Abstract
Background
Neuropathological damage due to chronic alcohol abuse often results in impairment of cognitive function. The damage is particularly marked in the frontal cortex. The 14-3-3 protein family consists of 7 proteins, β, γ, ε, ζ, η, θ and σ, encoded by 7 distinct genes. They are highly conserved molecular chaperones with roles in regulation of metabolism, signal transduction, cell-cycle control, protein trafficking, and apoptosis. They may also play an important role in neurodegeneration in chronic alcoholism.
Methods
We used Real-Time PCR to measure the expression of 14-3-3 mRNA transcripts in both the dorsolateral prefrontal cortex and motor cortex of human brains obtained at autopsy.
Results
We found significantly lower 14-3-3β, γ and θ expression in both cortical areas of alcoholics; but no difference in 14-3-3η expression, and higher expression of 14-3-3σ, in both areas. Levels of 14-3-3ζ and ε transcripts were significantly lower only in alcoholic motor cortex.
Conclusions
Altered 14-3-3 expression could contribute to synaptic dysfunction and altered neurotransmission in chronic alcohol misuse by human subjects.
Keywords: Alcohol abuse, gene expression, ethanol, Real-time PCR
Introduction
Chronic and excessive alcohol consumption has deleterious effects on the CNS, including damage to specific regions of the cerebral cortex (Kril et al., 1997). Chronic alcohol misuse can lead to impaired cognitive functions, including reduced ability to solve abstract problems, poorer memory, flawed fine motor skills, and reduced overall motor function (Harper and Matsumoto, 2005). Impairment of cognitive function correlates directly with the pattern of structural changes that occur in the brain, which includes reduced brain weight, reduction in white matter, and loss of neurons in specific cortical regions, primarily the dorsolateral prefrontal association cortex (prefrontal cortex, PFC; reviewed in Harper, 2009). Other changes include lower neuronal density (Kril et al., 1997), altered dendritic arbour and synaptic connectivity, and alterations to neurotransmitter systems (reviewed in Harper and Matsumoto, 2005). Comorbid diseases such as cirrhosis of the liver and Wernicke-Korsakoff Syndrome (WKS) exacerbate the neuropathological effects of chronic alcohol abuse (Kril et al., 1997).
The acute effects of alcohol are mediated by direct interaction with neurotransmitter and signalling systems, resulting in changes in the number and type of receptors, and in the amount of neurotransmitter released into the synapse. Continuous exposure to alcohol results in changes in neuronal function which are mediated, in part, by changes in gene expression (Nestler and Aghajanian, 1997) which are more extensive in the prefrontal cortex, emphasizing the importance of this region in compulsive alcohol misuse (Liu et al., 2004; 2006). Long-term exposure to cocaine and heroin results in patterns of brain plasticity similar to those induced by alcohol (Koob, 2004; Pignataro et al., 2009). It is likely that the remodelling of synaptic connections is dependent on changes in gene expression, which is responsible for these alterations in brain plasticity (reviewed in Pignataro et al., 2009).
cDNA Microarray and global proteomics studies have been used to identify genes and proteins that are differentially expressed in response to chronic alcohol misuse. Differentially expressed gene families include those involved in myelination, protein trafficking, ubiquitination, mitochondrial function, cell adhesion, neurogenesis, apoptosis, signal transduction and transcriptional control of gene expression (see Flatscher-Bader et al., 2005; Lewohl et al., 2000; Liu et al., 2004; 2006; Mayfield et al., 2002; Pignataro et al., 2009; Sokolov et al., 2003). These gene expression changes are consistent with theories of extensive alcohol-induced adaption of the CNS (reviewed in Crews et al., 2005; Lewohl et al., 2000; Liu et al., 2004; Pignataro et al., 2009; Sokolov et al., 2003).
The tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation proteins are a family of molecular chaperones commonly referred to as 14-3-3 proteins. The family consists of seven transcripts in mammals: 14-3-3β (YWHAB), 14-3-3γ (YWHAG), 14-3-3ε (YWHAE), 14-3-3ζ (YWHAZ), 14-3-3η (YWHAH), 14-3-3θ (YWHAQ) and 14-3-3σ also known as stratifin (SFN) (Berg et al., 2003; Fu et al., 2000; Wilker and Yaffe, 2004). Each protein is encoded by a distinct gene. The 14-3-3 family comprises highly conserved proteins with varying roles in regulation of metabolism, signal transduction, cell-cycle control, protein trafficking, transcription, stress response, apoptosis (Aitken, 2006; Dougherty and Morrison, 2004; Wilker and Yaffe, 2004) and dopamine synthesis (Wang et al., 2009). They are predominantly found in the brain and make up approximately 1% of total soluble brain protein (Fu et al., 2000; Ubl et al., 2002).
Previous studies have shown that members of the 14-3-3 family are differentially expressed in the brain of human alcoholics. However, the majority of these studies have used hybridization techniques (microarrays) or sequencing of protein fragments following two-dimensional gel electrophoresis, neither of which is ideally suited to measuring the isoforms of gene families with high degrees of sequence similarity. In a study using both cDNA and olignucleotide arrays, a 14-3-3 transcript with sequence homology to 14-3-3θ was increased in the prefrontal cortex of alcoholics when assayed by oligonucleotide array, but not by cDNA array (Lewohl et al., 2000). In a separate microarray study, 14-3-3η was reportedly upregulated in the nucleus accumbens of alcoholics (Flatscher-Bader et al., 2005). Global proteomics studies showed that four of the seven 14-3-3 proteins (14-3-3ε, ζ, η and γ) were down-regulated in the prefrontal cortex of alcoholics (Lewohl et al., 2004).
