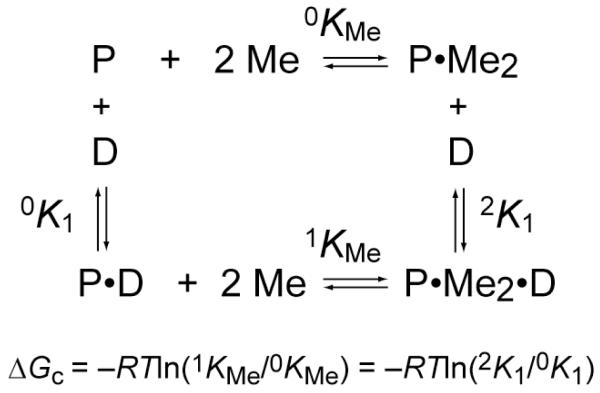Abstract
Prokaryotic organisms have evolved an impressive capacity to quickly adapt to a changing and challenging microenvironment in which the availability of both biologically required and non-essential transition metal ions can vary dramatically. In all bacteria, a panel of metalloregulatory proteins control the expression of genes encoding membrane transporters and metal trafficking proteins, that collectively manage metal homeostasis and resistance. These “metal sensors” are specialized allosteric proteins, in which the direct binding of a specific or small number of “cognate” metal ion(s) drives a conformational change in the regulator that allosterically activates or inhibits operator DNA binding, or alternatively, distorts the promoter structure thereby converting a poor promoter to a strong one. In this review, we discuss our current understanding of the features that control metal specificity of the allosteric response in these systems, and the role that structure, thermodynamics and conformational dynamics play in mediating allosteric activation or inhibition of DNA binding.
Keywords: transition metals, metal homeostasis, allostery, repressor, energetics, protein-DNA interactions
1. Transition metal biology and metal homeostasis
Transition metals are essential to a striking diversity of biological processes in the cell. The participation of a metal ion in a specific biological process is dictated by its intrinsic properties. These range from roles as essential cofactors for oxidation-reduction reactions, electron transfer, hydrolytic and acid-base chemistry, to structural centers that merely stabilize the protein fold. Zn(II) is unique among first row transition metals in that it possesses a dual role in the cell, as both a structural element and a catalytic cofactor. For example, Zn(II) stabilizes diverse “zinc-finger” proteins that mediate binding to DNA and RNA in eukaryotes [1, 2]. On the other hand, Zn(II) is an essential cofactor of many hydrolytic enzymes, including proteases, phosphatases, esterases and deacetylases, where it functions as a Lewis acid to activate a water molecule for catalysis [3]. Other transition metals are cofactors in catalases and superoxide dismutases (SODs) which enables resistance to cellular oxidative stress. SODs are ubiquitous enzymes found in virtually all cells and oxygen-tolerant organisms, the importance of which is highlighted by the wide array of metal centers, from Ni to Cu to Cu/Zn to Mn and Fe, used to catalyze the same reaction, sometimes in the same cell [4]. Ni is a cofactor for urease in the stomach pathogen H. pylori which allows this organism to colonize the acidic gastric lumen [5].
The other side of the “double-edged sword” of transition metal biology is that essential metal ions, notably iron and copper, can undergo facile oxidation-reduction and as a result, can catalyze the production of highly toxic reactive oxygen species by Fenton or Haber-Weiss chemistry from partially reduced forms of oxygen that form as by-products of aerobic respiration [6, 7]. In addition, heavy metals and metalloids that play no biological role, including mercury, cadmium, arsenic, lead and tin, are extremely toxic often as a result of forming strong coordinate covalent bonds with cellular thiols, or calcium-binding proteins in the case of Pb(II) [8-10]. Therefore, in order to utilize essential transition metal ions while detoxifying or effluxing nonessential ones, microorganisms and all cells have evolved mechanisms to regulate the intracellular metal concentration.
The remarkably precise control of cytoplasmic metal ion concentrations is termed metal homeostasis, in which metallochaperones, metal importers, and metal effluxing transporters each play vital roles in regulating metal bioavailability. The expression of genes encoding these proteins is controlled by a panel of specialized transcriptional regulators known as metalloregulatory proteins, or “metal sensor” proteins (see Section 2). These transcriptional regulators specifically sense one or a small number of metal ions and are classified into at least ten families on the basis of structural homology [11, 12]. Binding of the cognate metal to the metalloregulator activates or inhibits protein-DNA operator binding, which results in the transcriptional regulation of genes responsible for metal homeostasis [13, 14]. In addition to encoding proteins that traffic metal ions within or between cellular compartments, the regulon may also include genes that encode proteins with functionalities beyond metal homeostasis, including virulence determinants, an oxidative or nitrosative stress response, or precursors of enzymatic pathways [15-17]. Indeed, these operons are often vital for the survival of pathogenic bacteria in humans.
An increased level of complexity is introduced when metalloregulatory networks specific for one metal ion intersect with others in the cell. Structurally unrelated metallosensors with distinct metal specificities may interact within the same cytoplasm. For example, Ni(II) and Fe(II) metalloregulators NikR and FurR in H. pylori have overlapping DNA binding sites in the promoter regions of their regulons [18, 19], a finding consistent with numerous examples of Ni-Fe cross-talk in cells [20, 21]. In B. japonicum the Fur-family regulator Irr appears to be targeted for proteolytic degradation upon heme binding. Under conditions of low iron, heme is scarce and not sufficient to signal Irr for degradation; in addition, Mn(II) binds to Irr and further stabilizes it against denaturation. Therefore, when B. japonicum is grown under Mn(II) depleted conditions, Irr senses heme-Fe(II) at correspondingly lower concentrations, consistent with the hypothesis that Mn(II) and Fe(II) homeostasis are interconnected [22]. Non-cognate metal binding affects transition metal homeostasis as well. For example, in the human pathogen S. pneumoniae, Zn(II) stress induces a strong Mn(II) deprivation phenotype, thought to be caused by competition between Zn(II) and Mn(II) at the high affinity Mn(II) uptake system and possibly in the cytoplasmic Mn(II) sensor PsaR [23, 24].
Emerging evidence reveals that transition metal homeostasis influences many fundamental aspects of bacterial cell physiology and bacterial pathogenesis [25-27], and is therefore essential to understand the metal specificity and mechanisms of metalloregulation operative within the cell [11]. Important insights into what governs these processes come from biophysical and structural studies of purified metalloregulatory proteins. Herein, we review recent progress on two major regulatory aspects of bacterial metal homeostasis derived from physicochemical studies: 1) the coordination chemistry of metal coordination that governs the specificity of the response, and 2) structural, thermodynamic and dynamical mechanisms of allosteric regulation of operator-promoter binding by cognate vs. non-cognate metal ions.
2. Metal binding affinities of metal sensor proteins and control of metal homeostasis
Cellular homeostasis of total transition metal concentration is set within boundaries beyond which the cell experiences excess or deprivation, thereby inducing a cellular response. All cells actively concentrate cell-associated metal ions, to a degree that is more similar than different between cells, with total measurable zinc and iron in the 10−4-10−3 M range, manganese and copper approximately 10-fold lower, with nickel and cobalt another 10-fold lower [24, 28]. In prokaryotes, the cellular response to perturbations in metal homeostasis is nearly exclusively transcriptional. In many cases, a promoter exclusion model is operative where a transcriptional repressor binds to a specific DNA operator which sterically blocks the binding of RNA polymerase [29, 30]. A significant departure from this model is transcriptional activation by MerR-family regulators, where the metal-bound or otherwise “activated” form of the repressor allosterically changes the structure of the promoter from a poor promoter to a strong promoter, without protein dissociation [31-33].
In the former case, the simplest mechanism of allosteric regulation of operator binding requires that a metalloregulator (P) has at least one DNA binding site or domain (D) and one metal (M) regulatory site, often but not always located in structurally separated domains within a homooligomer (dimer or tetramer). The “ligand” binding sites can interact with each other in two distinct ways: If the binding of M stimulates the binding of D then this is termed positive allosteric regulation, and occurs most often in metal-mediated repression of uptake genes. On the contrary, if the binding of M antagonizes the binding of D, this manifests as negative heterotropic allosteric regulation, and it is this process that occurs most often in metal-mediated transcriptional derepression [12] (Fig. 1).
Figure 1.
A coupled equilibrium thermodynamic scheme that defines the relationship of all four allosteric states of the homodimer repressor (P) in equilibrium with DNA operator (D) and metal ions (Me), with a limiting stoichiometry of two metals per dimer. The allosteric coupling free energy, ΔGc, is defined as indicated.
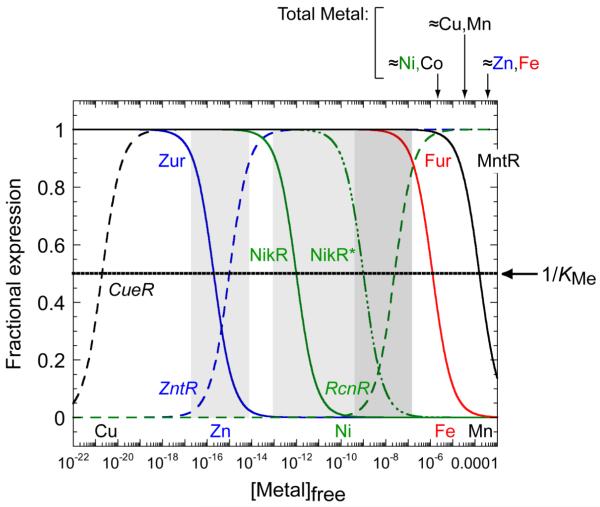
When the cytoplasm is experiencing metal excess, derepression of genes encoding for metal efflux or detoxification proteins, and concomitant repression of genes encoding metal uptake systems, will restore the cytoplasm to homeostasis. Thus, a long-standing hypothesis is that the intracellular metal activity or free metal concentration at which a metalloregulator is inhibited or activated to bind to its operator (or allosterically change the promoter structure) establishes the threshold or sensitivity of the transcriptional response [28]. This hypothesis holds that the reciprocal of the metal association equilibrium or metal affinity constant KMe, (1/KMe), defines the buffered or labile pool of metal at half-maximal activation or inhibition of the transcriptional response (Fig. 2).
Figure 2.
A graphical representation of the hypothesis that metal affinity for individual or pairs of metal sensor proteins (1/KMe) defines the ability of the cytoplasm to buffer biologically required transitions metal ions. Cu(I), black; Zn(II), blue; Ni(II), green; Fe(II), red; Mn(II), black. See Table 1 for KMe values for the regulators indicated. Efflux regulators are italicized, with transcriptional response curves (derepression or activation) represented by the dashed lines; uptake repressors are in straight font and transcriptional response curves (corepression) represented by the solid lines. NikR* represents filling of the secondary, low affinity sites which give rise to full repression of the high affinity nickel uptake system [48]. Approximate total cell-associated concentrations measured in E. coli under aerobic growth conditions on a minimal media are shown above the figure [28]; recent studies suggest that these concentrations are ≈5-fold larger in S. pneumoniae, which also exhibits a high Mn(II) quota relative to Fe(II) [24]. Note that this model is predicated on the simple notion that metal sensors are capable of “scanning” the cytoplasm for metal; this may not be the case [29, 139].
