Abstract
The social amoeba Dictyostelium discoideum is one of the leading model systems used to study how cells count themselves to determine the number and/or density of cells. In this review, we describe work on three different cell-density sensing systems used by Dictyostelium. The first involves a negative feedback loop in which two secreted signals inhibit cell proliferation during the growth phase. As the cell density increases, the concentrations of the secreted factors concomitantly increase, allowing the cells to sense their density. The two signals act as message authenticators for each other, and the existence of two different signals that require each other for activity may explain why previous efforts to identify autocrine proliferation-inhibiting signals in higher eukaryotes have generally failed. The second system involves a signal made by growing cells that is secreted only when they starve. This then allows cells to sense the density of just the starving cells, and is an example of a mechanism that allows cells in a tissue to sense the density of one specific cell type. The third cell density counting system involves cells in aggregation streams secreting a signal that limits the size of fruiting bodies. Computer simulations predicted, and experiments then showed, that the factor increases random cell motility and decreases cell-cell adhesion to cause streams to break up if there are too many cells in the stream. Together, studies on Dictyostelium cell density counting systems will help elucidate how higher eukaryotes regulate the size and composition of tissues.
Keywords: Dictyostelium, secreted, quorum sensing, chalone, cell density sensing, cell number counting
Introduction: Tumor dormancy
A major problem in treating cancer is the phenomenon of tumor dormancy: often, when a patient has a primary tumor and metastases, surgical removal of the primary tumor appears to stimulate cell proliferation in the metastatic foci (Demicheli, 2001, Guba et al., 2001, Peeters et al., 2008). Although the primary tumor appears to inhibit angiogenesis in the metastases (Holmgren et al., 1995), there is strong evidence that the primary tumor also inhibits the proliferation of single metastatic cells (Luzzi et al., 1998, Cameron et al., 2000, Guba et al., 2001). Endogenous angiogenesis inhibitors do not affect the proliferation of the single tumor cells (O’Reilly, 1997), leading to the hypothesis that the primary tumor secretes a factor that inhibits proliferation of the tumor cells, including cells in the metastatic foci (Guba et al., 2001). The ability to affect distant metastases suggests that the factor can circulate in the blood. Autocrine factors that inhibit cell proliferation as part of a negative feedback loop to regulate tissue size are called chalones. Identifying chalones and elucidating their signal transduction pathways could lead to the development of new therapies, such as simply injecting patients with the chalone, which might slow or stop the growth and proliferation of tumors and/ or metastases. However, despite good evidence indicating that chalones exist for a variety of tissues, in most cases the identity of these secreted factors is unknown (Gomer, 2001, Roisin-Bouffay & Gomer, 2004).
Secreted factors can be used to sense cell number
As a general principle, a secreted factor that can diffuse away from a group of cells can be used to sense the size of the group. As the number of cells in the group increases, the concentration of the factor increases (Figure 1). We have used diffusion calculations to show this formally for a variety of geometries; (Yuen & Gomer, 1994, Clarke & Gomer, 1995, Gomer, 2001, Roisin-Bouffay & Gomer, 2004). In addition, a diffusible secreted factor can be used to sense the number or density (with density being used to denote the number of cells/ ml) of cells of a specific type within a closed environment such as the body (Gomer, 2001, Roisin-Bouffay & Gomer, 2004). As the number of cells secreting the factor increases, the concentration of the factor in the closed system will also increase. To regulate the size of a tissue or group of cells, the cells can stop proliferating or undergo a morphogenetic rearrangement when the concentration of the factor indicates that a specific group size has been reached.
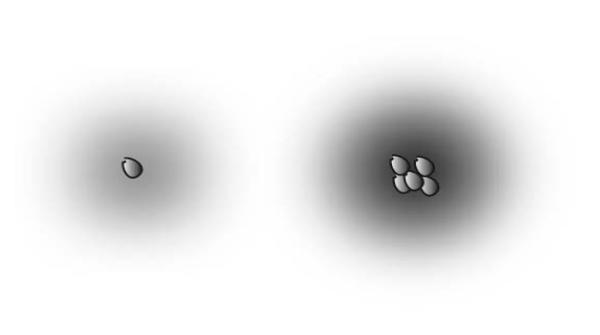
Figure 1.
A diffusible factor can be used by cells to sense the number of cells in a group. At left, a single cell secretes a diffusible factor; the intensity of the background represents the concentration of the factor. At right, a group of cells are all secreting the factor. Since the concentrations of the factor are effectively additive, the factor concentration in the vicinity of the cells is higher, allowing the cells to sense that there are other cells in the vicinity.
Dictyostelium cells secrete factors to communicate during growth and development
There are several signals secreted by Dictyostelium cells. During growth, at least two secreted polypeptide factors of unknown composition affect growth and gene expression (Clarke et al., 1987, Rathi et al., 1991, Rathi & Clarke, 1992, Whitbread et al., 1991, Iijima et al., 1995). DicA1, a cysteine-rich 80 kDa protein that is part of a ~400 kDa secreted complex, regulates gene expression during growth and development (Kolbinger et al., 2005). A ~450 kDa complex of unknown composition induces cells to begin development (Iijima et al., 1995). Aggregation and gene expression are mediated by the chemoattractant cAMP (Bonner, 1969, Barkley, 1969, Dottin et al., 1991, Milne et al., 1997, Saran et al., 2002). Other extracellular signals such as a chlorinated hydrocarbon called DIF (Brookman et al., 1987, Williams et al., 1987, Thompson et al., 2004), adenosine and ammonia (Gross et al., 1983, Schaap & Wang, 1986, Sternfeld, 1988, Kwong & Weeks, 1989, Xie et al., 1991), GABA, a steroid, and small peptides (Anjard et al., 1997, Anjard et al., 1998, Anjard & Loomis, 2006, Anjard et al., 2009) also play roles in development.
Identification of AprA, a secreted protein that regulates group size
While purifying a Dictyostelium secreted factor, we identified a 60 kDa contaminant protein we named AprA for autocrine proliferation repressor (Brock & Gomer, 2005). AprA has 28% identity to a Salmonella protein of undetermined function, and in a region of 37 amino acids has 37% identity and 59% similarity to a 100% conserved domain in a set of three different human putative proteins of unknown function. One (AAH35817) is expressed in lymphomas; one (BAA92109) is expressed in placenta, and the third (BAC04710) is expressed in liver.
To elucidate the function of AprA, we used homologous recombination to delete part of the aprA coding sequence to generate aprA− cells (Brock & Gomer, 2005). Developing aprA− cells form huge groups (Brock & Gomer, 2005). The aprA− phenotype was successfully rescued by expressing AprA under control of the Dictyostelium actin15 promoter in aprA− cells. Western blots stained with affinity-purified anti-AprA antibodies and size-exclusion gel chromatography indicated that AprA protein is part of a 138 kDa complex in the extracellular environment.
AprA represses proliferation
Proliferation curves (where proliferation refers to the increase in the number of cells, while growth refers to the increase in size and mass of the whole set of cells) for cells growing in liquid shaking culture indicated that aprA− cells proliferate faster than wild-type and reach stationary phase at a higher cell density, while aprA−/actin15::aprA cells proliferate slower and reach stationary phase at a lower density (Brock & Gomer, 2005). When grown on bacteria, aprA− cells also proliferated faster than wild-type, and aprA−/actin15::aprA cells were slower (Brock & Gomer, 2005). Videomicroscopy indicated that the cells were motile and thus viable, so that the differences in the proliferation of the cells were not due to differences in cell viability.
On a per nucleus basis, AprA does not affect growth
The growth (the increase in mass or protein per hour) and the proliferation (the increase in the number of cells per hour) of cells can be regulated independently (Gomer, 2001, Saucedo & Edgar, 2002, Dolznig et al., 2004, Jorgensen & Tyers, 2004). To determine if AprA regulates growth as well as proliferation, we measured the mass and protein content of populations of cells (Brock & Gomer, 2005). When the growth was calculated per nucleus, there was no significant difference in the mass accumulation rate between aprA− and wild type, while the mass accumulation rate was lower for aprA−/actin15::aprA. For all three cell lines, the protein accumulation rate per nucleus was approximately the same (Brock & Gomer, 2005). Together, the data suggested that although AprA inhibits proliferation, it does not affect growth in terms of mass or protein increase per nucleus per hour (Brock & Gomer, 2005).
