteins first discovered in yeast due to their role in the cell division cycle. Consistent with our findings, Sept9 expression has been reported in murine neural crest cells (Sorensen et al., 2002). Cathepsin B is a lysosomal cysteine proteinase involved in the effectiveness of invasive and metastatic cancers (Szpaderska and Frankfater, 2001) and cells appear to have differential abilities to migrate invasively based on cathepsin B expression both in vitro and in vivo (Szpaderska and Frankfater, 2001) (Lakka et al., 2004). Direct injections into tumor-containing animals of plasmids containing cathepsin B inhibited gliomal tumor growth and regressed pre-established tumor cells (Lakka et al., 2004).
Our screen revealed that the Neurofibromatosis 2 (NF2) gene, implicated in tumor production, is upregulated by neural crest induction. NF2 was previously shown to be expressed throughout development in the neural tube as well as in migrating neural crest cells and their derivatives (Akhmametyeva et al., 2006). The NF2 gene encodes the protein merlin, which is a member of the ezrin, radaxin, and moesin (ERM) family of membrane-cytoskeleton associated proteins. It is thought to have roles in cell adhesion, motility, and proliferation during development. Mutations in NF2 result in a predisposition to schwannomas, tumors of a neural crest origin. Familial schwannomas and meningiomas contain mutations that result in truncations of NF2. NF2 homozygous mutants are lethal at an early stage. However, conditional mutants targeted to Schwann and neural crest cells recapitulate the types of tumors seen in humans (Giovannini et al., 2000).
Two different type IV collagens, col4a5 and col4a6, were expressed by migratory neural crest and also have been implicated in malignant cell types. There are six collagen IV molecules that are arranged in a paired head-to-head fashion on three different chromosomes. Col4a5 and Col4a6 are the pair located on chromosome X and are found in patients with Alport syndrome associated with diffuse leiomyomatosis.
Other genes found in our screen, including PGK (Fig. 1), EWS, and HEPH, have transcripts that are expressed in both the premigratory and migratory neural crest cells and associated with various types of cancers. Phosphoglycerate kinase (PGK) is a glycolytic enzyme that catalyzes the reversible conversion of 1,3-diphosphoglycerate to 3-phosphoglycerate. It also is secreted by mouse fibrosarcoma tumor cells, and may participate in angiogenesis (Lay et al., 2000). EWS is a member of the TET family (TLS/FUS, EWS, TAFII68) that occurs in a translocation breakpoint associated with Ewing’s sarcoma, a primitive neuroectodermal tumor. The translocation generates a fusion with FLI-1, a member of the ETS family of transcription factors, and the fusion can function as a transcription factor (Ladomery and Dellaire, 2002). HEPH is involved in iron transport and shows altered expression in human colorectal carcinogenesis, resulting in increased intracellular iron that may induce proliferation and repress cell adhesion (Brookes et al., 2006).
Genes involved in wound healing and cell migration
Wound healing assays have long been used to look for cell migration defects. We found several genes in addition to those already described above as associated with wound healing and cell migration.
Col1a2 and Von Willebrand factor type A and cache domain containing 1 genes were expressed in the premigratory but not the migratory neural crest. Col1a2 is a major collagen subunit that plays a role in neural crest cell migration and may regulate cellular differentiation (He et al., 2005). In mouse, it is expressed in the head folds at e8.5–9.0. In adult tissues, it is upregulated in response to wounding, indicating that it may be involved in cell migration (Ponticos et al., 2004). Col1a2 also seems to mediate the adhesion and migration of germ cells during spermatogenesis in the mouse (He et al., 2005). Von Willebrand factor type A and cache domain containing 1 (VWCD1) is likely to be a voltage gated calcium channel or an ECM protein; genes containing similar domains participate in cell adhesion, migration, homing, pattern formation, and signal transduction (Colombatti et al., 1993).
