Abstract
HGAL is a germinal center (GC)-specific gene whose expression correlates with a favorable prognosis in patients with diffuse large B-cell and classicHodgkin lymphomas. HGAL is involved in negative regulation of lymphocyte motility. The movement of lymphocytes is directly driven by actin polymerization and actin-myosin interactions. We demonstrate that HGAL interacts directly and independently with both actin and myosin and delineate the HGAL and myosin domains responsible for the interaction. Furthermore, we show that HGAL increases the binding of myosin to F-actin and inhibits the ability of myosin to translocate actin by reducing the maximal velocity of myosin head/actin movement. No effects of HGAL on the actomyosin ATPase activity and on the rate of actin polymerization from G-actin to F-actin were observed. These findings reveal a new mechanism underlying the inhibitory effects of GC-specific HGAL protein on lymphocyte and lymphoma cell motility.
Keywords: HGAL, actin, myosin, motility, germinal center
Introduction
Lymphocyte migration to inflammatory sites and into and out of lymphoid organs is essential for organizing secondary immune anatomical structures such as lymphoid follicles and germinal centers (GC), permitting the appropriate development and propagation of immune responses. In GC, B-cell responses to antigens are amplified and refined in specificity[1]. GC B-lymphocytes are functionally and spatially segregated from extra-GC compartments due to limited inter-compartmental lymphocyte movement[2–6]. While restriction to the GC compartment may be necessary for successful completion of the GC reaction, the molecular mechanisms decreasing GC B-cell motility and GC exit are largely unknown.
We have recently cloned the Human Germinal Center–associated Lymphoma (HGAL) gene specifically expressed in GC B-lymphocytes[7]. HGAL is also expressed in GC-derived lymphomas and identifies biologically distinct subgroupsof diffuse large B-cell lymphomas (DLBCL) as well as classic Hodgkin lymphoma (cHL) associated with improved survivals[7–10]. Studies of HGAL murine homologue encoding the M17 protein using M17 knock-out mice revealed that this proteinis dispensable for GC formation, immunoglobulin somatic hypermutation, class-switch recombination, and mounting of T cell–dependent antibody responses[11]. However, M17-deficient mice exhibited reduced-sized Peyer’s patches[11]. We reported that HGAL inhibits migration of GC B-cells and HGAL-expressing lymphoma cells by activating the RhoA signaling pathway[12]. HGAL’s effect on RhoA is mediated by its direct interaction with RhoA-specific guanine nucleotide exchange factors (RhoGEFs) PDZ-RhoGEF and LARG that stimulate the GDP-GTP exchange rate. While this effect may underlie the inhibitory effects of HGAL on the motility of GC lymphocyte and GC-derived lymphoma cells, we also observed that HGAL coimmunoprecipitates and colocalizes with actin and myosin proteins[13], suggesting that additional molecular mechanisms may contribute to the inhibitory effect of HGAL on B-cell motility. Herein we demonstrate that HGAL protein can directly and independently bind to both cellular actin and myosin II proteins. We have also identified the regions within HGAL and myosin that mediate the interaction between HGAL and myosin II. Furthermore, we demonstrate that HGAL increases the binding of myosin to filamentous actin (F-actin) and inhibits the ability of myosin to translocate F-actin. These molecular mechanisms may further contribute to the inhibitory effects of HGAL on the motility of GC lymphocyte and GC-derived lymphoma cells.
Results
HGAL protein directly interacts with both F-actin and myosin
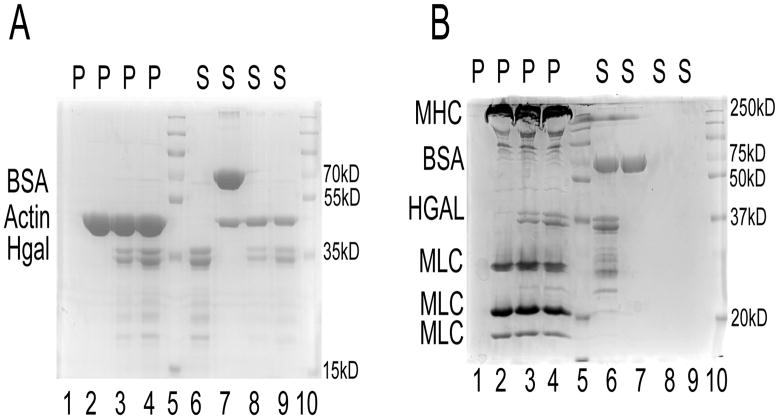
Using a co-immunoprecipitation assay and colocalization studies, we have previously shown that HGAL protein interacts with both actin and myosin II[13]. In order to determine whether the interaction is direct or indirect, we conducted the actin- HGAL and myosin-HGAL cosedimentation assays using purified recombinant HGAL and purified non-muscle F- actin and rabbit skeletal muscle myosin II proteins. As demonstrated in Figure 1A, after centrifugation, the majority of the HGAL protein was pulled down into the pellets with F-actin. Similarly, as pictured in Figure 1B, nearly all of the HGAL protein was precipitated with myosin. The binding of HGAL to F-actin and myosin was saturable and occurred at molar ratios of 5:1 and 2.5:1, respectively. Our results show that HGAL co-precipitates with both actin and myosin, suggesting that the interactions between HGAL and both actin and myosin are direct and independent.
Figure 1. HGAL protein directly interact with both F-actin and myosin.
(A). Coomassie-blue-stained gel showing a representative result of an F-actin co-sedimentation assay performed with recombinant HGAL protein. Pellet (lanes1–4) and Supernatant (lanes 6–9) fractions of co-sedimentation samples were resolved on a 10% SDS–PAGE gels. Lane 1,6: 0.01 mM HGAL; lanes 2,7: 0.05 mM F-actin and BSA; lanes 3,8: 0.05mM F-actin and 0.01mM HGAL; lanes 4,9: 0.05mM F-actin and 0.02mM HGAL; lanes 5,10: protein standard (Fermentas) (B) Coomassie-blue-stained gel showing a representative result of myosin HGAL co-sedimentation assay. Pellet (lanes1–4) and Supernatant (lanes 6–9) fractions of co-sedimentation samples were resolved on a 10% SDS–PAGE gels. Lane 1,6: 0.01 mM HGAL and BSA; lanes 2,7: 0.05 mM myosin and BSA; lanes 3,8: 0.05 mM myosin and 0.01mM HGAL; lanes 4,9: 0.05 mM myosin and 0.02mM HGAL; lanes 5,10: protein standard (Bio-rad). The experiments in A and B were repeated 3 times. MHC-myosin heavy chain; MLC-myosin light chains.