Because of the role of the 14-3-3 proteins in mediating cell damage and neurodegeneration, and the discrepancies between published studies, we measured the expression of all seven transcripts in the cerebral cortex of long-term alcohol misusers and healthy controls using Real-Time PCR. Expression was compared between the prefrontal cortex, a region of the brain that is particularly susceptible to the neuropathological effects of alcohol, and the primary motor cortex, which is relatively spared.
Methods
Tissue collection
Human autopsy brain tissue was collected by the Queensland Brain Bank, School of Chemistry and Molecular Biosciences, University of Queensland, and the NSW Tissue Resource Centre, University of Sydney, both of which are supported by the National Health and Medical Research Council (NH&MRC). Brain samples were collected by qualified pathologists under full ethical clearance and informed consent from the next of kin.
The cerebellum and brainstem were detached from the cerebrum by sectioning through the upper midbrain at the level of the superior colliculus. The cerebrum was hemisected, with one half being fixed in formalin for macro- and microscopic pathological examination. Cortical and sub-cortical pieces were dissected from the other hemisphere, immersed in ~10 vol of 0·32 M sucrose, and slowly frozen as previously described for optimal preservation (Dodd et al., 1986). Previous studies have shown that brain RNA is stable for up to 48 hours post mortem (Hynd et al., 2003) and can be used for PCR-based analysis (Lewohl et al., 1997). Frozen tissue pieces were stored at −80°C until required. Two regions from each brain were selected for analysis: dorsolateral prefrontal cortex (Brodmann areas 6 & 8) and the primary motor cortex (area 4).
Case Selection
Alcoholic and control groups were selected on the basis of alcohol consumption. Controls were defined as having consumed less than 20 g of ethanol per day, while alcoholics had consumed more than 80 g of ethanol per day, for most of their adult lives. Many alcoholics used in the study had consumed far greater daily quantities of alcohol than this, and had been drinking for more than 20 years. Alcoholics were further subdivided on the basis of comorbid disease. Uncomplicated alcoholics were differentiated from cirrhotic alcoholics on the basis of pathologically confirmed cirrhosis of the liver. Neither controls, uncomplicated alcoholics nor cirrhotic alcoholics had any other known disease complication at time of death. They were not drug users and had no psychiatric disorders requiring medication. Cases suffering other neurological diseases, such as Wernicke-Korsakoff syndrome or hepatic encephalopathy, were excluded.
RNA Isolation
Total RNA was extracted from the prefrontal cortex and primary motor cortex using TRIzol™ Reagent (Invitrogen, Mt Waverly, Vic, Australia) according to the manufacturer’s protocol. The frozen brain pieces were homogenized in 10 vol TRIzol™ Reagent using a polytron (Kinematika, Switzerland). Following ethanol precipitation the pellet was resuspended in DEPC-treated water. RNA quantity was measured by absorbance at 260 nm using a Nanodrop (Thermo Scientific, Waltham, MA, USA). Total RNA was stored in aliquots of 20 μL at −70°C until required.
Reverse transcription
Reverse transcription of samples was performed using an Oligo(dT)20mer. A mix consisting of 3.8 μM oligo(dT)20 (Sigma-Aldrich, Castle Hill, NSW, Australia), 2 μg Total RNA, 0.77 mM dNTPs (Promega, Annandale, NSW, Australia) in a final volume of 13 μL with DEPC-treated water was incubated at 65°C for 5 min and placed on ice for 1 min. 4 μL of 5x first-strand synthesis buffer, 50 mM dithiothreitol, 40 U RNaseOUT™ and 200 U Superscript™ III (Invitrogen) were added to the mix containing 2 μg RNA and incubated at 50°C for 1 h. The reaction was inactivated by incubation at 70°C for 15 min. Sample aliquots were stored at −70°C until required.
Primer Design
Primers were designed using Primer Express v1.5 Software (Applied Biosystems, Scoresby, Vic, Australia) and synthesised by Sigma-Aldrich. Primer specificity was verified using BLAST from the GenBank non-redundant nucleotide sequence database (NCBI; Altschul et al., 1997). Primer pairs were designed such that one of the pair spanned an exon boundary to eliminate amplification of gDNA. Primer sequences can be found in Table 1.
Table 1.
