Summary
Retinal photoreceptor phosphodiesterase (PDE6), a key enzyme for phototransduction, consists of a catalytic subunit complex (Pαβ) and two inhibitory subunits (Pγs). Pαβ has two non-catalytic cGMP-binding sites. Here, using bovine PDE preparations, we show the role of these cGMP-binding sites in PDE regulation. Pαβγγ and its transducin-activated form, Pαβγ, contain two and one cGMP, respectively. Only Pαβγ shows [3H]cGMP binding with a Kd ∼50 nM and Pγ inhibits the [3H]cGMP binding. Binding of cGMP to Pαβγ is suppressed during its formation, implying that cGMP binding is not involved in Pαβγγ activation. Once bound to Pαβγ [3H]cGMP is not dissociated even in the presence of a 1000-fold excess of unlabeled cGMP, binding of cGMP changes the apparent Stokes radius of Pαβγ, and the amount of [3H]cGMP-bound Pαβγ trapped by a filter is spontaneously increased during its incubation. These results suggest that Pαβγ slowly changes its conformation after cGMP binding, i.e., after formation of Pαβγ containing two cGMPs. Binding of Pγ greatly shortens the time to detect the increase in the filter-trapped level of [3H]cGMP-bound Pαβγ, but alters neither the level nor its Stokes radius. These results suggest that Pγ accelerates the conformational change, but does not add another change. These observations are consistent with the view that Pαβγ changes its conformation during its deactivation and that the binding of cGMP and Pγ is crucial for this change. These observations also imply that Pαβγγ changes its conformation during its activation and that release of Pγ and cGMP is essential for this change.
Keywords: PDE, PDE6, cGMP binding, GAF domains, transducin, retinal phototransduction, G protein-mediated signal transduction, cGMP-binding-dependent protein conformational change
Introduction
Cyclic GMP phosphodiesterase (EC 3.1.4.17), classified as PDE6 in the PDE family, is one of the key enzymes for phototransduction in retinal photoreceptor OS. Its activation is G protein-mediated: illuminated rhodopsin stimulates GTP/GDP exchange on Tα followed by dissociation of GTP-Tα from Tβγ. The GTP-Tα activates PDE, resulting in a decrease in the cytoplasmic [cGMP], closure of cGMP-gated channels, and hyperpolarization of plasma membranes (1-3).
The inactive form of rod PDE is composed of a catalytic subunit complex, Pαβ, and two inhibitory subunits, Pγs, i.e., Pαβγγ (4-10). A study using electron microscopy and image analysis of single particles (11) shows that bovine Pαβγγ, 150 × 108 × 60 Å, has the shape of a flattened bell with a handle-like protrusion (∼30 Å) and that the structure is divided into three distinct substructures by two holes. Except for the protrusion, the structure also appears to consist of two homologous structures arranged side by side. These characteristics are consistent with a model in which Pαβγγ's structure is determined by a dimer of homologous catalytic subunits consisting of two GAF regions and one catalytic region. Indeed, bovine Pαβγγ contains two cGMPs and these cGMPs bind tightly to substructures formed by GAF regions (12). These two substructures, called the non-catalytic cGMP-binding sites, are similar, but not identical, in shape and size (11). This implies that the manner of cGMP binding to each site and/or the role of cGMP binding to each site in PDE regulation, if present, may be different.
The current predominant model for PDE regulation is simple (13). For activation, GTP-Tα interacts with Pγ in Pαβγγ and the GTP-Tα·Pαβγγ complex, without altering the firm interaction between Pαβ and Pγ, expresses a high cGMP-hydrolytic activity. For deactivation, GTP in the GTP-Tα·Pαβγγ complex is hydrolyzed with help of RGS9 and accessory proteins, i.e., the GTP is hydrolyzed after formation of a huge complex, and then Pαβγγ is recovered after dissociation of various proteins including GDP-Tα. This model conveniently explains rapid activation and deactivation of PDE; however, there is no clear evidence to show a firm and continuous interaction between GTP-Tα and Pαβγγ during Pαβγγ activation as would be shown by the isolation of a complex of Pαβγγ with Tα containing a hydrolysis-resistant GTP analogue such as GTPγS. In addition, there is no definitive evidence to prove the formation of a GTP-Tα·Pαβγγ complex containing RGS9 and accessory proteins and its decomposition during deactivation of GTP-Tα-activated PDE.
Binding of cGMP to the non-catalytic site in Pαβ is believed to be involved in PDE regulation. Two models, the cGMP-regulated Pαβ-Pγ interaction model (14-18) and the cGMP-binding-direct regulation model (19), have been proposed to explain the role of cGMP-binding sites in PDE regulation. In the former model, the interaction between Pαβ and Pγ is dependent upon cGMP-presence at the non-catalytic site: When the non-catalytic sites of Pαβγγ are saturated with cGMP, GTP-Tα activates Pαβγγ without changing the tight interaction between Pαβ and Pγ, i.e., a GTP-Tα·Pαβγγ complex is formed and the complex expresses a high PDE activity. On the other hand, when the non-catalytic sites are not saturated, GTP-Tα activates Pαβγγ through dissociation of Pγ complexed with GTP-Tα, i.e., a Pγ-depleted PDE(s) is produced. Pγ in the GTP-Tα complex enhances GTPase activity of Tα, the resulted GDP-Tα instantly releases Pγ, and the released Pγ deactivates the GTP-Tα-activated PDE. In the latter model, binding of cGMP to the non-catalytic sites directly regulates PDE catalytic activity. These two models appear to explain some observations of cGMP binding to non-catalytic sites. However, as discussed later, these models have many ambiguous and controversial points. Thus, it is difficult to integrate these concepts smoothly into a coherent model for PDE regulation.
We have recently challenged the dominant model for PDE regulation by proposing a new and comprehensive model (11, 13, 20). In this model, GTP-Tα activates Pαβγγ by forming a complex with a Pγ thereby dissociating the Pγ·GTP-Tα complex. This occurs on membranes and is independent of the cytoplasmic [cGMP]. A significant portion of the Pγ·GTP-Tα complex is then released to the soluble fraction. Thus, Pαβγ is the GTP-Tα-activated PDE. After hydrolysis of GTP, both soluble and membranous Pγ·GDP-Tα complexes deactivate Pαβγ without liberating Pγ. These Pγ·GDP-Tα complexes appear to have a preferential order in deactivating Pαβγ. This new model is based on the following observations: 1) Pαβγ, but not Pαβ, is isolated only when OS homogenates are incubated with GTPγS; 2) The ratio of Pγ/Pαβ in Pαβγγ and Pαβγ is 2:1; 3) The enzymatic activity of Pαβγ is ∼12 times higher than that of Pαβγγ and inhibited by 30 nM Pγ; 4) The basic structure of these PDE species is not changed when Pαβγγ is shifted to Pαβγ; 5) Pγ·GTPγS-Tα is isolated from membranous and soluble fractions; 6) Both membranous and soluble Pγ·GDP-Tα complexes deactivate Pαβγ without liberating Pγ; 7) The membranous Pγ·GDP-Tα complex appears to be consumed earlier than the soluble Pγ·GDP-Tα complex; and 8) PDE regulatory mechanisms similar to this model are also found in mammalian and amphibian photoreceptors as well as in rods and cones. During these studies, we have also shown that 1) the interaction between Pαβγγ and GTPγS-Tα is short-lived, indicating that GTP-Tα·Pαβγγ is an intermediate, but not the GTP-Tα-activated PDE; 2) free Pγ is not detected in any preparations, implying that Pγ always forms complexes with other proteins; 3) Pαβγγδ and Pαβγδδ are formed when Pαβγγ and Pαβγ are solubilized with Pδ, a prenyl-binding protein; 4) the stoichiometry of Pαβγγδ suggests that only one lipid-moiety may be involved in the interaction of Pαβγγ with membranes; and 5) the stoichiometry of Pαβγδδ suggests that a lipid-moiety in Pαβ is also affected by Pγ dissociation.
In this study, we extend our model by integrating the role of cGMP binding to the non-catalytic site. We demonstrate that Pαβγγ and Pαβγ contain two and one cGMP, respectively, that only Pαβγ expresses [3H]cGMP-binding activity and that Pγ inhibits [3H]cGMP binding to Pαβγ. We also show that the cGMP binding to Pαβγ is suppressed during Pαβγγ activation, i.e., cGMP binding is not involved in Pαβγγ activation. We also suggest that cGMP binding to Pαβγ slowly changes its conformation and that binding of Pγ accelerates the conformational change. Based on these studies, we propose that binding of cGMP to Pαβγ is the first step in PDE deactivation.
Results
Binding of [3H]cGMP to OS membranes
Bovine OS membranes contain a [3H]cGMP-binding site(s) (Fig. 1A). Both GTPγS-treated and non-treated membranes showed [3H]cGMP-binding activities; however, the activity in GTPγS-treated membranes was much higher than that in GTPγS-non-treated membranes, indicating that GTPγS-Tα somehow enhances the [3H]cGMP-binding activity. On the contrary, the soluble fraction, whether obtained from GTPγS-treated or non-treated OS homogenates, showed only a negligible [3H]cGMP-binding activity (data not shown). This suggests that no protein in the soluble fraction contains the [3H]cGMP-binding site and/or expresses the [3H]cGMP-binding activity under our experimental conditions.
Figure 1.

Binding of [3H]cGMP to membranous PDE. A. Levels of [3H]cGMP binding to OS membranes treated with or without GTPγS. OS homogenates (27.5 mg protein) were suspended in 18.4 ml of Buffer A and divided into two portions. After incubation of a portion with 50 μM GTPγS overnight on ice, its membranes were washed with 5 ml Buffer A supplemented with 50 μM GTPγS (×2) and 5 ml Buffer A (×2) and suspended in 5 ml Buffer A. The other portion was treated in the same way but without GTPγS. Binding of [3H]cGMP to these suspensions (10 μl) was assayed using 1 μM [3H]cGMP. B and C. [3H]cGMP binding to proteins extracted from OS membranes treated with or without GTPγS. OS homogenates (27.7 mg protein) were suspended in 18 ml of Buffer A, divided into two portions and treated with or without GTPγS. Proteins were extracted from membranes with 3 ml Buffer B (×7), concentrated to ∼0.5 ml and applied to Bio-Gel A-0.5 m column. [3H]cGMP-binding activity (B) and PDE activity (not shown) were assayed using 60 and 5 μl of the fraction, respectively. Protein profiles in the fraction (90 μl) were analyzed by SDS-PAGE and staining with Coomassie blue (C). The left end lane shows Mr of standard proteins, 94, 67 and 43 kDa.
Solubilization and isolation of membranous proteins showed that a [3H]cGMP-binding activity (Fig. 1B) was detected only in the fraction containing a protein-double (Mr ∼88 kDa) (Fig. 1C) and that the activity appeared to be proportional to the level of the protein-doublet. These fractions also contained a PDE activity and the activity was proportional to the level of the protein-doublet (data not shown). The protein-doublet has been identified as Pαβ and 70-80% of Pαβ is extracted from membranes under these conditions (13, 20). These results suggest that the [3H]cGMP-binding activity in membranes is due to a Pαβ complex(s). This implies that cone PDEs, Pα′α′/Pγ′ complexes, are also present and that a Pα′α′/Pγ′ complex(s) expresses a [3H]cGMP-binding activity. However, neither Pα′ nor its [3H]cGMP-binding activity could be identified. These failures, we believe, are due to its small abundance in OS. The soluble fraction also contained a Pαβ/Pγ complex (peak b in Ref. 13); however, the complex showed only a negligible [3H]cGMP-binding activity (data not shown). This is consistent with the above-mentioned conclusion that [3H]cGMP-binding activity was not detected in the soluble fraction.
