Summary
Within the nucleus, the genome is spatially organized. Individual chromosomes are non-randomly positioned with respect to each other and with respect to nuclear landmarks [1,2]. Furthermore, the position of individual genes can reflect their expression. Here we discuss two well-characterized examples of gene relocalization associated with transcriptional activation: 1) developmentally regulated genes that move from the nuclear periphery to transcription factories in the nucleoplasm upon induction and 2) genes that are targeted from the nucleoplasm to the nuclear periphery, through interactions with the nuclear pore complex (NPC), upon activation. Finally, we speculate as to the mechanistic and functional commonalities of these phenomena.
Movement of developmentally regulated genes during differentiation
In differentiated metazoan cells, most heterochromatin localizes at the nuclear periphery [3,4]. Likewise, in budding yeast, silent subtelomeric genes localize at nuclear periphery [4,5,6]. These observations suggest that the nuclear periphery is a transcriptionally repressive environment. Consistent with this model, artificially tethering the yeast mating type locus to the nuclear envelope is sufficient to overcome loss of a cis-acting silencing element [7,8] and artificially tethering loci to the nuclear lamina in mammalian cells is sufficient to promote silencing [●9,●10]. This suggests that localization of genes at the nuclear periphery can promote transcriptional silencing.
Some genes localize to the nuclear periphery when repressed, but relocalize upon induction (Figure 1). A number of developmentally induced genes from different organisms and tissue types localize at the nuclear periphery in cells in which they are repressed and away from the nuclear periphery in cells in which they are expressed. This was first reported for the IgH and Igκ loci in mice [11], which localize to the nuclear periphery in hematopoietic progenitor cells and, after induction in pro-B cells, to the nuclear interior. In mice, several other loci relocalize from the nuclear periphery to the nuclear interior upon induction: the GFAP gene during astrocyte differentiation [12], the β-globin locus during erythroid development [13], the C-maf locus during T-cell development [14], the MyoD locus during myoblast development [15] and the Mash1 locus during neural development [16]. In humans, the CFTR gene moves away from the nuclear periphery in cells in which it is expressed [17]. This phenomenon has recently been observed for muscle-specific and gut-specific transgenes during development in C. elegans [●●18]. Thus, the movement of individual genes from the nuclear periphery to the nuclear interior upon differentiation is a common theme among developmentally induced genes.
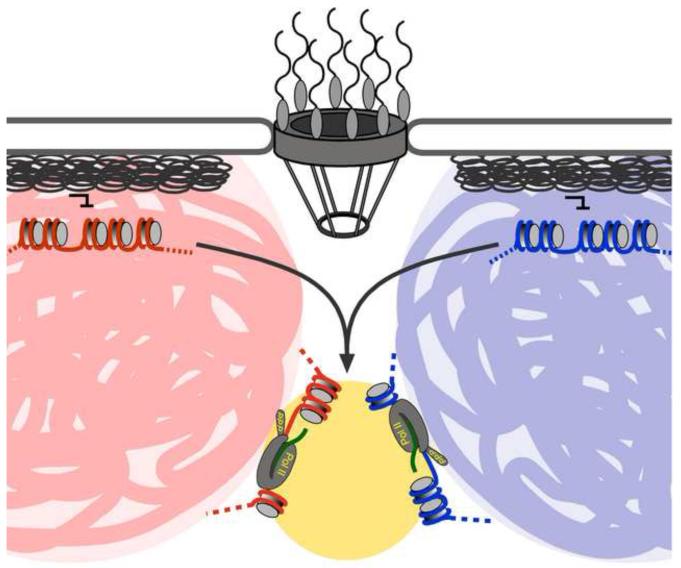
Figure 1. Relocalization of developmentally regulated genes.