Real-time PCR Primers
| Transcript | Primer Sequence | GenBank Accession Number |
Location |
|---|---|---|---|
| GAPDH | NM_002046 | ||
| Forward | 5′ – TGC ACC ACC AAC TGC TTA GC – 3′ | 529 | |
| Reverse | 5′ – GGC ATG GAC TGT GGT CAT GAG – 3′ | 595 | |
| 14-3-3β | NM_003404.3 | ||
| Forward | 5′ – GGC AAA GAG TAC CGT GAG AAG -3′ | 523 | |
| Reverse | 5′ – CTG GTT GTG TAG CAT TGG GAA TA -3′ | 625 | |
| 14-3-3γ | NM_012479.2 | ||
| Forward | 5′ – AGC CAC TGT CGA ATG AGG AAC – 3′ | 295 | |
| Reverse | 5′ – CTT CTG CTC AAT GCT ACT GAT GA – 3′ | 398 | |
| 14-3-3ε | NM_006761.3 | ||
| Forward | 5′ – AAT GAT TCG GGA ATA TCG GCA AA – 3′ | 353 | |
| Reverse | 5′ – ACT CGC CAG TGT TAG CTG C – 3′ | 462 | |
| 14-3-3ζ | NM_003406.2 | ||
| Forward | 5′ – CCT GCA TGA AGT CTG TAA CTG AG – 3′ | 155 | |
| Reverse | 5′ – GAC CTA CGG GCT CCT ACA ACA – 3′ | 254 | |
| 14-3-3η | NM_003405.3 | ||
| Forward | 5′ – CGA CCA CAC TCT TTT TCT TCT CC – 3′ | 683 | |
| Reverse | 5′ – TTC CAG TAT GAG AGC AAG GT – 3′ | 584 | |
| 14-3-3θ | NM_006826.2 | ||
| Forward | 5′ – AGC CAA TGC AAC TAA TCC AG – 3′ | 396 | |
| Reverse | 5′ – GTT TGT TTT CGA TCA TCA CCA – 3′ | 541 | |
| 14-3-3σ | NM_006142.3 | ||
| Forward | 5′ – AAG ATG AAG GGT GAC TAC TA – 3′ | 435 | |
| Reverse | 5′ – GAC CGG GCT GAG TCA ATG A – 3′ | 517 |
Real-Time PCR
Real-Time PCR was conducted using the Corbett RotorGene 3000 with SYBR® Green as the detection dye. Each PCR reaction on the 72 well rotor consisted of 5 ng of cDNA, 12.5 μL of SYBR Green PCR Master Mix (Applied Biosystems), 300 nM of each of the pair of oligonucleotide primers in a final reaction volume of 25 μL per well. No-template samples were included with each primer set to act as a negative control, while Universal Human Reference RNA (UHRR; Stratagene, La Jolla, CA) cDNA was used as a positive control to confirm amplification in each experiment for each primer pair. The endogenous control used for determining ΔCT values was glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Duplicate amplifications were conducted for each sample.
Threshold (T) was set above baseline in the exponential phase of the reaction using the plot of fluorescence vs. cycle number. To permit comparisons between experiments this threshold value was kept constant across experimental runs. Cycle threshold (CT) is defined as the number of cycles required for the fluorescence of an amplifying PCR product to cross the fixed threshold line. Assay variability (both intra- and inter-assay variability) was tested using cDNA serial dilutions generated from UHRR for each primer pair.
Differences between mean CT values of the duplicates (of each transcript) compared with the endogenous control gene were calculated using Microsoft Excel to give a ΔCT value. ΔCT values were used for statistical analysis and the means converted to 2−ΔCT for presentation as outlined in Livak and Schmittgen (2001). Relative expression was graphed using the means converted to 2−ΔCT for presentation using GraphPad version 5 (GraphPad Software Inc, La Jolla, CA, USA).
Statistical Analysis
Statistical Analysis was conducted using SPSS v16 (SPSS Inc, Chicago, IL, USA). Analysis of Variance (ANOVA) was followed by the Tukey hsd post hoc test where appropriate. The effects of age at death, post-mortem interval (PMI), brain weight (BWT) and gender were determined. Differences between the slopes of the regression lines were also tested. When regression was significant, covariance analysis (ANCOVA) was used to normalize the data and to produce adjusted least-squares mean and S.E.M. values.
Results
Case information is outlined in Table 2. Alcoholics and controls were matched as closely as possible for PMI and age at death. There was no significant difference in age between groups: controls 61 ± 3 y, uncomplicated alcoholics 52 ± 3 y, cirrhotic alcoholics 60 ± 4 y. There was no significant difference between groups for PMI: controls 30.6 ± 5.2 h, uncomplicated alcoholics 29.4 ± 3.3 h, cirrhotic alcoholics 24.2 ± 5.6 h. Mean brain weights were slightly lower in cirrhotic alcoholics (1249 ± 34 g) than in controls (1337 ± 25 g) and uncomplicated alcoholics (1334 ± 27 g) but this difference was not significant. However, females had significantly lower brain weight than their male counterparts, both overall and within each group. There were no significant differences across groups for either males or females in age, PMI or BWT.
Table 2.