The drawback to not having AprA is a decreased ability to form spores
A very puzzling aspect of the aprA gene is that disrupting it results in cells that proliferate faster. One would thus think that not having this gene would give an evolutionary advantage to cells. However, starvation is a selective pressure for Dictyostelium cells, and they have evolved to form spores when starved. We found that aprA− cells as well as aprA−/actin15::aprA cells formed approximately one sixth the number of viable spores compared to wild-type (Brock & Gomer, 2005). These data indicate that abnormally low or high levels of AprA reduce the ability of cells to form spores, and that cells thus need an optimal amount of AprA for efficient spore formation. In addition, aprA− cells die off relatively quickly after cells reach saturation. This suggests that Dictyostelium cells use AprA to slow proliferation at high cell density (when they are probably about to overgrow their food supply and starve) so that the cells will have more nutrient reserves.
AprA also helps to coordinate cytokinesis with mitosis
Counts of DAPI-stained cells showed that the aprA− population contained significantly more cells with three or more nuclei compared to wild-type cells, and showed that expression of AprA in the aprA− background rescues this defect (Brock & Gomer, 2005). It is interesting to note that several different human tumors involve multinucleate cells (Long & Aisenberg, 1975, Nonomura et al., 1995, Jayaram & Abdul Rahman, 1997, Ramos et al., 2002). One possibility is that as the population of proliferating cells reaches the density where AprA begins to slow cell proliferation, some nutrient becomes limiting, so that if the cells undergo mitosis too rapidly there is not enough time to completely assemble the cytokinesis machinery; another possibility is that AprA regulates a cell cycle checkpoint.
Verification that AprA is an extracellular signal that inhibits proliferation
Using known quantities of recombinant AprA (rAprA) as standards, and staining Western blots of conditioned growth medium and standards with affinity purified anti-AprA antibodies, we found that the extracellular concentration of AprA increases with cell density in the cultures, and at a density of 1.2 × 107 cells/ ml (where after one more round of cell division, cells will be at stationary phase), there was 300 ± 19 ng/ ml AprA (mean ± SEM, n=3) (Choe et al., 2009). rAprA at concentrations at and above 100 ng/ ml significantly slowed the proliferation of wild-type and aprA− cells. At concentrations above 4000 ng/ ml, the effect of rAprA on wild-type and aprA− cells is at a plateau, slowing rather than completely arresting proliferation. Comparing the bioactivity of different dilutions of conditioned growth medium (where we measured the concentration of AprA) to the bioactivity of different concentrations of rAprA, we observed that rAprA has roughly the same bioactivity as native AprA (Choe et al., 2009). These results show that AprA is an extracellular signal that inhibits proliferation.
Identification of CfaD as a second secreted protein that slows proliferation
We found that proliferating Dictyostelium cells secrete CfaD, a protein with similarity to mammalian cathepsins (Bakthavatsalam et al., 2008). Cathepsins are a family of lysosomal proteases (Nomura & Katunuma, 2005). Tumors often contain increased levels of cathepsins and, unlike most normal cells, secrete cathepsins (Jedeszko & Sloane, 2004, Gocheva & Joyce, 2007). Recombinant CfaD (rCfaD) has no protease activity, and both rCfaD and rCfaD with mutations in the putative cathepsin active site inhibit cell proliferation (Bakthavatsalam et al., 2008). This strongly suggests that CfaD acts as a proliferation-inhibiting signal despite its lack of detectable enzymatic activity. Interestingly, some of the mammalian cathepsins that CfaD has similarity to also have no detectable cathepsin activity, suggesting that these proteins also have some other function (Rawlings & Barrett, 1993). Like aprA− cells, cfaD− cells formed huge groups when starved, and expression of CfaD to generate cfaD−/CfaDOE cells rescued the phenotype.
As with AprA, the extracellular concentration of CfaD increases with cell density in the cultures (Bakthavatsalam et al., 2008). Physiological concentrations of rCfaD slow cell proliferation, and as with rAprA, high concentrations of rCfaD slow, but do not stop, proliferation (Bakthavatsalam et al., 2008). A possible explanation for this is that in the wild, a Dictyostelium cell might find itself in a small enclosed space where secreted factors might build up to very high concentrations, and having high concentrations of a secreted factor completely stop proliferation would be disadvantageous.
The cfaD− cells proliferate significantly faster and reach a significantly higher stationary phase density than wild-type or cfaD−/CfaDOE cells, while wild-type cells overexpressing CfaD proliferate slowly and reach stationary phase at a low cell density (Bakthavatsalam et al., 2008). After reaching stationary phase cell density, the cfaD− cells die off faster than wild-type or cfaD−/CfaDOE cells, while the cfaDOE cells were still alive when the wild type and cfaD−/cfaDOE cells had died. Like starved aprA− cells (Brock & Gomer, 2005), starved cfaD− cells form structures that have a reduced spore count and reduced spore viability. Expressing CfaD in the cfaD− background partially rescues both defects (Bakthavatsalam et al., 2008). Together, these data indicate that the evolutionary advantage for Dictyostelium to have AprA and CfaD appears to be that while both proteins slow proliferation, they both increase cell and spore viability (Bakthavatsalam et al., 2008, Brock & Gomer, 2005).
Like AprA, CfaD inhibits proliferation but not growth
Like aprA− cells, cfaD− cells tend to be multinucleate, and expression of CfaD in the cfaD− cells rescues this phenotype (Bakthavatsalam et al., 2008). We found that on a per nucleus basis, both AprA and CfaD do not affect growth, in terms of both cell mass accumulated per nucleus per hour, and protein accumulated per nucleus per hour (Bakthavatsalam et al., 2008, Choe et al., 2009).
AprA and CfaD form a complex in the extracellular environment
Size-exclusion gel chromatography indicated that in conditioned growth medium from wild-type cells, both AprA and CfaD are part of a ~138 kDa complex (Bakthavatsalam et al., 2008). In growth medium from aprA− cells, the CfaD-containing complex has a significantly smaller mass, while in growth medium from cfaD− cells, the AprA-containing complex has a smaller mass. In addition, pull-down assays showed that rCfaD added to wild type conditioned growth medium could pull down AprA, and that rAprA could pull down CfaD (Bakthavatsalam et al., 2008). These results suggest that in conditioned growth medium, CfaD and AprA interact with each other.
AprA is necessary for the effect of CfaD on proliferation, and vice versa
We found that rAprA had no observable effect on the proliferation of cfaD− cells (Choe et al., 2009), and rCfaD had no observable effect on the proliferation of aprA− cells (Bakthavatsalam et al., 2008). Adding mixtures of rCfaD and rAprA to cells showed that the presence of CfaD decreases the concentration of AprA needed to slow proliferation, and vice versa (Choe et al., 2009). These results suggest that two interacting proteins may function together as a chalone signal in a negative feedback loop that slows Dictyostelium cell proliferation. Having two different signals that are coupled to each other and which are both needed to activate a response is reminiscent of the phenomenon of T cell receptor activation in the immune system (Frauwirth & Thompson, 2002).
AprA and CrlA slow proliferation by lengthening G2
Compared to wild-type cells, aprA− and cfaD− cells have similar lengths of S and M phases, and shortened G2 phases (Hanson and Gomer, unpublished). This suggests that AprA and CfaD lengthen the cell cycle by lengthening the G2 phase.
AprA requires three G proteins for activity
Since many signal transduction pathways use G proteins, we examined the possibility that AprA and CfaD use G proteins in their signal transduction pathway. We found that like aprA− and cfaD− cells, Dictyostelium cells lacking the G protein components Gα8, Gα9, and Gβ proliferate faster and reach a higher stationary phase density than wild-type cells or cells lacking six other Gα subunits, despite secreting normal or high levels of AprA and CfaD (Bakthavatsalam et al., 2009). Like aprA− and cfaD− cells, gα8−, gα9− and gβ− cells die off faster than wild-type cells after reaching stationary phase (Bakthavatsalam et al., 2009). Compared to wild-type cells, the proliferation of gα8−, gα9− and gβ−cells are only weakly inhibited by rAprA. Like aprA− and cfaD− cells, gα8− and gβ− cells are multinucleate and have normal growth (the rate of increase in mass and protein per nucleus), whereas gα9− cells are not multinucleate and show increased growth (Bakthavatsalam et al., 2009). gα8− cells show normal cell-surface binding of rAprA, whereas gα9− and gβ− cells have fewer cell-surface rAprA binding sites, suggesting that Gα9 and Gβ regulate the synthesis or processing of the AprA receptor. Like other ligands that activate G proteins, rAprA induces the binding of [3H]GTP to membranes, and GTPγS inhibits the binding of rAprA to membranes. Both AprA-induced [3H]GTP binding and the GTPγS inhibition of rAprA binding require Gα8 and Gβ but not Gα9 (Bakthavatsalam et al., 2009). Like aprA− cells, ga8− cells have reduced spore viability (cells lacking Gβ do not aggregate or form fruiting bodies, so spore viability could not be tested in gβ− cells) (Bakthavatsalam et al., 2009). Together, the data showed that a chalone signal transduction pathway uses G proteins, and suggest that Gα8 and Gβ are part of the signal transduction pathway used by AprA to inhibit proliferation but not growth in Dictyostelium, whereas Gα9 is part of a different pathway that regulates both proliferation and growth.