Repressed genes often associate with the nuclear lamina at the nuclear periphery. Upon activation, these genes are often targeted to the nucleoplasm. Certain co-regulated genes, located on different chromosomes (chromosome territories represented as blue and pink zones) can colocalize with each other at transcription factories (yellow), located between territories. The colocalization of certain genes requires transcriptional activator (Klf1; green protein) that localizes to a subset of transcription factories [●●37]. Colocalization may promote expression of co-regulated genes by either concentrating factors that promote their expression or by allowing escape from repressive interactions with the nuclear lamina.
Several studies suggest that the interaction of genes with the nuclear lamina at the nuclear periphery promotes repression. Metazoan cells possess a lamina structure at the nuclear periphery, a fibrous mesh made up of lamins and lamin-associated proteins that colocalizes with heterochromatin [19,20]. Genome-wide studies in Drosophila show that much of the Drosophila genome interacts with lamins and that interaction with lamins correlates with transcriptional repression [21]. During astrocyte differentiation in mice, the association of the genome with the lamina changes in a cell type-specific manner, with genes that become active losing their association with the lamina [22,●●23]. Finally, artificially tethering mammalian genes to the nuclear lamina is sufficient to promote transcriptional repression of many neighboring genes [●9,●10,24]. These results suggest that interaction of genes with the nuclear lamina at the nuclear periphery promotes silencing.
How might interaction with the lamina promote repression? Recruitment of lamin A to promoters can repress transcription in both yeast and human cells, suggesting that lamins may directly inhibit transcription [25]. However, it is also possible that the mechanism is less direct. In mammals, histone deacetylases associated with repression interact with inner-nuclear-membrane (INM) proteins such as Emerin [26] and the lamin-associated protein LAP2β [27,28]. This may explain the concentration of hypoacetylated histones at the nuclear periphery [29,30] and the repression of genes artificially tethered to the nuclear lamina [●10]. Consistent with this model, transcriptional repression induced by tethering to the lamina can be relieved by treatment with tricostatin A, an HDAC inhibitor [●10]. This model is reminiscent of the mechanism by which subtelomeric genes are silenced in budding yeast. In yeast, the Sir proteins that catalyze deacetylation of histones at telomeres are concentrated at the nuclear periphery and anchoring of telomeres to the nuclear envelope seems to promote the establishment and fidelity of silencing of subtelomeric genes [6,31-35]. Thus, localization of genes at the nuclear periphery, coupled with a heterogeneous distribution of repressive factors, could promote repression. Furthermore, relocalization of genes from this environment to a more permissive environment might promote transcription.
After moving away from the nuclear periphery, some developmentally co-regulated genes colocalize, a phenomenon called gene “kissing” (Figure 1) [36,●●37]. Gene kissing can occur between genes on the same chromosome, often megabases apart, or between genes on different chromosomes. The genes colocalize either at foci of active RNA polymerase II called transcription factories [38] or near nuclear “speckles” [39]. Furthermore, the colocalization of genes on different chromosomes correlates with common translocation sites [40-44]. Gene kissing has been best characterized for genes induced in erythroid lineages in both humans and mice [36,45-47]. Colocalization of the active mouse Hbb and Hba globin genes with transcription factories has been demonstrated by both immuno-FISH and molecular techniques and requires Klf, a transcription factor that regulates their expression [●●37]. Because there seem to be a limited number of transcription factories per nucleus, kissing may concentrate factors that promote expression of related genes [●●37]. Thus, coupled with transcriptional regulation, certain genes can colocalize in association with subnuclear compartments.