Details of subjects
| Group | N | Age (y) | PMI (h) | BWT (g) (males) |
BWT (g) (females) |
|---|---|---|---|---|---|
| Control (11 female, 12 male) |
23 | 61.14 ± 2.86 |
30.64 ± 5.23 | 1394.08 ± 30.42 |
1265.64 ± 26.94 |
| Alcoholic (10 female, 15 male) |
25 | 52.45 ± 2.98 |
29.38 ± 3.31 | 1401.87 ± 30.85 |
1248.27 ± 27.53 |
| Cirrhotic (6 female, 7 male) |
13 | 59.54 ± 3.93 |
24.24 ± 5.60 | 1322.86 ± 33.57 |
1162.33 ± 38.47 |
PMI, post-mortem interval; BWT, brain weight
There was no significant difference between groups on any parameter
Age: F2,58 = 2.40, P = 0.10; PMI: F2, 58 = 0.41, P = 0.67; BWT: F2, 58 = 2.41, P = 0.10
Female: Age, F2,24 = 1.42, P = 0.26; PMI, F2,24 = 0.024, P = 0.98; BWT, F2,24 = 2.78, P = 0.08;
Male: Age, F2,31 = 1.542, P = 0.23; PMI, F2,31 = 0.10, P = 0.38; BWT, F2,31 = 1.41 , P = 0.26 Females have lower brain weight than their male counterparts in all groups (Overall, F1,59 = 33.65, P < 0.01; Controls, F1,21 = 10.43, P = 0.004; Uncomplicated Alcoholics, F1,23 = 15.64, P < 0.001; Cirrhotic Alcoholics, F1,11 = 9.98, P = 0.01)
To determine if the expression of any of the seven 14-3-3 isoforms was correlated with age at death, PMI or brain weight, linear regressions were performed on each data set. Regression analysis was conducted on all subjects combined. Where a significant correlation was found, further analysis on subjects separated according to alcoholic status (controls vs. combined alcoholics) was performed. There was no significant correlation between the expression of any of the 14-3-3 transcripts and age at death or PMI except for 14-3-3σ, which showed increasing expression levels with age, but only in the motor cortex (F1,21 = 5.93, P = 0.012). When case groups were analyzed separately, 14-3-3σ showed a significant correlation in controls (F1,21 = 5.93, P = 0.024) but there was no such correlation in combined alcoholic cases (F1,32 = 1.07, P = 0.31). ANCOVA on the expression of 14-3-3σ with age at death as the covariate did not alter the pattern of effects observed with ANOVA alone. Since the case groups did not differ significantly in age, a simple factorial ANOVA was appropriate to analyse the data.
Linear regressions on brain weight showed significant correlations for 14-3-3σ in both cortical regions (PFC: F1,57 = 7.35, P = 0.009; Motor, F1,55 = 9.5, P = 0.003) and for 14-3-3γ in motor cortex only (F1,53 = 5.23, P = 0.026). This correlation was not found for 14-3-3γ when case-groups were analysed separately. However, increasing 14-3-3σ expression was correlated with increasing brain weight in combined alcoholics, but not controls, in both cortical regions (combined alcoholics, PFC, F1,34 = 8.69, P = 0.006; Motor, F1,32 = 5.66, P = 0.024). ANCOVA of 14-3-3σ expression with brain weight as the covariate did not alter the pattern of significant differences in expression found with ANOVA. Since the case groups do not differ significantly in brain weight, a simple factorial ANOVA was appropriate to analyse the data.
Use of an appropriate endogenous control (housekeeping) gene as an internal reference is critical to the analysis of relative expression levels using Real-Time PCR. Expression of GAPDH was measured in each sample included in the study. Comprehensive statistical analysis was carried out on the raw CT values to determine differences in GAPDH expression (if any) between groups, genders and brain regions: no significant differences were observed. Transcript levels of all seven 14-3-3 genes were expressed relative to the CT of GAPDH in each sample (Pfaffl, 2001). Statistical tests were performed on the ΔCT values for each gene.
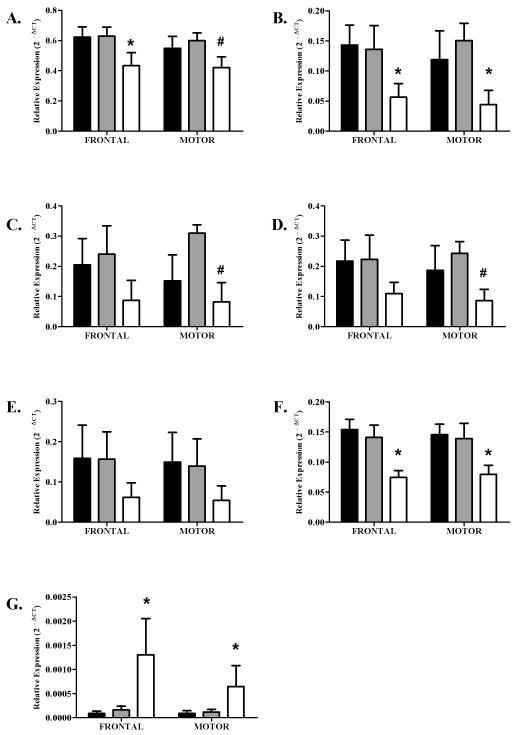
Initially, expression differences were analyzed across groups regardless of gender. Subsequent analysis addressed gender-specific expression levels of each transcript. Statistically significant differences in gene expression were observed for six of the seven 14-3-3 isoforms. In the prefrontal cortex, 14-3-3β RNA levels were significantly lower in cirrhotics compared with both controls and uncomplicated alcoholics, while in the motor cortex, RNA levels in cirrhotics were significantly lower than uncomplicated alcoholics (Figure 1, part A). Levels of 14-3-3γ and θ transcripts were significantly lower in both cortical regions of cirrhotic alcoholics compared with controls and uncomplicated alcoholics (Figure 1, part B & F). In contrast, 14-3-3σ expression was significantly greater in the prefrontal and motor cortices of cirrhotic alcoholics than in controls and uncomplicated alcoholics (Figure 1, part G). Expression levels of 14-3-3ε and ζ were significantly lower in cirrhotics compared to uncomplicated alcoholics, but only in the motor cortex (Figure 1, part C & D).