HGAL binds to the head and tail regions of myosin
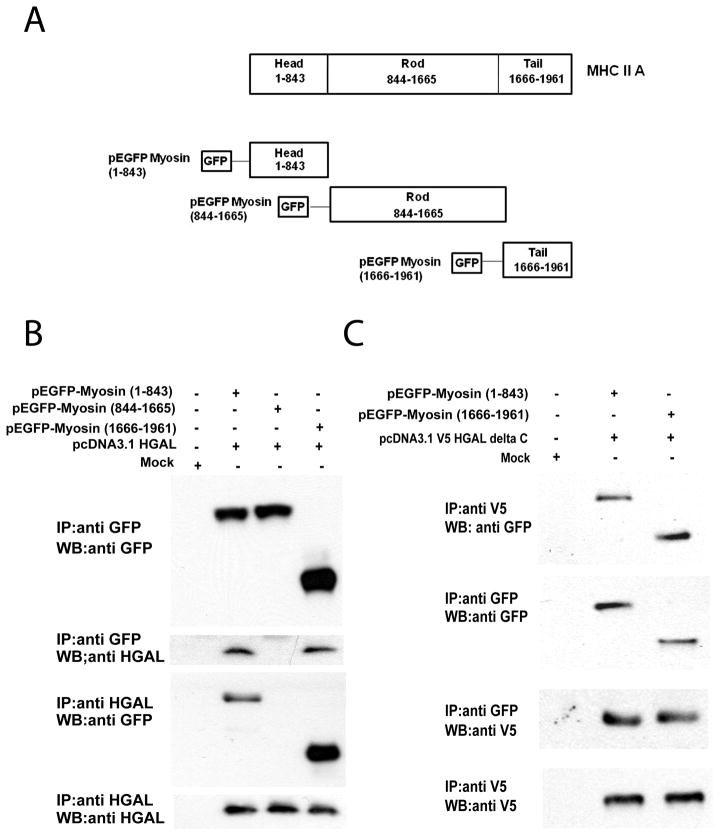
Since we have demonstrated a direct and independent binding of HGAL to myosin, we next carried out experiments to identify the domains of myosin where HGAL binds as well as the region of HGAL that interacts with myosin. We constructed plasmids that express the N-terminal portion of HGAL protein (pcDNA3.1V5 HGAL delta C encoding amino acids 1–118 of the HGAL protein) and plasmids that express the head region of myosin (sub-fragment 1 or S1), and the rod and tail regions. The myosin proteins pEGFP Myosin (1–843)-the head region, pEGFP Myosin (844–1665) - the rod region, and pEGFP Myosin (1666–1961) –the tail region can be expressed in 293Tcells (Figure 2A). Co-immunoprecipitation experiments demonstrated that HGAL interacts with the myosin head (S1) and tail regions but not with the rod region (Figures 2B and 2C). Search for potential HGAL consensus binding motif in the myosin head (S1) and tail regions failed to identify identical 5–8 amino acid sequences in these regions suggesting biding to different motifs. HGAL does not interact with the myosin light chains (data not shown). We further demonstrate that the N-terminal portion of the HGAL (aa1–118) is sufficient to allow its interaction with myosin (Figure 2C). Overall these studies demonstrate that the N-terminal portion of the HGAL protein (aa 1–118) can bind to the tail and head (S1) regions of myosin II, the latter containing the actin and ATP binding sites, but does not bind to the rod part of the myosin molecule that is responsible for filament formation.
Figure 2. HGAL binds to the head and tail regions of the myosin.
(A) Schematic structure of myosin and plasmid constructs used for experiments. (B) 293T cells were co-transfected with Mock empty vector(pcDNA3.1V5) or pcDNA3.1V5HGAL and pEGFP-Myosin(1–843), pEGFP-Myosin(844–1665) or pEGFP-Myosin (1666–1961). Cellular lysates were extracted after 48 hours and subjected to immunoprecipitation with anti–GFP or HGAL antibodies followed by anti-HGAL and anti–GFP Western immunoblotting, respectively. (C) 293T cells were co-transfected with Mock empty vector or pcDNA3.1V5 HGAL delta C encoding amino acids 1–118 of the HGAL protein and pEGFP-Myosin (1–843) or pEGFP-Myosin (1666–1961). Cellular lysates were extracted after 48 hours and subjected to immunoprecipitation with anti–GFP or HGAL antibodies followed by anti-HGAL and anti–GFP Western immunoblotting, respectively. The experiments in B and C were repeated three times. MHC-myosin heavy chain.
HGAL increases binding of myosin to F-actin
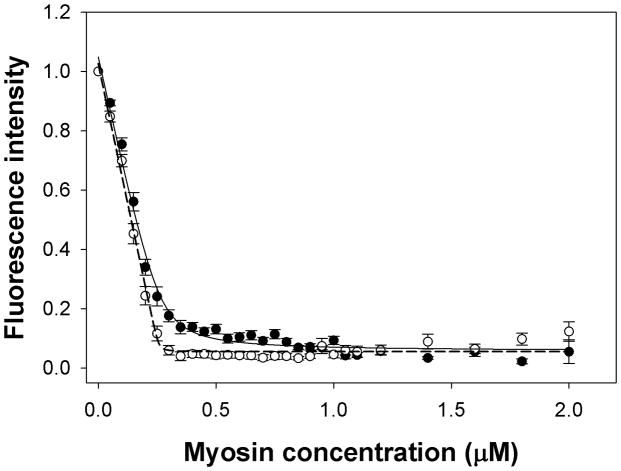
HGAL is involved in negative regulation of lymphocyte migration[13]. Cell movement appears to be driven directly by actin polymerization and by the actin-myosin interactions. The effect of HGAL on the binding of myosin to F-actin was examined using a fluorescence-based assay and pyrene labeled F-actin. As shown in Figure 3, the binding of myosin to F-actin was 54-fold stronger in the presence of HGAL compared to the absence of HGAL. The respective Kd values (in nM) were Kd=10.8±2.6 (n=11 independent experiments) in the absence of HGAL and Kd=0.2±0.5 in the presence of HGAL (n=10). The difference was statistically significant (p<0.01). Thus HGAL can increase the interaction between myosin II and F-actin. Negative control experiment showed no effect of BSA (bovine serum albumin) on binding of myosin to F-actin (data not shown) suggesting a specific HGAL-mediated effect on Kd.