Do these changes in gene positioning represent gene targeting to different subnuclear locations, or does gene positioning represent a downstream consequence of expression? The available data do not resolve this question. Consistent with the possibility that targeting might be specific and controlled by cis-acting information, promoters play an essential role in controlling gene positioning. The relocalization of the mouse β-globin locus to a transcription factory requires the Locus Control Region [13]. In C. elegans, transgenic promoters for housekeeping genes localize in the nucleoplasm in all cell types, whereas promoters from developmentally regulated genes localize in the nucleoplasm in cells in which they are expressed and at the nuclear periphery in cells in which they are not expressed [●●18]. Likewise, the colocalization of co-regulated genes in mouse erythroid cells requires the transcriptional activator Klf1 [●●37], but does not require ongoing transcription [48]. Consistent with the idea that gene kissing could concentrate factors that promote expression of related genes, Klf1 also localizes in a punctate pattern within the nucleus that overlaps with the transcription factories with which the genes interact (Figure 1) [●●37]. These results are consistent with the possibility that gene positioning and colocalization in the nucleoplasm may be controlled by cis-acting DNA elements in the promoters of these genes. However, it is not clear that localization to the nuclear lamina represents targeting. Association of genes with the nuclear lamina may represent a default state for silenced loci. If lamin-associated proteins both bind to hypoacetylated/heterochromatic loci and promote deacetylation/heterochromatinization, silencing might lead to peripheral localization and peripheral localization might stabilize silencing. If so, then this interaction might be blocked by transcription or by activator function. Indeed, when very large transgene arrays are integrated into the C. elegans genome they are silenced and their localization does not reflect the promoter sequences in the array. These heterochromatic arrays localize at the nuclear periphery, regardless of the promoters that they possess [●●18]. Likewise, a large array of lac repressor binding sites localizes at the nuclear periphery in hamster cells [49]. Tethering an activation domain to this array leads to relocalization of the array from the nuclear periphery to a more internal site [49]. Thus, although it is possible that active genes are targeted to particular sites during differentiation, it remains unclear if localization to the nuclear lamina represents targeting or a default destination for repressed loci.
Active genes at the nuclear periphery
Although heterochromatin and silenced genes localize at the nuclear periphery, localization at the nuclear periphery per se is not incompatible with transcription. Several of the genes that relocalize from the nuclear periphery to the nucleoplasm during differentiation are induced prior to relocalization [13,17,52]. Chromatin modifications associated with active transcription, as well as individual active genes have been observed at the nuclear periphery in mouse embryonic stem cells [●50,51]. Furthermore, even genes that are tethered directly to the nuclear lamina are not always repressed. Tethering of endogenous chromosomal loci to the lamina resulted in repression of some, but not all of the neighboring genes [●9,●10,24]. An artificial reporter gene tethered to the lamina is as inducible as the nucleoplasmic form of the gene [24]. Thus, transcription and localization to the nuclear periphery are not always mutually exclusive, suggesting that nuclear positioning can have different effects on different genes.
Some genes are targeted from the nucleoplasm to the nuclear periphery when activated (Figure 2). This phenomenon is best understood in budding yeast. Genome-wide chromatin immunoprecipitation microarray experiments against nuclear pore proteins demonstrated that hundreds of active genes interact with proteins of the nuclear pore complex (NPC) and localize at the nuclear periphery [53,54]. Inducible genes such as GAL1, INO1, GAL2, HSP104 and SUC2 localize in the nucleoplasm when repressed and relocalize to the nuclear periphery upon activation [53-58]. Localization at the nuclear periphery can promote transcription; tethering of INO1 [55,59] or HXK1 [57] to the nuclear envelope positively affects how fast or how robustly these genes are expressed and tethering of an artificial promoter to the NPC itself [60] is sufficient to induce transcription. Work from Drosophila [61] and mouse [39] raises the possibility that this phenomenon also occurs in metazoans (see below). Thus, some genes localize to the nuclear periphery when active and localization promotes transcription.