Figure 1.
Relative expression in both Prefrontal and Motor cortices. Data is presented as mean ΔCT converted to 2−ΔCT (with S.E.Ms). Asterixes denote significant changes between control and cirrhotic alcoholics, hashes denote significant changes between uncomplicated alcoholics and cirrhotic alcoholics (Tukey hsd P < 0.05). Solid bars represent Controls, lightly shaded bars represent Alcoholics without comorbid disease and open bars represent Cirrhotic Alcoholics. A significant reduction in gene expression was observed in both the prefrontal and motor cortices in 14-3-3β, γ and θ. In both 14-3-3 ε and ζ a significant reduction in expression was observed in the motor cortex only. There was no difference in either the prefrontal or the motor cortex for 14-3-3η. The only transcript to show a significant increase in expression was 14-3-3σ. This increase was seen in both the prefrontal and motor cortex.
A. 14-3-3β (PFC, F2,53 = 4.72, P = 0.01, Motor, F2,52 = 3.25, P < 0.05)
B. 14-3-3γ (PFC, F2,53 = 5.04, P < 0.01, Motor, F2,52 = 5.11, P < 0.01)
C. 14-3-3ε (PFC, F2,53 = 2.05, P = 0.14, Motor, F2,52 = 3.30, P = 0.05)
D. 14-3-3ζ (PFC, F2,53 = 2.10, P = 0.13, Motor, F2,52 = 3.62, P = 0.03)
E. 14-3-3η
F. 14-3-3θ (PFC, F2,55 = 13.09, P < 0.001, Motor, F2,55 = 6.45, P = 0.003)
G. 14-3-3σ (PFC, F2,56 = 10.57, P < 0.001, Motor, F2,54 = 4.83, P = 0.01)
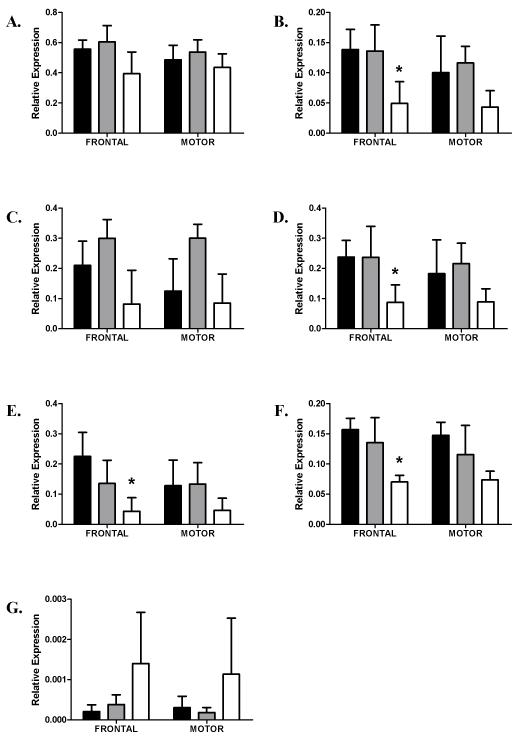
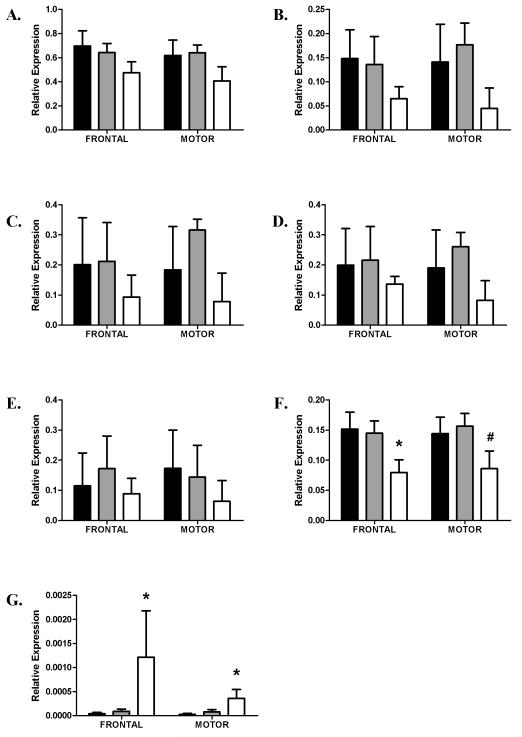
In female cases, significantly lower expression values were seen for 14-3-3γ, ζ, η and θ. For 14-3-3γ, and ζ expression levels were lower in the prefrontal cortex of cirrhotics compared with controls, while 14-3-3θ expression was lower in the prefrontal cortex of cirrhotic alcoholics compared with both controls and uncomplicated alcoholics (Figure 2, parts B, D, F). While no significant difference in the expression of 14-3-3η was found in either area of the combined case group, this expression of this isoform was significantly lower in the prefrontal cortex of female cirrhotics compared with controls (Figure 2, part E). In males, 14-3-3θ expression was significantly lower in cirrhotic alcoholics than in both controls and uncomplicated alcoholics in the prefrontal cortex. In motor cortex however, the expression of this isoform was lower in cirrhotics compared with uncomplicated alcoholics (Figure 3, part F). In the prefrontal cortex, 14-3-3σ expression was significantly higher in male cirrhotics compared with controls and uncomplicated alcoholics, while in motor cortex, 14-3-3σ was significantly higher in cirrhotics compared with controls (Figure 3, part G,).