CMF, a mechanism that senses the density of one type of cell
To attain efficient aggregation and spore dispersal, the amoebae need to coordinate their entry into the developmental pathway. Without such coordination, small cohorts of cells that happened to starve at the same time would form small, ineffectual fruiting bodies that would be unable to distribute spores far enough from the site of formation. This coordination is mediated by a mechanism that senses the density of starved cells and allows aggregation to occur only when there is a sufficiently high density of starved cells to form a properly sized fruiting body. Dictyostelium cells starved at low cell densities will aggregate and express prestalk and prespore markers only in starvation buffer previously conditioned by a high density of starved cells (Grabel & Loomis, 1978, Kay, 1982, Mehdy & Firtel, 1985, Gomer et al., 1986a, Gomer et al., 1986b). This indicated that starving Dictyostelium cells are able to sense each other, and are able to make a developmental decision based upon this information. This ability is not dependent upon cell-cell contact, but involves a secreted, soluble factor (conditioned medium factor, CMF) (Mehdy & Firtel, 1985). Using the prestalk and prespore gene expression assay described above, we purified CMF from conditioned starvation medium (Gomer et al., 1991). Using a CMF partial amino acid sequence, the cDNA coding for CMF was then isolated (Jain et al., 1992). CMF is an 80 kDa glycoprotein with optimal activity at 0.3 ng/ml (Jain et al., 1992). Cells lacking CMF fail to aggregate unless exogenous CMF is added (Jain et al., 1992). CMF is secreted at a constant rate from starving cells, and thus makes for an excellent quorum-sensing molecule, as its extracellular concentration is dependent upon the number of cells secreting it (Yuen & Gomer, 1994, Clarke et al., 1992, Clarke & Gomer, 1995).
Interestingly, there are two classes of CMF, the full length protein and a mixture of breakdown products ranging in size from 0.65 kDa to 6.5 kDa (Yuen et al., 1991). While full length CMF is produced and sequestered in vegetative cells, it is only secreted when cells begin to starve. In contrast, the small CMF products appear in the extracellular medium only during late development. The specific activity of these smaller CMF products is roughly 100-fold greater than that of full length CMF. One possible explanation for this difference is that the full-length CMF is used to coordinate aggregation in a population of cells, while the small breakdown products of CMF could allow cells which had wandered away from the main group of cells a chance to differentiate and potentially form spores even in the absence of fruiting body formation.
CMF regulates cAMP signal transduction
We found that CMF coordinates aggregation by regulating several aspects of cAMP signal transduction (Yuen et al., 1995). Dictyostelium cells respond to a pulse of cAMP in three ways. The cells move towards the source of cAMP, release a burst of cAMP themselves to relay the signal, and activate or deactivate expression of specific classes of genes (Mann & Firtel, 1989). The incoming cAMP pulse is detected by the cell surface cAMP receptor cAR1. The binding of cAMP to cAR1 causes the receptor to activate the heterotrimeric G protein Gα2βγ, which in turn activates many of the downstream responses associated with the activation of cAR1, including chemotaxis and gene expression (Kumagai et al., 1991 12352, Kumagai et al., 1989, Mann & Firtel, 1989). Activation of cAR1 and its associated G protein leads to transient activation of guanylyl cyclase and adenylyl cyclase. In addition, there is a cAMP-induced transient uptake of extracellular Ca++ and activation of development-specific genes (Milne & Devreotes, 1993, Kuwayama & van Haastert, 1998). CMF regulates these aspects of cAMP signaling as the activations of Ca++ influx, adenylyl cyclase, guanylyl cyclase, and gene expression in response to a pulse of cAMP are strongly inhibited in cells lacking CMF (Yuen et al., 1995).
CMF appears to regulate cAMP signaling through two separate pathways, using at least two different receptors (Jain & Gomer, 1994). CMF controls cAMP-dependent gene expression through the receptor CMFR1 (Deery & Gomer, 1999, Deery et al., 2002). Cells lacking this receptor exhibit a 50% decrease in CMF binding, suggesting that at least one other receptor for CMF exists. In addition, cmfR1− cells lack cAMP-dependent expression of prespore and prestalk genes, while retaining Ca++ influx, adenylyl cyclase, and guanylyl cyclase activiation. This suggests that a second receptor is responsible for CMF regulation of these specific effects.
The activations of Ca++ influx, adenylyl cyclase, and guanylyl cyclase in response to a pulse of cAMP are strongly inhibited in cells lacking CMF (Yuen et al., 1995). This inhibition does not occur at the level of cAR1 occupancy, as cmf− cells have normal levels of cAR1 expressed on the cell surface, and bind cAMP similarly to wild-type cells (Van Haastert et al., 1996). In addition, inhibition is not at the level of G protein activation, as the cAMP-stimulated GTP binding by Gα2, as measured by in vitro GTP binding, is not measurably affected by the presence or absence of CMF.
However, the deactivation of Gα2 is drastically altered by the presence of CMF. In vitro GTPase assays showed that CMF decreases the hydrolysis rate of the GTP associated with Gα2, and thus prolongs the length of time Gα2 remains in the GTP-bound, activated state (Brazill et al., 1997). Thus, CMF coordinates development by regulating cAMP signal transduction so that cells will not respond to cAMP and begin development until a sufficient number of cells are starving, as indicated to the cells by high levels of extracellular CMF.
The CMF signal transduction pathway
CMF exerts its effect on cAMP signaling through a G protein signaling pathway of its own. Specifically, CMF binds a G protein coupled receptor associated with Gα1. This pathway activates phospholipase C, an enzyme that converts the phosphoinositide PIP2 to inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (Brazill et al., 1998). CMF stimulates IP3 production, and this effect is lost in cells lacking phospholipase C, Gα1, or Gβ. RpkA, an unusual G-protein coupled receptor, appears to regulate Gα1 but may not be the CMF receptor per se (Bakthavatsalam et al., 2007). Interestingly, in the Gα1 null cells, the inositol 1,4,5-trisphosphate levels were higher than the levels seen in wild-type cells stimulated with CMF, suggesting that Gα1 is a negative regulator of phospholipase C. Conversely, in starving Gβ null cells, the inositol 1,4,5-trisphosphate levels were much lower than the levels seen in starving wild-type cells, suggesting that Gβ is a positive regulator of phospholipase C. Most importantly, phospholipase C is required for CMF to regulate the GTPase activity of Gα2, as well as for the activations of guanylyl and adenylyl cyclase. This pathway was further delineated when it was found that the phospholipase D orthologue, PldB, regulates CMF signaling. Cells lacking pldB aggregate at low cell density, as if the CMF signal transduction pathway is constitutively on. Conversely, cells overexpressing pldB do not aggregate at all, as though the CMF pathway is constitutively off (Chen et al., 2005). Both cell lines are insensitive to added CMF, suggesting that PldB is involved in the CMF quorum sensing pathway. Much like phospholipase C, PldB is required for CMF to regulate the GTPase activity of Gα2, as CMF can no longer decrease the GTPase activity of Gα2 in cells lacking PldB (Ray et al., 2010) (Figure 2).
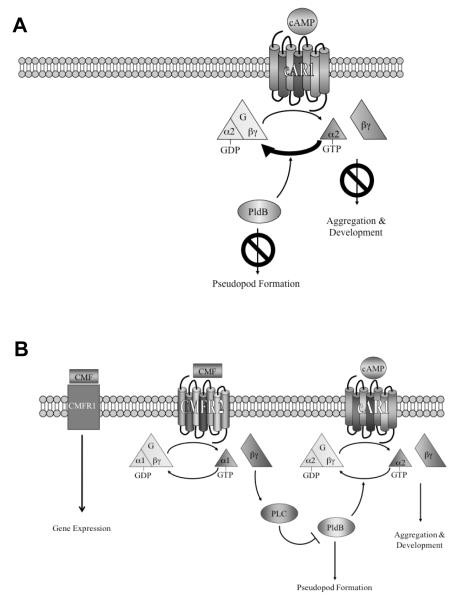
Figure 2.