To help illustrate this point, zinc homeostasis in E. coli is controlled by two metalloregulatory proteins, Zur and ZntR (Fig. 2). The Zn(II) sensor Zur is a member of the Fur family [34] that represses the expression of znuABC, which encodes a Zn(II) uptake ABC transporter in the presence of Zn(II). Conversely, ZntR is a MerR regulator that activates the transcription of zntA, a gene encoding a P-type ATPase that effluxes zinc from the cytoplasm to the periplasm [32, 35]. In vitro transcription experiments reveal that each metalloregulator competes for Zn(II) with an affinity constant (1/KMe) offset by approximately one log unit, resulting in an activation threshold positioned at approximately femtomolar (10−15 M) Zn(II) [28]. As shown in Table 1, other Zn(II) metalloregulatory proteins bind Zn(II) with log KMe of ≈12-13 at or above neutral pH; if this hypothesis is correct, then Zn(II) may well be buffered at somewhat higher concentration, 10−12-10−13 M in other microorganisms. In the eukaryotic cell, cytosolic Zn(II) is buffered at ≈10−9 M, with a labile pool of 0.2 μM in the mitochondrial matrix [36]. This suggests that the bacterial cytoplasm and mammalian cytosol possess different buffering capacities for zinc.
Table 1.
Metal binding affinities (KMe) for selected bacterial transition metal sensor proteins
| Regulator | Biological Processa |
Cognate metal |
Family | Species | Metal | log KMe |
pH | Reference |
|---|---|---|---|---|---|---|---|---|
| Zur | uptake | Zn | Fur | E. coli | Zn(II) | 15.7 | 7.6 | [28] |
| ZntR | efflux | Zn | MerR | E. coli | Zn(II) | 15.0 | 7.6 | [28] |
| CzrA | efflux | Zn | ArsR | S. aureus | Zn(II) | 12.4 b | 7.0 | [63] |
| Co(II) | 9.0 | 7.0 | ||||||
| SmtB | efflux/seq’n | Zn | ArsR | Synechococcus | Zn(II) | 11.3 c | 7.4 | [127] |
| Co(II) | 9.7 | 7.4 | ||||||
| AztR | efflux/seq’n | Zn/Cd | ArsR |
Anabaena
PCC7120 |
Zn(II) | >10 | 7.0 | [135] |
| Co(II) | ≥7.3 | 7.0 | ||||||
| Cd(II) | 7.3 | 7.0 | ||||||
| Pb(II) | 6.2 | 7.0 | ||||||
| BxmR | efflux/seq’n | Zn/Cd/Cu/Ag | ArsR | O. brevis | Zn(II) | 13.0 d | 6.3 | [136] |
| Cu(I) | 7.6 e | 6.3 | ||||||
| AdcR | uptake | Zn | MarR | S. pneumoniae | Zn(II) |
12.1
f
10.0 f |
8.0 6.0 |
[15] |
| Co(II) | 6.8 5.4 |
8.0 6.0 |
||||||
| Mn(II) | 5.1 | 8.0 | ||||||
| CueR | efflux | Cu | MerR | E. coli | Cu(I) | 20.7 | 8.0 | [43, 137] |
| Au(I) | 34.7 | 7.7 | ||||||
| CsoRBS | efflux | Cu | CsoR | B. subtilis | Zn(II) | 8.2 | 6.5 | [45] |
| Cu(I) | ≥19.0 | 6.5 | ||||||
| Ni(II) | 9.5 | 6.5 | ||||||
| Co(II) | ≤5.0 | 6.5 | ||||||
| CsoRMT | efflux | Cu | M. tuberculosis | Cu(I) | 18.0 | 7.0 | [44] | |
| CsoRSA | efflux | Cu | S. aureus | Cu(I) | 18.1 | 7.0 | [46] | |
| CupR | efflux | Au | MerR |
R.
metallidurans |
Au(I) | 34.1 | 7.7 | [137] |
| Cu(I) | 16.1, 18.2 |
7.7 | ||||||
| NikREC | uptake | Ni | NikR | E. coli | Zn(II) | >12.0 | 7.6 | [52] |
| Cu(II) | 16.9 | 7.6 | ||||||
| Ni(II) | 12.0 | 7.6 | ||||||
| Co(II) | 8.7 | 7.6 | ||||||
| Cd(II) | >9.0 | 7.6 | ||||||
| NikRHP | uptake | Ni | NikR | H. pylori | Ni(II) |
11.5
8.8 |
7.6 5.8 |
[138] |
| NmtR | efflux | Ni/Co | ArsR | M. tuberculosis | Zn(II) | ≥9.0 | 7.0 | [29] |
| Ni(II) | 10.0 | 7.0 | footnote g | |||||
| Co(II) | 5.9 | 7.0 | [29] | |||||
| RcnR | efflux | Ni/Co | CsoR | E. coli | Ni(II) | >7.6 | 7.0 | [54] |
| Co(II) | >8.3 | 7.0 | ||||||
| Fur | uptake | Fe | Fur | E. coli | Zn(II) | 5.9 | 7.0 | [47] |
| Co(II) | 6.8 | 7.0 | ||||||
| Fe(II) | 5.9 | 7.0 | ||||||
| Mn(II) | 4.6 | 7.0 | ||||||
| MntR | uptake | Mn | DtxR | B. subtilis | Zn(II) | 7.9 | 7.2 | [64] |
| Ni(II) | 5.7 | 7.2 | ||||||
| Co(II) | 5.3 | 7.2 | ||||||
| Mn(II) | 3.8 h | 7.2 | ||||||
| Cd(II) | 7.0 | 7.2 |
Indicated regulator controls the transcription of genes encoding proteins involved in metal uptake into the cytoplasm (uptake), extrusion from the cytoplasm (efflux), and of intracellular sequestration by metallothionens (seq’n).
First of two binding sites (KZn1) on the dimer (see text for details).
α5 site affinity.
α5 site affinity.
α3N site average affinity for each of two bound Cu(I) ions bound in a Cu2 cluster.
Highest affinity regulatory site; lower affinity sites not shown.
H. Reyes-Caballero, D. Giedroc, manuscript in preparation.
Determined by EPR spectroscopy.
B. anthracis AntR (an MntR homolog), log KMn=4.2 determined using EPR spectroscopy [134].
The Irving-Williams series of divalent metals ions establishes that Zn(II) and Cu(II) bind with the highest affinity to a model chelate, while Mn(II) and Fe(II) bind with the lowest affinity to the same chelate, Zn≤Cu>>Ni≥Co≥Fe≥Mn [11, 37]. Metalloregulatory proteins that have been extensively characterized largely follow this expected trend, although there are also some outliers (Table 1). The Irving-Williams series classifies Zn(II) as a highly “competitive” metal, i.e., Zn may be capable of binding as a non-cognate metal far more tightly than the corresponding cognate metal (KZn>>KMn for B. subtilis MntR, for example) (Table 1). As a result, the cytoplasm employs an overcapacity to chelate Zn(II), and it has been postulated that ribosomal proteins play an important role in this process [38-40]. The obligatory participation of specific metallochaperones that traffic Cu(I) to target proteins with KCu(I)≥1018 M−1 also likely reflects the need to maintain the bioavailable concentration of this metal under very tight control as well [41, 42]. As a result, E. coli CueR, a copper-selective member of the MerR family, is activated by Cu(I) in vivo and binds Cu(I) with an affinity of 1020.7 M−1 [43] (Table 1). Other copper sensors, e.g. CsoR, are characterized by a log KCu(I) of 18-19, in range of that of CueR [44-46]. Thus, this hypothesis suggests that Zn(II) and Cu(I) are buffered in cytoplasm in the picomolar-femtomolar and attomolar-zeptomolar range, respectively (Fig. 2). In contrast, E. coli Fur, an Fe(II)-dependent transcriptional repressor responds to the low micromolar range of iron (Table 1) [47], with binding affinities for Mn(II) regulators in the 103-105 M−1 range of KMe (Table 1). These affinity constants suggest that a correspondingly larger fraction of the Mn(II) and Fe(II) concentrations in the cell may be weakly bound or highly mobile with rapid off-rates [13] (Fig. 2).
The degree to which pairs of metalloregulators possess relatively well-matched metal affinities and specificities is not yet known. For example, a Cu(I) uptake regulator or an Fe or Mn efflux regulator have not yet been identified which suggests that “one-armed” transcriptional control (see Fig. 2) may be sufficient to maintain intracellular homeostasis of Cu(I) and Fe/Mn. The case of Ni(II) homeostasis in E. coli provides an interesting contrast to these systems. Like Zn(II), Ni(II) homeostasis in E. coli is controlled by a pair of regulators, in this case NikR and RcnR. NikR is a Ni(II)-specific D2-symmetric homotetrameric repressor [48, 49] that controls the transcription of nikABCD, a high affinity Ni(II) uptake transporter [50, 51]. NikR binds Ni(II) to the each C-terminal regulatory site at the tetrameric interface with affinity of 1012 M−1 [49, 52]. RcnR is a Ni(II) specific efflux regulator from the CsoR family that dissociates from the DNA operator sequence when Ni(II) binds, allowing the expression of rcnA, a gene that encodes for a Ni(II)/Co(II) transporter [53]. Ni(II) binds to RcnR with an affinity of ≈107.6 M−1 or approximately five orders of magnitude weaker than NikR at a similar pH (Table 1). Thus, NikR and RcnR possess poorly matched Ni(II) affinities and thus would appear to be in contrast to the zinc sensor pair in E. coli [54] (Fig. 2). However, NikR is known to possess an additional pair of low affinity Ni(II) regulatory sites (log KNi= 9) that are required for full NikR repression [55, 56]. This might suggest an ordered or step-wise regulatory response in the cell where uptake is first partially repressed, and then fully repressed, prior to Ni(II) efflux re-establishing Ni(II) homeostasis. Interestingly, two paralogous efflux regulators from the ArsR family [57] present in M. tuberculosis, NmtR and KmtR, may well collaborate to establish a similar two-step or graded repression response to Ni(II) toxicity as well, given log KNi values of 12 and 10, respectively [29, 58] (H. Reyes-Caballero and D. Giedroc, manuscript in preparation). The physiological importance of a potentially graded response to nickel toxicity in E. coli and M. tuberculosis has not yet been firmly established. The recent discovery of a two pairs of metalloregulatory sites within homodimeric S. coelicolor Zur [59], which appear to influence zinc-mediated repression at different promoters to different degrees, is also consistent with a graded response to zinc stress in that organism as well [60]. The sensitivity of the zinc efflux regulator has not yet been established in S. coelicolor.
Ongoing studies in our laboratory are focused on the control of Zn(II) homeostasis in S. pneumoniae, where two metal sensor proteins, AdcR and SczA, function to control the expression of genes that encode for Zn(II) uptake and efflux, respectively. AdcR is the first known metal-sensing MarR family member [61], and binds Zn(II) with an affinity of 1012 M−1 at pH 8.0 [15] as would be anticipated on the basis of KZn found for other Zn(II) metallosensors (Table 1). However, preliminary findings with SczA, a novel Zn(II) sensing TetR family member [62] reveals K of ≈ 108 M−1 Zn under the same conditions (K. Geiger and D. Giedroc, unpublished results), or about four orders of magnitude weaker than KZn for AdcR. This is consistent with expression analysis which suggests that under all conditions containing ≥20 μM total zinc in liquid media, the AdcR regulon is fully repressed and the cellular response to perturbation of zinc homeostasis is governed exclusively by SczA and metal efflux [15, 24].