Figure 3. HGAL increases binding of myosin to F-actin.
The effect of HGAL on the binding of myosin to F-actin was examined using a fluorescence-based assay. Pyrene-actin at 0.5 μM, was titrated with the increasing concentrations of myosin in the presence (open circle) or absence (closed circle) of HGAL protein. The fluorescence signal was read every 60 seconds until it reached a plateau. Kd= 10.8±2.6 nM for myosin alone (n=11), Kd= 0.2±0.5 nM for myosin and HGAL (n=10). p=0.0031.
HGAL inhibits the ability of myosin to translocate actin
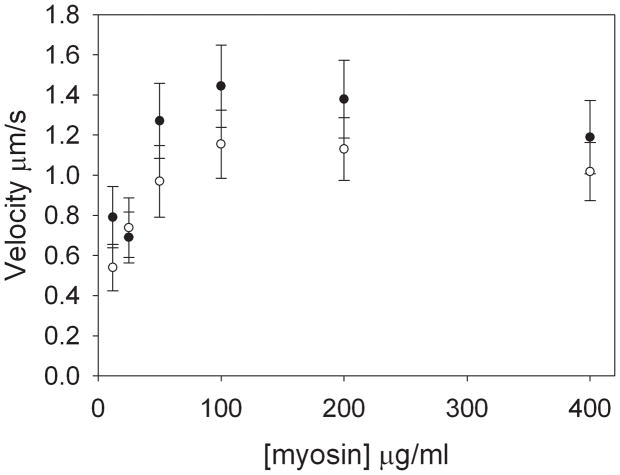
To determine whether HGAL affects the interaction between myosin and actin at the molecular level, we measured the ability of myosin to translocate actin filaments in the in vitro motility assay[14]. Unloaded actin filament velocity was measured over a range of myosin concentrations in the presence and absence of HGAL. Figure 4 shows that actin filament velocity was relatively insensitive to myosin concentrations over an broad range (100–400μg/ml) in both the presence and absence of HGAL. However, the velocity at which saturation occurred (Vsat) was reduced in the presence of HGAL (p=0.05).
Figure 4. HGAL inhibits the ability of myosin to translocate actin in In vitro Motility Assay.
Rabbit skeletal muscle myosin was introduced into the flow chamber in high salt buffer and allowed to bind to the nitrocellulose surface for 2 minutes. TRITC labeled F-actin filaments (~5 nM) in low salt buffer were added and allowed to bind to the myosin in the absence of ATP. HGAL was incubated for 5 minutes and allowed to bind to the surface immobilized myosin. The average velocity for a given filament was determined from the distance traveled by the filament between 10–12 consecutive video images taken at 1 second intervals using Retrac software. Closed circles- myosin alone; open circles-myosin with HGAL.
At myosin loading concentrations below the point of Vsat (<100 μg/ml), the number of filaments moving on the surface was reduced and the average velocity decreased. Actin filaments will move at maximum velocity in the motility assay when surface concentrations are sufficient to allow at least one myosin head to interact with the moving filament at all times. At concentrations below Vsat, velocity slows because there are prolonged periods of time where there are no myosin molecules bound to and moving the actin filament. Thus, the amount of myosin required to achieve maximal velocity gives a qualitative determination of the duty cycle, the fraction of myosin ATPase cycle where myosin is strongly bound to actin. The data show that actin filament velocity reaches maximum velocity at a similar myosin concentration in the presence and absence of HGAL. Therefore, although HGAL inhibits maximal velocity, the unloaded duty cycle of the isolated actomyosin is unaffected.
HGAL has no effect on actomyosin ATPase activity and actin polymerization
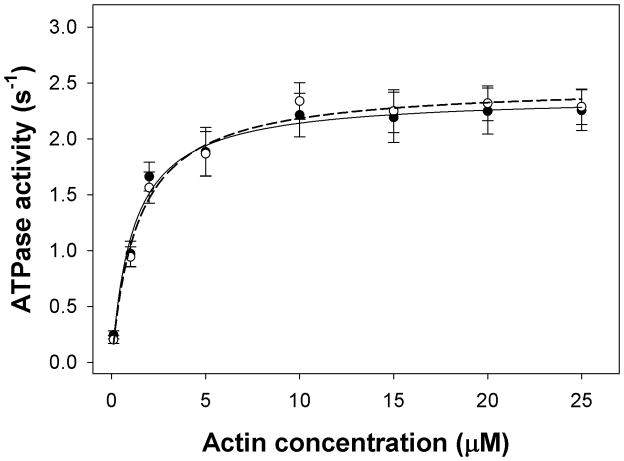
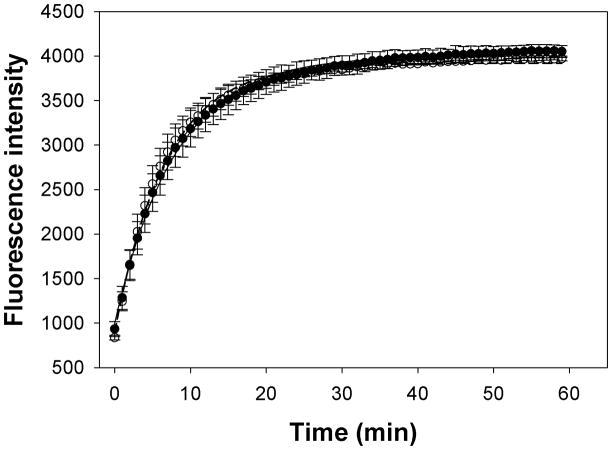
Actin polymerization and the actomyosin ATPase activity are required for chemotaxis and cytokinesis. Therefore we hypothesized that HGAL may affect actin polymerization and/or the actomyosin ATPase activity. As presented in Figure 5, the actin activated myosin ATPase activity was not affected by HGAL and no significant differences in the activity levels were observed in the presence of HGAL. Similarly, no effect of HGAL on the rate of actin polymerization was observed (Figure 6).