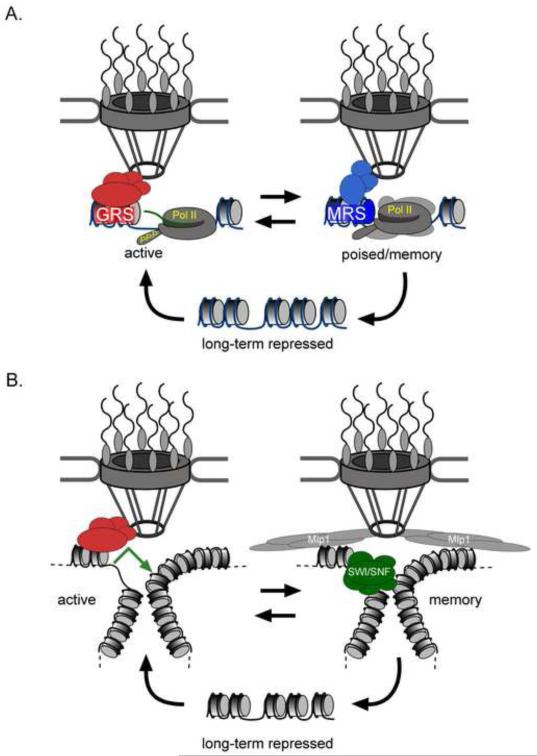
Figure 2. Genes associated with the Nuclear Pore Complex at the nuclear periphery.
A. The mechanism of INO1 targeting to the NPC. In yeast, active genes such as INO1 contain zip codes (Gene Recruitment Sequences, or GRSs) in their promoters that can localize at the nuclear periphery through interaction with the NPC, perhaps through the interaction adaptor proteins (X). After repression, some recently repressed genes remain associated with the NPC by a separate mechanism. Localization of the recently repressed form requires a different zip code (Memory Recruitment Sequence, or MRS), a different interaction with the NPC (perhaps through adaptor proteins such as Y) and leads to promoter poising in association with unphosphorylated RNA polymerase II. B. The mechanism of GAL gene transcriptional memory. After repression, GAL genes remain looped, with the 5′ and 3′ ends of the gene associated. This looping requires the Tpr homologue Mlp1 and, along with the SWI/SNF chromatin remodeler, promotes faster reactivation of the GAL genes [●74, ●75, 76].
Consistent with the physical association of genes with the NPC, a number of NPC proteins, mRNA transport factors or NPC-associated factors are required for peripheral targeting of INO1, GAL1 [59,62,●●63] GAL2 and HSP104 [58]. This suggested that the change in localization of these genes to the nuclear periphery might represent a consequence of transcription, perhaps through a bridging interaction of NPC-associated mRNA transport factors with the gene [54,58]. Consistent with this possibility, the interaction of some genes with the NPC is RNase sensitive [54], the targeting of HXK1 to the nuclear periphery requires the 3′UTR [57] and the targeting of GAL10 and HSP104 to the NPC requires the mRNA transport receptor Mex67 [58]. However, experiments with a temperature sensitive allele of RNA polymerase II showed that the interaction of GAL1 with the NPC [64] and the targeting of INO1 to the nuclear periphery [59] are independent of mRNA production. Therefore, the localization of some genes at the nuclear periphery is independent of transcription, suggesting that these genes might be targeted to the nuclear periphery in a manner that is coupled to transcription, but not dependent on transcription.
Consistent with this idea, genes possess cis-acting targeting elements that control localization. The targeting of the GAL2 gene to the nuclear periphery requires the promoter, but not the coding sequence or 3′UTR [58]. The targeting of INO1 to the nuclear pore complex is controlled by two cis-acting DNA sequences called Gene Recruitment Sequences (GRS I and GRS II) in its promoter (Figure 2) [●●63]. These sequences are distinct from the Upstream Activating Sequences that control INO1 transcription [65-67]. Importantly, these GRS elements function as “DNA zip codes”; when integrated at an ectopic locus, they are sufficient to confer both peripheral localization and a physical interaction with the NPC [●●63]. Mutations in the GRS elements block peripheral targeting of INO1 and another GRS-targeted gene, TSA2, leading to a defect in transcription [●●63]. This supports the idea that targeting to the NPC promotes transcription. Genome-wide, GRS I-containing promoters are enriched for genes that interact with the NPC and that are induced by protein folding stress [●●63]. Finally, the GRS I element, when introduced into the genome of the highly divergent yeast Schizosaccharomyces pombe, functions as a DNA zip code to confer peripheral localization [●●63]. This suggests that GRS I-mediated targeting to the NPC is an ancient mechanism, having been conserved for between 400 million and one billion years.