Paxillin, periplakin, and G-protein coupled receptor GPR144 are genes associated with wound healing and cell migration. They were distributed in the migrating neural crest but absent from premigratory neural crest. Paxillin expression was previously described in migrating mouse neural crest cells and derivatives such as dorsal root and trigeminal ganglia, and branchial arches. Loss of paxillin led to a decrease in focal adhesions and lamellipodia (Hagel et al., 2002). In Drosophila, overexpression of paxillin results in a ‘blistered wing’ phenotype considered characteristic of changes in cell adhesion. Paxillin also plays a role in the developing Drosophila eye (Chen et al., 2005). Plakins are proteins that make contact with intermediate filaments (Aho et al., 1998). Periplakin, a member of this family, localizes to the plasma membrane and is believed to play a major role in cell-cell adhesion (Nishimori et al., 2006). Plakins were shown by Aho et al. to be well expressed in the brain (Aho et al., 1998). G-protein coupled receptor GPR144 is a membrane bound adhesion protein with a GPS domain in the N-terminal region as well as a pentraxin domain (Bjarnadottir et al., 2004). Its developmental role is as yet unexplored.
Other genes previously implicated in wound healing and migration include TPM- γ, TRIO, MACF/ACF7, nonerythroid α-spectrin, PTK9L. Their mRNA transcripts were present in premigratory and migratory neural crest cells. Tropomyosins such as TPM-γ are components of the microfilament cytoskeleton that bind to the helical groove of the actin filament. Several tropomyosin isoforms have been shown to regulate cell contractility, morphology, migration and the organization of the cytoskeleton (Bryce et al., 2003). TRIO is a guanine nucleotide exchange factor (GEF) that functions in the activation of Rho GTPases such as RhoA, Rac1, and Cdc42 (Medley et al., 2003). It is essential for embryonic development and for the development of neural tissue in mouse (O’Brien et al., 2000). Furthermore, the TRIO-like unc-73 gene is essential for cell migration and axon guidance in C. elegans (Steven et al., 1998). MACF/ACF7 is from the family of cytoskeletal linkers and is the vertebrate homolog of Drosophila Shortstop/Kakapo. It binds both to microtubules through its GAR and GSR-repeat domains and to actin (Leung et al., 2001). In MACF-null cells, microtubules are destabilized and microtubule tracking along actin cables is perturbed (Kodama et al., 2003). It was previously shown to be expressed in the spinal cord and brain of the developing mouse as well as neural crest derivatives such as the dorsal root ganglia (Leung et al., 1999). Our data confirm that MACF1 is also expressed in the premigratory and migratory neural crest of chickens. Spectrins, including nonerythroid α-spectrin, are members of the cytoskeletal proteins that cross-link actin filaments or link actin to the cell membrane (Djinovic-Carugo et al., 2002). Anti-spectrin antibodies lead to the collapse of the intermediate filament network in cultured cells. Furthermore, depletion of alpha-spectrin in Drosophila results in the loss of cell-cell contact (Lee et al., 1993). Very little is known about PTK9L (protein tyrosine kinase 9-like or A6RP), but preliminary immunolocalization studies have previously indicated that it might be involved in focal adhesions (Rohwer et al., 1999).
Genes associated with human craniofacial and other birth abnormalities
Because it contributes to such a diversity of structures, the neural crest has been implicated in many birth abnormalities, including those that involve craniofacial malformations, intestinal innervation defects, and malformations of the cardiac system (Chai and Maxson, 2006) (Iwashita et al., 2003) (Wurdak et al., 2005). Several of the identified genes from this screen have been associated with birth abnormalities that are reminiscent of alterations in the normal course of neural crest cell development.
Nectins are molecules that are involved in adhesion, migration and polarization. We observed nectin-1 expression in migrating neural crest cells, but not in the dorsal neural tube, suggesting a possible role in cell movement and morphogenesis. Consistent with this, human mutations in nectin-1 result in various cleft lip/palate and ectodermal dysplasia disorders. Nectin functions as a cell adhesion molecule and are highly concentrated at adherens junctions. They signal through alpha-catenin, and are associated with F-actin bundles (Nakanishi and Takai, 2004) (Takai and Nakanishi, 2003). Trans-interactions of nectin induce the activation of Cdc42 and Rac small G proteins. The nectin-induced activation of Cdc42 increases the number of both filopodia and cell-cell contact sites while the activation of Rac induces formation of lamellipodia (Nakanishi and Takai, 2004). They also appear to form micro-clusters at sites of cell-cell contact (Takai et al., 2003).