Interestingly, the [3H]cGMP-binding activity in GTPγS-treated PDE was higher than that in GTPγS-non-treated PDE (Fig. 1B). When OS homogenates are incubated with GTPγS, the Pαβ content in membranes is increased 20-30% by binding of the Pαβ/Pγ complex existing in the soluble fraction (13). Therefore, binding of the Pαβ/Pγ complex to membranes and resulting expression of a [3H]cGMP-binding activity could increase the activity in membranes. However, the increase by GTPγS in the activity was much higher, ∼2.4 times (Fig. 1B). In addition, Pαβ in the Pαβ/Pγ complex has at most two cGMP-binding sites (12). Therefore, we conclude that even if the Pαβ/Pγ complex could express a [3H]cGMP-binding activity, the greater part of the increase is due to an increase in the activity of a Pαβ/Pγ complex(s) located on membranes. This is unexpected because previous studies using frog PDE/membranes (21, 22) showed that their [3H]cGMP-binding activity in GTP-non-treated PDE was much higher than that in GTP-treated PDE. We also note that this result, with the observation showing in Figure 1A, implies that [3H]cGMP binding to solubilized PDE species appears to be similar to that to membranous PDE species, i.e., properties of cGMP binding to membranous PDE species may be estimated by studying cGMP binding to solubilized PDE species.
Identification of PDE species expressing [3H]cGMP-binding activity
GTPγS-non-treated membranes contain Pαβγγ, and GTPγS-treated membranes have Pαβγγ and Pαβγ as major species and a Pαβ/Pγ complex as a minor species (20). These PDE species were extracted with a hypotonic buffer (Fig. 2A) or Pδ in an isotonic buffer (Fig. 2C) and their [3H]cGMP-binding activities were measured after isolation. The use of Pδ in an isotonic buffer may exclude a possible artifact(s) caused by the hypotonic extraction. OS homogenates were also treated with GTPγS in the presence of cGMP (GTPγS + cGMP), and after isolation of Pαβ/Pγ complexes, their [3H]cGMP-binding activities were measured (Fig. 2B). The result will be compared with the results shown in Figure 2A, as shown later.
Figure 2.

Binding of [3H]cGMP to PDE species extracted from OS membranes. A and B. PDE species extracted with a hypotonic buffer. Details of the procedure are in Experimental procedures. OS homogenates (50.4 mg protein) were suspended in 20 ml Buffer A and divided into three portions. After incubation with cGMP (A, upper panel), GTPγS (A, lower panel) or cGMP + GTPγS (B), proteins were extracted with Buffer B (a hypotonic buffer), applied to a TSK-DEAE 5PW column and eluted. Fractions containing PDE species were determined by SDS-PAGE and assaying PDE activity. Elution profiles of the 88-kDa-protein, Pαβ, are shown in each panel. Elution profile of other proteins was shown in Ref 20. PDE species were identified as described (20). Binding of [3H]cGMP to the fraction (60 μl) was measured with 0.5 μM [3H]cGMP. C. PDE species extracted with Pδ in an isotonic buffer. OS homogenates (12.4 mg) were suspended in 13 ml of Buffer A and divided into two portions. After incubation of a portion with GTPγS (50 μM) for 1 h on ice, membranes were washed with 2 ml of Buffer A containing GTPγS (50 μM) and 2 ml of Buffer A. The other portion was treated in the same way but without GTPγS. These membranes were suspended in 2.5 ml of Buffer D, incubated with Pδ (final 3 μM) overnight on ice, and washed with 2 ml of Buffer D (×2). All supernatants were collected and applied to a TSK-DEAE 5PW column. Rod and cone PDE species and their stoichiometry and transducin subunits were identified as described (20). Binding of [3H]cGMP to the fraction (50 μl) was measured with 0.5 μM [3H]cGMP (upper panel). Protein profiles in fractions (40 μl) were analyzed by SDS-PAGE and staining with Coomassie blue (lower panel). Due to the limited space, only results from GTPγS-treated membranes are shown. Profiles of PDE species from GTPγS-non-treated membranes are in Ref. 20.
Pαβγγ extracted by a hypotonic buffer. Pαβγγ was obtained from GTPγS-non-treated membranes (upper panel) and GTPγS-treated membranes (lower panel) (Fig. 2A). In the former preparation, the [3H]cGMP-binding activity appeared to be proportional to the level of Pαβ, implying that the Pαβγγ may express a [3H]cGMP-binding activity. However, the molecular ratio of [3H]cGMP/Pαβ was less than 0.01, indicating that only a negligible portion of the Pαβγγ expresses this activity. In the latter preparation, a small [3H]-radioactivity was detected in the fraction close to the Pαβγγ peak. However, the level of [3H]-radioactivity was not proportional to that of Pαβ in the Pαβγγ fraction, indicating that the [3H]-radioactivity is not attributable to [3H]cGMP bound to the Pαβγγ, i.e., the Pαβγγ does not show [3H]cGMP-binding activity and/or the Pαβγγ, when it exists with GTP-Tα, appears to lose a portion that may express the [3H]cGMP-binding activity (upper panel).
-
Pαβγγ extracted with Pδ in an isotonic buffer. The Pαβγγδ preparation was obtained from GTPγS-non-treated membranes (data not shown) and GTPγS-treated membranes (Fig. 2C). In the former preparation, the [3H]cGMP-binding activity appeared to be proportional to the level of Pαβ; however, the molecular ratio of [3H]cGMP/Pαβ in the Pαβγγδ was less than 0.01. These observations are identical to those for Pαβγγ extracted with a hypotonic buffer (Fig. 2A, upper panel). In the latter preparation, Pαβγγδ appeared to show a small [3H]cGMP-binding activity (Fig. 2C, upper panel). However, its amount was not exactly proportional to the Pαβ level in the fraction, indicating that the [3H]-radioactivity was not due to [3H]cGMP bound to the Pαβγγδ.
As shown later (Fig. 7), Pαβγγ can be trapped by the Millipore filter with a high efficiency, implying that the lack of [3H]cGMP-binding activity and/or the negligible level of [3H]cGMP-binding activity in Pαβγγ preparations are not due to the failure to trap the [3H]cGMP-bound Pαβγγ. Taken together, our results strongly suggest that Pαβγγ does not express [3H]cGMP-binding activity and that negligible activities occasionally detected in fractions containing Pαβγγ may be artifacts caused by experimental procedures. The level of [3H]-radioactivity was not proportional to that of Tα (Fig. 2C). This confirms that Tα has no cGMP-binding site (23). The amino-acid sequence of Tα also supports this notion. This is specifically noted here because we will use this information in a later discussion.
Pαβγ and Pαβ/Pγ. Whether extracted with the hypotonic buffer (Fig. 2A, lower panel) or with Pδ in the isotonic buffer (Fig. 2C), fractions containing these PDE species clearly showed [3H]cGMP-binding activities. In addition, the level of Pαβ was proportional to that of [3H]cGMP-binding activity in these fractions. These results indicate that both Pαβγ and Pαβ/Pγ express [3H]cGMP-binding activity.
Figure 7.
Pγ effect on the conformational change of Pαβγ by cGMP binding. A. The time to detect the increase in the level of [3H]cGMP-bound Pαβγ trapped by the filter. The experiment was carried out as a part of the study depicted in Figure 5A. Pαβγ (16.0 μg) was suspended in 640 μl of 55.5 mM Tris·HCl (pH 7.5) containing 4.44 mM EDTA and 1.11 mM IBMX, and [3H]cGMP-binding to the Pαβγ was initiated by adding 80 μl of 9 μM [3H]cGMP. After incubation for 30 min, an aliquot (72 μl) was withdrawn and applied to a Millipore filter, and its radioactivity was designated as the level at time 0. Simultaneously, a mixture (72 μl) of Pγ or its mutant (10 μM) with or without cGMP (10 mM) was added to the assay mixture: (●), with cGMP; and (○), without cGMP. After incubation for 0.25, 0.5, 0.75, 1, 2, 5, 10, and 20 min, an aliquot (80 μl) was withdrawn and applied to a Millipore filter, and its radioactivity was measured. The arrow (↑) indicates the addition of Pγ (or Pγ mutant) with or without cGMP. The arrow (⇦) indicates levels of [3H]cGMP-bound Pαβγ with (▲) or without (△) N18del. The 100% activity indicates that 1.32 pmol of [3H]cGMP was detected in 1.6 μg of Pαβγ (7.72 pmol). Data shown in Figure 5A was used as a control for this study. B. Elution profile of PDE species from a gel filtration column. Purified Pαβγ (70 μg) was incubated with unlabeled cGMP (0.5 mM) in 0.5 ml of 25 mM Tris·HCl, pH 7.5, 0.1 mM EDTA, and 1 mM IBMX for 30 min on ice and applied to a Superdex 200 HR column that had been equilibrated with Buffer E (black). The chromatography conditions are in Experimental Procedure. PDE activity (●) was assayed using 5 μl of the fraction. [3H]cGMP-binding activity (□) was measured using 50 μl of the fraction. Pαβγγ (90 μg) was also applied to the column and eluted in the same manner (red). PDE activity was assayed using 20 μl of the fraction (●). The 100% PDE activity indicates that 12.1 nmol cGMP hydrolyzed/min/tube. [3H]cGMP-binding activity was measured using 50 μl of the fraction (□). The 100% [3H]cGMP-binding activity indicates that 2.4 pmol of [3H]cGMP was detected in the assay mixture.
We emphasize that [3H]cGMP-binding activity in the fraction containing Pαβγδδ (Fig. 2C, upper panel) was similar to that in the fraction containing Pαβγ (Fig. 2A, lower panel), although these activities were apparently different due to use of different amounts of OS homogenates and different volumes of the fraction for the assay. We confirmed this observation by comparing the [3H]cGMP-binding activity of Pαβγ with that of Pαβγδδ (data not shown). These results indicate that Pδ binding to the lipid moiety of Pαβ does not affect the level of [3H]cGMP-binding activity in Pαβγ, implying that membrane binding of Pαβγ may not affect its cGMP-binding activity. This implication also supports our above-mentioned view that properties of cGMP binding to membranous PDE species may be estimated by studying the cGMP binding to solubilized PDE species. We also note that the NaCl-gradient in the study shown in Figure 2C was modified to collect both rod and cone PDEs with fraction numbers similar to those for rod PDEs (Fig. 2A). Therefore, their elution profile was slightly different from that shown in Figure 2A. We have already shown that the elution profile of PDE species containing Pδ is identical to that of PDE species without Pδ when the same NaCl-gradient was used (20). Comparison of the [3H]cGMP-binding activity of cone PDE with that of Pαβγ will be discussed later.
Contents of cGMP in Pαβγγ and Pαβγ
Pαβγγ and Pαβγ were purified from GTPγS-treated OS homogenates (Fig. 3A). These PDE species were clearly separated and characterization of these species including their specific activity and Pγ-sensitivity verified the clear separation (20). We also note that the level of protein staining with Coomassie blue is proportional to the Mr calculated based on its amino acid sequence under our staining conditions, i.e., the Pγ/Pαβ ratios also showed the clear separation (20). Molecular sieve chromatography of these PDE species also showed that the Pγ/Pαβ ratio in these PDE species was not changed during their storage.
Figure 3.

Levels of cGMP contained in Pαβγγ and Pαβγ. Pαβγγ (6.50 μg/20 μl) and Pαβγ (4.75 μg/50 μl) were purified from GTPγS-treated OS homogenates. A. Purity of these PDE preparations. Preparations of Pαβγγ (10 μl) and Pαβγ (25 μl) were applied to SDS-PAGE followed by staining with Coomassie blue. B. Levels of cGMP contained in these PDE species. Contents of cGMP were measured using a cGMP immunoassay kit.
We found that 3.0 pmol of the Pαβγγ contained ∼6.5 pmol of cGMP (Fig. 3B). This indicates that Pαβγγ contains 2 cGMPs. Pαβγγ isolated from GTPγS-non-treated OS homogenates also contained 2 cGMPs (data not shown). These results indicate that non-catalytic sites of Pαβγγ, whether located with or without GTP-Tα, are saturated by cGMP. These results also suggest that the saturation is a reason for the lack of [3H]cGMP-binding activity in Pαβγγ. These Pαβγγ preparations had been exposed to cGMP-free conditions for at least one week. This suggests that these cGMPs bind tightly to Pαβγγ, confirming a previous observation (12).