Several complementary studies examining the interaction of nucleoporins with the Drosophila melanogaster genome suggest that genes interact with nuclear pore proteins in flies [61,68,●●69,●●70]. Intriguingly, these studies identified at least two distinct types of genes: those that interact with the NPC at the nuclear periphery and those that interact with nucleoporins in the nucleoplasm. The genes that interact with nucleoporins in the nucleoplasm tended to be more active, developmentally important genes, whereas the NPC-associated genes were less active. Depletion of nucleoporins led to a defect in the transcription of the genes in the nucleoplasm [68,●●69,●●70]. This suggests that interactions with nucleoporins can also occur away from the pore and that interactions with nucleoporins in the nucleoplasm and at the NPC may have different effects on transcription.
Even in yeast, there are multiple mechanisms by which genes can be targeted to the NPC and these mechanisms that have distinct effects on transcription. Several genes that localize at the nuclear periphery when they are active remain at the nuclear periphery after they are repressed (Figure 2) [59]. In fact, the INO1 and GAL1 genes remain at the nuclear periphery in the population through several cell divisions [59]. Thus, localization of these repressed genes at the nuclear periphery represents an epigenetic form of “transcriptional memory”. Localization at the nuclear periphery correlates with a distinct mechanism of activation, suggesting that the function of this form of memory is to prime genes for reactivation [59,71]. In the case of INO1, the mechanism by which the recently repressed gene is localized at the nuclear periphery is distinct from the mechanism by which the active gene is localized at the nuclear periphery [72,●●73]. Whereas localization of active INO1 to the nuclear periphery requires the GRS DNA zip codes, localization of recently repressed INO1 to the nuclear periphery requires a DNA zip code called the Memory Recruitment Sequence (MRS; Figure 2). In the context of the INO1 promoter, the MRS only functions after INO1 has been repressed and the two targeting mechanisms are independent [●●73]. The targeting mediated by these two elements requires different NPC proteins, is regulated differently through the cell cycle [72] and leads to distinct biochemical interactions with the NPC [●●73]. Whereas GRS-mediated targeting of active INO1 to the NPC promotes robust transcription, MRS-mediated targeting of recently repressed INO1 to the NPC alters the chromatin state of the promoter and primes the gene for reactivation [●●73].
We do not understand how the interaction of genes with the NPC or nucleoporins promotes transcription or alters chromatin structure. In yeast, nucleoporins interact with the promoters of genes like GAL1 and INO1 and this interaction is important for transcription [●●63,64]. It is possible that, as with the Sir proteins, factors that promote transcription are concentrated at the NPC and targeting improves the efficiency of recruiting such factors. Alternatively, perhaps the NPC provides a stable surface on which three-dimensional events such as chromatin remodeling or gene looping are more efficient. Although there is no evidence that gene looping requires interaction with the NPC during transcription, the memory of recent GAL gene transcription involves a stable gene loop in association with the NPC (Figure 2B) [●74, ●75]. This interaction requires the Tpr homologue Mlp1 [●75] and is distinct from the mechanism used by the INO1 gene, requiring the SWI/SNF chromatin remodeling complex (Figure 2B) [76]. Because looping does not persist as long as GAL gene transcriptional memory, it remains to be seen if there are multiple mechanisms by which memory is conferred [71]. Finally, because nucleoporins can promote transcription in the nucleoplasm, it is also possible that these proteins have a novel, direct function in transcription. If so, then perhaps these functions could be carried out in association with the NPC in some organisms and in the nucleoplasm in other organisms.