Model of cAMP signaling in the absence (A) and presence (B) of CMF.
Interestingly, CMF, as a quorum sensing molecule, appears to not only influence cAMP signaling, but also seems to regulate cell shape (Yuen et al., 1995). Cells starved at high cell density have multiple ruffles and pseudopodia. Cells starved at low cell density are smooth and round. The addition of CMF to cells at low cell density causes them to develop pseudopodia and ruffles, much like cells at high cell density. Cells lacking CMF starved at high cell density are round and smooth. Thus CMF is both necessary and sufficient to induce drastic cell shape changes.
This same study demonstrated that cells starved in the presence of CMF extended pseudopodia 2.5 times more often than cells in the absence of CMF. The increased rate of pseudopod formation is likely a prelude to aggregation. To begin aggregating, cells need to become chemotactically responsive to cAMP. As the process of chemotaxis relies heavily on pseudopod formation, it makes sense that pseudopod extensions would be seen in starved cells. CMF appears to prepare the cells for chemotaxis by enabling them to create pseudopodia so that when the cell senses cAMP, it can extend a pseudopod towards the source of cAMP and thus initiate aggregation. Without CMF, pseudopod extension is suppressed, thus the cells are smooth and round. It is important to note that in these experiments, the changes in cell morphology induced by CMF occur at low cell densities, where no cAMP signaling can occur. Therefore, the effect of CMF on cell morphology appears to be a direct effect of CMF as opposed to a secondary effect from CMF regulation of cAMP signal transduction, suggesting that CMF regulates the cytoskeleton as part of its quorum sensing function.
A divergence of the signaling pathways that regulate the cytoskeleton and cAMP occurs after activation of Gα1 (Brazill et al., 1998). The point of divergence may be the phospholipase D orthologue, PldB, as it is involved in regulating both cAMP signaling and the cytoskeleton. Dictyostelium has two phospholipase D homologues. PldA is predominantly expressed in vegetative cells, whereas PldB is expressed during early development (Chen et al., 2005). The CMF signal transduction pathway appears to regulate PldB, which in turn produces phosphatidic acid which then regulates, through an unknown pathway, the GTPase activity of Gα2 (Ray et al., 2010).
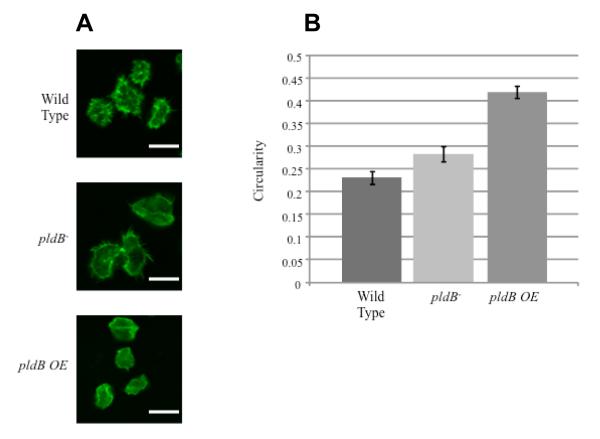
Phospholipase D also regulates actin localization (Zouwail et al., 2005). When pldB is overexpressed, actin is mislocalized and filopdial extensions, which require organized F-actin, are lost (Figure 3A). The amount of filopodia and pseudopods formed can be quantified by determining the circularity of a cell. Circularity is defined as 4π(area)/perimeter2. Thus, a cell whose perimeter forms a perfect circle has a circularity of 1. A cell with an increased number of extensions has an increased perimeter, which leads to a decreased circularity. Starved wild type and pldB− cells have circularities of 0.23 ± 0.01 and 0.28 ± 0.02 respectively. In contrast, starved cells overexpressing pldB have a circularity of 0.42 ± 0.02 (Figure 3B). Thus, much like cells in the absence of CMF, cells over expressing pldB suppress pseudopod formation. Interestingly, this effect is only seen in starved cells. Overexpression of PldB in vegetative cells has no effect on the actin cytoskeleton, suggesting that whatever PldB is acting through to affect actin is not present in vegetative cells.
Figure 3.
PldB regulates cell shape. A) Wild-type, pldB− and pldB overexpressing cells were starved, fixed and stained for F-actin with rhodamine-phalloidin. Bar is 10 μm. B) The circularity (4π(area)/perimeter2) of wild-type, pldB−, and pldB overexpressing cells was calculated using cells prepared as above.
CMF thus appears to control the initiation of development by regulating gene expression, the actin cytoskeleton, and cAMP signaling. Gene expression is regulated through a pathway involving CMFR1. The actin cytoskeleton and cAMP signaling are regulated through a single pathway which diverges after PldB, as shown in our model (Figure 2A). cAMP binds to the cAMP receptor cAR1, activating the associated G protein, Gα2βγ. The Gα2 subunit releases GDP and binds GTP, thus separating from Gβγ. Under low cell density conditions (Figure 2A), CMF levels are low, and therefore CMF signaling does not occur. This allows PldB activity to remain elevated, preventing pseudopod formation and allowing the intrinsic GTPase activity of Gα2 to rapidly hydrolyze the GTP back to GDP. This returns the G protein to its deactivated state, thus preventing aggregation. As more cells begin to starve, the levels of CMF increase until they reach a threshold level (Figure 2B). At this point, CMF activates its known receptor, CMFR1, allowing for the activation of gene expression. CMF also activates its presumed receptor, CMFR2, triggering a signaling cascade involving the activation of Gα1βγ and phospholipase C. This signaling leads to the inhibition of PldB activity, allowing pseudopod formation. In addition, decreased PldB activity supports dissociation of Gα2 and Gβγ by decreasing the GTPase activity of Gα2, allowing cAMP signaling to occur. Together, this supports aggregation and development.
CF regulates fruiting body size
A third secreted factor called counting factor (CF) is involved in sensing the number of cells in an aggregation stream, and causes the aggregation stream to break up if there are too many cells in the stream (Brock & Gomer, 1999). Because each aggregate undergoes morphogenesis to form a fruiting body, it is very important for Dictyostelium to modulate the size of aggregates during development. If an aggregate is too small, the resulting fruiting body will be too close to the ground for optimal spore dispersal. At the same time, if an aggregate is too big, the resulting fruiting body may topple over. Dictyostelium cells thus carefully regulate the size of aggregates.
The elucidation of the CF aggregation size regulation mechanism began with the identification of a mutant called smlAAS from a shotgun antisense mutagenesis screen (Spann et al., 1996). Developing smlAAS cells formed large numbers of abnormally small fruiting bodies (Spann et al., 1996, Brock et al., 1996). The antisense insert from the smlAAS cells encodes part of a novel protein which does not have any similarities to any known protein, and disruption of the smlA gene by homologous recombination caused the resulting smlA− cells to form large numbers of tiny fruiting bodies (Brock & Gomer, 1999). The addition of conditioned starvation medium harvested from developing smlA− cells to developing wild-type cells caused the wild-type cells to form tiny fruiting bodies, indicating that the smlA− cells were secreting a factor that reduces fruiting body size. Using the ability of the factor to cause wild-type cells to form tiny fruiting bodies as a bioassay, we purified the factor, and identified it as a ~450 kDa protein complex, which we named CF. CF is made up of at least 4 components: countin, CF45-1, CF50, and CF60, which are 40, 45, 50, and 60 kDa, respectively. Wild-type cells also secrete CF, indicating that the phenotype of smlA− cells is due to oversecretion of CF. Disruption of genes encoding each component of CF causes cells to secrete almost no detectable CF activity (Brock et al., 2002, Brock et al., 2003b, Brock et al., 2006). The aggregation streams seldom break in countin−, cf45-1−, and cf50− transformants, resulting in small numbers of huge aggregates. We were not able to obtain cf60− transformants, suggesting that cf60 may be essential (Brock et al., 2006).