Pαβγ, 6.0 pmol, contained ∼6.1 pmol of cGMP (Fig. 3B). This indicates that Pαβγ contains one cGMP, i.e., one of the non-catalytic sites in Pαβγ is empty. The possibility that cGMP existing in Pαβγ can be exchanged by [3H]cGMP during the assay of [3H]cGMP binding is quite low, as discussed later. Therefore, we conclude that the [3H]cGMP-binding activity in Pαβγ we observed is due to the binding of [3H]cGMP to the empty site, i.e., [3H]cGMP-bound Pαβγ contains one original cGMP and one [3H]cGMP. These results also indicate that GTP-Tα dissociates not only a single Pγ but also one cGMP from Pαβγγ during its activation. In other words, PDE activation is the mechanism to change Pαβγγ having two cGMPs to Pαβγ having one cGMP, and PDE deactivation is the mechanism to shift Pαβγ having one cGMP to Pαβγγ having two cGMPs. Pαβ/Pγ (Fig. 2A lower panel and 2C upper panel) is a minor species that is difficult to purify (20). Therefore, the content of cGMP in Pαβ/Pγ could not be measured.
Pαβγ was exposed to cGMP-free conditions for more than 3 days. Under these conditions, the molecular ratio of cGMP/Pαβ in Pαβγ is always ∼1.0 (Fig. 3B). This observation suggests that the affinity for cGMP is clearly different in Pαβγγ's two non-catalytic sites and that GTPγS-Tα (GTP-Tα) releases cGMP only from the same one site in Pαβγγ during its activation. This also implies that GTP-Tα dissociates Pγ from the same site in Pαβγγ during its activation.
Characterization of [3H]cGMP binding to Pαβγ
Purified Pαβγ showed a [3H]cGMP-binding activity (Fig. 4A). The level of [3H]cGMP binding reached a plateau as the [3H]cGMP concentration increased. Scatchard plotting of this saturable [3H]cGMP binding (Fig. 4A, Insert) indicates that Pαβγ has one kind of cGMP-binding site with Kd ∼50 nM. This is consistent with the above-mentioned view that [3H]cGMP binds to the same site in Pαβγ. The level of bound [3H]cGMP reached a plateau in less than 2 min under these conditions (Fig. 4B). Unlabeled cGMP, but not cAMP, competitively inhibited the [3H]cGMP binding (Fig. 4C). This indicates that the [3H]cGMP-binding site in Pαβγ is cGMP-specific.
Figure 4.
Binding of [3H]cGMP to Pαβγ. A. Concentration of [3H]cGMP. [3H]cGMP binding to Pαβγ (1.92 μg) was measured with indicated concentrations of [3H]cGMP. The [3H]cGMP-binding activity was analyzed by Scatchard plotting (Insert). B. Time-course. Pαβγ (17.3 μg) was incubated in 55 mM Tris·HCl, (pH 7.5) containing 4.4 mM EDTA and 1.1 mM IBMX (final volume, 720 μl) on ice for 10 min. The [3H]cGMP binding was initiated by adding 80 μl of 10 μM [3H]cGMP. After incubation for indicated periods, an aliquot (80 μl) was taken and applied to a Millipore filter. C. The cyclic nucleotide-specificity. After incubation of Pαβγ (1.92 μg) with indicated concentration of unlabeled cGMP (●) or cAMP (○) on ice for 10 min, [3H]cGMP binding was measured with 1 μM [3H]cGMP. The 100% activity indicates that 1.46 pmol [3H]cGMP bound to Pαβγ in tubes. D. Levels of [3H]cGMP-bound Pαβγ trapped by the filter. OS homogenates (18.9 mg protein) were suspended in 9.7 ml of Buffer A. After isolation by the TSK-DEAE 5PW column chromatography and concentration to 0.3 ml, the Pαβγ preparation (∼80 μg) was incubated with 1 μM [3H]cGMP for 30 min on ice and applied to a TSK 250 column that had been equilibrated with Buffer D. The level of [3H]cGMP bound to Pαβγ was calculated based on the [3H]-radioactivity in 70 μl of the fraction (●). The fraction (70 μl) was also applied to a Millipore filter and the [3H]-radioactivity on the filter was measured (□). Only fractions containing Pαβγ were shown. Insert: The rate of [3H]-radioactivity on the filter per the level of [3H]-radioactivity in the fraction. The 100% radioactivity indicates the [3H]-radioactivity detected in fraction 15. Fraction 15 (70 μl) contained 3.2 μg Pαβγ (15.5 pmol) and 13.1 pmol of [3H]cGMP.
Trapping of [3H]cGMP bound Pαβγ to a Millipore filter
After incubation with [3H]cGMP, Pαβγ was applied to a molecular sieve column and the amount of [3H]cGMP bound to Pαβγ was calculated based on the [3H]-radioactivity in the Pαβγ fraction (Fig. 4D). We found that 70 μl of fraction 15, the peak fraction, contained 3.2 μg Pαβγ (15.5 pmol) and 13.1 pmol of [3H]cGMP, i.e., ∼83% of Pαβγ in the fraction was occupied by [3H]cGMP. The average level of the occupation was ∼86% in three experiments. These results indicate that ∼100% of Pαβγ binds [3H]cGMP under these conditions. The result will also be confirmed later (Fig. 5). However, only ∼17% of the activity was detected when 70 μl of the fraction was applied to the filter and the [3H]cGMP-binding activity was obtained based on the [3H]-radioactivity trapped by the filter (Fig. 4D and its insert). This shows that the Millipore filter traps ∼17% of the [3H]cGMP-bound Pαβγ existing in the assay mixture. We could not get the result showing that 100% of Pαβγ expressed the [3H]cGMP-binding activity. We believe that this is due to an artifact caused by our experimental procedures, because the Pαβγ preparation we obtained appears to contain one kind of Pαβγ (20) and [3H]cGMP, once bound to Pαβγ, is not dissociated even in the presence of a 1000-fold excess of unlabeled cGMP (Fig. 5). In Figure 4D, fraction 15 apparently showed that 100% of Pαβγ binds [3H]cGMP. This is due to our intention to show the ratio of [3H]cGMP-binding activity measured by the filter. By the way, ∼18.2% of the [3H]cGMP-bound Pαβγ in the assay mixture was trapped by the filter in the result shown as Figure 4A. We emphasize that this low rate does not affect the properties shown in Figure 4A, 4B and 4C, because these properties are not affected by the low efficiency of the filter to trap [3H]cGMP-bound Pαβγ.
Figure 5.
Change of Pαβγ's characteristics by cGMP binding. A. Dissociation of [3H]cGMP bound to Pαβγ. Purified Pαβγ (16.0 μg) suspended in 640 μl of 55.5 mM Tris·HCl (pH 7.5) containing 4.44 mM EDTA and 1.11 mM IBMX, and [3H]cGMP binding was initiated by adding 80 μl of 9 μM [3H]cGMP. After incubation for 30 min on ice, an aliquot (72 μl) was withdrawn and applied to a Millipore filter, and its radioactivity was designated as the level at time 0. Simultaneously, 72 μl of 10 mM unlabeled cGMP (●) or water (○) was added to the assay mixture. After incubation for 0.25, 0.5, 0.75, 1, 2, 5, 10, and 20 min, an aliquot (80 μl) was withdrawn and applied to a Millipore filter, and its [3H]-radioactivity was measured. The arrow indicates the addition of cGMP or water. The 100% activity indicates that 1.32 pmol of [3H]cGMP was detected in 1.6 μg of Pαβγ (7.72 pmol). B. Elution profile of Pαβγ from a gel filtration column. Purified Pαβγ (70 μg) was incubated with (black) or without (red) unlabeled cGMP (0.5 mM) in 0.5 ml of 25 mM Tris·HCl, pH 7.5, 0.1 mM EDTA, and 1 mM IBMX for 30 min on ice and applied to a Superdex 200 HR column that had been equilibrated with Buffer E. Detailed conditions for this elution are in Experimental Procedure. PDE activity was assayed using 5 μl of the fraction (●). The 100% PDE activity indicates that 12.5 nmol cGMP hydrolyzed/min/tube. [3H]cGMP binding activity was measured using 50 μl of the fraction (□). The 100% activity indicates that 1.50 pmol of [3H]cGMP was detected in the assay mixture.
GTPγS-Tα-activated and Pδ-extracted cone PDE, Pα′α′γ′δδ (20), expressed a [3H]cGMP-binding activity (Fig. 2C, upper panel). We note that all Pα′α′γ′γ′ complexes present were activated to Pα′α′γ′ under our conditions (20). Interestingly, the level of [3H]cGMP-binding activity in fraction 13 was ∼ 5 times higher than that of fraction 27 (Fig. 2C, upper panel). A similar observation was also obtained when these PDEs were extracted with a hypotonic buffer (data not shown). These results indicate that ∼85% of [3H]cGMP-bound Pα′α′γ′ was trapped by the Millipore filter under the following assumptions: (a) Contents of Pαβ and Pα′α′ in these fractions are similar, (b) [3H]cGMP binds to all Pα′α′γ′ complexes, (c) Pα′α′γ′ has one cGMP binding site, (d) Pδ binding does not affect the level of [3H]cGMP-binding activity in Pα′α′γ′, and (e) ∼17% of [3H]cGMP-bound Pαβγ is trapped by the Millipore filter. We note that the level of protein staining with Coomassie blue is proportional to the Mr calculated based on its amino acid sequence under our staining conditions (20). Thus, amounts of Pαβ and Pα′α′ can be compared by comparing their staining levels in the same gel. We found that the stained level of Pαβ was similar to that of Pα′α′ (Fig. 2C, lower panel). This indicates that levels of Pαβ and Pα′α′ are similar, i.e., assumption a was proven. As described, we found that the [3H]cGMP-binding activity of Pα′α′γ′δδ was similar to that of Pα′α′γ′, i.e., assumption d was proven. Assumption e was also proven, as described above. Assumptions b and c are not yet proven; however, these assumptions are reasonable if characteristics of the [3H]cGMP binding to Pαβγ are taken into consideration. Therefore, we conclude that the low trapping rate is specific to [3H]cGMP-bound Pαβγ.
Conformational change of Pαβγ by cGMP binding
After incubation with [3H]cGMP for 30-min (i.e., after binding of [3H]cGMP to ∼100% of Pαβγ), dissociation of [3H]cGMP bound to Pαβγ was followed with or without 1 mM unlabeled cGMP (Fig. 5A). We found that the level of [3H]cGMP binding to Pαβγ was not changed even in the presence of 1 mM unlabeled cGMP at least for the first 5 min. Under the similar conditions, [3H]cGMP binding to Pαβγ reached the maximum in less than 2 min (Fig. 4B), indicating that the 5-min incubation was enough to chase [3H]cGMP bound to Pαβγ if indeed the [3H]cGMP could be chased. Therefore, this observation indicates that [3H]cGMP, once bound to Pαβγ, cannot be dissociated. This strongly suggests that Pαβγ, after binding of [3H]cGMP, changes its conformation, especially the conformation of its non-catalytic site and/or a region(s) near the non-catalytic site, and that the Pαβγ, after changing its conformation, firmly holds the [3H]cGMP. A large conformational change initiated by cGMP binding has been reported in one of GAF domains in cone PDE (24). A similar conformational change may occur when [3H]cGMP binds to Pαβγ, i.e., when Pαβγ having one cGMP is shifted to Pαβγ having two cGMPs.