Figure 5. HGAL has no effect on actomyosin ATPase activity.
Rabbit skeletal myosin at the concentration of 1.9μM was titrated with the increasing concentrations of rabbit skeletal F-actin (in μM): 0.1, 1, 2, 5, 10, 15, 20 and 25. The assays were performed in triplicates on 96-well microplates in a 120-μl reaction volume. Actin activated ATPase activity for myosin alone (closed circle): Vmax = 2.39 s−1, KM=1.15 μM; for myosin and HGAL (open circle) Vmax = 2.49 s−1, KM=1.39 μM; p>0.05. The experiment was repeated five times.
Figure 6. HGAL does not affect actin polymerization.
Pyrene-actin was added into the G buffer and mixed with either HGAL or just HGAL buffer. The samples were read on a Spectra Max Gemini EM fluorescence microplate reader at 60 second interval for 120 cycles with excitation at 368 nm and emission at 430nm. Rate of polymerization for actin alone (closed circle) is 0.125 min−1, for actin + HGAL (open circle) is 0.149 min−1 (for the curves above). Averaged results (n=6): rate of polymerization for actin alone is 0.118 ± 0.045 min−1, for actin + HGAL is 0.111 ± 0.022 min−1.
Discussion
Lymphocyte motility and migration are critical features of various physiological and pathological processes, including immune response and lymphoma dissemination. Lymphocyte motility is a multi-step process that results from a coordinated cytoskeletal remodeling primarily mediated by cyclic interactions between the myosin head domains projecting from the myosin filaments and polymerized F-actin in the presence of ATP. Hydrolysis of the ATP by myosin releases free energy utilized to generate mechanical force for directed actin-based movement. In the case of skeletal muscle, this complex process is regulated by the tropomyosin-troponin regulatory proteins that control the actin-myosin interaction depending on calcium ion concentrations in the cell. In non-muscle cells, other distinct proteins may control actin-myosin interaction.
The regulatory processes controlling lymphocytes motility, especially the motility of GC lymphocytes and lymphoma cells are not fully elucidated. In the GC, B-lymphocytes are functionally and spatially segregated from extra-GC compartments. Some of the GC B-lymphocytes demonstrate restricted motility between the light and dark zones of the GC[3], while other B cells are stationary throughout these zones. Additionally, in contrast to largely spherical naïve and memory B lymphocytes, GC B-cells frequently exhibit irregular contours with shifting prominent cytoplasmic processes resulting in polarized shapes[2–5]. These observations may suggest the existence of GC B-cell unique cytoskeletal regulatory proteins. Indeed, HGAL (also named germinal center-expressed transcript 2-GCET2) is specifically expressed in GC B-lymphocytes and GC-derived lymphomas and decreases lymphocyte motility[9, 12, 13]. We have demonstrated that the inhibitory effect of HGAL on lymphocyte motility may in part be attributed to the activation of the RhoA signaling pathway[12]. HGAL’s effect on RhoA is mediated by its direct interaction with RhoA-specific guanine nucleotide exchange factors (RhoGEFs) PDZ-RhoGEF and LARG that stimulate the GDP-GTP exchange rate and activation of RhoA downstream effectors that regulate cytoskeletal remodeling. However, we previously also observed that HGAL colocalizes and interacts with actin and myosin II, as shown by co-immunoprecipitation experiments[13], suggesting additional mechanisms that may contribute to HGAL’s inhibitory effect on cell motility. Herein, by using recombinant purified HGAL and purified F- actin and myosin proteins in cosedimentation assays, we demonstrate that HGAL interacts directly and independently with both actin and myosin. These findings corroborate our previous observation that inhibiting actin polymerization and disrupting microfilament organization with cytochalasin D or latrunculin B did not affect HGAL-myosin interactions[13]. We show that the N–terminal portion of the HGAL protein can interact with two myosin regions, the head (S1) and tail, but not with the rod region. While the tail domain generally mediates interaction with the “cargo” and/or other myosin molecules, the head domain is involved in binding to F-actin, which is coupled to steps in the hydrolysis of ATP and transduction of free energy into directed actin-based movement. As we demonstrate here, HGAL does not affect the actomyosin ATPase activity, but significantly increases the strength of actin-myosin binding. This may lead to prolongation of the myosin-actin interaction and contribute to a decreased ability of myosin to translocate actin. An HGAL-induced decrease in actin filament sliding velocity shown by the in vitro motility assays, support this idea. Alternatively, but not mutually exclusive, HGAL binding to myosin and/or actin may serve as a “load” that resists actin filament motion, thus leading to the same effects. One can hypothesize that HGAL functions as a molecular tether between the thick myosin-containing filaments and filamentous actin and regulates the kinetics of acto-myosin interactions and thus cellular motility. The HGAL-mediated bridging properties are similar to those imposed by other myosin binding protein, myosin-binding protein C (MyBP-C) shown to form a linkage between the thick and thin filaments of cardiac muscle and regulate cardiac contractility[15, 16]. Like HGAL, MyBP-C can bind to two different regions in the myosin molecule and affects actomyosin motility[17]. No change in the actomyosin ATPase activity suggests that HGAL does not affect the number or the rate of the myosin cross-bridges undergoing a transition from the weakly to strongly bound state and generation of force. Overall these findings demonstrate that HGAL interaction with actin and myosin may contribute to the decreased motility of GC lymphocytes expressing HGAL protein.
Previous studies in mice identified regulators of actin dynamics and cell motility as key determinants of in vivo lymphoma progression[18]. Specifically, inhibition of cytoskeletal remodeling resulted in decreased lymphoma dissemination and progression. HGAL expression in human DLBCL tumors was associated with limited stage and prolonged survival[7, 19]. HGAL regulation of actin dynamics via RhoA activation and direct effects on actin-myosin interactions may explain the association between HGAL expression and favorable outcome of DLBCL and classical HL patients whose tumors express this protein, consistent with observations in mice. Since HGAL is specifically expressed only in GC lymphocytes and GC-derived lymphomas, further studies investigating the unique role of HGAL-mediated inhibition of lymphocyte motility on immune response and in lymphomagenesis are needed.