Computer simulations predicted that CF regulates adhesion and motility
To gain insight into how a secreted factor can regulate group size, we performed mathematical modeling (Roisin-Bouffay et al., 2000, Dallon et al., 2006, Brock et al., 2003b, Tang et al., 2002, Jang & Gomer, 2008). After testing many different parameters, we found that the local concentration of a secreted factor such as CF can modulate group size by altering cell-cell adhesion and random motility in an adhesion stream (Roisin-Bouffay et al., 2000). In addition, computer simulations showed that it is not cell-cell adhesion or random motility per se, but the ratio of cell-cell adhesion and random motility that determines the group size (Dallon et al., 2006). The essential prediction from the computer simulations was that if a stream has too many cells in it, as indicated to the cells by high levels of CF, the cells can disrupt the integrity of the stream by decreasing cell-cell adhesion and increasing random cell motility, causing streams to physically break. In addition, a virtual mutant created by computer simulations predicted that cells can regulate group size by changing the frequency of cell reorientation. Experiments then confirmed that, as predicted, CF decreases cell-cell adhesion and increases cell motility, and that altering cell-cell adhesion or cell motility qualitatively affects group size in accordance with the above prediction (Roisin-Bouffay et al., 2000, Tang et al., 2002).
Different components of CF have different activities
The components of CF complex have distinct and overlapping functions. For example, the aggregation streams in transformants with disruptions of countin, cf45-1, or cf50 gene rarely break, so that large but few aggregates are formed (Brock & Gomer, 1999, Brock et al., 2003b, Brock et al., 2002). In cf50− transformants, we observed some degradation in secreted countin, suggesting that the role of CF50 may be to protect countin (Brock et al., 2002). Even though cf50− cells form large groups, the developing structures of cf50− cells are more aberrant than countin− cells, suggesting that CF50 may play a role in cell-type differentiation. In countin− and cf50− cells, we observed a large amount of CF45-1 accumulation during development (Brock et al., 2003b). The addition of recombinant countin, CF45-1, CF50, or CF60 to wild-type cells reduced group size (Gao et al., 2002, Brock et al., 2003b, Brock et al., 2003a, Brock et al., 2006). Even though recombinant CF45-1 reduces group size when added to wild-type and cf45-1− cells, it has very little effect when added to countin− or cf50− cells, suggesting that CF45-1 requires countin and CF50 to affect group size (Brock et al., 2003b). Similarly, recombinant CF60 reduces group size when added to wild-type, countin−, or cf45-1− cells, but does not affect group size when added to cf50− cells, suggesting that the effect of CF60 on group size requires CF50 (Brock et al., 2006).
CF regulates key aspects of the cAMP signal transduction pathway
During aggregation, the CF signal transduction pathway seems to adopt the cAMP and cGMP signaling pathways to mediate group size determination (Tang et al., 2001). In developing Dictyostelium cells, the cAMP and cGMP signaling pathways regulate many phenomena including cell-cell adhesion and motility (Mato et al., 1977, Wurster et al., 1977, Verkerke-van Wijk & Schaap, 1997, Bosgraaf & van Haastert, 2002, Bosgraaf & van Haastert, 2006). CF upregulates the size of the cAMP-induced cAMP pulse and downregulates the cAMP-induced cGMP pulse, and regulates key components of the cAMP and cGMP signal transduction pathway (Tang et al., 2001, Brock et al., 2002). For instance, countin regulates cAMP-induced Akt/PKB translocation and activity (Gao et al., 2004). Individual CF components however have different effects on these pathways. Countin increases cAMP-induced cAMP pulses whereas CF50 decreases cAMP-induced cAMP pulses. In addition, countin and CF50 have opposite effects on cAMP-stimulated erk2 activation (Brock et al., 2003a).
CF regulates glucose to regulate fruiting body size
In addition to cAMP and cGMP signaling pathways, glucose is also implicated in CF-mediated group size determination (Jang et al., 2002, Jang & Gomer, 2005, Jang & Gomer, 2006, Jang et al., 2009). Transformants lacking bioactive CF (countin−, cf45-1−, and cf50−), as well as wild-type cells treated with antibodies to deplete secreted CF, have high internal glucose levels, while smlA− cells (which oversecrete CF) and wild-type cells treated with recombinant components of CF have low internal glucose levels (Jang et al., 2002). The addition of 1 mM glucose to starving cells increases internal glucose levels, negates the effect of high levels of CF, and mimics the effect of depleting CF on group size, cell-cell adhesion, and random motility (Jang et al., 2002). As a part of the signal transduction pathway whereby CF decreases internal glucose levels, CF decreases the activity of glucose-6-phosphatase by increasing the Km to inhibit the last step of gluconeogenesis (Jang & Gomer, 2005, Jang & Gomer, 2006). CF also increases the levels of glucose-6-phosphate and fructose-6-phosphate and decreases the levels of fructose-1,6-bisphosphate, pyruvate, lactate, ATP, and the rate of oxygen consumption (Jang & Gomer, 2008). We hypothesized that there exists a primordial response in cells that decreases adhesion and increases motility when cells are in the presence of low nutrients (as sensed by low internal glucose levels), and that CF modulates this mechanism to regulate adhesion and motility.
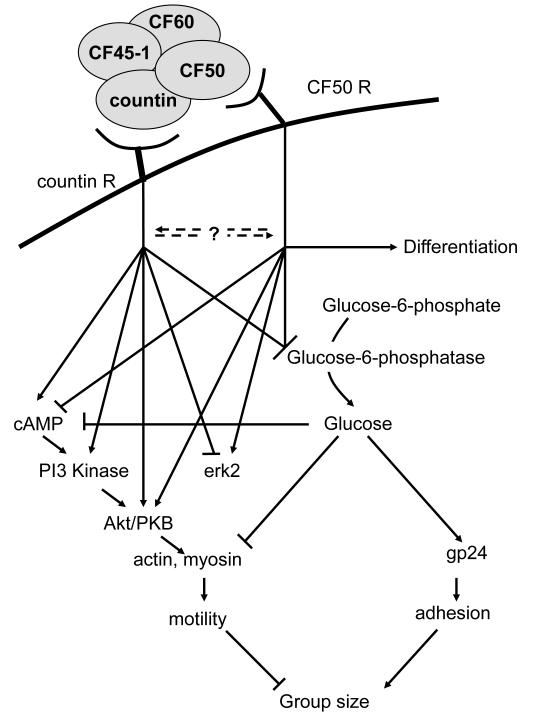
Finally, there seems to be at least two separate receptors (a countin receptor and a CF50 receptor) for components of the CF complex, and thus at least two separate pathways which can be distinguished by their effects on cAMP signal transduction and cell-type differentiation (Gao et al., 2002, Brock et al., 2003a). Our working hypothesis is that countin and CF50 bind to their receptors and activate their own signal transduction pathways (Figure 4). At the same time, one signal transduction pathway may modulate the other signaling pathway. The two pathways then converge to regulate cell-cell adhesion and random motility, and finally group size. Having separate signaling pathways for the different components of CF complex may allow cells to more precisely modulate cell-cell adhesion and random motility to regulate group size.
Figure 4.
Summary of the CF signal transduction pathway. Our current model is that countin and CF50 bind to separate surface receptors and trigger separate signaling pathways distinguished by their effects on cAMP signal transduction and cell-type differentiation.
Summary
In this review, we have summarized what is known about three different systems that Dictyostelium uses for cell density sensing. In the AprA/CfaD system, we found a chalone pathway in Dictyostelium where the chalone signal has two components that need each other for activity and thus act as message authenticators for each other. CMF represents a system that uses a secreted signal to sense the composition of a group of cells. Finally, CF represents a system where cells undergo a morphogenetic rearrangement to form groups of cells of a defined size. This work will hopefully help us to elucidate the physics and biochemistry of development in higher eukaryotes. For instance, this work has already led to the identification of a human blood serum protein that appears to play a role in wound healing and fibrosing diseases such as cardiac fibrosis, pulmonary fibrosis, and end-stage kidney disease (Pilling et al., 2003, Pilling et al., 2007, Haudek et al., 2006, Naik-Mathuria et al., 2008, Gomer et al., 2009), and clinical trials using this secreted factor are currently underway (Duffield & Lupher, 2010).
Acknowledgements and disclosure
This work was supported by NIH grants GM074990, S06-GM606564, and RR03037, NSF grant 0346975, and Dongguk University research fund DRIMS 2006-1094-Z. We thank Sarah Herlihy and Jonathan Phillips for comments on the manuscript. Dr. Gomer is a Science Advisory Board member for Trellis Bioscience, and has received royalties and stock options from Trellis. He is also a cofounder and Science Advisory Board members for Promedior, and has received royalties and stock options from Promedior.