To further prove that binding of cGMP changes Pαβγ's conformation, we directly compared the relative compactness (Stokes radius) of cGMP-treated Pαβγ with that of cGMP-non-treated Pαβγ (Fig. 5B). This method has been used to show a conformational change by cGMP binding in PDE5 (25-27). After incubation of Pαβγ with or without cGMP for 30 min on ice, these Pαβγs were applied to a gel filtration column and PDE activity was measured to identify the fraction containing Pαβγ. The cGMP-non-treated Pαβγ, as expected, was eluted as a single peak with the peak activity in fraction 38. The [3H]cGMP-binding activity was also observed in these fractions. However, the cGMP-treated Pαβγ was eluted as two peaks, the major peak in fraction 34 and the minor peak in fraction 38, and only the Pαβγ in fraction 38 showed the [3H]cGMP-binding activity. These observations indicate that the apparent Stokes radius of cGMP-treated Pαβγ was 4-7 Å larger than that of cGMP-non-treated Pαβγ, i.e., the Stokes radius of Pαβγ appears to be increased when Pαβγ having one cGMP is shifted to Pαβγ having two cGMPs. We note that the difference in the Stokes radius was observed in the Tris buffer; however, the difference was less clear in a phosphate buffer (data not shown). This may be due to a tendency of Pαβγ to change its structure in a Tris buffer (11). We also note that 50 μl of the peak fraction of the cGMP-non-treated Pαβγ contained 2.4 μg Pαβγ (11.6 pmol of Pαβγ) and bound 9.90 pmol [3H]cGMP. This indicates that ∼85% of the Pαβγ expressed [3H]cGMP-binding activity, confirming that almost all Pαβγ complexes show the [3H]cGMP-binding activity (Fig. 4D). We also note that the major peak of the cGMP-treated Pαβγ showed no ability to bind [3H]cGMP, confirming that cGMP, once bound to Pαβγ, is not dissociated (Fig. 5A).
Rate of the conformational change in Pαβγ
The level of [3H]cGMP-binding abruptly increased after a 10-min incubation (Fig. 5A). The level was increased ∼3 times the level at time 0 after 20-min (Fig. 5A) and ∼ 4 times after 40-min (data not shown). Since ∼100% of Pαβγ present bound [3H]cGMP during the pre-incubation, these observations indicate that the amount of [3H]cGMP-bound Pαβγ trapped by the filter abruptly increased.
The incubation of [3H]cGMP-bound Pαβγ was initiated by addition of unlabeled cGMP or water (Fig. 5A). The increase in the trapped level of [3H]cGMP-bound Pαβγ was observed after addition of 1 mM unlabeled cGMP, indicating that the increase is not due to new binding of [3H]cGMP to Pαβγ. The increase was also detected after addition of water, implying that the unlabelled cGMP is not involved in this increase. Addition of unlabeled cGMP or water slightly diluted the mixture, ∼10%; however, it is unlikely that such a small dilution could cause this increase. Modification of the Pαβγ during the incubation could also be ignored because the Pαβγ was pure (Fig. 3A) and the incubation was carried out on ice. Taken together, these observations deny the possibility that the increase is attributed to a reaction occurred during the incubation.
During the pre-incubation, [3H]cGMP bound to Pαβγ. As another important change during the pre-incubation, the buffer in the Pαβγ preparation, a phosphate buffer containing Mg2+, was changed to a Tris buffer containing IBMX but not Mg2+. Pαβγ appears to have a tendency to change its structure in a Tris buffer, but not in a phosphate buffer (11), and Mg2+ binds to Pαβ (28, 29). IBMX may also increase the cGMP affinity of non-catalytic sites, as discussed later. Therefore, these changes might affect Pαβγ's properties and this property change might cause to increase the level of [3H]cGMP-bound Pαβγ trapped by the filter. However, this increase was observed with either Pαβγ stored in the original buffer or in the pre-incubation buffer, a Tris buffer without Mg2+ (data not shown). The increase was also detected with or without IBMX (data not shown). Therefore, these explanations may be disregarded. Modification of Pαβγ during the pre-incubation could also be ignored, as described above. Taken together, these observations strongly suggest that [3H]cGMP binding to Pαβγ during the pre-incubation is the sole reason for the increase in the filter-trapping level of [3H]cGMP-bound Pαβγ, i.e., the increase appears to be caused by a conformational change on Pαβγ upon binding of [3H]cGMP.
This increase in the filter-trapping level of [3H]cGMP-bound Pαβγ was observed only after ∼10-min incubation, i.e., ∼40-min appeared to be required to detect the increase (Fig. 5A). Why is the increase detected after such a long incubation if the increase is due to [3H]cGMP binding? We believe that the conformational change caused by [3H]cGMP binding progresses consistently, but slowly, and that the increase is detected only after the Pαβγ having the altered conformation accumulates to a certain level. In other words, there is a threshold to trap the [3H]cGMP-bound Pαβγ. We emphasize that a mechanism to accelerate the conformational change should be present if this conformational change is indeed involved in PDE regulation.
Suppression of cGMP binding during activation of Pαβγγ
Two possible stages for cGMP binding to Pαβγ are expected in PDE regulation: during Pαβγγ activation to Pαβγ and during Pαβγ deactivation to Pαβγγ. First we investigate whether cGMP binds to Pαβγ during the activation of Pαβγγ to Pαβγ. After incubation of OS homogenates with GTPγS in the presence (Fig, 2B) or absence (Fig. 2A, lower panel) of cGMP, PDE species were extracted with Buffer B and applied to a TSK-DEAE column, and their [3H]cGMP-binding activities were measured. Both OS homogenates were incubated in the presence of IBMX and more than 20% of added cGMP remained in the cGMP-added homogenate when membranes were isolated. We found that the [3H]cGMP binding activity of cGMP-treated Pαβγ appeared to be slightly higher than that of cGMP-non-treated Pαβγ. However, the difference was unclear in other two studies. Therefore, we conclude that cGMP-incubated Pαβγ has the ability to bind [3H]cGMP similar to that in Pαβγ obtained without cGMP. The same result was obtained when Pαβγ was extracted with Pδ in a isotonic buffer (data not shown). Pαβγ, once it binds cGMP, holds the cGMP and cannot accept [3H]cGMP (Fig. 5B). Therefore, the [3H]cGMP-binding activity we observed (Fig. 2B) indicates that Pαβγ cannot bind cGMP during activation of Pαβγγ to Pαβγ.
Pαβ/Pγ, the minor GTPγS-Tα-activated PDE (Fig. 2), lost its [3H]cGMP-binding activity when the fraction containing Pαβ/Pγ was pre-treated with cGMP (data not shown). However, Pαβ/Pγ obtained from cGMP-treated OS homogenates showed a [3H]cGMP-binding activity (Fig. 2B) similar to that of Pαβ/Pγ obtained from cGMP-non-treated homogenates (Figs. 2A, lower panel). This suggests that binding of cGMP to Pαβ/Pγ is suppressed during its formation. Together, our observations indicate that the cGMP-binding activity of GTP-Tα-activated PDE species is suppressed during its formation. This, we believe, is a critical finding to identify the function of cGMP binding in PDE regulation. We note that the Pαβ/Pγ was eluted slightly earlier when OS homogenates were incubated with cGMP, as previously shown (20). The presence of cGMP may be crucial for the early elution; however, the real reason is unknown.
Binding of cGMP during deactivation of Pαβγ
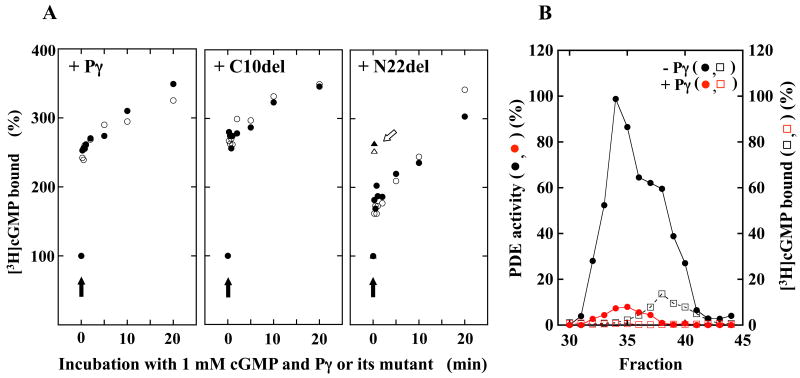
Next, we investigated whether cGMP binds to Pαβγ during deactivation of Pαβγ to Pαβγγ. Binding of cGMP may be involved in Pαβγ deactivation in two ways: after interaction with Pγ and before interaction with Pγ. First, we study whether cGMP binds to Pαβγ after Pγ binding to Pαβγ. We assayed [3H]cGMP-binding activity of Pαβγ after incubation of Pαβγ with Pγ or its mutants (Fig. 6). Here these Pαβγ complexes are termed Pαβγ·Pγ or Pαβγ·Pγ-mutant to emphasize that [3H]cGMP-binding activity is assayed after formation of these complexes. Pγ·GDP-Tα, instead of Pγ, should be used, because Pγ·GDP-Tα, but not free Pγ, is the inhibitor of Pαβγ (13, 20). However, it is unknown whether Pγ mutants we used form a complex with GDP-Tα. Therefore, free Pγ was used in this study.
Figure 6.
Effects of Pγ and its mutants on [3H]cGMP binding to Pαβγ. A. The effect on the level of [3H]cGMP binding. After incubation or Pαβγ (1.92 μg) with various concentrations of Pγ or its mutants, the [3H]cGMP-binding activity was measured. The 100% activity indicates that 1.46 pmol [3H]cGMP bound to Pαβγ in tubes. Following Pγ and its mutants were used: ●, wild type Pγ; □, N18del; △, N22del; ▲, C18Sub, and ▼, C10del. B. The effect on the time-course of [3H]cGMP-binding. Pαβγ (17.3 μg) was incubated with 1.11 μM Pγ or its mutants in 55 mM Tris·HCl, (pH 7.5) containing 4.4 mM EDTA and 1.1 mM IBMX (final volume, 720 μl) on ice for 30 min. The [3H]cGMP binding was initiated by adding 80 μl of 10 μM [3H]cGMP. After incubation for indicated periods on ice, an aliquot (80 μl) was taken and applied to a Millipore filter. Following Pγ and its mutants were used: ○, control; ●, wild type Pγ; △, N22del; and ▲, C18Sub. C. The effect on the Scatchard plot. Pαβγ (1.92 μg) was incubated with 1 mM of wild type Pγ (●) or N22del (△). As a control, Pαβγ alone was incubated (○). Then, [3H]cGMP binding was initiated by adding indicated concentrations of [3H]cGMP (C-1). The [3H]cGMP binding in C-1 is analyzed by Scatchard plotting (C-2).
[3H]cGMP binding to Pαβγ (control). The level of [3H]cGMP binding to Pαβγ reached a plateau in less than 2 min and was not changed during the incubation period of at least 40 min (Figs. 4B and 6B). After reaching to the plateau, ∼100% of Pαβγ bound [3H]cGMP in the mixture. However, the plateau indicates the level of [3H]cGMP-bound Pαβγ trapped by the filter. In this case, the filter trapped ∼16 % of the Pαβγ existing in the mixture.
[3H]cGMP binding to Pαβγ·Pγ. The level of bound [3H]cGMP was reduced when Pαβγ·Pγ was formed (Fig. 6A). A reason for the reduction is that binding of [3H]cGMP to Pαβγ·Pγ was slow and, even after 30 min incubation, did not reach the level that Pαβγ could reach in 2 min (Fig. 6B). The Kd for cGMP in Pαβγ/Pγ is ∼0.33 μM (Fig. 6C), indicating that the binding of Pγ to Pαβγ reduces its affinity for cGMP ∼6.5 times. This reduction may be a reason for the slow binding of [3H]cGMP to Pαβγ·Pγ. The efficiency of Millipore filter for trapping of [3H]cGMP-bound Pαβγ was increased when Pγ binds to [3H]cGMP-bound Pαβγ, as shown below (Fig. 7A). Therefore, the reduction in the level of [3H]cGMP-binding to Pαβγ·Pγ is not due to the reduction of the Millipore filter's ability to trap the [3H]cGMP-bound Pαβγ·Pγ.