GARS-AIRS-GART, and WBSCR20A (present in the region critical for the Williams-Beuren syndrome) have transcripts in both premigratory and migratory neural crest. Human GARS-AIRS-GART encodes two different proteins that are differentially expressed during human brain development but down-regulated after birth. They continue to be expressed postnatally in the cerebellum of patients with Down syndrome and have been suggested to contribute to this syndrome (Brodsky et al., 1997). Williams-Beuren syndrome displays many features consistent with neural crest pathology, including congenital heart and vascular disease, dental abnormalities, dysmorphic facial features, and mental retardation. One of the genes that are present in the region critical for this syndrome is WBSCR20A (Nol1, p120), a 120 kDA protein that was found to be expressed in the human fetal brain by RT-PCR (Merla et al., 2002).
Cell cycle genes
Neural crest cells actively proliferate as they delaminate and migrate. Thus, it is not surprising that numerous genes identified in our screen were associated with the cell cycle including Lamin B2 (Fig. 1), Cyclin B2, Cyclin B3, FMS Interacting Protein, Mcm2 and Mcm5. All of these were expressed in both premigratory and migratory neural crest cells in early development. Lamin proteins are filaments that form a mesh that lines the inner side of the nuclear envelope (Lehner et al., 1987). Lamin B2 is an essential gene that is associated with spindles in mitosis and deficit results in spindle defects (Harborth et al., 2001; Tsai et al., 2006). Cyclin B2 has been shown to be essential for the control of the cell cycle at the G2/M transition and, in embryonic heart cells undergoing EMT, appears to be regulated by RhoA (Tavares et al., 2006). Cyclin B3 is localized to the nucleus and is degraded during anaphase entry. Its prolonged expression interferes with progression through G1 and entry into S-phase (Nguyen et al., 2002). Human Mcm2 and Mcm5 proteins form a complex that localizes to the nucleus. Blocking Mcm2 inhibits DNA synthesis, indicating that these proteins may play a role in DNA replication and regulation of the cell cycle (Tsuruga et al., 1997). Finally, FMIP (FMS Interacting Protein) has been shown to control differentiation of granulocytes and macrophages (Tamura et al., 1999).
Neural crest cells are induced at the border of the neural plate and the non-neural ectoderm, migrate through stereotyped pathways, and finally form such disparate cell types as melanocytes and cartilage. Identifying the different gene cascades that are active in these cells is critical to a greater understanding of neural crest cells as well as the many disorders in which they are implicated. With the results from this screen, we expand the known genes, including transcription factors, involved in neural crest development. These data provide new players to be included in existing gene transduction networks and further our understanding of the development of neural crest cells.
References
- Aho S, McLean WH, Li K, Uitto J. cDNA cloning, mRNA expression, and chromosomal mapping of human and mouse periplakin genes. Genomics. 1998;48:242–247. doi: 10.1006/geno.1997.5188. [DOI] [PubMed] [Google Scholar]
- Akhmametyeva EM, Mihaylova MM, Luo H, Kharzai S, Welling DB, Chang LS. Regulation of the Neurofibromatosis 2 gene promoter expression during embryonic development. Dev Dyn. 2006;235:2771–2785. doi: 10.1002/dvdy.20883. [DOI] [PubMed] [Google Scholar]
- Alberti S, Krause SM, Kretz O, Philippar U, Lemberger T, Casanova E, Wiebel FF, Schwarz H, Frotscher M, Schutz G, Nordheim A. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc Natl Acad Sci U S A. 2005;102:6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ. Stem cells and transcription factors in the development of the mammalian neural crest. Faseb J. 1994;8:707–713. doi: 10.1096/fasebj.8.10.8050669. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, Credidio C, Saint-Jeannet JP. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev Biol. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- Ayadi A, Zheng H, Sobieszczuk P, Buchwalter G, Moerman P, Alitalo K, Wasylyk B. Net-targeted mutant mice develop a vascular phenotype and up-regulate egr-1. Embo J. 2001;20:5139–5152. doi: 10.1093/emboj/20.18.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Reintsch W, Kaufmann C, Dreyer C. The function of Xenopus germ cell nuclear factor (xGCNF) in morphogenetic movements during neurulation. Dev Biol. 2003;257:329–342. doi: 10.1016/s0012-1606(03)00109-x. [DOI] [PubMed] [Google Scholar]
- Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in xenopus. Dev Cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Ben-Tabou de-Leon S, Davidson EH. Gene regulation: gene control network in development. Annu Rev Biophys Biomol Struct. 2007;36:191. doi: 10.1146/annurev.biophys.35.040405.102002. [DOI] [PubMed] [Google Scholar]
- Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends Genet. 2005;21:188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Bjarnadottir TK, Fredriksson R, Hoglund PJ, Gloriam DE, Lagerstrom MC, Schioth HB. The human and mouse repertoire of the adhesion family of G-protein-coupled receptors. Genomics. 2004;84:23–33. doi: 10.1016/j.ygeno.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Brodsky G, Barnes T, Bleskan J, Becker L, Cox M, Patterson D. The human GARS-AIRS-GART gene encodes two proteins which are differentially expressed during human brain development and temporally overexpressed in cerebellum of individuals with Down syndrome. Hum Mol Genet. 1997;6:2043–2050. doi: 10.1093/hmg/6.12.2043. [DOI] [PubMed] [Google Scholar]
- Brookes MJ, Hughes S, Turner FE, Reynolds G, Sharma N, Ismail T, Berx G, McKie AT, Hotchin N, Anderson GJ, Iqbal T, Tselepis C. Modulation of iron transport proteins in human colorectal carcinogenesis. Gut. 2006;55:1449–1460. doi: 10.1136/gut.2006.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, Bamburg JR, Jeffrey PL, Hardeman EC, Gunning P, Weinberger RP. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell. 2003;14:1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas FW, Lee K, Edelhoff S, Disteche C, Braun RE. Cloning and characterization of the mouse interleukin enhancer binding factor 3 (Ilf3) homolog in a screen for RNA binding proteins. Mamm Genome. 1999;10:451–456. doi: 10.1007/s003359901022. [DOI] [PubMed] [Google Scholar]
- Buchwalter G, Gross C, Wasylyk B. The ternary complex factor Net regulates cell migration through inhibition of PAI-1 expression. Mol Cell Biol. 2005;25:10853–10862. doi: 10.1128/MCB.25.24.10853-10862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke LJ, Hollemann T, Pieler T, Renkawitz R. Molecular cloning and expression of the chromatin insulator protein CTCF in Xenopus laevis. Mech Dev. 2002;113:95–98. doi: 10.1016/s0925-4773(02)00005-9. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Chen GC, Turano B, Ruest PJ, Hagel M, Settleman J, Thomas SM. Regulation of Rho and Rac signaling to the actin cytoskeleton by paxillin during Drosophila development. Mol Cell Biol. 2005;25:979–987. doi: 10.1128/MCB.25.3.979-987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Coles EG, Gammill LS, Miner JH, Bronner-Fraser M. Abnormalities in neural crest cell migration in laminin alpha5 mutant mice. Dev Biol. 2006;289:218–228. doi: 10.1016/j.ydbio.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Colombatti A, Bonaldo P, Doliana R. Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix. 1993;13:297–306. doi: 10.1016/s0934-8832(11)80025-9. [DOI] [PubMed] [Google Scholar]
- Davidson EH. The regulatory genome : gene regulatory networks in development and evolution. xi. Amsterdam ; Boston: Elsevier/Academic Press; 2006. p. 289. [Google Scholar]
- Dickinson ME, Selleck MA, McMahon AP, Bronner-Fraser M. Dorsalization of the neural tube by the non-neural ectoderm. Development. 1995;121:2099–2106. doi: 10.1242/dev.121.7.2099. [DOI] [PubMed] [Google Scholar]