Figure 2.
Relative expression in Prefrontal and Motor cortices in females. Data is presented as mean ΔCT converted to 2−ΔCT (with S.E.Ms). Asterixes denote significant changes between control and cirrhotic alcoholics (Tukey hsd P < 0.05). Key as for Fig. 1. Significantly lower expression of 14-3-3 γ, ζ, η, and θ was seen in the prefrontal cortex only of female cirrhotics compared with controls.
A. 14-3-3β
B. 14-3-3γ (PFC, F2,22 =3.93, P = 0.035, Motor, F2,22 = 1.61, P = 0.22)
C. 14-3-3ε
D. 14-3-3ζ (PFC, F2,22 =3.51, P < 0.05, Motor, F2,22 = 1.26, P = 0.30)
E. 14-3-3η (PFC, F2,23 =3.68, P = 0.04, Motor, F2,23 = 1.28, P = 0.30)
F. 14-3-3θ (PFC, F2,23 =6.8, P = 0.005, Motor, F2,23 = 2.78, P = 0.08)
G. 14-3-3σ
Figure 3.
Relative expression in Prefrontal and Motor cortices in males. Data is presented as mean ΔCT converted to 2−ΔCT (with S.E.Ms). Asterixes denote significant changes between control and cirrhotic alcoholics, hashes denote significant changes between uncomplicated alcoholics and cirrhotic alcoholics (Tukey hsd P < 0.05). Key as for Fig. 1. The expression of 14-3-3θ in prefrontal and motor cortices was significantly lower in male cirrhotics than in either controls or non-cirrhotic alcoholics. This was true in motor cortex when male cirrhotic alcoholics were compared with male non-cirrhotic alcoholics. Expression of 14-3-3σ in prefrontal and motor cortices was significantly higher in male cirrhotic than in controls.
A. 14-3-3β
B. 14-3-3γ
C. 14-3-3ε
D. 14-3-3ζ
E. 14-3-3η
F. 14-3-3θ (PFC, F2,29 =5.58, P = 0.009, Motor, F2,29 = 4.16, P = 0.026)
G. 14-3-3σ (PFC, F2,29 =10.764, P < 0.001, Motor, F2,29 = 5.90, P = 0.007)
Discussion
Previous microarray and global proteomic studies have shown that 14-3-3 mRNA transcripts and proteins may be differentially expressed in the prefrontal cortex of chronic alcoholics (2004; Lewohl et al., 2000). However, due to the nature of microarray and proteomic studies, distinguishing the expression profiles of specific isoforms, which share regions of high homology, is very difficult. Due to the proposed role of 14-3-3 isoforms in a number of neurodegenerative disorders (Mackie and Aitken, 2005), and the conflicting results of previous studies, we quantified the mRNA expression of all seven human 14-3-3 isoforms in the prefrontal and motor cortices of alcoholics and controls using isoform-specific primers and Real-Time PCR.
Six isoforms demonstrated statistically significant changes in expression in the brain. Five of these isoforms showed lower expression in cirrhotic alcoholics and one, 14-3-3σ showed higher expression in cirrhotic alcoholics than controls. There were also differences in the expression profile of these isoforms between the two cortical regions studied. The expression of 14-3-3γ and θ were lower in cirrhotic alcoholics than in controls in both cortical regions whereas 14-3-3β and 14-3-3ζ were differentially expressed only in the motor cortex of cirrhotic alcoholics.. In addition, female cirrhotics showed lower expression of 14-3-3γ, ζ, η and θ in the prefrontal cortex only, while in male cirrhotics, 14-3-3θ expression was lower and 14-3-3σ expression higher in both prefrontal and motor cortices. The difference in the expression profile of 14-3-3 isoforms with disease state, between cortical regions and between males and females suggests that these isoforms are regulated by distinct mechanisms. Although the isoform-specific regulation of expression of these isoforms has not been studied in detail, there is emerging evidence that the expression of 14-3-3σ is mediated via DNA methylation (Vera et al. 2010) and that 14-3-3ζ is regulated by at least one microRNA, miR-451 (Yu et al. 2010). The role of DNA methylation or microRNA regulation of the other isoforms remains to be elucidated.
14-3-3 proteins are abundantly expressed in the brain and are associated with several neurological diseases, including status epilepticus, Creutzfeldt-Jakob disease, Parkinson disease, and Alzheimer’s disease (Reviewed in Berg et al., 2003). Isoforms of this highly conserved group are enriched in areas of the brain that are of particular importance in neuroplasticity and higher brain functions such as the prefrontal cortex (Skoulakis and Davis, 1998). These proteins are involved in a range of different cellular processes, from cellular proliferation and signal transduction to apoptosis and cell-cycle control (Dean et al., 2007; Dougherty and Morrison, 2004; Fu et al., 2000; Xiao et al., 1995). Their involvement in these processes is through phosphorylation-dependent interaction with protein targets, which include a variety of signalling molecules, transcription factors, cytoskeletal proteins, biosynthetic enzymes, apoptotic factors, tumour suppressors, and ion channels (Dougherty and Morrison, 2004). Direct binding of 14-3-3s to their protein targets increases enzyme activity and controls the subcellular localization of target proteins (Berg et al., 2003). To date, very few isoform-specific binding partners have been identified. However, it is likely that each of the seven isoforms exhibit distinct expression profiles and specificity of binding partners.