Furthermore, it is of note that both RhoA GTPase and actin cytoskeleton have been implicated also in B-cell receptor (BCR) signaling. Saci et al showed that RhoA is activated in response to BCR stimulation and affects BCR-dependent calcium flux and cell proliferation[20]. In addition, it was demonstrated that BCR stimulation induces rapid global actin depolymerization in a BCR signal strength-dependent manner followed by polarized actin repolymerization and actin depolymerization enhanced BCR signaling[21]. Since HGAL can regulate the activation of RhoA and affects actomyosin cytoskeleton, it is possible that HGAL may also contribute to regulation of the BCR signaling. These studies are currently ongoing in our laboratory.
Experimental Procedures
Cells, cell culture, plasmids and transfection
HEK (human embryonic kidney) 293 cells were cultured at 37 °C and 5% CO2 in Dulbecco’s Modification of Eagle’s Medium (DMEM) (Mediatech, Manassas, VA), supplemented with 10% fetal bovine serum (FBS) (Hyclone Logan, UT), 2mM glutamine and 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen-GIBCO, Grand Island, NY).
The full length wild-type human non-muscle myosin IIA heavy chain plasmid pEGFP-myosin IIA and its truncated mutant pEGFP-myosin II ARF (1666–1961)[22, 23] were generous gifts from Dr. Masayuki Takahashi (Division of Chemistry, Graduate School of Science, Hokkaido University, Sapporo, 060–0810 Japan). Using standard cloning techniques, truncated mutants encoding the head portion of myosin IIA linked to EGFP (pEGFP1-843) and the rod part of myosin IIA linked to EGFP (pEGFP844-1665) were generated from the pEGFP-myosin IIA plasmid that was used as a template. The vectors pcDNA3.1V5HGAL and pcDNA3.1V5 HGAL delta C encoding amino acids 1–118 of the HGAL protein were previously reported.[13]
Polyfect transfection reagent (Qiagen, Valencia, CA) was used for transfection of plasmids into HEK 293 cells according to the manufacturer’s instructions. Briefly, cells were plated at 2 × 106 cells in 100-mm culture dishes (Nalge Nunc International, Rochester, NY) in 10 mL DMEM and grown overnight at 37°C and 5% CO2. Plasmid DNA (8μg), diluted and mixed in 300 μl of serum-free DMEM was incubated for 5 min at room temperature. Polyfect transfection reagent was added to the mixture, which was incubated for additional 10 min at room temperature and then added to the cells in a drop-wise manner. The cells were incubated for 48 h before proceeding with further experiments.
Antibodies, Western blot analysis and immunoprecipitation
Mouse monoclonal anti-HGAL antibody was generated in our laboratory, as reported previously[9]. Mouse monoclonal anti GFP antibody was from Santa Cruz Biotechnology Inc. (Santa Cruz, CA) and mouse monoclonal anti V5 antibody was from Invitrogen Inc. (Carlsbad, CA) Western blotting and immunoprecipitation were performed as previously reported[24]. Briefly, whole-cell extracts were preparedby lysing 5 × 106 cells in radioimmunoprecipitation assay (RIPA)buffer (1x phosphate-buffered saline, 1% Nonidet P-40 [NP-40],0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS],10 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 100 mM sodium orthovanadate) on ice for 30 min. Cell lysate was centrifuged at 14,000 g for 10 minutes at 4°C. Protein concentration of lysates was determined using Coomassie protein assay reagent (Thermo Scientific, Rockford, IL) and samples were adjusted to equal protein concentration. For immunoprecipitation (IP), 400 μg of protein was precipitated for 1 to 2 hours with the indicated antibody at 4°C with rotation. Protein G-agarose (Invitrogen, CA) or TrueBlotTM anti-Mouse Ig IP Beads (eBioscinece, CA) were added and the mixture was rotated for an additional 1 hour. Precipitated complexes were washed four times in RIPA buffer, boiled in Laemmli buffer (2 x concentrate: 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue, 0.125 M Tris HCl (pH 6.8)), separated on 12% SDS-PAGE gel and immunoblotted with the indicated antibodies.
Expression and purification of recombinant HGAL protein
For expression and purification of Trx-HGAL fusion protein, BL21 (DE3) cells were transformed with pTRX-HGAL plasmid. Individual clones were grown at 37 °C in liquid broth containing 100 μg/ml ampicillin until A600 reached 0.6, followed by addition of isopropyl β-D-1-thiogalactopyranoside (IPTG, final concentration 0.1 mM) for 3 h. The bacteria were collected and re-suspended in a sonication buffer (10mM Tris–HCl, 150 mM NaCl, pH8.0) and lysed by sonication on ice. Following 20 min centrifugation at 12,000 g, the supernatant was applied to Ni2+ chelating Sepharose column (50/40 cm, Amersham–Pharmacia Biotech; Sunnyvale, CA) pre-equilibrated with starting buffer (50mM Tris–HCl, 150 mM NaCl, and 10mM imidazole, pH8.0), washed with washing buffer (50mM Tris–HCl, 150 mM NaCl, and 80mM imidazole, pH7.0) and fusion protein was eluted using elution buffer (50mM Tris–HCl, 150 mM NaCl, and 200 mM imidazole, pH7.0). The buffer of the eluate was changed to the strong anion ion exchange chromatography starting buffer (50 mM Tris–HCl, 20 mM NaCl, pH8.0) and the solution was applied to Q.F.F. sepharose column (16/20, GE Healthcare Bio-Science Corp; Piscataway, NJ). The proteins were eluted from the column with linear gradient of 1M NaCl in starting buffer at 5 ml/min constant speed for 100 min. Fractions containing the Trx-Src I fusion protein were pooled together.