3. Metal selectivity, ligand donor sets and coordination geometry
A large number of metal sensor proteins have now been structurally characterized by x-ray crystallography, NMR spectroscopy and other spectroscopic methods. As a result, it is now possible to identify common features of metal sensing sites specific for a particular metal ion (Fig. 3). Transition metal ions become more polarizable or “soft” as the number of d orbital electrons increases. Although the first row transition metal ions Fe(II), Co(II), Ni(II), Cu(II) and Zn(II) are considered as a group to be borderline hard/soft, subtle ligand preferences are apparent as Mn(II) is d5 and is considered “hard” relative to d10 Zn(II) which is correspondingly “soft”. Likewise, Cu(I) is more polarizable than Cu(II) since a decrease in valence means an additional d-electron has been added. Soft metals prefer soft ligands, with the trend in polarizability decreasing from Cys to His/Met to Asp/Glu.
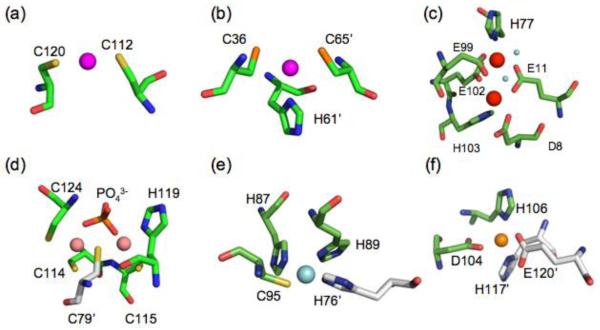
Figure 3.
Native metal coordination geometries for selected bacterial metalloregulatory proteins. A colored sphere represent the metal ion unless is otherwise indicated. (a) E. coli CueR, Cu(I) linear (PDB ID:1Q05), (b) M. tuberculosis CsoR, trigonal Cu(I) (PDB ID: 2HH7), (c) B. subtilis MntR, penta-coordinate dinuclear Mn(II), H2O molecules are represented as cyan spheres (PDB ID: 1ON1), (d) E. coli ZntR, tetrahedral dinuclear Zn(II) (PDB ID: 1Q08), (e) E. coli NikR, square planar Ni(II) (PDB ID: 2HZV), (f) S. elongatus PC7942 SmtB, Zn(II) tetrahedral (PDB ID: 1R22). Graphical representation was created using PyMol [140].
These trends in metal-ligand polarizabilities from small molecule coordination chemistry are largely recapitulated in metal ligand donor sets in metal sensor proteins. This has been most extensively established in the ArsR/SmtB (or ArsR) family of metalloregulatory proteins, widely represented across bacterial species [14, 57, 58]. ArsR family proteins regulate the expression of genes that encode for proteins responsible for metal ion detoxification, sequestration and cytoplasmic efflux, as well as many other processes not yet discovered [58]. The metalloregulators of this family have collectively evolved an impressive range of regulatory metal binding sites on a relatively unchanging protein scaffold with clearly distinct metal specificities [12]. Soft metal ion sensors from the ArsR family, such as Cd(II)/Pb(II) sensor S. aureus CadC, the Cd(II)/Pb(II) sensor M. tuberculosis CmtR, As(III)/Sb(III) sensor E. coli ArsR, and Cu(I) sensor O. brevis BxmR, all harbor metal regulatory sites that are rich in cysteine residues. In contrast, the Zn(II)/Co(II) sensor S. aureus CzrA [63] and the Ni(II)/Co(II) M. tuberculosis sensor NmtR bind the metal with more electronegative and less polarizable “hard” donor ligands including glutamate and aspartate residues (Fig. 3).
B. subtilis MntR is a Mn(II)-dependent repressor of the transcription of mntABCD which encodes a high affinity Mn(II) importer. MntR binds Mn(II) weakly with KMn in the order of 105 M−1 [64] (Table 1), yet is Mn(II)-specific among first-row transition metal ions in vivo [65]. Structurally MntR is similar to the Fe(II) sensor DtxR/IdeR [66, 67], but is a stable dimer that binds one to two metal ions per subunit depending on solution conditions [68]. Among non-cognate metals, Cd(II) activates MntR in vitro to the same or greater degree while Zn(II), Ni(II), Fe(II), and Cu(II), activate DNA binding with low efficacy [69]. A similar metal specificity profile characterizes DtxR-like family repressors PsaR and ScaR from Streptococcus spp. [24, 70]. Selective allosteric switching by Mn(II) over Fe(II) appears to be largely based on ligand preferences of Mn(II) vs. Fe(II) [71]. The metal regulatory sites in MntR and DtxR/IdeR are structurally equivalent yet differ in the nature of the coordinating ligands. Here, the thioether sulfur of Met10 in the α1 helix and thiolate sulfur of Cys102 in the regulatory domain found in the Fe(II) selective repressor DtxR/IdeR are replaced by Asp8 and Glu99, respectively, in Mn(II)-specific MntR [71]. Inspection of the structural database confirms that nature selects against coordination of Mn(II) by cysteine [72] in contrast to the well known propensity of Fe(II) to form iron-sulfur clusters of varying nuclearities [71]. On the other hand, Mn(II) selects against Zn(II) on the basis of distinct coordination geometries of each, since the tetrahedral coordination of Zn(II) differs from the native more highly coordinated hexa- or heptavalent coordination geometry observed for Mn(II) and Cd(II), respectively [64, 73].
The S. aureus Zn(II) metalloregulator CzrA of the ArsR family represses the transcription of the cation diffusion facilitator (CDF) CzrB in the absence of metal stress [14]. CzrA is denoted an “α5” ArsR family sensor as it binds Zn(II) with negative homotropic cooperativity at two regulatory sites positioned between the pair of α5 helices at the dimer interface with step-wise binding affinities KZn1 and KZn2 of ≈1012 M−1 and ≈1010 M−1, respectively, at pH 7.0 (Table 1) [63, 74]. Functional characterization of metal ligands in the α5 helix shows a tolerance for variation at some liganding positions, while other metal ligands are absolutely essential for allosteric regulation of DNA binding [63]. In contrast, the mutagenesis of any of the α5 ligands in the homologous Ni(II)/Co(II) sensor M. tuberculosis NmtR abrogates allosteric inhibition of DNA binding (H. Reyes-Caballero and D. Giedroc, manuscript in preparation). NmtR binds Ni(II) in an octahedral coordination geometry, but shares an analogous core of four α5 ligands with CzrA [75].
Comparative structural and spectroscopic studies of CzrA and the related zinc sensor, Synechococcus SmtB with NmtR reveals a key aspect of regulation by “α5” ArsR family sensors. These studies establish that Zn(II) binds in a tetrahedral or distorted tetrahedral coordination geometry to CzrA and SmtB [63, 76] while Ni(II) binds to NmtR in an octahedral coordination geometry [75]. These coordination geometries parallel the natural preferences of each metal [77-80]. However, a non-cognate metal ion will often bind to the same site in such a way to force a new non-native coordination geometry, consistent with intrinsic preferences of the non-cognate metal [75]. However, formation of a non-native coordination geometry always results in a weaker or abrogated allosteric response, relative to the cognate metal, and thus a poorer ability to sense these metals in the cell. For example, in the functional analysis of the α5 metal ligand chelate of CzrA, only those substitutions that preserved a native tetrahedral Zn(II) coordination geometry were capable of allosteric regulation of DNA binding in vitro and could adopt an allosterically inhibited conformational state, irrespective of KZn [63]. These findings on CzrA support the hypothesis that the structure of the first coordination shell around the cognate metal is a major determinant of biological specificity [63].
Detailed studies with other regulators are largely consistent with this hypothesis. The M. tuberculosis Cu(I) dependent repressor, CsoRMT is a founding member of a large family of bacterial regulatory proteins [46, 54, 81]. M. tuberculosis CsoR controls the expression of ctpV, a gene that encodes a predicted copper ATPase that effluxes Cu(I) from the cytoplasm [81]. CsoR binds to Cu(I) with an association equilibrium constant in the order of 1018 M−1 [44, 45]. CsoRs from Bacillus subtilis (CsoRBS) and S. aureus (CsoRSA) are homologous to CsoRMT and each adopts a S2N coordination complex that is structurally identical to that of CsoRMT [45, 46] (Fig. 3). CsoRBS also binds Zn(II) and Ni(II) tightly in what appear to be non-native tetrahedral and square planar geometries, respectively, but these metals are quantitatively less capable than trigonal Cu(I) of driving dissociation of CsoRBS from the DNA operator [45]. In contrast, the CsoR-family Ni(II) efflux regulator E. coli RcnR [12] adopts an octahedral coordination geometry upon binding cognate metals Ni(II) and Co(II) by recruiting additional metal ligands from the N-terminal region that are not found in the copper sensor CsoR; this results in an S(N/O)5 coordination sphere [54]. The metal specificity of CsoR and RcnR for Cu(I) or Ni(II) is therefore dictated by the coordination geometry which may or may not track with metal affinity [82], analogous to the conclusions reached with the ArsR family of regulators.
Full repression by the Ni(II) sensor E. coli NikR is also observed in vivo only when cognate metal Ni(II) binds in a square planar coordination geometry to the C-terminal regulatory sites. Cu(II) forms a complex that is isostructural with that of Ni(II), but will not be regulatory in vivo due to the vanishingly small concentrations of Cu(II) expected to be present in the reducing bacterial cytoplasm (see Fig. 2) [54]. However, a clearly non-native coordination geometry is adopted by all other non-repressing metals; Zn(II) adopts a tetrahedral coordination geometry and Co(II) an octahedral complex. As expected, both metals poorly activate DNA operator binding by NikR [50, 55, 83, 84].
This close correspondence of functional metal selectivity and coordination geometry also characterizes other classes of metalloregulatory proteins, including the transcriptional activators of the MerR family from E. coli, CueR and ZntR. In MerR family proteins, metal ions typically bind to a C-terminal loop which is packed against the N-terminal DNA binding domain. CueR binds Cu(I) in a characteristic linear Cu(I) bis-thiolato coordination geometry [85], a coordination structure also observed for CueR in complex with Ag(I) and Au(I) [43] (Fig. 3). In contrast, Zn(II) binds to ZntR to form a phosphate-bridged binuclear Zn(II) center, with each Zn(II) ion adopting a tetrahedral coordination geometry [43] (Fig. 3). Finally, the Fur family Ni(II) sensor Nur [86] has been proposed to discriminate against other metals by harboring an octahedral coordination site not found in other Fur family repressors, with an additional site analogous to the regulatory Fe(II) binding sites in the Fe(II)-specific regulator, Fur [87]. In contrast, the regulatory site(s) in the Zn(II)-specific Fur family regulator, Zur, clearly adopt four-coordinate tetrahedral complexes [60, 88], fully consistent with nearly all other Zn(II) regulators, with the exception of AdcR [15]. In no case, however, have systematic quantitative studies been carried out to assess the degree to which the coordination geometry of the cognate or regulatory metal differs from that of a non-cognate metal in allosteric activation of DNA binding.