[3H]cGMP binding to Pαβγ·C18del and Pαβγ·C10del. Both C18sub and C10del drastically reduced the level of [3H]cGMP bound (Fig. 6A and B). A simple explanation for the reduction is that Pγ mutants lacking the C-terminus interrupt the entry of [3H]cGMP to a non-catalytic site in Pαβγ. This explanation appears to be correct because Pγ's domains other than the C-terminus, especially its N-terminus, are located in the vicinity of GAF domains (30-32). However, the reduction was much larger than that caused by Pγ (Fig. 6A), suggesting that the way for binding of these Pγ mutants to Pαβγ is slightly different from that of Pγ and that the inhibitory effect is neutralized, although partially, by its C-terminus. Alternatively, all of the Pαβγ·C-terminal-mutant complexes would bind [3H]cGMP; however, only the filter might trap a negligible amount of these complexes. However, the level trapped by the filter was increased when C10del was added to [3H]cGMP-bound Pαβγ (Fig. 7A). Therefore, this scenario is unlikely.
[3H]cGMP binding to Pαβγ·N18del and Pαβγ·N22del. The apparent level of [3H]cGMP binding was increased when Pαβγ formed a complex with N18del and N22del (Figs. 6A, 6B and 6C). There are two possible explanations for this observation; 1) the level of [3H]cGMP binding to these Pαβγ·N-terminal-mutant complexes was really increased, and 2) the level trapped by the filter of [3H]cGMP-bound Pαβγ was increased. We compared [3H]cGMP binding to Pαβγ with that to Pαβγ·N22del (and Pαβγ·N18del). We found that 1) both Pαβγ (Figs. 4A and 6C) and Pαβγ·N22del (Fig. 6C) had one kind of [3H]cGMP-binding site; 2) the rate of [3H]cGMP binding to Pαβγ·N22del appeared to be similar or slightly faster than that to Pαβγ (Fig. 6B); and 3) the affinity of Pαβγ·N22del for [3H]cGMP was higher than that of Pαβγ (Fig. 6C). These findings indicate that Pαβγ·N22del binds [3H]cGMP more effectively than Pαβγ does. As described, ∼100% of Pαβγ rapidly binds [3H]cGMP under these conditions. Therefore, we conclude that ∼100% of Pαβγ·N22del also rapidly binds [3H]cGMP and that the apparent increase in the level of [3H]cGMP-bound Pαβγ·N22del is due to the effective trapping by the filter of [3H]cGMP-bound Pαβγ·N22del. In other words, explanation 2, but not 1, is appropriate. The small difference in levels of [3H]cGMP binding to Pαβγ·N22del and Pαβγ·N18del (Fig. 7A) may also be caused by the difference in levels trapped by the filter of [3H]cGMP-bound Pαβγ·N22del and Pαβγ·N18del.
In conclusion, Pγ appears to suppress [3H]cGMP binding to Pαβγ by interrupting the entry of [3H]cGMP to a non-catalytic site in Pαβγ. This indicates that the scheme in which Pγ binds to Pαβγ first and then cGMP binds to the Pαβγγ is unlikely. This also implies that Pγ in Pαβγγ interferes with cGMP release. This may be a reason for the observations that cGMPs are tightly bound to Pαβγγ (Fig. 3 and Ref. 12) and that release of cGMP from Pαβγγ is coupled with Pγ dissociation (Fig. 2).
Effect of Pγ on cGMP-bound Pαβγ
Next we study how Pγ affects [3H]cGMP-bound Pαβγ, i.e., Pαβγ having two cGMPs. Since ∼100% of Pαβγ binds [3H]cGMP and the Pαβγ firmly folds [3H]cGMP, we investigate the effect of Pγ on the filter-trapping level of [3H]cGMP-bound Pαβγ (Fig. 7A). The study is based on the result shown in Figure 5: The increase in the filter-trapping level of [3H]cGMP-bound Pαβγ is an indicator for Pαβγ's conformational change by cGMP binding. Pγ·GDP-Tα, instead of Pγ, should be used in this study because Pγ·GDP-Tα, but not free Pγ, is the inhibitor of Pαβγ (13, 20). However, as described above, it is unknown whether Pγ mutants we used form a complex with GDP-Tα. Therefore, free Pγ was used in this study.
When Pγ was added to [3H]cGMP-bound Pαβγ exactly in the same way as in the study shown in Figure 5A, the level trapped by the filter of [3H]cGMP-bound Pαβγ was increased in less than 15 seconds, but the trapping level of [3H]cGMP-bound Pαβγ was not changed (Fig. 7A). A simple explanation for these phenomena is that Pγ shortens the time required to detect the slow increase, i.e., Pγ accelerates the conformational change of [3H]cGMP-bound Pαβγ.
To strengthen the above-mentioned conclusion, the relative compactness, the Stokes radius, of cGMP-pretreated Pαβγ, Pαβγ having two cGMPs, was also compared with or without Pγ (Fig. 7B). Purified Pαβγγ was used as Pαβγγ having two cGMPs because the Pαβγγ has two cGMPs (Fig. 3). As expected, the cGMP-pretreated Pαβγ eluted as two peaks, the major peak of the Pαβγ was eluted in fraction 34, and the fraction showed no [3H]cGMP-binding activity. The important point is that Pαβγγ was also eluted in fraction 34 (Fig. 7B). This strongly suggests that Pγ does not change the relative compactness of Pαβγ having two cGMPs, i.e., binding of Pγ does not change the conformation of Pαβγ having two cGMPs. Together with the observation that cGMP-binding to Pαβγ slowly changes its conformation (Fig. 5) and that Pγ shortens the time required to detect the slow increase (Fig. 7A), the result strongly suggests that binding of Pγ accelerates cGMP-dependent conformational change on Pαβγ.
The rapid increase by Pγ in the filter-trapping level of [3H]cGMP-bound Pαβγ (Fig. 7A) might be due to a change by Pγ in its surface and/or total charge of Pαβγ. However, in such cases, the slow increase (Fig. 5A) could also be detected. However, the slow increase was indeed lacking. Therefore, this possibility is unlikely. We also note that the result (Fig. 7B) does not eliminate the possibility that Pγ binding causes a small and/or localized conformational change on the [3H]cGMP-bound Pαβγ. However, the clear change in its Stokes radius (Figs. 5B) suggests that this kind of conformational change is not involved.
When C10del was added, the level trapped by the filter of [3H]cGMP-bound Pαβγ was also rapidly increased and the increase by a longer incubation (Fig. 5A) disappeared (Fig. 7A). Similar observations were also observed when N22del was added (Fig. 7A). These results suggest that binding of C10del or N22del, similar to Pγ binding, accelerates the conformational change on [3H]cGMP-bound Pαβγ and that a Pγ domain(s) located between N22 and C10 is involved in this acceleration. We note that the level increased by N22del of [3H]cGMP-bound Pαβγ was smaller than that increased by Pγ or C10del; however, the level increased by N18del was similar to that by Pγ or C10del (Fig. 7A). The level of [3H]cGMP binding to Pαβγ·N18del was also consistently higher than that to Pαβγ·N22del (Fig. 6A). These results suggest that all or some of 4 amino-acid residues between 19-22 in Pγ's amino-acid-sequence may be crucial for the acceleration.
In conclusion, these results strongly suggest that Pγ binding accelerates the cGMP-binding-initiated conformational change on Pαβγ. Together with the result showing that Pγ inhibits cGMP-binding to Pαβγ (Fig. 6), these results also imply that the scheme in which cGMP binds to Pαβγ first and then Pγ binds to the Pαβγ is appropriate for Pαβγ deactivation, the process to shift the Pαβγ having one cGMP to the Pαβγγ having two cGMPs (Fig. 8). These results also imply a large conformational change during Pαβγγ activation, the process to shift the Pαβγγ having two cGMPs to the Pαβγ having one cGMP, although its direction is opposite to that for Pαβγ deactivation (Fig. 8).
Figure 8.
Role of the non-catalytic cGMP-binding site in PDE regulation. Activation. GTP-Tα activates Pαβγγ/2cGMPs (Pαβγγ having two cGMPs) to Pαβγ/cGMP (Pαβγ having one cGMP). At the initial stage, cGMP is present in OS; however, binding of cGMP to the empty site on Pαβγ/cGMP is suppressed. Deactivation. After hydrolysis of cGMP, retinal guanylate cyclase initiates to produce cGMP from GTP. When the [cGMP] in OS is increased to ∼50 nM, the cGMP binds to Pαβγ/cGMP and Pαβγ/2cGMPs is formed. Then, the PDE species slowly changes its conformation. Interaction with Pγ·GDP-Tα accelerates the conformational change and swiftly establishes the inactive form of PDE (Pαβγγ/2cGMPs).
Discussion
Using bovine PDE preparations, we have recently proposed a new and comprehensive model for PDE regulation (13, 20). In this study, we try to integrate the role of non-catalytic and cGMP-specific binding sites in Pαβ in this new model. We show that Pαβγγ, the inactive form, and Pαβγ, the GTP-Tα-activated form, contain two and one cGMP, respectively, and that only Pαβγ shows [3H]cGMP binding. We also show that Pαβγ's ability to bind cGMP is suppressed during formation of Pαβγ. We also strongly suggest that the cGMP binding slowly changes the conformation of Pαβγ and that Pγ binding accelerates this conformational change. These findings are consistent with the view that Pαβγ rapidly changes its conformation during its deactivation and that binding of cGMP and Pγ plays crucial roles in the conformational change (Fig. 8). These findings also imply that Pαβγγ rapidly changes its conformation during its activation and that the release of Pγ and cGMP plays important roles for the conformational change (Fig. 8). This is, to our understanding, the first model that the role of non-catalytic binding sites is smoothly integrated in PDE regulation.
Pαβγ is the PDE species expressing [3H]cGMP-binding activity
Identification of a PDE species expressing [3H]cGMP-binding activity is the first step in the exploration of the role of cGMP-binding to the non-catalytic site in PDE regulation. We have shown that the GTP-Tα-activated PDE in membranes has a high [3H]cGMP-binding activity (Fig. 1), that Pαβγ contains one cGMP, i.e., Pαβγ has one empty non-catalytic site, (Fig. 3), and that cGMP binds to Pαβγ (Figs. 2, 3 and 4) with Kd ∼50 nM (Figs. 4A and 6C). Three points related to these issues need discussion. 1) In the currently dominant model, Pαβγγ complexed with GTP-Tα is believed to be the GTP-Tα-activated PDE. Some groups also suggest that Pγ-free Pαβ is a GTP-Tα-activated PDE. However, neither the GTPγS-Tα·Pαβγγ complex nor the Pγ-free Pαβ was detected in any OS homogenates (13, 20). Therefore, in their absence, we could not characterize the [3H]cGMP-binding activities of these Pαβ complexes. We believe that this failure may not be crucial in exploring the role of cGMP binding in PDE regulation. 2) When [3H]cGMP binding to Pαβγ was assayed, cGMP present originally in Pαβγ could be exchanged with [3H]cGMP. However, based on the following two reasons, we conclude that the cGMP/[3H]cGMP exchange does not occur and that the [3H]-radioactivity we detected is only attributed to the [3H]cGMP bound to the empty site in Pαβγ: 1) Pαβγ had been exposed to the cGMP-free conditions more than 5 days before the final step for purification; however, the purified Pαβγ contains exactly one cGMP (Fig. 3). This result suggests that Pαβγ firmly holds the cGMP, i.e., the cGMP is hardly exchanged with [3H]cGMP. This is true especially when the concentration of [3H]cGMP is as low as 1 μM. 2) Fractions containing Pαβγ did not show any [3H]cGMP-binding activity when the Pαβγ was pre-treated with unlabeled cGMP (Figs. 5B and 7B). This observation indicates that neither the originally bound cGMP nor the newly bound cGMP on Pαβγ can be exchanged with [3H]cGMP in the assay mixture. We emphasize that the exchange did not occur even with 1 mM cGMP (Figs. 5A and 7A). 3) We added IBMX to suppress hydrolysis of [3H]cGMP when [3H]cGMP binding to Pαβγ was assayed. We found that the affinity for cGMP of a PDE preparation was increased when 1 mM IBMX was present in the assay mixture (data not shown). Under these conditions, a large part of the [3H]cGMP was not yet hydrolyzed even in the absence of IBMX, suggesting that IBMX itself affects the affinity of the PDE preparation for cGMP. IBMX also enhances the cGMP affinity of frog PDE (23). Therefore, in OS, the Pαβγ's affinity for cGMP may be lower than that we observed. We note that the assay mixture for [3H]cGMP binding did not contain Mg2+ to suppress hydrolysis of [3H]cGMP. We found that both the amount of [3H]cGMP bound to Pαβγ and the affinity of Pαβγ for [3H]cGMP were not notably changed in the presence or absence of 5 mM MgCl2 (data not shown). Thus, we believe that the lack of Mg2+ in the assay mixture does not affect the affinity of Pαβγ for cGMP.