References
- Anjard C, Loomis WF. GABA induces terminal differentiation of Dictyostelium through a GABAB receptor. Development. 2006;133:2253–2261. doi: 10.1242/dev.02399. [DOI] [PubMed] [Google Scholar]
- Anjard C, Su Y, Loomis WF. Steroids initiate a signaling cascade that triggers rapid sporulation in Dictyostelium. Development. 2009;136:803–812. doi: 10.1242/dev.032607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjard C, Van Bemmelen M, Veron M, Reymond CD. A new spore differentiation factor (SDF) secreted by Dictyostelium cells is phosphorylated by the cAMP dependent protein kinase. Differentiation. 1997;62:43–49. doi: 10.1046/j.1432-0436.1997.6210043.x. [DOI] [PubMed] [Google Scholar]
- Anjard C, Zeng C, Loomis WF, Nellen W. Signal transduction pathways leading to spore differentiation in Dictyostelium discoideum. Dev. Biol. 1998;193:146–155. doi: 10.1006/dbio.1997.8804. [DOI] [PubMed] [Google Scholar]
- Bakthavatsalam D, Brazill D, Gomer RH, Eichinger L, Rivero F, Noegel AA. A G protein-coupled receptor with a lipid kinase domain is involved in cell-density sensing. Curr Biol. 2007;17:892–897. doi: 10.1016/j.cub.2007.04.029. [DOI] [PubMed] [Google Scholar]
- Bakthavatsalam D, Brock DA, Nikravan NN, Houston KD, Hatton RD, Gomer RH. The secreted Dictyostelium protein CfaD is a chalone. J. Cell Science. 2008;121:2473–2480. doi: 10.1242/jcs.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavatsalam D, Choe JM, Hanson NE, Gomer RH. A Dictyostelium chalone uses G proteins to regulate proliferation. BMC Biol. 2009;7:44. doi: 10.1186/1741-7007-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley DS. Adenosine-3′,5′-phosphate: Identification as acrasine in a species of cellular slime mould. Science. 1969;165:1133–1134. doi: 10.1126/science.165.3898.1133. [DOI] [PubMed] [Google Scholar]
- Bonner JT. Hormones in social ameba and mammals. Sci. Amer. 1969;220:78–91. doi: 10.1038/scientificamerican0669-78. [DOI] [PubMed] [Google Scholar]
- Bosgraaf L, Van Haastert PJ. The regulation of myosin II in Dictyostelium. Eur J Cell Biol. 2006;85:969–979. doi: 10.1016/j.ejcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Bosgraaf L, Van Haastert PJM. A model for cGMP signal transduction in Dictyostelium in perspective of 25 years of cGMP research. J. Muscle Res. Cell Motil. 2002;23:781–791. doi: 10.1023/a:1024431813040. [DOI] [PubMed] [Google Scholar]
- Brazill DT, Gundersen R, Gomer RH. A cell-density sensing factor regulates the lifetime of a chemoattractant-induced Gα-GTP conformation. FEBS Letters. 1997;404:100–104. doi: 10.1016/s0014-5793(97)00104-x. [DOI] [PubMed] [Google Scholar]
- Brazill DT, Lindsey DF, Bishop JD, Gomer RH. Cell density sensing mediated by a G protein-coupled receptor activating phospholipase C. J. Biol. Chem. 1998;273:8161–8168. doi: 10.1074/jbc.273.14.8161. [DOI] [PubMed] [Google Scholar]
- Brock DA, Buczynski F, Spann TP, Wood SA, Cardelli J, Gomer RH. A Dictyostelium mutant with defective aggregate size determination. Development. 1996;122:2569–2578. doi: 10.1242/dev.122.9.2569. [DOI] [PubMed] [Google Scholar]
- Brock DA, Ehrenman K, Ammann R, Tang Y, Gomer RH. Two components of a secreted cell-number counting factor bind to cells and have opposing effects on cAMP signal transduction in Dictyostelium. J Biol Chem. 2003a;278:52262–52272. doi: 10.1074/jbc.M309101200. [DOI] [PubMed] [Google Scholar]
- Brock DA, Gomer RH. A cell-counting factor regulating structure size in Dictyostelium. Genes Dev. 1999;13:1960–1969. doi: 10.1101/gad.13.15.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock DA, Gomer RH. A secreted factor represses cell proliferation in Dictyostelium. Development. 2005;132:4553–4562. doi: 10.1242/dev.02032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock DA, Hatton RD, Giurgiutiu D-V, Scott B, Ammann R, Gomer RH. The different components of a multisubunit cell number-counting factor have both unique and overlapping functions. Development. 2002;129:3657–3668. doi: 10.1242/dev.129.15.3657. [DOI] [PubMed] [Google Scholar]
- Brock DA, Hatton RD, Giurgiutiu D-V, et al. CF45-1, a secreted protein which participates in group size regulation in Dictyostelium. Eukaryotic Cell. 2003b;2:788–797. doi: 10.1128/EC.2.4.788-797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock DA, Van Egmond WN, Shamoo Y, Hatton RD, Gomer RH. A 60-kilodalton protein component of the counting factor complex regulates group size in Dictyostelium discoideum. Eukaryot Cell. 2006;5:1532–1538. doi: 10.1128/EC.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookman JJ, Jermyn KA, Kay RR. Nature and distribution of the morphogen DIF in the Dictyostelium slug. Development. 1987;100:119–124. doi: 10.1242/dev.100.1.119. [DOI] [PubMed] [Google Scholar]
- Cameron MD, Schmidt EE, Kerkvliet N, et al. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res. 2000;60:2541–2546. [PubMed] [Google Scholar]
- Chen Y, Rodrick V, Yan Y, Brazill D. PldB, a putative phospholipase D homologue in Dictyostelium discoideum mediates quorum sensing during development. Eukaryot Cell. 2005;4:694–702. doi: 10.1128/EC.4.4.694-702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe JM, Bakthavatsalam D, Phillips JE, Gomer RH. Dictyostelium cells bind a secreted autocrine factor that represses cell proliferation. BMC Biochem. 2009;10:4. doi: 10.1186/1471-2091-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Dominguez N, Yuen IS, Gomer RH. Growing and starving Dictyostelium cells produce distinct density-sensing factors. Dev. Biol. 1992;152:403–406. doi: 10.1016/0012-1606(92)90147-9. [DOI] [PubMed] [Google Scholar]
- Clarke M, Gomer RH. PSF and CMF, autocrine factors that regulate gene expression during growth and early development of Dictyostelium. Experientia. 1995;51:1124–1134. doi: 10.1007/BF01944730. [DOI] [PubMed] [Google Scholar]
- Clarke M, Kayman SC, Riley K. Density-dependent induction of discoidin-I synthesis in exponentially growing cells of Dictyostelium discoideum. Differentiation. 1987;34:79–87. doi: 10.1111/j.1432-0436.1987.tb00053.x. [DOI] [PubMed] [Google Scholar]
- Dallon J, Jang W, Gomer RH. Mathematically modelling the effects of counting factor in Dictyostelium discoideum. Math Med Biol. 2006;23:45–62. doi: 10.1093/imammb/dqi016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deery WJ, Gao T, Ammann R, Gomer RH. A single cell-density sensing factor stimulates distinct signal transduction pathways through two different receptors. J. Biol. Chem. 2002;277:31972–31979. doi: 10.1074/jbc.M204539200. [DOI] [PubMed] [Google Scholar]
- Deery WJ, Gomer RH. A putative receptor mediating cell-density sensing in Dictyostelium. J. Biol. Chem. 1999;274:34476–34482. doi: 10.1074/jbc.274.48.34476. [DOI] [PubMed] [Google Scholar]
- Demicheli R. Tumour dormancy: findings and hypotheses from clinical research on breast cancer. Semin Cancer Biol. 2001;11:297–306. doi: 10.1006/scbi.2001.0385. [DOI] [PubMed] [Google Scholar]
- Dolznig H, Grebien F, Sauer T, Beug H, Mullner E. Evidence for a size-sensing mechanism in animal cells. Nat. Cell Biol. 2004;6:899–905. doi: 10.1038/ncb1166. [DOI] [PubMed] [Google Scholar]
- Dottin RP, Bodduluri SR, Doody JF, Haribabu B. Signal transduction and gene expression in Dictyostelium discoideum. Dev. Genetics. 1991;12:2–5. doi: 10.1002/dvg.1020120103. [DOI] [PubMed] [Google Scholar]