- Djinovic-Carugo K, Gautel M, Ylanne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513:119–123. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- Eroglu B, Wang G, Tu N, Sun X, Mivechi NF. Critical role of Brg1 member of the SWI/SNF chromatin remodeling complex during neurogenesis and neural crest induction in zebrafish. Dev Dyn. 2006;235:2722–2735. doi: 10.1002/dvdy.20911. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M. Genomic analysis of neural crest induction. Development. 2002;129:5731–5741. doi: 10.1242/dev.00175. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Gonzalez C, Gu C, Bronner-Fraser M. Guidance of trunk neural crest migration requires neuropilin 2/semaphorin 3F signaling. Development. 2006;133:99–106. doi: 10.1242/dev.02187. [DOI] [PubMed] [Google Scholar]
- Gessert S, Maurus D, Rossner A, Kuhl M. Pescadillo is required for Xenopus laevis eye development and neural crest migration. Dev Biol. 2007;310:99–112. doi: 10.1016/j.ydbio.2007.07.037. [DOI] [PubMed] [Google Scholar]
- Giovannini M, Robanus-Maandag E, van der Valk M, Niwa-Kawakita M, Abramowski V, Goutebroze L, Woodruff JM, Berns A, Thomas G. Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 2000;14:1617–1630. [PMC free article] [PubMed] [Google Scholar]
- Hagel M, George EL, Kim A, Tamimi R, Opitz SL, Turner CE, Imamoto A, Thomas SM. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol Cell Biol. 2002;22:901–915. doi: 10.1128/MCB.22.3.901-915.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- He Z, Feng L, Zhang X, Geng Y, Parodi DA, Suarez-Quian C, Dym M. Expression of Col1a1, Col1a2 and procollagen I in germ cells of immature and adult mouse testis. Reproduction. 2005;130:333–341. doi: 10.1530/rep.1.00694. [DOI] [PubMed] [Google Scholar]
- Hirota Y, Katsumata A, Takeya T. Nucleotide and deduced amino acid sequences of chicken lactate dehydrogenase-A. Nucleic Acids Res. 1990;18:6432. doi: 10.1093/nar/18.21.6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, van der Vliet PC, Timmers HT. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. Embo J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- Honore SM, Aybar MJ, Mayor R. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev Biol. 2003;260:79–96. doi: 10.1016/s0012-1606(03)00247-1. [DOI] [PubMed] [Google Scholar]
- Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, Wang X. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci U S A. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashita T, Kruger GM, Pardal R, Kiel MJ, Morrison SJ. Hirschsprung disease is linked to defects in neural crest stem cell function. Science. 2003;301:972–976. doi: 10.1126/science.1085649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joos TO, David R, Dreyer C. xGCNF, a nuclear orphan receptor is expressed during neurulation in Xenopus laevis. Mech Dev. 1996;60:45–57. doi: 10.1016/s0925-4773(96)00599-0. [DOI] [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. ACF7: an essential integrator of microtubule dynamics. Cell. 2003;115:343–354. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- Ladomery M, Dellaire G. Multifunctional zinc finger proteins in development and disease. Ann Hum Genet. 2002;66:331–342. doi: 10.1017/S0003480002001215. [DOI] [PubMed] [Google Scholar]
- Lakka SS, Gondi CS, Yanamandra N, Olivero WC, Dinh DH, Gujrati M, Rao JS. Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene. 2004;23:4681–4689. doi: 10.1038/sj.onc.1207616. [DOI] [PubMed] [Google Scholar]
- Lay AJ, Jiang XM, Kisker O, Flynn E, Underwood A, Condron R, Hogg PJ. Phosphoglycerate kinase acts in tumour angiogenesis as a disulphide reductase. Nature. 2000;408:869–873. doi: 10.1038/35048596. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Brito JM, Creuzet S. Role of the neural crest in face and brain development. Brain Res Rev. 2007;55:237–247. doi: 10.1016/j.brainresrev.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Lee JK, Coyne RS, Dubreuil RR, Goldstein LS, Branton D. Cell shape and interaction defects in alpha-spectrin mutants of Drosophila melanogaster. J Cell Biol. 1993;123:1797–1809. doi: 10.1083/jcb.123.6.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF, Stick R, Eppenberger HM, Nigg EA. Differential expression of nuclear lamin proteins during chicken development. J Cell Biol. 1987;105:577–587. doi: 10.1083/jcb.105.1.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CL, Liem RK, Parry DA, Green KJ. The plakin family. J Cell Sci. 2001;114:3409–3410. doi: 10.1242/jcs.114.19.3409. [DOI] [PubMed] [Google Scholar]