Binding of cGMP to Pαβγ is not involved PDE activation
Binding of [3H]cGMP to Pαβγ implies that the cGMP binding may be involved in the activation of Pαβγγ to Pαβγ and/or in the deactivation of Pαβγ to Pαβγγ. Here we show that the ability of Pαβγ to bind cGMP is suppressed during the formation of Pαβγ by GTPγS-Tα in OS homogenates (Fig. 2). This indicates that the cGMP binding is not involved in the activation of Pαβγγ to Pαβγ. It is unknown now how this property of Pαβγ is suppressed in OS homogenates containing GTPγS-Tα. However, we emphasize that either membrane-bound Pαβγ (Fig. 1A) or Pαβγ purified from membranes (Fig. 4A) expresses a [3H]cGMP-binding activity. This suggests that a soluble factor in the OS homogenates is involved in the suppression of cGMP binding. This factor may be GTPγS-Tα, because GTPγS-Tα, by interacting with Pαβ (33, 34), may inhibit the entry of cGMP to the non-catalytic site in Pαβγ. Alternatively, the interaction with GTPγS-Tα may compel Pαβγ to change its conformation and the Pαβγ having the altered conformation may not be able to bind cGMP. We have shown that GTPγS-Tα releases not only one Pγ but also one cGMP from Pαβγγ when Pαβγγ is activated (Fig. 3). The GTPγS-Tα releases Pγ by forming a complex with Pγ (13, 20); however, the mechanism to release cGMP is unknown because neither GTPγS-Tα nor Pγ can bind [3H]cGMP (Fig. 2 and Ref. 23). It is possible that this alternative mechanism may also be involved in the release of cGMP.
Binding of cGMP to Pαβγ may be a mechanism for light adaptation
Our results strongly suggest that binding of cGMP, i.e., formation of Pαβγ having two cGMPs from Pαβγ having one cGMP, changes the conformation of Pαβγ. The important points are that this Pαβγ's conformational change is slow (Fig. 5A); however, binding of Pγ to Pαβγ having two cGMPs accelerates its conformational change (Fig. 7). Overall, these findings indicate that cGMP binds to Pαβγ first and then Pγ binds to the Pαβγ in Pαβγ deactivation (Fig. 8). In this scheme, the residual [cGMP] is crucial for the rate of Pαβγ deactivation. The residual [cGMP] in OS is dependent upon the level of illumination: If illumination is low, the residual [cGMP] is higher than the Pαβγ's Kd and the cGMP-binding-dependent conformational change may be initiated on Pαβγ immediately after disappearance of GTP-Tα. Thus, Pαβγ will be rapidly deactivated. If illumination is high, the residual [cGMP] is lower than the Kd and the conformational change may be delayed. Therefore, Pαβγ will be slowly deactivated. This may be a mechanism for light adaptation. The important point for this argument is whether the residual [cGMP] in OS can be increased to the Kd before deactivation of Pαβγ. The Kd of Pαβγ for cGMP we measured is ∼50 nM (Figs. 4A and 6C), and the Kd in OS may be higher than ∼50 nM, as described above. However, the Kd in OS should be much lower than the dark level of cGMP, 4-5 μM (2, 3). Therefore, a partial recovery of the [cGMP] may be enough to initiate the Pαβγ deactivation. The [cGMP] in OS is recovered by retinal guanylate cyclase. We have recently shown that retinal guanylate cyclase is activated by a light-initiated, ATP-stimulated and Ca2+-sensitive mechanism (35-37) and that this activity is much higher than that expected by the current model. Indeed, it has been reported that the in vivo activation of retinal guanylate cyclase is much higher than that shown based on the current model (38-40). Therefore, the [cGMP] in OS may be partially recovered before complete shut down of Pαβγ and the [cGMP], after partially recovered, may be enough to initiate the cGMP-binding-dependent conformational change on Pαβγ.
Using N18del and N22del, we suggest that all or some of 4 amino-acid residues between 19-22 in the Pγ amino-acid-sequence may be involved in the Pγ-dependent acceleration of Pαβγ's conformational change (Fig. 7A). This sequence includes a part of the Pro20-Xaa21-Thr22-Pro23-Arg24 sequence, the sequence essential for phosphorylation of Thr22 by cyclin-dependent protein kinase 5 (41-45). It is unknown now whether Thr22 phosphorylation affects the Pγ-dependent acceleration of Pαβγ's conformational change; however, this study is very interesting. Comparison with PDE5 (25-27) will also be of great interest for future studies.
Previous models for the role of cGMP binding in PDE regulation
It should be emphasized that all previous studies, including our studies, did not identify PDE species expressing cGMP-binding activity. For frog PDE, PDE species exiting in OS membranes (14-16, 21, 22, 33) or PDE species treated with trypsin (15) were used. For bovine PDE, PDE species treated with trypsin were used (17, 18). Therefore, the cGMP-binding activity reported might be the activity ascribed by the mixture of PDE species and/or affected by other PDE species. Moreover, the cGMP-binding activity obtained might be affected by different rates of filter trapping. Therefore, conclusions in previous studies may not be correct. For example, previous studies suggested that Pγ stimulated cGMP binding to GTP-Tα-activated frog PDE/membranes (21, 22). We propose to reevaluate this conclusion. Trypsin-treated PDE species have other problems: Trypsin digests one PDE species in various ways, and the trypsin-treated PDE appears to lose not only all Pγs but also all cGMPs (17, 18). Presence of such PDE species in OS is doubtful (13, 20).
Two previous models to explain the role of non-catalytic site in PDE regulation, the cGMP-regulated Pαβ-Pγ interaction model (14-18) and the cGMP-binding-direct regulation model (19), were also based on unclear identification of the PDE species expressing cGMP-binding activity. For example, the former model implies the presence of GTP-Tα·Pαβγγ complex containing two cGMPs or the presence of GTP-Tα·Pαβγγ complex containing one or no cGMP and cGMP-binding to the complex. However, neither the presence of these complexes nor cGMP binding to the complex have been verified. In addition, the mechanism by which GTP-Tα activates Pαβγγ without changing the interaction between Pαβ and Pγ has not been shown. The former model also implies that GTP-Tα releases Pγ when the [cGMP] in OS is low or absent and that the released Pγ accelerates deactivation of GTP-Tα-activated PDE. These implications are also problematical; 1) Pγ is released from Pαβγγ to accelerate PDE deactivation. This appears to be a self-contradiction; 2) cGMP is released from Pαβγγ when [cGMP] is low or absent. However, as shown previously (12) and now, this is not the case in bovine PDE; 3) Pγ is released from GDP-Tα. However, Pγ forms a tight complex with GDP-Tα (33, 46, 47), and the mechanism to release Pγ from the complex is unknown {We have shown that Pγ·GDP-Tα, without Pγ liberation, inhibits Pαβγ (13)}; and 4) they indicate, to our understanding, that the released Pγ enhances GTPase activity of the Tα that is already complexed with Pγ in Pαβγγ. However, this mechanism is unknown. In the latter model, there is no data showing which PDE species binds cGMP, how the cGMP-binding changes the PDE catalytic activity and how GTP-Tα is involved in the mechanism. Therefore, it is difficult to integrate these models in PDE regulation. We and another group (48-50) have also proposed a model in which the non-catalytic sites serve as a cytoplasmic buffer for cGMP. This model appears not to be directly related to PDE regulation, and thus we do not discuss here.
Experimental procedures
Materials
Dark-adapted frozen bovine retinas were obtained from J. A. Lawson Co. (Lincoln NE). [3H]cGMP was from PerkinElmer Life Sciences. Other chemical reagents and materials were purchased from various sources described in previous articles (13, 20).
Preparation of OS membranes and isolation of PDE species
Membranes were prepared from OS homogenates as described (13). PDE species were solubilized with a hypotonic buffer or Pδ in an isotonic buffer and purified by using DEAE and molecular sieve columns (13, 20). In each study, protein-contents in OS homogenates and volumes of buffers used to wash membranes or to extract PDE species were slightly different. These differences have been shown not to be critical (13, 20). 1) Use of a hypotonic buffer. Details have been published (20). Here, as an example, the procedure for the experiment shown in Figure 2 is described: OS homogenates (∼50 mg protein) were suspended in 20 ml Buffer A (20 mM HEPES, pH 7.5, 5 mM DTT, 5 mM MgCl2, 5 μM leupeptin, 5 μM pepstatin A, 0.1 mM PMSF, 1 mM benzamidine, and 150 mM NaCl) and divided into three portions. To each portion, cGMP (final 1 mM), GTPγS (final 50 μM), or cGMP + GTPγS was added. After overnight incubation on ice, membranes were isolated and washed (×2) with 7 ml Buffer A supplemented with cGMP (1 mM), GTPγS (50 μM), or cGMP + GTPγS. Finally, all membranes were washed with 7 ml Buffer A (×2). PDE species were extracted from these membranes with 7 ml Buffer B (5 mM Tris·HCl, pH 7.5, 5 mM DTT, 0.5 mM MgCl2, 5 μM leupeptin, 5 μM pepstatin A, 0.1 mM PMSF and 1 mM benzamidine) (×7) and applied to a TSK-DEAE 5PW column. After assaying PDE activity and identifying a 88-kDa-protein-doublet by SDS-PAGE, fractions containing PDE species were concentrated to ∼0.5 ml and applied to a Superdex 200 HR column. The 88-kDa-doublet has been identified as Pαβ (13, 20). When cGMP was added, IBMX (final 1 mM) was also added to all portions. 2) Use of Pδ in an isotonic buffer. Details of this procedure have been reported (20). After incubation of OS homogenates with or without GTPγS, membranes were isolated and washed as described above. These membranes were suspended in Buffer C (10 mM Na-phosphate, pH 6.8, 5 mM DTT, 5 mM MgCl2, 5 μM leupeptin, 5 μM pepstatin A, 0.1 mM PMSF, 1 mM benzamidine, 100 mM NaCl), incubated with Pδ (final 3 μM) overnight on ice, and washed with Buffer C (×2) (13, 20). All supernatants were combined together and applied to a TSK-DEAE 5PW column. Pαβγγ and Pαβγ are isolated as Pαβγγδ and Pαβγδδ, respectively (20). We also obtained a PDE preparation using a Bio-Gel A-0.5 m column (9 × 600 mm) that had been equilibrated with Buffer D (10 mM Na-phosphate, pH 6.8, 1 mM DTT, 2 mM MgCl2, 5 μM leupeptin, 5 μM pepstatin A, 0.1 mM PMSF, 1 mM benzamidine, 150 mM NaCl and 15% glycerol). Proteins were eluted with Buffer D (Fig. 1B and C). The chromatography conditions were: flow rate, 0.8 ml/13 min; and fraction volume, 0.7 ml. The resin was chosen because the resin does not have any charge and only a small amount of proteins may stick to the resin.