- Duffield JS, Lupher ML., Jr. PRM-151 (recombinant human serum amyloid P/pentraxin 2) for the treatment of fibrosis. Drug News Perspect. 2010;23:305–315. doi: 10.1358/dnp.2010.23.5.1444206. [DOI] [PubMed] [Google Scholar]
- Frauwirth KA, Thompson CB. Activation and inhibition of lymphocytes by costimulation. J Clin Invest. 2002;109:295–299. doi: 10.1172/JCI14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Ehrenman K, Tang L, Leippe M, Brock DA, Gomer RH. Cells respond to and bind countin, a component of a multisubunit cell number counting factor. J Biol Chem. 2002;277:32596–32605. doi: 10.1074/jbc.M203075200. [DOI] [PubMed] [Google Scholar]
- Gao T, Knecht D, Tang L, Hatton RD, Gomer R. A cell number counting factor regulates Akt/Protein Kinase B to regulate group size in Dictyostelium discoideum group size. Eukaryotic Cell. 2004;3:1176–1184. doi: 10.1128/EC.3.5.1176-1184.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocheva V, Joyce JA. Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle. 2007;6:60–64. doi: 10.4161/cc.6.1.3669. [DOI] [PubMed] [Google Scholar]
- Gomer RH. Not being the wrong size. Nat. Rev. Mol. Cell Biol. 2001;2:48–54. doi: 10.1038/35048058. [DOI] [PubMed] [Google Scholar]
- Gomer RH, Armstrong D, Leichtling BH, Firtel RA. cAMP induction of prespore and prestalk gene expression in Dictyostelium is mediated by the cell-surface cAMP receptor. Proc. Natl. Acad. Sci. USA. 1986a;83:8624–8628. doi: 10.1073/pnas.83.22.8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomer RH, Datta S, Firtel RA. Cellular and subcellular distribution of a cAMP-regulated prestalk protein and prespore protein in Dictyostelium discoideum: A study on the ontogeny of prestalk and prespore cells. J.Cell Biol. 1986b;103:1999–2015. doi: 10.1083/jcb.103.5.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomer RH, Pilling D, Kauvar LM, et al. A serum amyloid P-binding hydrogel speeds healing of partial thickness wounds in pigs. Wound Repair Regen. 2009;17:397–404. doi: 10.1111/j.1524-475X.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomer RH, Yuen IS, Firtel RA. A secreted 80×103 Mr protein mediates sensing of cell density and the onset of development in Dictyostelium. Development. 1991;112:269–278. doi: 10.1242/dev.112.1.269. [DOI] [PubMed] [Google Scholar]
- Grabel L, Loomis WF. Effector controlling accumulation of N-acteylglucosaminidase during development of Dictyostelium discoideum. Dev. Biol. 1978;64:203–209. doi: 10.1016/0012-1606(78)90072-6. [DOI] [PubMed] [Google Scholar]
- Gross JD, Bradbury J, Kay RR, Peacey MJ. Intracellular pH and the control of cell differentiation in Dictyostelium discoideum. Nature. 1983;303:244–245. doi: 10.1038/303244a0. [DOI] [PubMed] [Google Scholar]
- Guba M, Cernaianu G, Koehl G, et al. A primary tumor promotes dormancy of solitary tumor cells before inhibiting angiogenesis. Cancer Res. 2001;61:5575–5579. [PubMed] [Google Scholar]
- Haudek SB, Xia Y, Huebener P, et al. Bone marrow-derived fibroblast precursors mediate ischemic cardiomyopathy in mice. Proc Natl Acad Sci U S A. 2006;103:18284–18289. doi: 10.1073/pnas.0608799103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren L, O’reilly MS, Folkman J. Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat. Med. 1995;1:149–153. doi: 10.1038/nm0295-149. [DOI] [PubMed] [Google Scholar]
- Iijima N, Takagi T, Maeda Y. A proteinous factor mediating intercellular communication during the transition of Dictyostelium cells from growth to differentiation. Zool. Sci. 1995;12:61–69. doi: 10.2108/zsj.12.61. [DOI] [PubMed] [Google Scholar]
- Jain R, Gomer RH. A developmentally regulated cell surface receptor for a density-sensing factor in Dictyostelium. J. Biol. Chem. 1994;269:9128–9136. [PubMed] [Google Scholar]
- Jain R, Yuen IS, Taphouse CR, Gomer RH. A density-sensing factor controls development in Dictyostelium. Genes Dev. 1992;6:390–400. doi: 10.1101/gad.6.3.390. [DOI] [PubMed] [Google Scholar]
- Jang W, Chiem B, Gomer RH. A secreted cell-number counting factor represses intracellular glucose levels to regulate group size in Dictyostelium. J Biol Chem. 2002;277:39202–39208. doi: 10.1074/jbc.M205635200. [DOI] [PubMed] [Google Scholar]
- Jang W, Gomer RH. Exposure of cells to a cell-number counting factor decreases the activity of glucose-6-phosphatase to decrease intracellular glucose levels in Dictyostelium. Eukaryotic Cell. 2005;4:991–998. doi: 10.1128/EC.4.1.72-81.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W, Gomer RH. A protein in crude cytosol regulates glucose-6-phosphatase activity in crude microsomes to regulate group size in Dictyostelium. J Biol Chem. 2006;281:16377–16383. doi: 10.1074/jbc.M509995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W, Gomer RH. Combining experiments and modelling to understand size regulation in Dictyostelium discoideum. J R Soc Interface. 2008;5(Suppl 1):S49–58. doi: 10.1098/rsif.2008.0067.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W, Schwartz OG, Gomer RH. A cell number counting factor alters cell metabolism. Commun Integr Biol. 2009;2:293–297. doi: 10.4161/cib.2.4.8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram G, Rahman N. Abdul. Cytology of Ki-1-positive anaplastic large cell lymphoma. A report of two cases. 1997;41:1253–1260. doi: 10.1159/000333484. [DOI] [PubMed] [Google Scholar]
- Jedeszko C, Sloane BF. Cysteine cathepsins in human cancer. Biol Chem. 2004;385:1017–1027. doi: 10.1515/BC.2004.132. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Kay RR. cAMP and spore differentiation in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA. 1982;79:3228–3231. doi: 10.1073/pnas.79.10.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbinger A, Gao T, Brock D, et al. A cysteine-rich extracellular protein containing a PA14 domain mediates quorum sensing in Dictyostelium discoideum. Eukaryot Cell. 2005;4:991–998. doi: 10.1128/EC.4.6.991-998.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Hadwiger JA, Pupillo M, Firtel RA. Molecular genetic analysis of two Galpha protein subunits in Dictyostelium. J. Biol. Chem. 1991;266:1220–1228. [PubMed] [Google Scholar]
- Kumagai A, Pupillo M, Gundersen R, Miake-Lye R, Devreotes PN, Firtel RA. Regulation and function of Gα protein subunits in Dictyostelium. Cell. 1989;57:265–275. doi: 10.1016/0092-8674(89)90964-1. [DOI] [PubMed] [Google Scholar]
- Kuwayama H, Van Haastert PJM. Chemotactic and osmotic signals share a cGMP transduction pathway in Dictyostelium discoideum. FEBS Lett. 1998;424:248–252. doi: 10.1016/s0014-5793(98)00183-5. [DOI] [PubMed] [Google Scholar]
- Kwong L, Weeks G. Studies on the accumulation of the differentiation-inducing factor (DIF) in high-cell-density monolayers of Dictyostelium discoideum. Dev. Biol. 1989;132:554–558. doi: 10.1016/0012-1606(89)90250-9. [DOI] [PubMed] [Google Scholar]
- Long J, Aisenberg A. Richter’s syndrome. A terminal complication of chronic lymphocytic leukemia with distinct clinicopathologic features. Am J Clin Pathol. 1975;63:786–795. doi: 10.1093/ajcp/63.6.786. [DOI] [PubMed] [Google Scholar]