Rabbit skeletal muscle myosin
Rabbit skeletal muscle myosin II was isolated and purified as described in Greenberg et al[25]. Briefly, minced rabbit skeletal muscle was extracted in 3 volumes of ice cold Guba Straub buffer (0.1 M KH2PO4, 0.05 M K2HPO4, and 0.3 M KCl, pH 6.5) with constant stirring. The muscle mince was pelleted by centrifugation at 12,000 × g for 30 minutes at 4°C. The myosin containing supernatant was decanted and the myosin was precipitated using 13 volumes of ice-cold 1mM EDTA. Precipitated myosin was collected by serial centrifugation at 8,000 × g for 10 minutes at 4°C. The final spin was for 10 minutes at 12,000 × g. The supernatant was removed and discarded. The myosin pellets were then dissolved in cold 20 mM MOPS, pH 7, 1 mM DTT buffer and KCl to a final concentration of 0.5 M. Resuspended myosin was then ultracentrifuged at 125,000 × g for 1.5 h at 4°C. The myosin containing supernatant was then removed and precipitated a second time using 14 volumes of ice cold water. Myosin was collected by centrifugation at 4,500 × g for 10 minutes at 4°C. Following centrifugation the supernatant was removed and discarded. Myosin pellets were left with a minimal volume of 1mM DTT/water and stored overnight on ice. The following morning myosin pellets were resuspended as previously in 20 mM MOPS, pH 7, 1 mM DTT and 0.5 M KCl and ultracentrifuged again. The supernatant containing myosin was removed, mixed with glycerol in a 1:1 by volume ratio and stored at −20°C until needed for experiments.
Muscle F-actin
Rabbit skeletal muscle actin was prepared according to Pardee and Spudich[26] with modifications. Briefly, rabbit skeletal acetone powder was extracted with a G-actin buffer consisting of 2 mM Tris-HCl (pH 8.0), 0.2 mM Na2ATP, 0.5 mM β-mercaptoethanol, 0.2 mM CaCl2, and 0.0005% NaN3 at a ratio of 20 ml/g for 30 min with stirring on ice. The extract was centrifuged at 7,000 × g at 4 °C for 1 h to clarify and the tissue pellet was discarded. The supernatant was removed and adjusted to a final concentration of 40 mM KCl, 2 mM MgCl2 and 1 mM Na2ATP (pH 8.0). The F-actin was allowed to polymerize for 2 h at 4°C. The KCl concentration was then increased again very slowly to a final concentration of 0.6 M and the solution was stirred slowly on ice for 30 min. This step was necessary to remove possible traces of tropomyosin-troponin. The F-actin was then collected by ultracentrifugation at 200,000 × g at 4 °C for 1.5 h. The supernatant was discarded and the F-actin pellets were re-dissolved in a buffer consisting of 10 mM Mops (pH 7.0) and 40 mM KCl.
Labeling of F-actin with pyrene
For fluorescence assays rabbit skeletal actin was labeled with Pyrene lodoacetamide (PIA) by the method of Cooper et al[27]. Briefly 20–40 μM F-actin was incubated at room temperature, in the dark, for 16 hours with a 1.5 molar excess of N-(1-pyrene) iodoacetamide (Invitrogen, Molecular Probes) in a buffer containing 10 mM MOPS, pH 7.0 and 40 mM KCl. Then the reaction was quenched with 1mM DTT and the preparation was centrifuged at 200,000 × g for 1.5 hours. Then F-actin was dialyzed against G-actin buffer (5 mM Tris-HCl pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP) to remove excess of pyrene and DTT and polymerized overnight to form F-actin. The resulting molar ratio of pyrene/F-actin was 0.8 determined using the molar extinction coefficient, ε344(pyrene)=22,000 M−1cm−1.
Actin cosedimentation assay
Non-muscle human actin (Cytoskeleton Inc, Denver, CO) was diluted to 1mg/ml in a buffer containing 5 mM Tris pH 8, 0.2 mM CaCl2, 0.2 mM ATP and 0.5 mM DTT, centrifuged at 40,000 × g for 10 minutes at 4°C. The supernatant was used to induce actin polymerization by adding 50 mM KCl, 1 mM ATP and 2 mM MgCl2. The polymerization occurred at room temperature for 1 hour. Recombinant HGAL protein was spun down at 100,000 × g in Beckman ultracentrifuge for 20 minutes at 22°C. HGAL protein in the supernatant (0.01mM and 0.02mM) and F-actin (0.05mM) were incubated in the buffer containing 10 mM Tris pH 7.0, 1 mM ATP, 0.2 mM DTT, 1 mM EGTA, 0.1 mM CaCl2 and 2 mM MgCl2 for 1 hour at room temperature, then spun at 100,000 × g at 22°C. The supernatants were carefully removed and 5 X Laemmli SDS-PAGE sample buffer was added; 1 X Laemmli SDS-PAGE sample buffer was added to the pellets. The pellets and supernatants were analyzed following their separation by SDS-PAGE and Coomassie Blue staining of the gel.
Myosin-HGAL cosedimentation assay
Rabbit skeletal muscle myosin was precipitated with 13 volumes of ice cold 1mM DTT. Myosin was then collected by centrifugation at 8,000 × g for 10 minutes. The pellet was resuspended in a buffer containing 0.4M KCl, 1mM DTT, and 10mM MOPS at pH 7 and dialyzed overnight. Myosin was diluted at 1:11 ratio with 1mM DTT and put on ice for one hour. Recombinant HGAL protein was spun down at 100,000 × g in a Beckman ultracentrifuge for 20 minutes at 22°C. HGAL in the supernatant (0.01mM and 0.02mM) and myosin (0.05mM) were mixed and incubated on ice for 30 minutes and then spun down at 100,000 × g in Beckman ultracentrifuge for 30 minutes at 22°C. The supernatants and pellets were carefully collected and analyzed following their separation by SDS-PAGE and Coomassie Blue staining.