In the brain, 14-3-3 proteins play a pivotal role in the regulation of neurotransmission and have the capability to bind to more than 100 neuronal proteins. One of the key neuronal protein targets is tyrosine hydroxlyase (TH), which catalyses the rate limiting step in dopamine synthesis. 14-3-3 proteins bind directly with TH and this interaction is required for its optimal activation and stabilization (Obsilova et al., 2008; Wang et al., 2009). A reduction of 14-3-3 transcripts in the prefrontal cortex of human alcoholics may therefore result in a decrease in dopamine synthesis and a reduction in dopaminergic neurotransmission.
14-3-3 proteins also bind to and mediate the subcellular localization of many proteins, including neurotransmitter receptors. One such interaction is the binding of 14-3-3η and ζ to the GABAB receptor. These isoforms bind to the C-terminus of the GABAB-R1 subunit and control the assembly of functional receptors by preventing the dimerization of GABAB-R1 with GABABR2 (Couve et al., 2001). Thus, reduced expression of 14-3-3 isoforms in the prefrontal cortex may alter GABA neurotransmission by altering the number of functional GABAB receptors in the neuronal membrane.
The 14-3-3 family of proteins has also been implicated in the regulation of apoptosis. Selected 14-3-3 isoforms bind to and sequester specific pro-apoptotic proteins. For example, 14-3-3θ binds to Bad (Zha et al., 1996) and Bcl2-associated X protein (Bax) (Nomura et al., 2003) and 14-3-3ζ binds to apoptosis signal-regulating kinase 1 (Zhang et al., 1999). The binding of 14-3-3s to these pro-apoptotic proteins prevents their function and inhibits apoptosis. A loss of functional 14-3-3 proteins may therefore result in increased free pro-apoptotic proteins in the cytoplasm and ultimately cell death (Ubl et al., 2002). Alcoholics show neuronal loss, particularly in the prefrontal cortex (Harper and Matsumoto, 2005). Deregulation of neuronal survival mechanisms by 14-3-3 proteins may contribute to alcohol-related neuronal loss.
In the brain, the acute effects of alcohol, mediated by interactions with various neurotransmitter and signalling systems, result in the modified release of neurotransmitters from presynaptic terminals. The 14-3-3 protein isoforms may be important contributors to neuronal loss and may therefore play a critical role in the development and progression of the damage that results from chronic alcoholism. This study has identified changes in 14-3-3 gene expression levels that implicate these proteins in the brain’s response to alcohol and will aid in elucidating the mechanisms and pathways by which alcohol affects the brain.
Acknowledgments
We would like to acknowledge the Queensland Brain Bank, the Australian Brain Bank Network and the NSW Tissue Resource Centre who provided the alcoholic and control brain tissue for analysis. Allison Eckert and Donna Sheedy provided detailed information on each of the cases used. We thank the next of kin for providing informed written consent for the studies. Financial support was provided by the National Institute of Alcoholism and Alcohol Abuse (USA, NIH AA12404) (R01 AA012725-04.
Financial Support:
Financial support was provided by the National Institute of Alcoholism and Alcohol Abuse (USA, NIH AA12404) (R01 AA012725-04.
Footnotes
There was no conflict of interest.
References
- Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16(3):162–72. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D, Holzmann C, Riess O. 14-3-3 proteins in the nervous system. Nat Rev Neurosci. 2003;4(9):752–62. doi: 10.1038/nrn1197. [DOI] [PubMed] [Google Scholar]
- Couve A, Kittler JT, Uren JM, Calver AR, Pangalos MN, Walsh FS, Moss SJ. Association of GABA(B) receptors and members of the 14-3-3 family of signaling proteins. Mol Cell Neurosci. 2001;17(2):317–28. doi: 10.1006/mcne.2000.0938. [DOI] [PubMed] [Google Scholar]
- Crews FT, Buckley T, Dodd PR, Ende G, Foley N, Harper C, He J, Innes D, Loh EW, Pfefferbaum A, Zou J, Sullivan EV. Alcoholic neurobiology: changes in dependence and recovery. Alcohol Clin Exp Res. 2005;29(8):1504–1513. doi: 10.1097/01.alc.0000175013.50644.61. [DOI] [PubMed] [Google Scholar]
- Dean B, Boer SA, Mackinnon A, Berk M. CNS 14-3-3zeta: changes with sex but not psychiatric diagnoses or psychotropic drug treatment. Schizophr Res. 2007;93(1-3):51–7. doi: 10.1016/j.schres.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Dodd PR, Hardy JA, Baig EB, Kidd AM, Bird ED, Watson WE, Johnston GA. Optimization of freezing, storage, and thawing conditions for the preparation of metabolically active synaptosomes from frozen rat and human brain. Neurochem Pathol. 1986;4(3):177–98. doi: 10.1007/BF02834357. [DOI] [PubMed] [Google Scholar]