4. Allosteric signal propagation
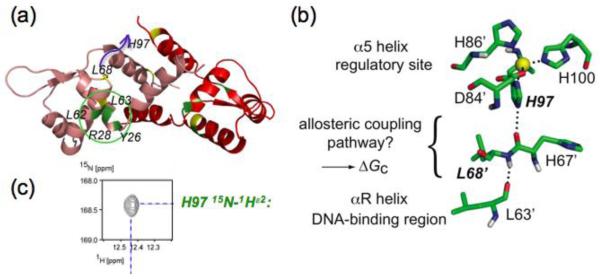
The studies summarized above establish that the structure of the first coordination shell adopted by cognate vs. non-cognate metal ions is a major determinant for biological metal specificity. Recent structural studies are beginning to reveal how formation of a native coordination geometry is tied to structural changes in a repressor [89, 90]. Early crystallographic structures of the apo- and Zn(II) forms of Synechoccocus SmtB and S. aureus CzrA provided the first insights into a possible structural coupling mechanism [91]. Two hydrogen bonds appeared to form a pathway that connected the α5 metal center and the α4 DNA recognition helix on the opposite protomer through the αR’-β1 loop upon metal binding (Fig. 4). The non-chelating nitrogen (Nε2) of the essential ligand His117 (H97 in CzrA) forms a key second coordination shell hydrogen bond and is therefore predicted to be responsible for triggering the allosteric response upon Zn(II) binding [91]. Support for this model came from the observation of an intense Nε2-Hε2 correlation from His117 in the 1H-15N HSQC spectrum, which is lost upon metal dissociation [91] (Fig. 4). Analogous observations are found for His97 in the related Zn(II) sensor CzrA [91, 92] (Fig. 4).
Figure 4.
Illustration of a potential hydrogen bonding pathway in CzrA that links the metal binding and DNA binding sites. (a) Ribbon representation of the solution structure of CzrA bound to DNA with key residues highlighted [89] The blue arrow defines the distance (≈10 Å) between the L68′ and H97 on opposite protomers that are proposed to hydrogen bond in the allosterically inhibited Zn2-bound state (see panel b). (b) Two hydrogen bonds are proposed to link the NHε2 of His97 with the main chain C=O of L63′ in the DNA binding αR helix [91]. (c) 15N-1H correlation for the HNε2 group of H97 observed in a simple 15N-1H HSQC spectrum of Zn2-CzrA, indicative of slow exchange with solvent in the allosterically inhibited metal-bound state, consistent with a hydrogen bonding interaction. This correlation is found in all first coordination shell mutants of CzrA that are capable of negatively regulating DNA binding upon Zn(II) binding [63], and is lost in apo-CzrA [91].
Semisynthetic native chemical ligation is currently being used to directly test this allosteric coupling model in CzrA by site-specifically incorporating unnatural His97 analogs designed to maintain an Nδ1-Zn(II) coordination bond, but lack the ability to donate a hydrogen bond on the other side of the imidazole ring (Z. Ma., Y. Fu and D. Giedroc, manuscript in preparation) (see Fig. 4(b)). Such an approach was recently employed on the Cu(I) sensor M. tuberculosis CsoR in which it was established that the nonliganding Nε2 face of His61 mediates a hydrogen bonding network to Tyr35′ and Glu81 across the subunit interface to stabilize the low DNA binding affinity of the tetramer [44]. Although the precise structural mechanism of Cu(I) regulation in CsoR is not completely understood, these second coordination shell interactions seem to be required for allosterically coupling cognate metal binding and DNA binding sites. The neutral charge of the histidine side chain at physiological pH and the frequency of events of this kind found in structural databases [93, 94] reveal that nature exploits a recurring mechanism to enable allosteric signal propagation in metal sensor proteins. Current efforts are underway to identify other allosteric residues in CzrA.
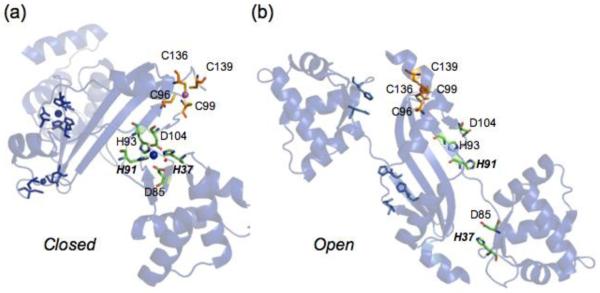
The mechanism of activation of DNA binding by Fur family transcriptional repressors is based largely on studies of the hydrogen peroxide sensor PerR, which employs an iron chelate to sense oxidative stress in B. subtilis. Here, the metal ligand His37 from the N-terminal winged helix DNA binding domain effectively couples the N-terminal and C-terminal domain which stabilizes a “closed” Fe(II)- bound conformation that activates binding of PerR to operator sites upstream of oxidative stress sensing genes (Fig. 4) [95]. Mechanistically, PerR uses an open coordination site on Fe(II) to bind H2O2, which results in oxidation of Fe(II) to Fe(III) and creating OH• which in turn leads to oxidation of two metal liganding histidines, His37 and His91, to 2-oxo-histidine [96]. This is thought to lower the affinity of PerR for Fe(III), allowing for metal dissociation, and triggering a quaternary structural change leading to an “open” apo-like structure of lower DNA binding affinity, as predicted by the crystal structure of inactivated (apo) PerR (Fig. 5) [95, 97, 98]. All ligands in the regulatory site are essential for peroxide sensing in vivo, but each plays a distinct function. His37 appears to function in a key allosteric role, but this time via ligand oxidation and perturbation of the first coordination shell; this effectively uncouples the DNA binding domain from the regulatory domain. His91, on the other hand, is predicted to play an important role in stabilizing the Fe(II) complex; oxidation therefore simply increases dissociation of the metal, which drives “open” the molecule [99, 100] (Fig. 5).
Figure 5.
The first coordination shell of a liganding histidine as an allosteric trigger in moving from the (a) “closed” PerRMnZn (3F89) and (b) “open” apo-Zn (2FE3) structures. The structure shows two metal binding sites occupied by Mn(II) (blue) and Zn(II) (magenta). Mn(II) binds to the regulatory site in a distorted square pyramidal geometry involving ligands H37 and H91 and in the Fe(II) complex, an open coordination site that accommodate H2O2. Zn(II) binds to a thiolate rich site that plays no role in the catalysis [95, 96], but is essential for structural integrity of the dimer [99].
Other Fur family repressors bind metal ions to activate DNA binding to Fur-boxes upstream of genes that often encode for metal ion uptake systems [101]. In the available structures of Fur homologs that are activated to bind DNA by divalent metal ions, e.g,. zinc-sensing M. tuberculosis and S. coelicolor Zur [60, 88, 102], P. aeuroginosa, V. colera and H. pylori Fe(II) sensors Fur [72, 103, 104] and the Ni(II) sensor Streptomyces coelicolor Nur [86], all possess metal binding sites that are structurally analogous to the Fe(II) site in PerR. In most of these cases, it has been hypothesized that coordination of metal ions anchored by a conserved pair of His residues analogous to His37 and His91 in B. subtilis PerR stabilizes a “closed” conformation that is competent to bind operator DNA [72, 95] (Fig. 5).
5. Thermodynamics of allosteric regulation by metals
An emerging topic in the study of allosteric regulation in metal sensor proteins is the measurement of the energetics and conformational dynamics that underlie the mechanism of metal activation or inhibition of DNA operator binding. These studies are motivated in part by a lack of a dramatic structural change observed when comparing the metal-bound vs. metal-free crystal structures of ArsR/SmtB sensors, for example [91]. This picture differs dramatically from the large conformational change in PerR and perhaps other Fur family regulators as well as in NikR [90] discussed above (see Fig. 5). In MerR family activators, there is little understanding of this process due to incomplete structural characterization of all relevant allosteric states, e.g., apo-, ligand-bound, DNA-bound ligand activated, within a single MerR regulator [33, 105] Indeed, accumulating evidence suggests that allosteric proteins can clearly function in the absence of large structural changes, mediated instead by changes in conformational dynamics, e.g., stiffening or enhanced mobility, that can be detected by residue-specific NMR methods (see Section 5) or globally through thermodynamic methods [106-108].
Although biological systems are open systems that operate far from equilibrium [109], the study of metalloregulatory proteins in equilibrium with its two ligands, DNA operator and metal ion, represents a powerful tool that has enabled the discovery of many fundamental aspects of metalloregulation [13, 74]. This coupled equilibrium can be cast in terms of a simple thermodynamic cycle (Fig. 1; Fig. 6) [13]. The magnitude of the thermodynamic linkage, ΔGtC, defines the efficacy of the allosteric ligand (metal) to activate or inhibit DNA operator binding. Such a formalism facilitates quantitative comparisons of different metal sensor proteins with different metal ions, but more importantly provides a thermodynamic construct with which to identify key residues that are involved in the allosteric switching mechanism, independent of their effect on binding affinity for either ligand [13, 44, 110].
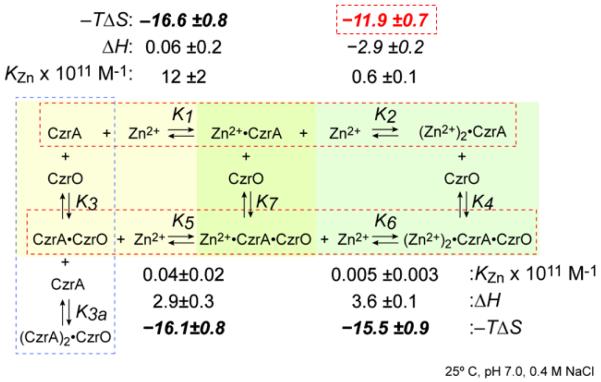
Figure 6.
Global energetics of the step-wise coupled equilibrium that describes the allosteric effect of Zn(II) binding to CzrA free in solution and when bound to DNA. Thermodynamic parameters determined by isothermal titration calorimetry (ITC) are shown [74] at pH 7.0, 0.4 M NaCl. 25.0 °C.. This is an expanded thermodynamic linkage scheme relative to that outlined in Fig. 1. CzrO, czr DNA operator. K1, K2, K5 and K6 are KZni from each step in the equilbrium where ΔGi = −RTlnKZni. Allosteric (heterotropic) coupling free energies, ΔGci are obtained as follows: ΔGc1 = ΔG5 − ΔG1 (defined by the yellow box); ΔGc2 = ΔG6 − ΔG2 (defined by the green box); ΔGct = (ΔG5 + ΔG6) − (ΔG1 + ΔG2) = ΔGc1 + ΔGc2 = ΔG4 − ΔG3 (overall linkage). See text for other details.
Isothermal titration calorimetry (ITC) has recently been used to measure a complete set of thermodynamic parameters for the binding of Zn(II) to CzrA homodimer in the free form and in the protein-DNA complex. This approach has allowed extraction of the homotropic coupling free energies within the two states and ultimately the step-wise heterotropic coupling free energies, ΔGci (Fig. 6) [74]. The Zn(II) binding isotherms were readily fit to a two-step binding model where the energetics associated with formation of the Zn1-CzrA and Zn2-CzrA complexes could be measured in the free and the DNA bound complex (top, bottom horizontal equilibria, Fig. 6). The first Zn(II) binding event to the apoprotein is entropically driven (−TΔS1 = −16.6 kcal mol−1), with a small opposing enthalpy term (ΔH1 = 0.06 kcal mol−1) [74]. Structurally, this Zn1 state is characterized by a loss of symmetry detectable in an 1H-15N HSQC spectrum, representing a superposition of apo and metallated states, consistent with strong negative cooperativity of metal binding [63, 111]. The magnitude of the heat capacity change (ΔCp1 ≈−230 cal mol K−1) may be reporting on solvent reorganization as a result of restructuring the dimer interface [74]. In striking contrast, binding of the second Zn(II) to Zn1-CzrA is entropically less favorable by ≈5 kcal mol−1 (−TΔS2 =−11.9 kcal mol−1) relative to the first binding event (Fig. 6). This is thought to be the energetic origin of negative homotropic cooperativity of Zn(II) binding, where the second site globally quenches the short timescale dynamics [74].