Actin-activated myosin ATPase activity in the presence or absence of HGAL
The kinetics of the actomyosin interaction in the presence or absence of HGAL protein was measured using actin-activated myosin ATPase assays. Rabbit skeletal muscle myosin at the concentration of 1.9 μM was titrated with the increasing concentrations of rabbit skeletal F-actin (in μM): 0.1, 1, 2, 5, 10, 15, 20 and 25. The assays were performed it triplicates on 96-well microplates in a 120-μl reaction volume containing 25 mM imidazole (pH 7.0), 4 mM MgCl2, 1 mM EGTA and 1 mM DTT. The final salt (KCl) concentration was 0.107 M. The reactions were initiated with the addition of 2.5 mM ATP with mixing in a Jitterbug incubator shaker (Boekel) and allowed to proceed for 5 minutes at 30 °C and then terminated by the addition of 4% trichloroacetic acid. Samples were then centrifuged at 3720 × g for 20 minutes and 50 μl of supernatants were transferred to the microplate for determination of inorganic phosphate by the Fiske–Subbarow method[28]. Data were analyzed using the Michaelis–Menten equation, yielding the Vmax and KM parameters. In control experiments, pyrene-actin was titrated with myosin containing either HGAL-buffer with no HGAL added or BSA (bovine serum albumin) included in each titration point with myosin.
The effect of HGAL on the binding of myosin to F-actin
We used a fluorescence-based assay to examine the effect of HGAL on the binding of rabbit skeletal muscle myosin to pyrene-labeled F-actin. The measurements were performed in a 2ml cuvette in a buffer containing 10 mM MOPS, 0.4M KCl, pH 7 using a JASCO FP-6500 Fluorometer (Jasco, USA). Pyrene-labeled F-actin at 0.5 μM, was titrated with the increasing concentrations of rabbit skeletal muscle myosin in the presence or absence of HGAL protein. Fluorescence was recorded at 408 nm with excitation wavelength of 340 nm. The data were collected using the computer program Felix (Photon Technology International) and fitted to a nonlinear binding model yielding apparent dissociation constants Kd[29].
The effect of HGAL on actin polymerization
Actin polymerization assay was done using the Actin Polymerication Biochem Kit (Cytoskeleton Inc, Denver, CO) according to the manufacturer’s instructions. Briefly, pyrene-actin was added into the G-actin buffer (5 mM Tris-HCl, 0.2 mM CaCl2, 0.2 mM ATP, pH 8.0) and mixed with HGAL or just HGAL buffer (10 mM MOPS, 40 mM KCl, 1mM DTT, pH 7.0). The samples were read on a Spectra Max Gemini EM fluorescence microplate reader (Molecular Devices) at 60 second interval for 120 cycles with excitation at 368 nm and emission at 430nm.
In vitro motility assay
To examine the effect of HGAL on the interaction of actin and myosin at the molecular level we have utilized the in vitro motility assays, as described previously. Briefly, [14] a flow chamber was formed between a nitrocellulose coated coverslip and a standard glass slide with 3M double stick tape at the boundaries. Rabbit skeletal muscle myosin was introduced into the chamber in high salt buffer (300 mM KCl, 25 mM imidazole, 1 mM EGTA, 4 mM MgCl2, 1 mM DTT) and allowed to bind to the nitrocellulose surface for 2 minutes. Sixtyμl of 0.5 mg/ml BSA in high salt buffer was then flowed through the chamber to remove any unbound myosin and block the remaining surface sites to avoid nonspecific attachment of actin filaments or HGAL. After blocking, 60μl of low salt buffer (25 mM KCl, 25 mM imidazole, 1mM EGTA, 4 mM MgCl2, 1 mM DTT) was flowed through the chamber to remove any unbound BSA. Tetramethylrhodamine isothiocyanate (TRITC) labeled F-actin filaments (~5 nM) in low salt buffer were added and allowed to bind to the myosin in the absence of ATP. Unbound actin was removed by washing with low salt buffer and movement initiated by the addition of low salt buffer with 1mM ATP, scavenger (glucose oxidase, catalase, and dextrose) and 0.5% methylcellulose added. For some experiments 30 μl of 400 μg/ml HGAL was incubated for 5 minutes after the BSA blocking step and allowed to bind to the surface immobilized myosin.
Data was collected for several different loading concentrations of myosin. Filament movement was observed at 25oC with an ICCD camera model IC200 (PTI, Birmingham, NJ). Video sequences were captured using Scion image and an AG-5 image grabber (Scion Corp, Frederick, MD). The average velocity for a given filament was determined from the distance traveled by the filament between 10–12 consecutive video images taken at 1 second intervals using Retrac, the freeware program written by Dr. Nick Carter. 15–25 filaments from each video segment were averaged and at least two video segments were obtained per flow cell.
Statistical analysis
Data are expressed as the average of n experiments ± SEM (standard error of the mean). Comparisons between groups were performed using an unpaired Student’s t-test (Sigma Plot 11; Systat Software, Inc., San Jose, CA, USA). The significance was defined as p< 0.05.