- Leung CL, Sun D, Zheng M, Knowles DR, Liem RK. Microtubule actin cross-linking factor (MACF): a hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J Cell Biol. 1999;147:1275–1286. doi: 10.1083/jcb.147.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BC, Prescott JE, Campbell SE, Shim H, Orlowski RZ, Dang CV. Tumor induction by the c-Myc target genes rcl and lactate dehydrogenase A. Cancer Res. 2000;60:6178–6183. [PubMed] [Google Scholar]
- Luo T, Lee YH, Saint-Jeannet JP, Sargent TD. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proc Natl Acad Sci U S A. 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R, Aybar MJ. Induction and development of neural crest in Xenopus laevis. Cell Tissue Res. 2001;305:203–209. doi: 10.1007/s004410100369. [DOI] [PubMed] [Google Scholar]
- Mayor R, Morgan R, Sargent MG. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- McDonald ER, 3rd, El-Deiry WS. Suppression of caspase-8- and -10-associated RING proteins results in sensitization to death ligands and inhibition of tumor cell growth. Proc Natl Acad Sci U S A. 2004;101:6170–6175. doi: 10.1073/pnas.0307459101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medley QG, Buchbinder EG, Tachibana K, Ngo H, Serra-Pages C, Streuli M. Signaling between focal adhesion kinase and trio. J Biol Chem. 2003;278:13265–13270. doi: 10.1074/jbc.M300277200. [DOI] [PubMed] [Google Scholar]
- Merla G, Ucla C, Guipponi M, Reymond A. Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum Genet. 2002;110:429–438. doi: 10.1007/s00439-002-0710-x. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Takai Y. Roles of nectins in cell adhesion, migration and polarization. Biol Chem. 2004;385:885–892. doi: 10.1515/BC.2004.116. [DOI] [PubMed] [Google Scholar]
- Nguyen TB, Manova K, Capodieci P, Lindon C, Bottega S, Wang XY, Refik-Rogers J, Pines J, Wolgemuth DJ, Koff A. Characterization and expression of mammalian cyclin b3, a prepachytene meiotic cyclin. J Biol Chem. 2002;277:41960–41969. doi: 10.1074/jbc.M203951200. [DOI] [PubMed] [Google Scholar]
- Nishimori T, Tomonaga T, Matsushita K, Oh-Ishi M, Kodera Y, Maeda T, Nomura F, Matsubara H, Shimada H, Ochiai T. Proteomic analysis of primary esophageal squamous cell carcinoma reveals downregulation of a cell adhesion protein, periplakin. Proteomics. 2006;6:1011–1018. doi: 10.1002/pmic.200500262. [DOI] [PubMed] [Google Scholar]
- O’Brien SP, Seipel K, Medley QG, Bronson R, Segal R, Streuli M. Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc Natl Acad Sci U S A. 2000;97:12074–12078. doi: 10.1073/pnas.97.22.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticos M, Abraham D, Alexakis C, Lu QL, Black C, Partridge T, Bou-Gharios G. Col1a2 enhancer regulates collagen activity during development and in adult tissue repair. Matrix Biol. 2004;22:619–628. doi: 10.1016/j.matbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Pugacheva EM, Kwon YW, Hukriede NA, Pack S, Flanagan PT, Ahn JC, Park JA, Choi KS, Kim KW, Loukinov D, Dawid IB, Lobanenkov VV. Cloning and characterization of zebrafish CTCF: Developmental expression patterns, regulation of the promoter region, and evolutionary aspects of gene organization. Gene. 2006;375:26–36. doi: 10.1016/j.gene.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Reichman TW, Muniz LC, Mathews MB. The RNA binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol Cell Biol. 2002;22:343–356. doi: 10.1128/MCB.22.1.343-356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekdal C, Sjottem E, Johansen T. The nuclear factor SPBP contains different functional domains and stimulates the activity of various transcriptional activators. J Biol Chem. 2000;275:40288–40300. doi: 10.1074/jbc.M006978200. [DOI] [PubMed] [Google Scholar]
- Rishi AK, Zhang L, Yu Y, Jiang Y, Nautiyal J, Wali A, Fontana JA, Levi E, Majumdar AP. Cell cycle- and apoptosis-regulatory protein-1 is involved in apoptosis signaling by epidermal growth factor receptor. J Biol Chem. 2006;281:13188–13198. doi: 10.1074/jbc.M512279200. [DOI] [PubMed] [Google Scholar]