Concluding remarks
The two phenomena highlighted in this brief review represent the best-characterized changes in gene positioning associated with changes in gene expression. Given the lack of true compartmentalization of the nucleus, we propose that the spatial organization of the nucleus is achieved by: 1) protein-protein interactions that create spatial heterogeneities within the nucleus (eg. Sir protein foci that promote silencing of subtelomeric genes or transcription factories that could promote transcription of coregulated genes; Figure 1), 2) the folding of chromosomes such that individual genes can access the appropriate subnuclear compartments (eg. Figure 1), 3) the stabilization of both chromatin conformation and subnuclear compartments through interaction with surfaces such as the nuclear envelope (eg. the lamina or the NPC) and 4) molecular mechanisms that allow regulated movement of genes from one compartment to another. The two phenomena discussed here illustrate these ideas. If the nuclear lamina both concentrates repressive factors and interacts with heterochromatin, it could create a positive feedback mechanism to both package and repress large parts of the genome (Figure 1). The NPC and transcription factories may represent stable sites to which co-regulated genes are targeted and achieve more robust transcription through colocalization and concentration of shared factors (Figures 1 & 2). Thus, subnuclear compartments need not always be pre-existing entities but might be produced through conditional interactions among genes. A better understanding of the molecular basis for localization to these sites, and how localization impacts transcription, will allow us to answer a number of fascinating questions. How do DNA elements confer conditional localization to particular subnuclear sites? Why is the expression of different genes affected differently by localization to the same subnuclear compartment? To what extent is localization really important? In other words, are genes targeted to particular places/factories within the nucleus and, if so, does this matter for their proper expression? The answers to these questions will provide important insights into how genomes function within cells.
Acknowledgements
We thank Sara Ahmed, Will Light, Lauren Meldi and Donna Garvey Brickner for helpful comments on the manuscript. J.H.B is supported by NIH grant GM080484 and a W.M. Keck Young Scholars in Medical Research Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- 3.Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- 4.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 5.Taddei A, Hediger F, Neumann FR, Gasser SM. The function of nuclear architecture: a genetic approach. Annu Rev Genet. 2004;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- 6.Taddei A, Van Houwe G, Nagai S, Erb I, van Nimwegen E, Gasser SM. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res. 2009;19:611–625. doi: 10.1101/gr.083881.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 8.Andrulis ED, Zappulla DC, Ansari A, Perrod S, Laiosa CV, Gartenberg MR, Sternglanz R. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol Cell Biol. 2002;22:8292–8301. doi: 10.1128/MCB.22.23.8292-8301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●9.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. In this study, the authors show that artificially tethering immunoglobulin genes to the inner nuclear membrane leads to transcriptional repression. Furthermore, they show that these loci interact with the nuclear lamina in cells in which they are repressed.
- ●10.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. This study shows that artificially tethering human genes to the nuclear membrane suppresses expression of some, but not all, of the genes nearby. Repression was dependent on histone deacetylase activity.
- 11.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 12.Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 2008;22:489–498. doi: 10.1101/gad.1634608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hewitt SL, High FA, Reiner SL, Fisher AG, Merkenschlager M. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur J Immunol. 2004;34:3604–3613. doi: 10.1002/eji.200425469. [DOI] [PubMed] [Google Scholar]
- 15.Lee H, Quinn JC, Prasanth KV, Swiss VA, Economides KD, Camacho MM, Spector DL, Abate-Shen C. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 2006;20:784–794. doi: 10.1101/gad.1392006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 17.Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●18.Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 2010;24:766–782. doi: 10.1101/gad.559610. This study establishes a system for observing the localization of developmentally regulated transgenes during differentiation in a living animal. In developing worms, promoters of tissue-specific genes localize to the nuclear interior in cells in which they are expressed and the nuclear periphery in cells in which they are repressed. Furthermore, large arrays of transgenes become heterochromatic and associate with the nuclear envelope, regardless of the promoters in the array.
- 19.Belmont AS. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 22.Kind J, van Steensel B. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol. 2010;22:320–325. doi: 10.1016/j.ceb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- ●●23.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. Using genome-wide DAM ID, the authors determine the spatial behavior of the mouse genome in mouse embryonic stem cells and through differentiation. They show that hundreds of sites in the genome, including both individual genes and blocks of genes change their association with the lamina during differentiation. As in other studies, the association with the nuclear lamina was correlated with transcriptional silencing.