Preparation of Pγ and its mutants
Bovine Pγ and its mutants, N18del (a Pγ mutant in which 18 amino-acids in the N-terminus were deleted), N22del (a Pγ mutant in which 22 amino-acids in the N-terminus were deleted), C18Sub (a Pγ mutant in which 18 amino-acids in the C-terminus were substituted with a frame-shift mutation), and C10del (a Pγ mutant in which 10 amino-acids in the C-terminus were deleted), were used. Oligonucleotides (49) and procedures to express and purify Pγ and these Pγ mutants (51) have been reported.
Contents of cGMP in Pαβγγ and Pαβγ
Contents of cGMP bound to purified Pαβγγ and Pαβγ were measured using a cGMP immunoassay kit, the Correlate-EIA Direct Cyclic GMP Enzyme Immunoassay Kit (Assay Designs, Ann Arbor, MI). After incubation of 100 μl of these preparations with 900 μl 0.1 M HCl for 2 h on ice, supernatants were obtained by centrifugation (350,000 × g, 20 min, 4 °C) and diluted with 0.1 M HCl. The amount of cGMP in these mixtures (100 μl) was measured as the instruction of the kit indicated. Apparent Mrs of Pαβγγ (216,894) and Pαβγ (207,226) were used for the calculation of cGMP concentrations.
Determination of [3H]cGMP-binding activity in PDE
Two methods were used. 1) Filter assay. This assay was performed as described (23, 30). Typically, Pαβγ (∼15 μg before purification, and ∼2 μg, after purification) was incubated with 0.5 or 1 μM [3H]cGMP (1 mCi/ml) in the assay medium (final volume, 100 μl) containing 25 mM Tris·HCl, pH 7.5, 0.1 mM EDTA, and 1 mM IBMX for 30 min on ice, and 80 μl aliquots were applied to a Millipore filter (HA, pore size 0.45 μm) that had been wetted with 20 mM sodium phosphate buffer (pH 6.5). The filter was washed with 3 ml of phosphate buffer (×3) and dissolved in 4 ml of scintillation cocktail. When 1 μM [3H]cGMP was used, the maximum level (∼1.7% of the added counts) was reached in 2 min and the binding was stable for at least 40 min (Fig. 4B). Addition of a 1000-fold excess of unlabeled cGMP to the [3H]cGMP inhibited [3H]cGMP-binding by greater than 99%. A linear relationship existed between the level trapped by the filter of [3H]cGMP-bound Pαβγ and the protein level. 2) Gel filtration. This method was used to estimate the efficiency of the Millipore filter to trap [3H]cGMP-bound Pαβγ. After incubation with 1 μM [3H]cGMP in 0.5 ml of 25 mM Tris·HCl, pH 7.5, containing 0.1 mM EDTA and 1 mM IBMX for 30 min on ice, Pαβγ (40-90 μg) was applied to a Superdex 200 HR column that had been equilibrated with Buffer D. The chromatography conditions were: room temperature, 0.3-0.5 ml/min flow rate and 0.3-0.5 ml fraction volume. The level of [3H]cGMP bound to Pαβγ was calculated based on the [3H]-radioactivity in 50-70 μl of the fraction. The fraction (50-70 μl) was also applied to a Millipore filter and the [3H]-radioactivity on the filter was measured. The efficiency of the filter's ability to trap [3H]cGMP-bound Pαβγ was estimated by comparison of these [3H]-radioactivities.
Determination of Stokes radii of Pαβγ treated with or without cGMP
Purified Pαβγ (70 μg) was incubated with or without unlabeled cGMP (0.5 mM) in 0.5 ml of 25 mM Tris·HCl, pH 7.5, 0.1 mM EDTA, and 1 mM IBMX for 30 min on ice. After concentration to ∼0.25 ml, the mixture was applied to a Superdex 200 HR column which had been equilibrated with Buffer E (10 mM Tris·HCl, pH 7.5, 1 mM DTT, 2 mM MgCl2, 5 μM leupeptin, 5 μM pepstatin A, 0.1 mM PMSF, 1 mM benzamidine, 100 mM NaCl and 10% glycerol). The chromatography conditions were: room temperature, 0.25 ml/min flow rate and 0.25 ml fraction volume. PDE activity was assayed using 5-20 μl of the fraction. [3H]cGMP-binding activity was measured using 50 μl of the fraction. Neither IBMX nor cGMP was added to the elution buffer, Buffer E, because IBMX may change Pαβγ's affinity for cGMP, as discussed in this study, and cGMP, once bound to Pαβγ, is not dissociated. The apparent Stokes radii of these PDE species were calculated as described (25-27). The following proteins were used to standardize the column: ovalbumin, 30.5 Å; conalubumin, 40.4 Å; aldolase, 48.1 Å; and thyroglobulin, 85.0 Å.
Analytical procedures
PDE activity was assayed as described (30). Protein concentration was measured with bovine serum albumin as the standard (52). SDS-PAGE using 8-16% gradient gels was carried out as described (53). Protein staining with Coomassie blue was also performed as described (54). The advantage of this staining method has been described in previous reports (13, 20). The method was also used to estimate roughly the content of Pαβ in gels. In our results, individual points represent the average values of duplicate assays. All experiments were carried out at least three times and the results were similar. The data shown are representative of these experiments.
Acknowledgments
We especially thank Dr. Richard Needleman, Wayne State University, for critical reading of the manuscript. This work was supported in part by National Institute of Health Grants EY07546 and EY09631, Jules and Doris Stein Professorship and an unrestricted grant from Research to Prevent Blindness, and an unrestricted grant from College of Osteopathic Medicine, Touro University.
Abbreviation
- PDE
cGMP phosphodiesterase
- OS
outer segments of retinal photoreceptors
- Pα and Pβ
rod PDE catalytic subunits
- Pγ
rod PDE inhibitory subunit
- Pα′
cone PDE catalytic subunit
- Pγ′
cone PDE inhibitory subunit
- Pαβ/Pγ
Pαβ complexes having an unknown number of Pγ
- Pδ
a prenyl-binding protein
- Mr
molecular weight
- Tα
transducin α subunit
- Tβγ
transducin β and γ subunits
- GTP-Tα
GTP-bound Tα
- GDP-Tα
GDP-bound Tα
- GTPγS
guanosine 5′-O-(3-thiotriphosphate)
- GTPγS-Tα
GTPγS-bound Tα
- PAGE
polyacryamide gel electrophoresis
- PMSF
phenylmethylsulfonyl fluoride
- IBMX
1-methyl-3-isobutylxanthine
References
- 1.Hurley JB. Molecular properties of the cGMP cascade of vertebrate photoreceptors. Annu Rev Physiol. 1987;49:793–812. doi: 10.1146/annurev.ph.49.030187.004045. [DOI] [PubMed] [Google Scholar]
- 2.Yau KW, Baylor DA. Cyclic GMP-activated conductance of retinal photoreceptor cells. Annu Rev Neurosci. 1989;12:289–327. doi: 10.1146/annurev.ne.12.030189.001445. [DOI] [PubMed] [Google Scholar]
- 3.Miller WH. Dark mimic. Invest Ophthalmol Vis Sci. 1990;31:1659–1673. [PubMed] [Google Scholar]
- 4.Miki N, Baraban JM, Keirns JJ, Boyce JJ, Bitensky MW. Purification and properties of the light-activated cyclic nucleotide phosphodiesterase of rod outer segments. J Biol Chem. 1975;250:6320–6327. [PubMed] [Google Scholar]
- 5.Baehr W, Devlin MJ, Applebury ML. Isolation and characterization of cGMP phosphodiesterase from bovine rod outer segments. J Biol Chem. 1979;254:11669–11677. [PubMed] [Google Scholar]
- 6.Hurley JB, Stryer L. Purification and characterization of the gamma regulatory subunit of the cyclic GMP phosphodiesterase from retinal rod outer segments. J Biol Chem. 1982;257:11094–11099. [PubMed] [Google Scholar]
- 7.Deterre P, Bigay J, Forquet F, Robert M, Chabre M. cGMP phosphodiesterase of retinal rods is regulated by two inhibitory subunits. Proc Natl Acad Sci U S A. 1988;85:2424–2428. doi: 10.1073/pnas.85.8.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fung BK, Young JH, Yamane HK, Griswold-Prenner I. Subunit stoichiometry of retinal rod cGMP phosphodiesterase. Biochemistry. 1990;29:2657–2664. doi: 10.1021/bi00463a006. [DOI] [PubMed] [Google Scholar]
- 9.Ovchinnikov YA, Lipkin VM, Kumarev VP, Gubanov VV, Khramtsov NV, Akhmedov NB, Zagranichny VE, Muradov KG. Cyclic GMP phosphodiesterase from cattle retina. Amino acid sequence of the gamma-subunit and nucleotide sequence of the corresponding cDNA. FEBS Lett. 1986;204:288–292. doi: 10.1016/0014-5793(86)80830-4. [DOI] [PubMed] [Google Scholar]
- 10.Lipkin VM, Khramtsov NV, Vasilevskaya IA, Atabekova NV, Muradov KG, Gubanov VV, Li T, Johnston JP, Volpp KJ, Applebury ML. Beta-subunit of bovine rod photoreceptor cGMP phosphodiesterase. Comparison with the phosphodiesterase family. J Biol Chem. 1990;265:12955–12959. [PubMed] [Google Scholar]
- 11.Kajimura N, Yamazaki M, Morikawa K, Yamazaki A, Mayanagi K. Three-dimensional structure of non-activated cGMP phosphodiesterase 6 and comparison of its image with those of activated forms. J Struct Biol. 2002;139:27–38. doi: 10.1016/s1047-8477(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 12.Gillespie PG, Beavo JA. cGMP is tightly bound to bovine retinal rod phosphodiesterase. Proc Natl Acad Sci U S A. 1989;86:4311–4315. doi: 10.1073/pnas.86.11.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamazaki A, Bondarenko VA, Matsuura I, Tatsumi M, Kurono S, Komori N, Matsumoto H, Hayashi F, Yamazaki RK, Usukura J. Mechanism for the regulation of mammalian cGMP phosphodiesterase6. 1: identification of its inhibitory subunit complexes and their roles. Mol Cell Biochem. 2010;339:215–233. doi: 10.1007/s11010-010-0387-8. [DOI] [PubMed] [Google Scholar]
- 14.Arshavsky VY, Dumke CL, Bownds MD. Noncatalytic cGMP-binding sites of amphibian rod cGMP phosphodiesterase control interaction with its inhibitory gamma-subunits. A putative regulatory mechanism of the rod photoresponse. J Biol Chem. 1992;267:24501–24507. [PubMed] [Google Scholar]
- 15.D'Amours MR, Cote RH. Regulation of photoreceptor phosphodiesterase catalysis by its non-catalytic cGMP-binding sites. Biochem J. 1999;340:863–869. [PMC free article] [PubMed] [Google Scholar]
- 16.Norton AW, D'Amours MR, Grazio HJ, Hebert TL, Cote RH. Mechanism of transducin activation of frog rod photoreceptor phosphodiesterase. Allosteric interaction between the inhibitory gamma subunit and the noncatalytic cGMP-binding sites. J Biol Chem. 2000;275:38611–38619. doi: 10.1074/jbc.M004606200. [DOI] [PubMed] [Google Scholar]
- 17.Mou H, Grazio HJ, 3rd, Cook TA, Beavo JA, Cote RH. cGMP binding to noncatalytic sites on mammalian rod photoreceptor phosphodiesterase is regulated by binding of its gamma and delta subunits. J Biol Chem. 1999;274:18813–18820. doi: 10.1074/jbc.274.26.18813. [DOI] [PubMed] [Google Scholar]