- Rohwer A, Kittstein W, Marks F, Gschwendt M. Cloning, expression and characterization of an A6-related protein. Eur J Biochem. 1999;263:518–525. doi: 10.1046/j.1432-1327.1999.00537.x. [DOI] [PubMed] [Google Scholar]
- Sasai N, Mizuseki K, Sasai Y. Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development. 2001;128:2525–2536. doi: 10.1242/dev.128.13.2525. [DOI] [PubMed] [Google Scholar]
- Schratt G, Philippar U, Berger J, Schwarz H, Heidenreich O, Nordheim A. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck MA, Bronner-Fraser M. The genesis of avian neural crest cells: a classic embryonic induction. Proc Natl Acad Sci U S A. 1996;93:9352–9357. doi: 10.1073/pnas.93.18.9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen AB, Warming S, Fuchtbauer EM, Pedersen FS. Alternative splicing, expression, and gene structure of the septin-like putative proto-oncogene Sint1. Gene. 2002;285:79–89. doi: 10.1016/s0378-1119(02)00406-7. [DOI] [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet JP. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development. 2002;129:421–432. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- Stec I, van Ommen GJ, den Dunnen JT. WHSC1L1, on human chromosome 8p11.2, closely resembles WHSC1 and maps to a duplicated region shared with 4p16.3. Genomics. 2001;76:5–8. doi: 10.1006/geno.2001.6581. [DOI] [PubMed] [Google Scholar]
- Steven R, Kubiseski TJ, Zheng H, Kulkarni S, Mancillas J, Ruiz Morales A, Hogue CW, Pawson T, Culotti J. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- Steventon B, Carmona-Fontaine C, Mayor R. Genetic network during neural crest induction: from cell specification to cell survival. Semin Cell Dev Biol. 2005;16:647–654. doi: 10.1016/j.semcdb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Sakai D, Osumi N, Wada H, Wakamatsu Y. Sox genes regulate type 2 collagen expression in avian neural crest cells. Dev Growth Differ. 2006;48:477–486. doi: 10.1111/j.1440-169X.2006.00886.x. [DOI] [PubMed] [Google Scholar]
- Szpaderska AM, Frankfater A. An intracellular form of cathepsin B contributes to invasiveness in cancer. Cancer Res. 2001;61:3493–3500. [PubMed] [Google Scholar]
- Takai Y, Irie K, Shimizu K, Sakisaka T, Ikeda W. Nectins and nectin-like molecules: roles in cell adhesion, migration, and polarization. Cancer Sci. 2003;94:655–667. doi: 10.1111/j.1349-7006.2003.tb01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y, Nakanishi H. Nectin and afadin: novel organizers of intercellular junctions. J Cell Sci. 2003;116:17–27. doi: 10.1242/jcs.00167. [DOI] [PubMed] [Google Scholar]
- Tamura T, Mancini A, Joos H, Koch A, Hakim C, Dumanski J, Weidner KM, Niemann H. FMIP, a novel Fms-interacting protein, affects granulocyte/macrophage differentiation. Oncogene. 1999;18:6488–6495. doi: 10.1038/sj.onc.1203062. [DOI] [PubMed] [Google Scholar]
- Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134:1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares AL, Mercado-Pimentel ME, Runyan RB, Kitten GT. TGFbeta-mediated RhoA expression is necessary for epithelial-mesenchymal transition in the embryonic chick heart. Dev Dyn. 2006;235:1589–1598. doi: 10.1002/dvdy.20771. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Tsuruga H, Yabuta N, Hashizume K, Ikeda M, Endo Y, Nojima H. Expression, nuclear localization and interactions of human MCM/P1 proteins. Biochem Biophys Res Commun. 1997;236:118–125. doi: 10.1006/bbrc.1997.6865. [DOI] [PubMed] [Google Scholar]
- Wurdak H, Ittner LM, Lang KS, Leveen P, Suter U, Fischer JA, Karlsson S, Born W, Sommer L. Inactivation of TGFbeta signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 2005;19:530–535. doi: 10.1101/gad.317405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu QaW, DG, editors. In situ hybridization: A practical approach. Oxford University Press; 1992. [Google Scholar]
- Zhao G, Shi L, Qiu D, Hu H, Kao PN. NF45/ILF2 tissue expression, promoter analysis, and interleukin-2 transactivating function. Exp Cell Res. 2005;305:312–323. doi: 10.1016/j.yexcr.2004.12.030. [DOI] [PubMed] [Google Scholar]