These thermodynamics of Zn(II) binding contrast sharply with those obtained with the repressing apo-CzrA-DNA complex. Here, the entropic penalty for the binding of the second Zn(II) relative to the first is far smaller (Δ(−TΔS)’=0.6 kcal mol−1) suggesting that the Zn2-CzrA-DNA complex is far more dynamic than unbound Zn2 CzrA [74]. Parallel differences are observed in the sign and magnitude of the enthalpy term, which while overall favorable in the unbound state, is unfavorable or near zero (ΔH2’ − ΔH1’ ≈0.7 kcal mol−1) in the DNA complex; this is consistent with the greatly reduced Δ(−TΔS’). Thus, although metal binding to the protein-DNA complex is also negatively cooperative (ΔΔG=1.3 kcal mol−1), the underlying energetics are completely different. This in turn drives strong negative allosteric (heterotropic) cooperativity in this system, ΔGct, which is sizable at 6.3 kcal mol−1. In experiments that employed a covalently fused CzrA dimer, the intermediate Zn1-CzrA state shows metal dependent inhibition of DNA binding in vitro to 70% of the fully metallated Zn2-state [111]. In contrast, the stepwise coupling free energies, ΔGc1, ΔGc2, were found to make approximately equal contributions to ΔGct [74] (Fig. 6). The origin of this discrepancy is unknown but it may lie in the covalent coupling of the C-terminus of one protomer to the N-terminus of the other, since the linker length was found to have a profound influence on the stoichiometry and cooperativity of Zn(II) binding to various fused CzrAs [111].
It is interesting to note that studies like these have the potential to systematically access all four allosteric “end-states,” including the ternary CzrA-Zn2-DNA complex (P•Me2•D in Fig. 1) [112]. This complex is likely a transiently formed intermediate in the cell since Zn(II) binding by the repressing CzrA-DNA complex will quickly lead to disassembly of the complex. As might be expected, qualitative inspection of the 1H-15N TROSY spectrum of this intermediate reveals that CzrA adopts a “hybrid” conformation as it attempts to optimize interactions with both metal and DNA ligands (A. Arunkumar and D. Giedroc, unpublished results). There are a number of residues for which backbone amide and side chain methyl chemical shifts are distinguishable in the CzrA-Zn2-DNA complex relative to the apo-CzrA-DNA and allosterically inhibited Zn2 states. These residues are strong candidates for playing a key role in structural coupling between metal and DNA binding sites [112], which may buttress the hydrogen bonding pathway shown (Fig, 4). In any case, when NMR studies like these are combined with a multiple sequence alignment of ArsR family regulators of known distinct metal binding sites [14, 58], this approach can be used to identify candidate residues for substitution and quantification of ΔGc [63, 74].
6. Conformational dynamics in allosteric regulation by metals
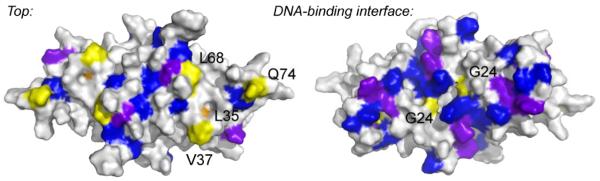
These global thermodynamics are in general consistent with insights gleaned from residue-specific conformational dynamics, readily measured by a variety of NMR approaches. For example, the fully metallated Zn2 state of CzrA, relative to apo-CzrA, reveals that Zn(II) binding rigidifies the quaternary structure of the molecule [89, 91]. The rate of hydrogen-deuterium solvent exchange of backbone amides is globally quenched [91] and investigation of the short timescale (ps-ns) dynamics and qualitative insights into intermediate (μs) timescale dynamics using the same methods are consistent with dynamical quenching of not only the α5 and α1 helical core that comprises much of the dimer interface, but also into the more peripheral α4 (DNA recognition) helices (Fig. 7). The β-hairpin is structurally unaffected, which is conformationally dynamic in both apo and metallated states [89].
Figure 7.
Zinc binds to apo-CzrA homodimer and quenches the conformational dynamics in the core α5, α1 helical regions but also in more peripheral DNA-binding αR helices of the molecule [89]. Spacefilling models of Zn2 CzrA are shown (left, α5 helices on top; right, view from the DNA-binding interface) with residues shaded according to a specific change in dynamics relative to apo-CzrA. Blue, residues for which the order parameter S2 increases (≥0.02); purple, residues for which significant chemical exchange broadening (Rex) is lost on Zn(II) binding; beige, residues for which Rex becomes measurable upon Zn(II) binding; yellow, residues for which S2 decreases on Zn(II) binding. This picture is consistent with significant quenching of the conformational dynamics (indicated by blue, purple residues) far from the allosteric zinc binding sites, relative to those residues which increase their motional disorder (beige, yellow); these may define “hinge” regions in the allosterically inhibited Zn(II) conformational ensemble [90].
This picture of the allosterically inhibited Zn2 state contrasts sharply with that of repressing apo-CzrA-DNA complex relative to same apo-CzrA reference state [89]. Here, the expected stabilization of the protein-DNA interfacial region is observed; however, the metal binding sites in the α5 helix become highly mobile as evidenced by rapid H-D exchange rates and increases in backbone dynamical disorder in the ns to ps time scale, which are small but extend into the core of the protein [89]. These dynamical findings are consistent with the solution structure of the DNA-bound CzrA. This structure reveals that while the global fold of each protomer is unchanged relative to apo-CzrA, the quaternary structure differs dramatically, with the DNA recognition helices (α4) directed away from the core and pointing in a direction that allows for favorable contacts with successive major grooves of the DNA operator (see Fig. 4(a)). In order for this to occur, the α5 helices pull apart from one another and expose the core of the dimer, thus strongly enhancing conformational mobility in the allosteric sites. Enhanced dynamics in this region may well increase the kinetics of metal association and thus drive subsequent dissociation of the Zn(II)-bound repressor from the DNA [113].
Although studies of the conformational dynamics have only been reported for a few other metal sensor proteins [114, 115], functionally important stabilization of the native structure upon binding metals may be a recurring theme. This can result from metal binding to a structural site that plays no direct role in regulation or to the regulatory site(s) directly, similar to that which occurs in CzrA. For example in H. pylori FurHP [19, 116-120], Zn(II) binds to a tetrathiolate, distorted tetrahedral site found in a C-terminal regulatory domain involving two C-terminal Cys residues that are necessary to stabilize the dimer, but play no direct role in metalloregulation [121]. Hence, formation of this structural site is a necessary prerequisite of metal binding to the regulatory sites [121]. This S4 structural site is found in many, e.g., Zur [60, 88], Nur [86] and PerR [95], but not all Fur family sensors. Some Fur family dimers contain a third pair of “secondary” metal binding sites, whose coordination structure and function seems to vary from the one to another regulator [34, 40, 60, 88, 104, 122].
Similarly, DtxR/IdeR regulators often require metal occupancy of what is thought to be a secondary structural site that stabilizes the quaternary structure by increasing the stability of the dimer [70, 123-125]. This may be a necessary prerequisite for a second metal to bind to the pair of regulatory sites in the dimer [126]. In the ArsR/SmtB family, the α5 metal site that plays a regulatory role in CzrA (see Fig. 4) and in SmtB [127] is also present in many (but not all) Cd(II)/Pb(II) sensing CadCs but plays no role in regulation; it may stabilize the dimer and function as a necessary prerequisite for Cd(II) regulation in the more peripheral α3N sensing sites [128, 129]. In E. coli NikR, the binding of cognate metal to the C-terminal domain nucleates a hydrogen bonding network that likely stabilizes the native structure against thermal denaturation; in contrast, non-cognate metals that adopt distinct coordination structures do not [49, 52, 130]. Structural stabilization of NikR allows formation of a pair of lower affinity Ni(II) sites that are essential for full activation of nik operator binding [56].
The crystallographic structures of C. diphtherium DtxR, M. tuberculosis IdeR and B. subtilis MntR in the metal-free and allosterically activated, metal-bound states reveal only small differences that bring the pair of DNA binding domains into a more “closed” conformation which is thought to directly stabilize protein-DNA interactions [73, 126, 131-133]. An alternative interpretation of these small structural changes is that the binding of activating metals functions largely to quench conformational dynamics which “locks in” or “freezes” a high DNA-binding affinity conformation. Hydrogen-deuterium exchange mass spectrometry carried out with Mn(II)-MntR [115] and EPR spectroscopy of the structural homolog B. anthracis AntR [114] are generally consistent with this picture. In AntR, the DNA binding domain is characterized by an ensemble of different conformational states in rapid exchange on the ns timescale that are narrowed by non-cognate Zn(II) binding [114, 134]. The degree to which cognate metal Mn(II) narrows the distribution relative to Zn(II) is unknown but of interest since Zn(II) is a poor activator of operator binding by MntR and other MntR-like repressors including S. pneumoniae MntR and PsaR [23, 24] and S. gordonii ScaR [70]. A subject of ongoing work in our laboratory is to determine the degree to which allosteric inhibition and activation may possess a common origin in “stiffening” or rigidifying the metal-bound conformation [89].
7. Concluding remarks
In this review, we discuss three major emerging themes that are predicted to govern functional metal selectivity in bacterial transition metal homeostasis: 1) Metal sensor specificity for one or a few closely related metals from different structural families seem to exhibit characteristic trends in metal affinity when measured under similar solution conditions. It is these affinities that may well govern the intracellular sensitivity of these allosteric switches in the cell, thus establishing the limits of the “buffered” concentration of metal, beyond which mediates a transcriptional response. Cu(I) and Zn(II) are highly competitive metals and thus their bioavailability is tightly controlled intracellularly by the mechanisms discussed here. 2) Cognate metal complexes in metal sensor proteins are characterized by a coordination geometry that optimizes subsequent noncovalent interactions that drive a structural or dynamical change in the oligomer that leads to allosteric signal propagation. This sometimes occurs through the “second” coordination shell of the inducing metal complex. 3) Allosteric switching by metal sensor proteins involves changes in structure and/or dynamics driven by cognate metal binding and to a far lesser extent, by non-cognate metal binding. Future challenges are to determine the degree to these rules derived from a detailed analysis of only a few systems characterize other metal sensor proteins, and how nature re-engineers an existing allosteric metal site and evolves the ability to respond to other stresses, namely oxidative and nitrosative stress. How multiple parallel pathways of transition metal homeostasis and oxidative stress resistance intersect in the cell is also an important topic. Careful investigations of the quantitative biology of these fascinating allosteric proteins will continue to provide new insights into the perturbation and adaptability of the metallome in cells.