- Dougherty MK, Morrison DK. Unlocking the code of 14-3-3. J Cell Sci. 2004;117(Pt 10):1875–84. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93(2):359–70. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–47. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Harper C. The neuropathology of alcohol-related brain damage. Alcohol Alcohol. 2009;44(2):136–40. doi: 10.1093/alcalc/agn102. [DOI] [PubMed] [Google Scholar]
- Harper C, Matsumoto I. Ethanol and brain damage. Curr Opin Pharmacol. 2005;5(1):73–8. doi: 10.1016/j.coph.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Hynd MR, Lewohl JM, Scott HL, Dodd PR. Biochemical and molecular studies using human autopsy brain tissue. J Neurochem. 2003;85(3):543–62. doi: 10.1046/j.1471-4159.2003.01747.x. [DOI] [PubMed] [Google Scholar]
- Koob GF. A role for GABA mechanisms in the motivational effects of alcohol. Biochem Pharmacol. 2004;68(8):1515–25. doi: 10.1016/j.bcp.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Kril JJ, Halliday GM, Svoboda MD, Cartwright H. The cerebral cortex is damaged in chronic alcoholics. Neuroscience. 1997;79(4):983–98. doi: 10.1016/s0306-4522(97)00083-3. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Crane DI, Dodd PR. A method for the quantitation of the alpha 1, alpha 2, and alpha 3 isoforms of the GABAA receptor in human brain using competitive PCR. Brain Res Brain Res Protoc. 1997;1(4):347–56. doi: 10.1016/s1385-299x(97)00009-3. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Van Dyk DD, Craft GE, Innes DJ, Mayfield RD, Cobon G, Harris RA, Dodd PR. The application of proteomics to the human alcoholic brain. Ann N Y Acad Sci. 2004;1025:14–26. doi: 10.1196/annals.1316.002. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24(12):1873–82. [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem. 2004;90(5):1050–8. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31(7):1574–82. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mackie S, Aitken A. Novel brain 14-3-3 interacting proteins involved in neurodegenerative disease. FEBS J. 2005;272(16):4202–10. doi: 10.1111/j.1742-4658.2005.04832.x. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81(4):802–13. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Aghajanian GK. Molecular and cellular basis of addiction. Science. 1997;278(5335):58–63. doi: 10.1126/science.278.5335.58. [DOI] [PubMed] [Google Scholar]
- Nomura M, Shimizu S, Sugiyama T, Narita M, Ito T, Matsuda H, Tsujimoto Y. 14-3-3 Interacts directly with and negatively regulates pro-apoptotic Bax. J Biol Chem. 2003;278(3):2058–65. doi: 10.1074/jbc.M207880200. [DOI] [PubMed] [Google Scholar]
- Obsilova V, Silhan J, Boura E, Teisinger J, Obsil T. 14-3-3 proteins: a family of versatile molecular regulators. Physiol Res. 2008;57(Suppl 3):S11–21. doi: 10.33549/physiolres.931598. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignataro L, Varodayan FP, Tannenholz LE, Harrison NL. The regulation of neuronal gene expression by alcohol. Pharmacol Ther. 2009 doi: 10.1016/j.pharmthera.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulakis EM, Davis RL. 14-3-3 proteins in neuronal development and function. Mol Neurobiol. 1998;16(3):269–84. doi: 10.1007/BF02741386. [DOI] [PubMed] [Google Scholar]
- Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J Neurosci Res. 2003;72(6):756–67. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- Ubl A, Berg D, Holzmann C, Kruger R, Berger K, Arzberger T, Bornemann A, Riess O. 14-3-3 protein is a component of Lewy bodies in Parkinson’s disease-mutation analysis and association studies of 14-3-3 eta. Brain Res Mol Brain Res. 2002;108(1-2):33–9. doi: 10.1016/s0169-328x(02)00510-7. [DOI] [PubMed] [Google Scholar]
- Vera J, Schultz J, Ibrahim S, Raatz Y, Wolkenhauer O, Kunz M. Dynamical effects of epigenetic silencing of 14-3-3sigma expression. Mol Biosyst. 2010;6(1):264–73. doi: 10.1039/b907863k. [DOI] [PubMed] [Google Scholar]
- Wang J, Lou H, Pedersen CJ, Smith AD, Perez RG. 14-3-3zeta contributes to tyrosine hydroxylase activity in MN9D cells: localization of dopamine regulatory proteins to mitochondria. J Biol Chem. 2009;284(21):14011–9. doi: 10.1074/jbc.M901310200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker E, Yaffe MB. 14-3-3 Proteins--a focus on cancer and human disease. J Mol Cell Cardiol. 2004;37(3):633–42. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Xiao B, Smerdon SJ, Jones DH, Dodson GG, Soneji Y, Aitken A, Gamblin SJ. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376(6536):188–91. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- Yu D, dos Santos CO, Zhao G, Jiang J, Amigo JD, Khandros E, Dore LC, Yao Y, D’Souza J, Zhang Z, Ghaffari S, Choi J, Friend S, Tong W, Orange JS, Paw BH, Weiss MJ. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 24(15):1620–33. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87(4):619–28. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chen J, Fu H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc Natl Acad Sci U S A. 1999;96(15):8511–5. doi: 10.1073/pnas.96.15.8511. [DOI] [PMC free article] [PubMed] [Google Scholar]