- 24.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee DC, Welton KL, Smith ED, Kennedy BK. A-type nuclear lamins act as transcriptional repressors when targeted to promoters. Exp Cell Res. 2009;315:996–1007. doi: 10.1016/j.yexcr.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holaska JM, Wilson KL. An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry. 2007;46:8897–8908. doi: 10.1021/bi602636m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Somech R, Shaklai S, Geller O, Amariglio N, Simon AJ, Rechavi G, Gal-Yam EN. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J Cell Sci. 2005;118:4017–4025. doi: 10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- 28.Nili E, Cojocaru GS, Kalma Y, Ginsberg D, Copeland NG, Gilbert DJ, Jenkins NA, Berger R, Shaklai S, Amariglio N, et al. Nuclear membrane protein LAP2beta mediates transcriptional repression alone and together with its binding partner GCL (germ-cell-less) J Cell Sci. 2001;114:3297–3307. doi: 10.1242/jcs.114.18.3297. [DOI] [PubMed] [Google Scholar]
- 29.Sadoni N, Langer S, Fauth C, Bernardi G, Cremer T, Turner BM, Zink D. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J Cell Biol. 1999;146:1211–1226. doi: 10.1083/jcb.146.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilchrist S, Gilbert N, Perry P, Bickmore WA. Nuclear organization of centromeric domains is not perturbed by inhibition of histone deacetylases. Chromosome Res. 2004;12:505–516. doi: 10.1023/B:CHRO.0000034892.64739.ff. [DOI] [PubMed] [Google Scholar]
- 31.Taddei A, Gasser SM. Multiple pathways for telomere tethering: functional implications of subnuclear position for heterochromatin formation. Biochim Biophys Acta. 2004;1677:120–128. doi: 10.1016/j.bbaexp.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. Embo J. 2004;23:1301–1312. doi: 10.1038/sj.emboj.7600144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taddei A, Gartenberg MR, Neumann FR, Hediger F, Gasser SM. Multiple pathways tether telomeres and silent chromatin at the nuclear periphery: functional implications for sir-mediated repression; Novartis Found Symp; 2005; pp. 140–156. discussion 156-165, 227-130. [PubMed] [Google Scholar]
- 34.Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- 35.Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 2004;119:955–967. doi: 10.1016/j.cell.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●37.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. This study provides a genome-wide analysis within mouse erythroid cells, revealing intra- and inter- chromosomal associations between the active mouse globin genes and numerous other co-regulated genes. The genes colocalize with transcription factories and colocalization requires the Klf transcription factor.
- 38.Xu M, Cook PR. The role of specialized transcription factories in chromosome pairing. Biochim Biophys Acta. 2008;1783:2155–2160. doi: 10.1016/j.bbamcr.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 39.Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozubek S, Lukasova E, Ryznar L, Kozubek M, Liskova A, Govorun RD, Krasavin EA, Horneck G. Distribution of ABL and BCR genes in cell nuclei of normal and irradiated lymphocytes. Blood. 1997;89:4537–4545. [PubMed] [Google Scholar]
- 41.Kozubek S, Lukasova E, Mareckova A, Skalnikova M, Kozubek M, Bartova E, Kroha V, Krahulcova E, Slotova J. The topological organization of chromosomes 9 and 22 in cell nuclei has a determinative role in the induction of t(9,22) translocations and in the pathogenesis of t(9,22) leukemias. Chromosoma. 1999;108:426–435. doi: 10.1007/s004120050394. [DOI] [PubMed] [Google Scholar]
- 42.Neves H, Ramos C, da Silva MG, Parreira A, Parreira L. The nuclear topography of ABL, BCR, PML, and RARalpha genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood. 1999;93:1197–1207. [PubMed] [Google Scholar]
- 43.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- 44.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 46.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 47.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palstra FP, O’Connell MF, Ruzzante DE. Population structure and gene flow reversals in Atlantic salmon (Salmo salar) over contemporary and long-term temporal scales: effects of population size and life history. Mol Ecol. 2007;16:4504–4522. doi: 10.1111/j.1365-294X.2007.03541.x. [DOI] [PubMed] [Google Scholar]
- 49.Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- ●50.Luo L, Gassman KL, Petell LM, Wilson CL, Bewersdorf J, Shopland LS. The nuclear periphery of embryonic stem cells is a transcriptionally permissive and repressive compartment. J Cell Sci. 2009;122:3729–3737. doi: 10.1242/jcs.052555. Using quantitative immunofluorescence to localize chromatin modifications associated with active and repressed genes as well as FISH to localize individual genes, the authors find that both repressed and active genes localize to the nuclear periphery in mouse embryonic stem cells.