Acknowledgements
We gratefully acknowledge the US National Institutes of Health (GM042569) for financial support of this project and members of the Giedroc laboratory for their comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Berg JM, Godwin HA. Lessons from zinc-binding peptides. Annu Rev Biophys Biomol Struct. 1997;26:357–371. doi: 10.1146/annurev.biophys.26.1.357. [DOI] [PubMed] [Google Scholar]
- [2].Seneque O, Bonnet E, Joumas FL, Latour JM. Cooperative Metal Binding and Helical Folding in Model Peptides of Treble-Clef Zinc Fingers. Chemistry. 2009;15:4798–4810. doi: 10.1002/chem.200900147. [DOI] [PubMed] [Google Scholar]
- [3].Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- [4].Perry JJP, Shin DS, Getzoff ED, Tainer JA. The structural biochemistry of the superoxide dismutases. Biochim Biophys Acta. 2010;1804:245–262. doi: 10.1016/j.bbapap.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. Active sites of transition-metal enzymes with a focus on nickel. Curr Opin Struct Biol. 1998;8:749–758. doi: 10.1016/s0959-440x(98)80095-x. [DOI] [PubMed] [Google Scholar]
- [6].Arredondo M, Nunez MT. Iron and copper metabolism. Mol Aspects Med. 2005;26:313–327. doi: 10.1016/j.mam.2005.07.010. [DOI] [PubMed] [Google Scholar]
- [7].Singleton C, Le Brun N. Atx1-like chaperones and their cognate P-type ATPases: copper-binding and transfer. Biometals. 2007;20:275–289. doi: 10.1007/s10534-006-9068-1. [DOI] [PubMed] [Google Scholar]
- [8].Hartwig A. Zinc Finger Proteins as Potential Targets for Toxic Metal Ions: Differential Effects on Structure and Function. Antiox Redox Signal. 2001;3:625–634. doi: 10.1089/15230860152542970. [DOI] [PubMed] [Google Scholar]
- [9].Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- [10].Godwin HA. The biological chemistry of lead. Curr Opin Chem Biol. 2001;5:223–227. doi: 10.1016/s1367-5931(00)00194-0. [DOI] [PubMed] [Google Scholar]
- [11].Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- [12].Ma Z, Jacobsen FE, Giedroc DP. Coordination Chemistry of Bacterial Metal Transport and Sensing. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Giedroc DP, Arunkumar AI. Metal sensor proteins: nature’s metalloregulated allosteric switches. Dalton Trans. 2007;7:3107–3120. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- [14].Busenlehner LS, Pennella MA, Giedroc DP. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol Rev. 2003;27:131–143. doi: 10.1016/S0168-6445(03)00054-8. [DOI] [PubMed] [Google Scholar]
- [15].Reyes-Caballero H, Guerra AJ, Jacobsen FE, Kazmierczak KM, Cowart D, Koppolu UMK, Scott RA, Winkler ME, Giedroc DP. The Metalloregulatory Zinc Site in Streptococcus pneumoniae AdcR, a Zinc-activated MarR Family Repressor. J Mol Biol. 2010;403:197–216. doi: 10.1016/j.jmb.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen H, Wu R, Xu G, Fang X, Qiu X, Guo H, Tian B, Hua Y. DR2539 is a novel DtxR-like regulator of Mn/Fe ion homeostasis and antioxidant enzyme in Deinococcus radiodurans. Biochem Biophys Res Comm. 2010;396:413–418. doi: 10.1016/j.bbrc.2010.04.106. [DOI] [PubMed] [Google Scholar]
- [17].Ernst FD, Kuipers EJ, Heijens A, Sarwari R, Stoof J, Penn CW, Kusters JG, van Vliet AHM. The Nickel-Responsive Regulator NikR Controls Activation and Repression of Gene Transcription in Helicobacter pylori. Infect. Immun. 2005;73:7252–7258. doi: 10.1128/IAI.73.11.7252-7258.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Danielli A, Scarlato V. Regulatory circuits in Helicobacter pylori : network motifs and regulators involved in metal-dependent responses. FEMS Microbiol Rev. 2010;34:738–752. doi: 10.1111/j.1574-6976.2010.00233.x. [DOI] [PubMed] [Google Scholar]
- [19].Danielli A, Roncarati D, Delany I, Chiarini V, Rappuoli R, Scarlato V. In Vivo Dissection of the Helicobacter pylori Fur Regulatory Circuit by Genome-Wide Location Analysis. J Bacteriol. 2006;188:4654–4662. doi: 10.1128/JB.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ahn B-E, Cha J, Lee E-J, Han A-R, Thompson CJ, Roe J-H. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol Microbiol. 2006;59:1848–1858. doi: 10.1111/j.1365-2958.2006.05065.x. [DOI] [PubMed] [Google Scholar]
- [21].Wang S, Wu Y, Outten FW. Fur and the novel regulator YqjI control transcription of the ferric reductase gene yqjH in Escherichia coli. J Bacteriol. 2011;193:563–574. doi: 10.1128/JB.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Puri S, Hohle TH, O’Brian MR. Control of bacterial iron homeostasis by manganese. Proc Natl Acad Sci U S A. 2010;107:10691–10695. doi: 10.1073/pnas.1002342107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kloosterman TG, Witwicki R, Pol M, Bijlsma J, Kuipers E. Opposite effects of Mn2+ and Zn2+ on the PsaR-mediated expression of the virulence genes pcpA, prtA and psaB(CA) of Streptococcus pneumoniae. Mol Microbiol. 2008 doi: 10.1128/JB.00307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jacobsen FE, Kazmierczak KM, Lisher JP, Winkler ME, Giedroc DP. Interplay between manganese and zinc homeostasis in the human pathogen Streptococcus pneumoniae. Metallomics. 2011;3:38–41. doi: 10.1039/c0mt00050g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol Microbiolbiol. 2010;77:1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Festa RA, Jones MB, Butler-Wu S, Sinsimer D, Gerads R, Bishai WR, Peterson SN, Darwin KH. A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol Microbiolbiol. 2011;79:133–148. doi: 10.1111/j.1365-2958.2010.07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2011;108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Outten CE, O’Halloran TV. Femtomolar Sensitivity of Metalloregulatory Proteins Controlling Zinc Homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- [29].Cavet JS, Meng W, Pennella MA, Appelhoff RJ, Giedroc DP, Robinson NJ. A nickel-cobalt-sensing ArsR-SmtB family repressor. Contributions of cytosol and effector binding sites to metal selectivity. J Biol Chem. 2002;277:38441–38448. doi: 10.1074/jbc.M207677200. [DOI] [PubMed] [Google Scholar]
- [30].Chauhan S, Kumar A, Singhal A, Tyagi JS, Krishna Prasad H. CmtR, a cadmium-sensing ArsR–SmtB repressor, cooperatively interacts with multiple operator sites to autorepress its transcription in Mycobacterium tuberculosis. FEBS Journal. 2009;276:3428–3439. doi: 10.1111/j.1742-4658.2009.07066.x. [DOI] [PubMed] [Google Scholar]
- [31].O’Halloran TV, Frantz B, Shin MK, Ralston DM, Wright JG. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989;56:119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- [32].Outten CE, Outten FW, O’Halloran TV. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J Biol Chem. 1999;274:37517–37524. doi: 10.1074/jbc.274.53.37517. [DOI] [PubMed] [Google Scholar]
- [33].Lee PE, Demple B, Barton JK. DNA-mediated redox signaling for transcriptional activation of SoxR. Proc Natl Acad Sci U S A. 2009;106:13164–13168. doi: 10.1073/pnas.0906429106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jin-Won L, John DH. Functional specialization within the Fur family of metalloregulators. BioMetals. 2007 doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- [35].Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- [36].Dittmer PJ, Miranda JG, Gorski JA, Palmer AE. Genetically encoded sensors to elucidate spatial distribution of cellular zinc. J Biol Chem. 2009;284:16289–16297. doi: 10.1074/jbc.M900501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Irving H, Williams RJP. Order of Stability of Metal Complexes. Nature. 1948;162:746–747. [Google Scholar]
- [38].Panina EM, Mironov AA, Gelfand MS. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc Natl Acad Sci U S A. 2003;100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gabriel SE, Helmann JD. Contributions of Zur-Controlled Ribosomal Proteins to Growth under Zinc Starvation Conditions. J Bacteriol. 2009;191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dian C, Vitale S, Leonard GA, Bahlawane C, Fauquant C, Leduc D, Muller C, de Reuse H, Michaud-Soret I, Terradot L. The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Mol Microbiolbiol. 2011;79:1260–1275. doi: 10.1111/j.1365-2958.2010.07517.x. [DOI] [PubMed] [Google Scholar]
- [41].Robinson NJ, Winge DR. Copper Metallochaperones. Annual Review of Biochemistry. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dainty SJ, Patterson CJ, Waldron KJ, Robinson NJ. Interaction between cyanobacterial copper chaperone Atx1 and zinc homeostasis. J Biol Inorg Chem. 2009;15:77–85. doi: 10.1007/s00775-009-0555-z. [DOI] [PubMed] [Google Scholar]
- [43].Changela A, Chen K, Xue Y, Holschen J, Outten CE, O’Halloran TV, Mondragon A. Molecular Basis of Metal-Ion Selectivity and Zeptomolar Sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- [44].Ma Z, Cowart DM, Ward BP, Arnold RJ, DiMarchi RD, Zhang L, George GN, Scott RA, Giedroc DP. Unnatural Amino Acid Substitution as a Probe of the Allosteric Coupling Pathway in a Mycobacterial Cu(I) Sensor. J Am Chem Soc. 2009;131:18044–18045. doi: 10.1021/ja908372b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ma Z, Cowart DM, Scott RA, Giedroc DP. Molecular Insights into the Metal Selectivity of the Copper(I)-Sensing Repressor CsoR from Bacillus subtilis. Biochemistry. 2009;48:3325–3334. doi: 10.1021/bi900115w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Grossoehme N, Kehl-Fie TE, Ma Z, Adams KW, Cowart DM, Scott RA, Skaar EP, Giedroc DP. Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J Biol Chem. 2011 doi: 10.1074/jbc.M111.220012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mills SA, Marletta MA. Metal Binding Characteristics and Role of Iron Oxidation in the Ferric Uptake Regulator from Escherichia coli. Biochemistry. 2005;44:13553–13559. doi: 10.1021/bi0507579. [DOI] [PubMed] [Google Scholar]
- [48].Chivers PT, Sauer RT. NikR repressor: High-affinity nickel binding to the C-terminal domain regulates binding to operator DNA. Chemistry & Biology. 2002;9:1141–1148. doi: 10.1016/s1074-5521(02)00241-7. [DOI] [PubMed] [Google Scholar]
- [49].Schreiter ER, Sintchak MD, Guo Y, Chivers PT, Sauer RT, Drennan CL. Crystal structure of the nickel-responsive transcription factor NikR. Nat Struct Mol Biol. 2003;10:794–799. doi: 10.1038/nsb985. [DOI] [PubMed] [Google Scholar]