- 18.Mou H, Cote RH. The catalytic and GAF domains of the rod cGMP phosphodiesterase (PDE6) heterodimer are regulated by distinct regions of its inhibitory gamma subunit. J Biol Chem. 2001;276:27527–27534. doi: 10.1074/jbc.M103316200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang XJ, Cahill KB, Elfenbein A, Arshavsky VY, Cote RH. Direct allosteric regulation between the GAF domain and catalytic domain of photoreceptor phosphodiesterase PDE6. J Biol Chem. 2008;283:29699–29705. doi: 10.1074/jbc.M803948200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki A, Tatsumi M, Bondarenko VA, Kurono S, Komori N, Matsumoto H, Matsuura I, Hayashi F, Yamazaki RK, Usukura J. Mechanism for the regulation of mammalian cGMP phosphodiesterase6. 2: isolation and characterization of the transducin-activated form. Mol Cell Biochem. 2010;339:235–251. doi: 10.1007/s11010-010-0404-y. [DOI] [PubMed] [Google Scholar]
- 21.Yamazaki A, Bartucca F, Ting A, Bitensky MW. Reciprocal effects of an inhibitory factor on catalytic activity and noncatalytic cGMP binding sites of rod phosphodiesterase. Proc Natl Acad Sci U S A. 1982;79:3702–3706. doi: 10.1073/pnas.79.12.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cote RH, Bownds MD, Arshavsky VY. cGMP binding sites on photoreceptor phosphodiesterase: role in feedback regulation of visual transduction. Proc Natl Acad Sci U S A. 1994;91:4845–4849. doi: 10.1073/pnas.91.11.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamazaki A, Sen I, Bitensky MW, Casnellie JE, Greengard P. Cyclic GMP-specific, high affinity, noncatalytic binding sites on light-activated phosphodiesterase. J Biol Chem. 1980;255:11619–11624. [PubMed] [Google Scholar]
- 24.Martinez SE, Heikaus CC, Klevit RE, Beavo JA. The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change. J Biol Chem. 2008;283:25913–25919. doi: 10.1074/jbc.M802891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu DM, Corbin JD, Grimes KA, Francis SH. Activation by cyclic GMP binding causes an apparent conformational change in cGMP-dependent protein kinase. J Biol Chem. 1997;272:31922–31928. doi: 10.1074/jbc.272.50.31922. [DOI] [PubMed] [Google Scholar]
- 26.Francis SH, Chu DM, Thomas MK, Beasley A, Grimes K, Busch JL, Turko IV, Haik TL, Corbin JD. Ligand-induced conformational changes in cyclic nucleotide phosphodiesterases and cyclic nucleotide-dependent protein kinases. Methods. 1998;14:81–92. doi: 10.1006/meth.1997.0567. [DOI] [PubMed] [Google Scholar]
- 27.Zoraghi R, Bessay EP, Corbin JD, Francis SH. Structural and functional features in human PDE5A1 regulatory domain that provide for allosteric cGMP binding, dimerization, and regulation. J Biol Chem. 2005;280:12051–12063. doi: 10.1074/jbc.M413611200. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava D, Fox DA, Hurwitz RL. Effects of magnesium on cyclic GMP hydrolysis by the bovine retinal rod cyclic GMP phosphodiesterase. Biochem J. 1995;308:653–658. doi: 10.1042/bj3080653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He F, Seryshev AB, Cowan CW, Wensel TG. Multiple zinc binding sites in retinal rod cGMP phosphodiesterase, PDE6alpha beta. J Biol Chem. 2000;275:20572–20577. doi: 10.1074/jbc.M000440200. [DOI] [PubMed] [Google Scholar]
- 30.Yamazaki M, Li N, Bondarenko VA, Yamazaki RK, Baehr W, Yamazaki A. Binding of cGMP to GAF domains in amphibian rod photoreceptor cGMP phosphodiesterase (PDE). Identification of GAF domains in PDE alphabeta subunits and distinct domains in the PDE gamma subunit involved in stimulation of cGMP binding to GAF domains. J Biol Chem. 2002;277:40675–40686. doi: 10.1074/jbc.M203469200. [DOI] [PubMed] [Google Scholar]
- 31.Guo LW, Muradov H, Hajipour AR, Sievert MK, Artemyev NO, Ruoho AE. The inhibitory gamma subunit of the rod cGMP phosphodiesterase binds the catalytic subunits in an extended linear structure. J Biol Chem. 2006;281:15412–15422. doi: 10.1074/jbc.M600595200. [DOI] [PubMed] [Google Scholar]
- 32.Song J, Guo LW, Muradov H, Artemyev NO, Ruoho AE, Markley JL. Intrinsically disordered gamma-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure. Proc Natl Acad Sci U S A. 2008;105:1505–1510. doi: 10.1073/pnas.0709558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamazaki A, Yamazaki M, Tsuboi S, Kishigami A, Umbarger KO, Hutson LD, Madland WT, Hayashi F. Regulation of G protein function by an effector in GTP-dependent signal transduction. An inhibitory subunit of cGMP phosphodiesterase inhibits GTP hydrolysis by transducin in vertebrate rod photoreceptors. J Biol Chem. 1993;268:8899–8907. [PubMed] [Google Scholar]
- 34.Liu W, Clark WA, Sharma P, Northup JK. Mechanism of allosteric regulation of the rod cGMP phosphodiesterase activity by the helical domain of transducin alpha subunit. J Biol Chem. 1998;273:34284–34292. doi: 10.1074/jbc.273.51.34284. [DOI] [PubMed] [Google Scholar]
- 35.Yamazaki A, Yu H, Yamazaki M, Honkawa H, Matsuura I, Usukura J, Yamazaki RK. A critical role for ATP in the stimulation of retinal guanylyl cyclase by guanylyl cyclase-activating proteins. J Biol Chem. 2003;278:33150–33160. doi: 10.1074/jbc.M303678200. [DOI] [PubMed] [Google Scholar]
- 36.Yamazaki A, Yamazaki M, Yamazaki RK, Usukura J. Illuminated rhodopsin is required for strong activation of retinal guanylate cyclase by guanylate cyclase-activating proteins. Biochemistry. 2006;45:1899–1909. doi: 10.1021/bi0519396. [DOI] [PubMed] [Google Scholar]
- 37.Bondarenko VA, Hayashi F, Usukura J, Yamazaki A. Involvement of rhodopsin and ATP in the activation of membranous guanylate cyclase in retinal photoreceptor outer segments (ROS-GC) by GC-activating proteins (GCAPs): a new model for ROS-GC activation and its link to retinal diseases. Mol Cell Biochem. 334:125–139. doi: 10.1007/s11010-009-0323-y. [DOI] [PubMed] [Google Scholar]
- 38.Cornwall MC, Fain GL. Bleached pigment activates transduction in isolated rods of the salamander retina. J Physiol. 1994;480(Pt 2):261–279. doi: 10.1113/jphysiol.1994.sp020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koutalos Y, Nakatani K, Tamura T, Yau KW. Characterization of guanylate cyclase activity in single retinal rod outer segments. J Gen Physiol. 1995;106:863–890. doi: 10.1085/jgp.106.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Detwiler P. Open the loop: dissecting feedback regulation of a second messenger transduction cascade. Neuron. 2002;36:3–4. doi: 10.1016/s0896-6273(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 41.Tsuboi S, Matsumoto H, Yamazaki A. Phosphorylation of an inhibitory subunit of cGMP phosphodiesterase in Rana catesbeiana rod photoreceptors. II. A possible mechanism for the turnoff of cGMP phosphodiesterase without GTP hydrolysis. J Biol Chem. 1994;269:15016–15023. [PubMed] [Google Scholar]
- 42.Tsuboi S, Matsumoto H, Jackson KW, Tsujimoto K, Williams T, Yamazaki A. Phosphorylation of an inhibitory subunit of cGMP phosphodiesterase in Rana catesbeiana rod photoreceptors. I. Characterization of the phosphorylation. J Biol Chem. 1994;269:15024–15029. [PubMed] [Google Scholar]
- 43.Matsuura I, Bondarenko VA, Maeda T, Kachi S, Yamazaki M, Usukura J, Hayashi F, Yamazaki A. Phosphorylation by cyclin-dependent protein kinase 5 of the regulatory subunit of retinal cGMP phosphodiesterase. I. Identification of the kinase and its role in the turnoff of phosphodiesterase in vitro. J Biol Chem. 2000;275:32950–32957. doi: 10.1074/jbc.M000702200. [DOI] [PubMed] [Google Scholar]
- 44.Hayashi F, Matsuura I, Kachi S, Maeda T, Yamamoto M, Fujii Y, Liu H, Yamazaki M, Usukura J, Yamazaki A. Phosphorylation by cyclin-dependent protein kinase 5 of the regulatory subunit of retinal cGMP phosphodiesterase. II. Its role in the turnoff of phosphodiesterase in vivo. J Biol Chem. 2000;275:32958–32965. doi: 10.1074/jbc.M000703200. [DOI] [PubMed] [Google Scholar]
- 45.Yamazaki A, Moskvin O, Yamazaki RK. Phosphorylation by cyclin-dependent protein kinase 5 of the regulatory subunit (Pγ) of retinal cGMP phosphodiesterase (PDE6): its implications in phototransduction. Adv Exp Med Biol. 2002;514:131–153. doi: 10.1007/978-1-4615-0121-3_9. [DOI] [PubMed] [Google Scholar]
- 46.Yamazaki A, Hayashi F, Tatsumi M, Bitensky MW, George JS. Interactions between the subunits of transducin and cyclic GMP phosphodiesterase in Rana catesbeiana rod photoreceptors. J Biol Chem. 1990;265:11539–11548. [PubMed] [Google Scholar]
- 47.Otto-Bruc A, Antonny B, Vuong TM, Chardin P, Chabre M. Interaction between the retinal cyclic GMP phosphodiesterase inhibitor and transducin. Kinetics and affinity studies. Biochemistry. 1993;32:8636–8645. doi: 10.1021/bi00084a035. [DOI] [PubMed] [Google Scholar]
- 48.Yamazaki A, Yamazaki M, Bondarenko VA, Matsumoto H. Discrimination of two functions of photoreceptor cGMP phosphodiesterase gamma subunit. Biochem Biophys Res Commun. 1996;222:488–493. doi: 10.1006/bbrc.1996.0771. [DOI] [PubMed] [Google Scholar]
- 49.Yamazaki A, Bondarenko VA, Dua S, Yamazaki M, Usukura J, Hayashi F. Possible stimulation of retinal rod recovery to dark state by cGMP release from a cGMP phosphodiesterase noncatalytic site. J Biol Chem. 1996;271:32495–32498. doi: 10.1074/jbc.271.51.32495. [DOI] [PubMed] [Google Scholar]
- 50.Cote RH, Brunnock MA. Intracellular cGMP concentration in rod photoreceptors is regulated by binding to high and moderate affinity cGMP binding sites. J Biol Chem. 1993;268:17190–17198. [PubMed] [Google Scholar]
- 51.Bondarenko VA, Desai M, Dua S, Yamazaki M, Amin RH, Yousif KK, Kinumi T, Ohashi M, Komori N, Matsumoto H, Jackson KW, Hayashi F, Usukura J, Lipkin VM, Yamazaki A. Residues within the polycationic region of cGMP phosphodiesterase gamma subunit crucial for the interaction with transducin alpha subunit. Identification by endogenous ADP-ribosylation and site-directed mutagenesis. J Biol Chem. 1997;272:15856–15864. doi: 10.1074/jbc.272.25.15856. [DOI] [PubMed] [Google Scholar]
- 52.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 53.Yamazaki A, Tatsumi M, Torney DC, Bitensky MW. The GTP-binding protein of rod outer segments. I. Role of each subunit in the GTP hydrolytic cycle. J Biol Chem. 1987;262:9316–9323. [PubMed] [Google Scholar]
- 54.Fairbanks G, Steck TL, Wallach DF. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971;10:2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]