Acknowledgments
I.S.L. is supported by National Institutes of Health (NIH) grantsNIH CA109335 and NIH CA122105, and the Dwoskin Family and Fidelity Foundations. D.S-C. is supported by NIH grants HL071778 and HL090786. JM is supported by HL077280 and HL086655. D.M.H. is supported by the WCU Program through the National Research Foundation of Korea funded by the Ministry of Education Science and Technology (R31-2008-000-10071-0)
Abbreviations
- HGAL
human Germinal Center Associated Lymphoma
- GC
germinal center
- DLBCL
diffuse large B-cell lymphoma
- cHL
classic Hodgkin lymphoma
- RhoGEFs
RhoA-specific guanine nucleotide exchange factors
References
- 1.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 2.Hauser AE, Shlomchik MJ, Haberman AM. In vivo imaging studies shed light on germinal-centre development. Nat Rev Immunol. 2007;7:499–504. doi: 10.1038/nri2120. [DOI] [PubMed] [Google Scholar]
- 3.Hauser AE, Junt T, Mempel TR, Sneddon MW, Kleinstein SH, Henrickson SE, von Andrian UH, Shlomchik MJ, Haberman AM. Definition of germinal-center B cell migration in vivo reveals predominant intrazonal circulation patterns. Immunity. 2007;26:655–667. doi: 10.1016/j.immuni.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 5.Schwickert TA, Lindquist RL, Shakhar G, Livshits G, Skokos D, Kosco-Vilbois MH, Dustin ML, Nussenzweig MC. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 2007;446:83–87. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- 6.Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lossos IS, Alizadeh AA, Rajapaksa R, Tibshirani R, Levy R. HGAL is a novel interleukin-4-inducible gene that strongly predicts survival in diffuse large B-cell lymphoma. Blood. 2003;101:433–440. doi: 10.1182/blood-2002-06-1931. [DOI] [PubMed] [Google Scholar]
- 8.Natkunam Y, Hsi ED, Aoun P, Zhao S, Elson P, Pohlman B, Naushad H, Bast M, Levy R, Lossos IS. Expression of the human germinal center-associated lymphoma (HGAL) protein identifies a subset of classic Hodgkin lymphoma of germinal center derivation and improved survival. Blood. 2007;109:298–305. doi: 10.1182/blood-2006-04-014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Natkunam Y, Lossos IS, Taidi B, Zhao S, Lu X, Ding F, Hammer AS, Marafioti T, Byrne GE, Jr, Levy S, Warnke R, Levy R. Expression of the human germinal center-associated lymphoma (HGAL) protein, a new marker of germinal center B-cell derivation. Blood. 2005;105:3979–3986. doi: 10.1182/blood-2004-08-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azambuja D, Lossos IS, Biasoli I, Morais JC, Britto L, Scheliga A, Pulcheri W, Natkunam Y, Spector N. Human germinal center-associated lymphoma protein expression is associated with improved failure-free survival in Brazilian patients with classical Hodgkin lymphoma. Leuk Lymphoma. 2009;50:1830–1836. doi: 10.3109/10428190903242628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schenten D, Egert A, Pasparakis M, Rajewsky K. M17, a gene specific for germinal center (GC) B cells and a prognostic marker for GC B-cell lymphomas, is dispensable for the GC reaction in mice. Blood. 2006;107:4849–4856. doi: 10.1182/blood-2005-10-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang X, Lu X, McNamara G, Liu X, Cubedo E, Sarosiek KA, Sanchez-Garcia I, Helfman DM, Lossos IS. HGAL, a germinal center specific protein, decreases lymphoma cell motility by modulation of the RhoA signaling pathway. Blood. 2010;116:5217–5227. doi: 10.1182/blood-2010-04-281568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Chen J, Malumbres R, Cubedo Gil E, Helfman DM, Lossos IS. HGAL, a lymphoma prognostic biomarker, interacts with the cytoskeleton and mediates the effects of IL-6 on cell migration. Blood. 2007;110:4268–4277. doi: 10.1182/blood-2007-04-087775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg MJ, Kazmierczak K, Szczesna-Cordary D, Moore JR. Cardiomyopathy-linked myosin regulatory light chain mutations disrupt myosin strain-dependent biochemistry. Proc Natl Acad Sci U S A. 2010;107:17403–17408. doi: 10.1073/pnas.1009619107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kensler RW, Shaffer JF, Harris SP. Binding of the N-terminal fragment C0-C2 of cardiac MyBP-C to cardiac F-actin. J Struct Biol. 2010;174:44–51. doi: 10.1016/j.jsb.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winegrad S. Myosin binding protein C, a potential regulator of cardiac contractility. Circ Res. 2000;86:6–7. doi: 10.1161/01.res.86.1.6. [DOI] [PubMed] [Google Scholar]
- 17.Saber W, Begin KJ, Warshaw DM, VanBuren P. Cardiac myosin binding protein-C modulates actomyosin binding and kinetics in the in vitro motility assay. J Mol Cell Cardiol. 2008;44:1053–1061. doi: 10.1016/j.yjmcc.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meacham CE, Ho EE, Dubrovsky E, Gertler FB, Hemann MT. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat Genet. 2009;41:1133–1137. doi: 10.1038/ng.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baecklund E, Natkunam Y, Backlin C, Iliadou A, Askling J, Ekbom A, Feltelius N, Klareskog L, Enblad G, Lossos IS, Levy R, Sundstrom C, Rosenquist R. Expression of the human germinal-center-associated lymphoma (HGAL) protein in diffuse large B-cell lymphomas in patients with rheumatoid arthritis. British Journal of Hematology. 2008;141:69–72. doi: 10.1111/j.1365-2141.2008.07011.x. [DOI] [PubMed] [Google Scholar]
- 20.Saci A, Carpenter CL. RhoA GTPase regulates B cell receptor signaling. Mol Cell. 2005;17:205–214. doi: 10.1016/j.molcel.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Hao S, August A. Actin depolymerization transduces the strength of B-cell receptor stimulation. Mol Biol Cell. 2005;16:2275–2284. doi: 10.1091/mbc.E04-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Ya’acov A, Ravid S. Epidermal growth factor-mediated transient phosphorylation and membrane localization of myosin II-B are required for efficient chemotaxis. J Biol Chem. 2003;278:40032–40040. doi: 10.1074/jbc.M306948200. [DOI] [PubMed] [Google Scholar]
- 23.Sato MK, Takahashi M, Yazawa M. Two regions of the tail are necessary for the isoform-specific functions of nonmuscle myosin IIB. Mol Biol Cell. 2007;18:1009–1017. doi: 10.1091/mbc.E06-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Chen J, Sasmono RT, Hsi ED, Sarosiek KA, Tiganis T, Lossos IS. T-cell protein tyrosine phosphatase, distinctively expressed in activated-B-cell-like diffuse large B-cell lymphomas, is the nuclear phosphatase of STAT6. Mol Cell Biol. 2007;27:2166–2179. doi: 10.1128/MCB.01234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenberg MJ, Mealy TR, Watt JD, Jones M, Szczesna-Cordary D, Moore JR. The molecular effects of skeletal muscle myosin regulatory light chain phosphorylation. Am J Physiol Regul Integr Comp Physiol. 2009;297:R265–274. doi: 10.1152/ajpregu.00171.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 27.Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 28.Fiske C, Subbarow Y. The Colorimetric Determination of Phosphorus. J Biol Chem. 1925;66:375–400. [Google Scholar]
- 29.Pant K, Watt J, Greenberg M, Jones M, Szczesna-Cordary D, Moore JR. Removal of the cardiac myosin regulatory light chain increases isometric force production. FASEB J. 2009;23:3571–3580. doi: 10.1096/fj.08-126672. [DOI] [PMC free article] [PubMed] [Google Scholar]