- [50].De Pina K, Desjardin V, Mandrand-Berthelot M-A, Giordano G, Wu L-F. Isolation and Characterization of the nikR Gene Encoding a Nickel-Responsive Regulator in Escherichia coli. J Bacteriol. 1999;181:670–674. doi: 10.1128/jb.181.2.670-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dosanjh NS, Michel SLJ. Microbial nickel metalloregulation: NikRs for nickel ions. Curr Opin Chem Biol. 2006;10:123–130. doi: 10.1016/j.cbpa.2006.02.011. [DOI] [PubMed] [Google Scholar]
- [52].Wang SC, Dias AV, Bloom SL, Zamble DB. Selectivity of metal binding and metal-induced stability of Escherichia coli NikR. Biochemistry. 2004;43:10018–10028. doi: 10.1021/bi049405c. [DOI] [PubMed] [Google Scholar]
- [53].Iwig JS, Rowe JL, Chivers PT. Nickel homeostasis in Escherichia coli - the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol Microbiol. 2006;62:252–262. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- [54].Iwig JS, Leitch S, Herbst RW, Maroney MJ, Chivers PT. Ni(II) and Co(II) Sensing by Escherichia coli RcnR. J Am Chem Soc. 2008;130:7592–7606. doi: 10.1021/ja710067d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wang SC, Dias AV, Zamble DB. The “metallo-specific” response of proteins: a perspective based on the Escherichia coli transcriptional regulator NikR. Dalton Trans. 2009:2459–2466. doi: 10.1039/b818167p. [DOI] [PubMed] [Google Scholar]
- [56].Wang SC, Li Y, Ho M, Bernal M-E, Sydor AM, Kagzi WR, Zamble DB. The Response of Escherichia coli NikR to Nickel: A Second Nickel-Binding Site. Biochemistry. 2010;49:6635–6645. doi: 10.1021/bi100685k. [DOI] [PubMed] [Google Scholar]
- [57].Osman D, Cavet JS. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat Prod Rep. 2010;27:668–680. doi: 10.1039/b906682a. [DOI] [PubMed] [Google Scholar]
- [58].Campbell DR, Chapman KE, Waldron KJ, Tottey S, Kendall S, Cavallaro G, Andreini C, Hinds J, Stoker NG, Robinson NJ, Cavet JS. Mycobacterial cells have dual nickel-cobalt sensors: sequence relationships and metal sites of metal-responsive repressors are not congruent. J Biol Chem. 2007;282:32298–32310. doi: 10.1074/jbc.M703451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shin JH, Oh SY, Kim SJ, Roe JH. The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2) J Bacteriol. 2007;189:4070–4077. doi: 10.1128/JB.01851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Shin JH, Jung HJ, An YJ, Cho YB, Cha SS, Roe JH. Graded expression of zinc-responsive genes through two regulatory binding sites in Zur. Proc Natl Acad Sci U S A. 2011;108 doi: 10.1073/pnas.1017744108. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol. 2006;8:51–62. [PubMed] [Google Scholar]
- [62].Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJE, Kuipers OP. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol Microbiolbiol. 2007;65:1049–1063. doi: 10.1111/j.1365-2958.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- [63].Pennella MA, Arunkumar AI, Giedroc DP. Individual Metal Ligands Play Distinct Functional Roles in the Zinc Sensor Staphylococcus aureus CzrA. J Mol Biol. 2006;356:1124–1136. doi: 10.1016/j.jmb.2005.12.019. [DOI] [PubMed] [Google Scholar]
- [64].Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. Metal binding studies and EPR spectroscopy of the manganese transport regulator MntR. Biochemistry. 2006;45:15359–15372. doi: 10.1021/bi0607406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Schmitt MP. Analysis of a DtxR-like metalloregulatory protein, MntR, from Corynebacterium diphtheriae that controls expression of an ABC metal transporter by an Mn2+-dependent mechanism. J Bacteriol. 2002;184:6882–6892. doi: 10.1128/JB.184.24.6882-6892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Que Q, Helmann JD. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol. 2000;35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- [67].Glasfeld A, Guedon E, Helmann JD, Brennan RG. Structure of the manganese-bound manganese transport regulator of Bacillus subtilis. Nat Struct Biol. 2003;10:652–657. doi: 10.1038/nsb951. [DOI] [PubMed] [Google Scholar]
- [68].Lieser SA, Davis TC, Helmann JD, Cohen SM. DNA-binding and oligomerization studies of the manganese(II) metalloregulatory protein MntR from Bacillus subtilis. Biochemistry. 2003;42:12634–12642. doi: 10.1021/bi0350248. [DOI] [PubMed] [Google Scholar]
- [69].Golynskiy MV, Davis TC, Helmann JD, Cohen SM. Metal-induced structural organization and stabilization of the metalloregulatory protein MntR. Biochemistry. 2005;44:3380–3389. doi: 10.1021/bi0480741. [DOI] [PubMed] [Google Scholar]
- [70].Stoll KE, Draper WE, Kliegman JI, Golynskiy MV, Brew-Appiah RAT, Phillips RK, Brown HK, Breyer WA, Jakubovics NS, Jenkinson HF, Brennan RG, Cohen SM, Glasfeld A. Characterization and Structure of the Manganese-Responsive Transcriptional Regulator ScaR. Biochemistry. 2009;48:10308–10320. doi: 10.1021/bi900980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Guedon E, Helmann JD. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol Microbiol. 2003;48:495–506. doi: 10.1046/j.1365-2958.2003.03445.x. [DOI] [PubMed] [Google Scholar]
- [72].Pearson RG. Hard and soft acids and bases, HSAB, part 1: Fundamental principles. J Chem Educ. 1968;45:581. [Google Scholar]
- [73].Kliegman JI, Griner SL, Helmann JD, Brennan RG, Glasfeld A. Structural basis for the metal-selective activation of the manganese transport regulator of Bacillus subtilis. Biochemistry. 2006;45:3493–3505. doi: 10.1021/bi0524215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Grossoehme NE, Giedroc DP. Energetics of Allosteric Negative Coupling in the Zinc Sensor S. aureus CzrA. J Am Chem Soc. 2009;131:17860–17870. doi: 10.1021/ja906131b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Pennella MA, Shokes JE, Cosper NJ, Scott RA, Giedroc DP. Structural elements of metal selectivity in metal sensor proteins. Proc Natl Acad Sci U S A. 2003;100:3713–3718. doi: 10.1073/pnas.0636943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].VanZile ML, Cosper NJ, Scott RA, Giedroc DP. The zinc metalloregulatory protein Synechococcus PCC7942 SmtB binds a single zinc ion per monomer with high affinity in a tetrahedral coordination geometry. Biochemistry. 2000;39:11818–11829. doi: 10.1021/bi001140o. [DOI] [PubMed] [Google Scholar]
- [77].Maret W, Li Y. Coordination Dynamics of Zinc in Proteins. Chem Rev. 2009;109:4682–4707. doi: 10.1021/cr800556u. [DOI] [PubMed] [Google Scholar]
- [78].Patel K, Kumar A, Durani S. Analysis of the structural consensus of the zinc coordination centers of metalloprotein structures. Biochim Biophys Acta. 2007;1774:1247–1253. doi: 10.1016/j.bbapap.2007.07.010. [DOI] [PubMed] [Google Scholar]
- [79].Dudev T, Lim C. Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem Rev. 2003;103:773–787. doi: 10.1021/cr020467n. [DOI] [PubMed] [Google Scholar]
- [80].Kuppuraj G, Dudev M, Lim C. Factors Governing Metal-Ligand Distances and Coordination Geometries of Metal Complexes. J Phys Chem B. 2009;113:2952–2960. doi: 10.1021/jp807972e. [DOI] [PubMed] [Google Scholar]
- [81].Liu T, Ramesh A, Ma Z, Ward SK, Zhang LM, George GN, Talaat AM, Sacchettini JC, Giedroc DP. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- [82].Chivers PT. Coordinating intracellular nickel-metal-site structure-function relationships and the NikR and RcnR repressors. Nat Prod Rep. 2010;27:658–667. doi: 10.1039/b906683g. [DOI] [PubMed] [Google Scholar]
- [83].Leitch S, Bradley MJ, Rowe JL, Chivers PT, Maroney MJ. Nickel-Specific Response in the Transcriptional Regulator, Escherichia coli NikR. J Am Chem Soc. 2007;129:5085–5095. doi: 10.1021/ja068505y. [DOI] [PubMed] [Google Scholar]
- [84].Bloom SL, Zamble DB. Metal-Selective DNA-Binding Response of Escherichia coli NikR. Biochemistry. 2004;43:10029–10038. doi: 10.1021/bi049404k. [DOI] [PubMed] [Google Scholar]
- [85].Chen K, Yuldasheva S, Penner-Hahn JE, O’Halloran TV. An Atypical Linear Cu(I)-S2 Center Constitutes the High-Affinity Metal-Sensing Site in the CueR Metalloregulatory Protein. J Am Chem Soc. 2003;125:12088–12089. doi: 10.1021/ja036070y. [DOI] [PubMed] [Google Scholar]
- [86].An YJ, Ahn BE, Han AR, Kim HM, Chung KM, Shin JH, Cho YB, Roe JH, Cha SS. Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucl Acids Res. 2009;37:3442–3451. doi: 10.1093/nar/gkp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ahmad R, Brandsdal BO, Michaud-Soret I, Willassen NP. Ferric uptake regulator protein: Binding free energy calculations and per-residue free energy decomposition. Proteins. 2009;75:373–386. doi: 10.1002/prot.22247. [DOI] [PubMed] [Google Scholar]
- [88].Lucarelli D, Russo S, Garman E, Milano A, Meyer-Klaucke W, Pohl E. Crystal Structure and Function of the Zinc Uptake Regulator FurB from Mycobacterium tuberculosis. J Biol Chem. 2007;282:9914–9922. doi: 10.1074/jbc.M609974200. [DOI] [PubMed] [Google Scholar]
- [89].Arunkumar AI, Campanello GC, Giedroc DP. Solution structure of a paradigm ArsR family zinc sensor in the DNA-bound state. Proc Natl Acad Sci U S A. 2009;106:18177–18182. doi: 10.1073/pnas.0905558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Schreiter ER, Wang SC, Zamble DB, Drennan CL. NikR-operator complex structure and the mechanism of repressor activation by metal ions. Proc Natl Acad Sci U S A. 2006;103:13676–13681. doi: 10.1073/pnas.0606247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol. 2003;333:683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- [92].Arunkumar AI, Pennella MA, Kong XM, Giedroc DP. Resonance assignments of the metal sensor CzrA in the apo-, Zn2- and DNA-bound (42 kDa) states. Biomol NMR Assign. 2007;1:99–101. doi: 10.1007/s12104-007-9027-y. [DOI] [PubMed] [Google Scholar]
- [93].Chakrabarti P. Geometry of interaction of metal-ions with histidine-residues in protein structures. Protein Engin. 1990;4:57–63. doi: 10.1093/protein/4.1.57. [DOI] [PubMed] [Google Scholar]
- [94].Alberts IL, Nadassy K, Wodak SJ. Analysis of zinc binding sites in protein crystal structures. Protein Science. 1998;7:1700–1716. doi: 10.1002/pro.5560070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Jacquamet L, Traore DAK, Ferrer JL, Proux O, Testemale D, Hazemann JL, Nazarenko E, El Ghazouani A, Caux-Thang C, Duarte V, Latour JM. Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol Microbiol. 2009;73:20–31. doi: 10.1111/j.1365-2958.2009.06753.x. [DOI] [PubMed] [Google Scholar]