- 51.Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J, Stein S, Vincent S, Molloy KR, Kreth G, et al. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol. 2006;174:27–38. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malhas A, Lee CF, Sanders R, Saunders NJ, Vaux DJ. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J Cell Biol. 2007;176:593–603. doi: 10.1083/jcb.200607054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 54.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarma NJ, Haley TM, Barbara KE, Buford TD, Willis KA, Santangelo GM. Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics. 2007;175:1127–1135. doi: 10.1534/genetics.106.068932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 58.Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brickner DG, Cajigas IC, Fondufe-Mittendorf Y, Ahmed S, Lee P-C, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci U S A. 2005;102:5749–5754. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. Embo J. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- ●●63.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. This study identifies two Gene Recruitment Sequences (GRSs) within the promoter regions of certain genes in yeast that act as “zip codes” in targeting them to the nuclear periphery upon activation. Targeting requires interaction with the NPC, and full transcriptional activation of GRS-containing genes requires targeting to the NPC.
- 64.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 65.Bachhawat N, Ouyang Q, Henry SA. Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J Biol Chem. 1995;270:25087–25095. doi: 10.1074/jbc.270.42.25087. [DOI] [PubMed] [Google Scholar]
- 66.Ambroziak J, Henry SA. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- 67.Schwank S, Ebbert R, Rautenstrauss K, Schweizer E, Schuller HJ. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:230–237. doi: 10.1093/nar/23.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●69.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- ●●70.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. In these two studies, the authors discovered that, in Drosophila, nucleoporins can interact with genes both at the nuclear pore complex and in the nucleoplasm. These two classes of genes are distinct in terms of their function and their expression. They also find that interaction of nucleoporins with certain genes is important for their proper expression.
- 71.Brickner JH. Transcriptional memory at the nuclear periphery. Curr Opin Cell Biol. 2009;21:127–133. doi: 10.1016/j.ceb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brickner DG, Brickner JH. Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Mol Biol Cell. 2010;21:3421–3432. doi: 10.1091/mbc.E10-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●73.Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell. 2010;40:112–125. doi: 10.1016/j.molcel.2010.09.007. This study reports that the INO1 gene employs two distinct mechanisms of targeting to the nuclear pore complex. When active, INO1 is targeted to the NPC via GRSs in its promoter. After repressed, INO1 remains associated with the NPC via a Memory Recruitment Sequence (MRS). This interaction alters the chromatin structure of the INO1 promoter, allowing RNA polymerase II to bind to the repressed promoter and priming the gene for faster reactivation.
- ●74.Philippe J-P, Singh BN, Krishnamurthy S, Hampsey M. A physiologcal role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●75.Tan-Wong SM, Wijayatilake HD, Proudfoot N. Gene loops function ot maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. These two studies show that GAL genes form loops that remain intact after repression by a mechanism involving the NPC-associated protein Mlp1. Looping is required for faster reactivation of the GAL genes.
- 76.Kundu S, Horn PJ, Peterson CL. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]