- [96].Lee J-W, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- [97].Traoré DAK, El Ghazouani A, Ilango S, Dupuy J, Jacquamet L, Ferrer JL, Caux-Thang C, Duarte V, Latour JM. Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol Microbiol. 2006;61:1211–1219. doi: 10.1111/j.1365-2958.2006.05313.x. [DOI] [PubMed] [Google Scholar]
- [98].Traoré DAK, Ghazouani AE, Jacquamet L, Borel F, Ferrer J-L, Lascoux D, Ravanat J-L, Jaquinod M, Blondin G, Caux-Thang C, Duarte V, Latour J-M. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat Chem Biol. 2009;5:53–59. doi: 10.1038/nchembio.133. [DOI] [PubMed] [Google Scholar]
- [99].Won YB, Ji CJ, Cho JH, Lee JW. Mutational Analysis of the Metal-binding Sites of Peroxide Sensor PerR. Bull Kor Chem Soc. 2010;31:1573–1576. [Google Scholar]
- [100].Giedroc DP. Hydrogen peroxide sensing in Bacillus subtilis : it is all about the (metallo)regulator. Mol Microbiol. 2009;73:1–4. doi: 10.1111/j.1365-2958.2009.06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- [102].Lucarelli D, Vasil ML, Meyer-Klaucke W, Pohl E. The metal-dependent regulators FurA and FurB from Mycobacterium tuberculosis. Internat J Mol Sci. 2008;9:1548–1560. doi: 10.3390/ijms9081548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol. 2003;47:903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- [104].Sheikh A, Taylor GL. Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol Microbiol. 2009;72:1208–1220. doi: 10.1111/j.1365-2958.2009.06718.x. [DOI] [PubMed] [Google Scholar]
- [105].Changela A, Chen K, Xue Y, Holschen J, Outten C, O’Halloran T, Mondragon A. Molecular Basis of Metal-Ion Selectivity and Zeptomolar Sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- [106].Kern D, Zuiderweg ER. The role of dynamics in allosteric regulation. Curr Opin Struct Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- [107].Popovych N, Sun S, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nat Struct Mol Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Tsai C-J, del Sol A, Nussinov R. Allostery: Absence of a Change in Shape Does Not Imply that Allostery Is Not at Play. J Mol Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Zotin AI. Thermodynamic aspects of developmental biology. Mono Dev Biol. 1972;5:i–viii. 1–159. [PubMed] [Google Scholar]
- [110].Reinhart GD. Quantitative analysis and interpretation of allosteric behavior. Methods Enzymol. 2004;380:187–203. doi: 10.1016/S0076-6879(04)80009-0. [DOI] [PubMed] [Google Scholar]
- [111].Lee S, Arunkumar AI, Chen X, Giedroc DP. Structural Insights into Homo- and Heterotropic Allosteric Coupling in the Zinc Sensor S. aureus CzrA from Covalently Fused Dimers. J Am Chem Soc. 2006;128:1937–1947. doi: 10.1021/ja0546828. [DOI] [PubMed] [Google Scholar]
- [112].Fenton AW. Allostery: an illustrated definition for the ‘second secret of life’. Trends Biochem Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci U S A. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sen KI, Logan TM, Fajer PG. Protein dynamics and monomer-monomer interactions in AntR activation by electron paramagnetic resonance and double electron-electron resonance. Biochemistry. 2007;46:11639–11649. doi: 10.1021/bi700859p. [DOI] [PubMed] [Google Scholar]
- [115].Golynskiy M, Li S, Woods VL, Jr., Cohen SM. Conformational studies of the manganese transport regulator (MntR) from Bacillus subtilis using deuterium exchange mass spectrometry. J Biol Inorg Chem. 2007;12:699–709. doi: 10.1007/s00775-007-0216-z. [DOI] [PubMed] [Google Scholar]
- [116].Delany I, Spohn G, Rappuoli R, Scarlato V. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol Microbiolbiol. 2001;42:1297–1309. doi: 10.1046/j.1365-2958.2001.02696.x. [DOI] [PubMed] [Google Scholar]
- [117].Delany I, Pacheco ABF, Spohn G, Rappuoli R, Scarlato V. Iron-Dependent Transcription of the frpB Gene of Helicobacter pylori Is Controlled by the Fur Repressor Protein. J Bacteriol. 2001;183:4932–4937. doi: 10.1128/JB.183.16.4932-4937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Harris AG, Hinds FE, Beckhouse AG, Kolesnikow T, Hazell SL. Resistance to hydrogen peroxide in Helicobacter pylori: role of catalase (KatA) and Fur, and functional analysis of a novel gene product designated ‘KatA-associated protein’, KapA (HP0874) Microbiology. 2002;148:3813–3825. doi: 10.1099/00221287-148-12-3813. [DOI] [PubMed] [Google Scholar]
- [119].Ernst FD, Bereswill S, Waidner B, Stoof J, Mader U, Kusters JG, Kuipers EJ, Kist M, van Vliet AHM, Homuth G. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology. 2005;151:533–546. doi: 10.1099/mic.0.27404-0. [DOI] [PubMed] [Google Scholar]
- [120].Ernst FD, Homuth G, Stoof J, Mader U, Waidner B, Kuipers EJ, Kist M, Kusters JG, Bereswill S, van Vliet AHM. Iron-Responsive Regulation of the Helicobacter pylori Iron-Cofactored Superoxide Dismutase SodB Is Mediated by Fur. J. Bacteriol. 2005;187:3687–3692. doi: 10.1128/JB.187.11.3687-3692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Vitale S, Fauquant C, Lascoux D, Schauer K, Saint-Pierre C, Michaud-Soret I. A ZnS4 Structural Zinc Site in the Helicobacter pylori Ferric Uptake Regulator. Biochemistry. 2009;48:5582–5591. doi: 10.1021/bi9004396. [DOI] [PubMed] [Google Scholar]
- [122].Carpenter BM, Gancz H, Benoit SL, Evans S, Olsen CH, Michel SLJ, Maier RJ, Merrell DS. Mutagenesis of Conserved Amino Acids of Helicobacter pylori Fur Reveals Residues Important for Function. J. Bacteriol. 2010;192:5037–5052. doi: 10.1128/JB.00198-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Tao X, Zeng HY, Murphy JR. Transition metal ion activation of DNA binding by the diphtheria tox repressor requires the formation of stable homodimers. Proc Natl Acad Sci U S A. 1995;92:6803–6807. doi: 10.1073/pnas.92.15.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Spiering MM, Ringe D, Murphy JR, Marletta MA. Metal stoichiometry and functional studies of the diphtheria toxin repressor. Proc Natl Acad Sci U S A. 2003;100:3808–3813. doi: 10.1073/pnas.0737977100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Love JF, vanderSpek JC, Marin V, Guerrero L, Logan TM, Murphy JR. Genetic and biophysical studies of diphtheria toxin repressor (DtxR) and the hyperactive mutant DtxR(E175K) support a multistep model of activation. Proc Natl Acad Sci U S A. 2004;101:2506–2511. doi: 10.1073/pnas.0303794101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].D’Aquino JA, Tetenbaum-Novatt J, White A, Berkovitch F, Ringe D. Mechanism of metal ion activation of the diphtheria toxin repressor DtxR. Proc Natl Acad Sci U S A. 2005;102:18408–18413. doi: 10.1073/pnas.0500908102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].VanZile ML, Chen X, Giedroc DP. Structural characterization of distinct α3N and α5 metal sites in the cyanobacterial zinc sensor SmtB. Biochemistry. 2002;41:9765–9775. doi: 10.1021/bi0201771. [DOI] [PubMed] [Google Scholar]
- [128].Kandegedara A, Thiyagarajan S, Kondapalli KC, Stemmler TL, Rosen BP. Role of Bound Zn(II) in the CadC Cd(II)/Pb(II)/Zn(II)-responsive Repressor. J Biol Chem. 2009;284:14958–14965. doi: 10.1074/jbc.M809179200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Busenlehner LS, Weng TC, Penner-Hahn JE, Giedroc DP. Elucidation of primary (α3N) and vestigial (α5) heavy metal-binding sites in Staphylococcus aureus pI258 CadC: Evolutionary implications for metal ion selectivity of ArsR/SmtB metal sensor proteins. J Mol Biol. 2002;319:685–701. doi: 10.1016/S0022-2836(02)00299-1. [DOI] [PubMed] [Google Scholar]
- [130].Phillips CM, Schreiter ER, Guo Y, Wang SC, Zamble DB, Drennan CL. Structural basis of the metal specificity for nickel regulatory protein NikR. Biochemistry. 2008;47:1938–1946. doi: 10.1021/bi702006h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].DeWitt MA, Kliegman JI, Helmann JD, Brennan RG, Farrens DL, Glasfeld A. The conformations of the manganese transport regulator of Bacillus subtilis in its metal-free state. J Mol Biol. 2007;365:1257–1265. doi: 10.1016/j.jmb.2006.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Tanaka T, Shinkai A, Bessho Y, Kumarevel T, Yokoyama S. Crystal structure of the manganese transport regulatory protein from Escherichia coli. Proteins. 2009;77:741–746. doi: 10.1002/prot.22541. [DOI] [PubMed] [Google Scholar]
- [133].Wisedchaisri G, Chou CJ, Wu MT, Roach C, Rice AE, Holmes RK, Beeson C, Hol WGJ. Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis. Biochemistry. 2007;46:436–447. doi: 10.1021/bi0609826. [DOI] [PubMed] [Google Scholar]
- [134].Sen KI, Sienkiewicz A, Love JF, vanderSpek JC, Fajer PG, Logan TM. Mn(II) binding by the anthracis repressor from Bacillus anthracis. Biochemistry. 2006;45:4295–4303. doi: 10.1021/bi052288g. [DOI] [PubMed] [Google Scholar]
- [135].Liu T, Golden JW, Giedroc DP. A zinc(II)/lead(II)/cadmium(II)-inducible operon from the Cyanobacterium anabaena is regulated by AztR, an α3N ArsR/SmtB metalloregulator. Biochemistry. 2005;44:8673–8683. doi: 10.1021/bi050450+. [DOI] [PubMed] [Google Scholar]
- [136].Liu T, Chen XH, Ma Z, Shokes J, Hemmingsen L, Scott RA, Giedroc DP. A Cu-I-sensing ArsR family metal sensor protein with a relaxed metal selectivity profile. Biochemistry. 2008;47:10564–10575. doi: 10.1021/bi801313y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Jian X, Wasinger EC, Lockard JV, Chen LX, He C. Highly Sensitive and Selective Gold(I) Recognition by a Metalloregulator in Ralstonia metallidurans. J Am Chem Soc. 2009;131:10869–10871. doi: 10.1021/ja904279n. [DOI] [PubMed] [Google Scholar]
- [138].Li YJ, Zamble DB. pH-Responsive DNA-Binding Activity of Helicobacter pylori NikR. Biochemistry. 2009;48:2486–2496. doi: 10.1021/bi801742r. [DOI] [PubMed] [Google Scholar]
- [139].Rowe JL, Starnes GL, Chivers PT. Complex Transcriptional Control Links NikABCDE-Dependent Nickel Transport with Hydrogenase Expression in Escherichia coli. J Bacteriol. 2005;187:6317–6323. doi: 10.1128/JB.187.18.6317-6323.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].DeLano WL. The PyMol molecular graphics system, Version 1.2r3pre. Schrödinger; LLC, San Carlos: 2004. [Google Scholar]