- Luzzi KJ, Macdonald IC, Schmidt EE, et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am. J. Pathol. 1998;153:865–873. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SK, Firtel RA. Two-phase regulatory pathway controls cAMP receptor-mediated expression of early genes in Dictyostelium. Proc Natl Acad Sci USA. 1989;86:1924–1928. doi: 10.1073/pnas.86.6.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato JM, Krens FA, Van Haastert PJM, Konijn TM. 3′:5′-Cyclic AMP-dependent 3′:5′-cyclic GMP accumulation in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA. 1977;74:2348–2351. doi: 10.1073/pnas.74.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdy MC, Firtel RA. A secreted factor and cyclic AMP jointly regulate cell-type-specific gene expression in Dictyostelium discoideum. Mol. Cell. Biol. 1985;5:705–713. doi: 10.1128/mcb.5.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JL, Devreotes PN. The surface cyclic AMP receptors, cAR1, cAR2, and cAR3, promote Ca2+ influx in Dictyostelium discoideum by a G alpha-2-independent mechanism. Mol Biol Cell. 1993;4:283–292. doi: 10.1091/mbc.4.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JLS, Caterina MJ, Devreotes PN. Random mutagenesis of the cAMP chemoattractant receptor, cAR1, of Dictyostelium - Evidence for multiple states of activation. J. Biol. Chem. 1997;272:2069–2076. doi: 10.1074/jbc.272.4.2069. [DOI] [PubMed] [Google Scholar]
- Naik-Mathuria B, Pilling D, Crawford JR, et al. Serum amyloid P inhibits dermal wound healing. Wound Repair Regen. 2008;16:266–273. doi: 10.1111/j.1524-475X.2008.00366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Katunuma N. Involvement of cathepsins in the invasion, metastasis and proliferation of cancer cells. J Med Invest. 2005;52:1–9. doi: 10.2152/jmi.52.1. [DOI] [PubMed] [Google Scholar]
- Nonomura A, Mizukami Y, Shimizu J, et al. Small giant cell carcinoma of the lung diagnosed preoperatively by transthoracic aspiration cytology. A case report. Acta Cytol. 1995;39:129–133. [PubMed] [Google Scholar]
- O’reilly MS. Angiostatin: an endogenous inhibitor of angiogenesis and of tumor growth. EXS. 1997;79:273–294. [PubMed] [Google Scholar]
- Peeters CF, De Waal RM, Wobbes T, Ruers TJ. Metastatic dormancy imposed by the primary tumor: does it exist in humans? Ann Surg Oncol. 2008;15:3308–3315. doi: 10.1245/s10434-008-0029-5. [DOI] [PubMed] [Google Scholar]
- Pilling D, Buckley CD, Salmon M, Gomer RH. Inhibition of fibrocyte differentiation by serum amyloid P. J Immunol. 2003;171:5537–5546. doi: 10.4049/jimmunol.171.10.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling D, Roife D, Wang M, et al. Reduction of bleomycin-induced pulmonary fibrosis by serum amyloid P. J Immunol. 2007;179:4035–4044. doi: 10.4049/jimmunol.179.6.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos D, Monteagudo C, Carda C, Ramon D, Gonzalez-Devesa M, Llombart-Bosch A. Ultrastructural and immunohistochemical characterization of the so-called giant multinucleate cells in cutaneous collagenomas. Histopathology. 2002;41:134–143. doi: 10.1046/j.1365-2559.2002.01438.x. [DOI] [PubMed] [Google Scholar]
- Rathi A, Clarke M. Expression of early developmental genes in Dictyostelium discoideum is initiated during exponential growth by an autocrine-dependent mechanism. Mech. Dev. 1992;36:173–182. doi: 10.1016/0925-4773(92)90068-u. [DOI] [PubMed] [Google Scholar]
- Rathi A, Kayman SC, Clarke M. Induction of gene expression in Dictyostelium by prestarvation factor, a factor secreted by growing cells. Dev. Genetics. 1991;12:82–87. doi: 10.1002/dvg.1020120115. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem J. 1993;290(Pt 1):205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Chen Y, Ayoung J, Hanna R, Brazill D. Phospholipase D controls Dictyostelium development by regulating G protein signaling. Cell Signal. 2010 doi: 10.1016/j.cellsig.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roisin-Bouffay C, Gomer RH. How to reach the right size? Med Sci (Paris) 2004;20:219–224. doi: 10.1051/medsci/2004202219. [DOI] [PubMed] [Google Scholar]
- Roisin-Bouffay C, Jang W, Gomer RH. A precise group size in Dictyostelium is generated by a cell-counting factor modulating cell-cell adhesion. Mol. Cell. 2000;6:953–959. [PubMed] [Google Scholar]
- Saran S, Meima ME, Alvarez-Curto E, Weening KE, Rozen DE, Schaap P. cAMP signaling in Dictyostelium - Complexity of cAMP synthesis, degradation and detection. J. Muscle Res. Cell Motil. 2002;23:793–802. doi: 10.1023/a:1024483829878. [DOI] [PubMed] [Google Scholar]
- Saucedo L, Edgar B. Why size matters: altering cell size. Curr. Opin. Genet. Devel. 2002;12:565–571. doi: 10.1016/s0959-437x(02)00341-6. [DOI] [PubMed] [Google Scholar]
- Schaap P, Wang M. Interactions between adenosine and oscillatory cAMP signaling regulate size and pattern in Dictyostelium. Cell. 1986;45:137–144. doi: 10.1016/0092-8674(86)90545-3. [DOI] [PubMed] [Google Scholar]
- Spann TP, Brock DA, Lindsey DF, Wood SA, Gomer RH. Mutagenesis and gene identification in Dictyostelium by shotgun antisense. Proc. Natl. Acad. Sci. USA. 1996;93:5003–5007. doi: 10.1073/pnas.93.10.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternfeld J. Proportion regulation in Dictyostelium is altered by oxygen. Differentiation. 1988;37:173–179. [Google Scholar]
- Tang L, Ammann R, Gao T, Gomer RH. A cell number-counting factor regulates group size in Dictyostelium by differentially modulating cAMP-induced cAMP and cGMP pulse sizes. J. Biol. Chem. 2001;276:27663–27669. doi: 10.1074/jbc.M102205200. [DOI] [PubMed] [Google Scholar]
- Tang L, Gao T, Mccollum C, et al. A cell number-counting factor regulates the cytoskeleton and cell motility in Dictyostelium. Proc. Natl. Acad. Sci., USA. 2002;99:1371–1376. doi: 10.1073/pnas.022516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C, Fu Q, Buhay C, Kay R, Shaulsky G. A bZIP/bRLZ transcription factor required for DIF signaling in Dictyostelium. Development. 2004;131:513–523. doi: 10.1242/dev.00939. [DOI] [PubMed] [Google Scholar]
- Van Haastert PJM, Bishop JD, Gomer RH. The cell density factor CMF regulates the chemoattractant receptor cAR1 in Dictyostelium. J. Cell Biol. 1996;134:1543–1549. doi: 10.1083/jcb.134.6.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerke-Van Wijk I, Schaap P. cAMP, a signal for survival. In: Maeda Y, Inouye K, Takeuchi I, editors. Dictyostelium - A model system for cell and developmental biology. Universal Academy Press; Tokyo, Japan: 1997. pp. 145–162. [Google Scholar]
- Whitbread JA, Sims M, Katz ER. Evidence for the Presence of a Growth Factor in Dictyostelium discoideum. Dev. Genetics. 1991;12:78–81. doi: 10.1002/dvg.1020120114. [DOI] [PubMed] [Google Scholar]
- Williams JG, Ceccarelli A, Mcrobbie S, et al. Direct induction of Dictyostelium prestalk gene expression by DIF provides evidence that DIF is a morphogen. Cell. 1987;49:185–192. doi: 10.1016/0092-8674(87)90559-9. [DOI] [PubMed] [Google Scholar]
- Wurster B, Schubiger K, Wick U, Gerisch G. Cyclic GMP in Dictyostelium discoideum; Oscillations and pulses in response to folic acid and cyclic AMP signals. FEBS Lett. 1977;76:141–144. doi: 10.1016/0014-5793(77)80139-7. [DOI] [PubMed] [Google Scholar]
- Xie YJ, Kwong L, Weeks G. A possible role for DIF-2 in the formation of stalk cells during Dictyostelium development. Dev. Biol. 1991;145:195–200. doi: 10.1016/0012-1606(91)90225-r. [DOI] [PubMed] [Google Scholar]
- Yuen IS, Gomer RH. Cell density-sensing in Dictyostelium by means of the accumulation rate, diffusion coefficient and activity threshold of a protein secreted by starved cells. J. Theor. Biol. 1994;167:273–282. doi: 10.1006/jtbi.1994.1069. [DOI] [PubMed] [Google Scholar]
- Yuen IS, Jain R, Bishop JD, et al. A density-sensing factor regulates signal transduction in Dictyostelium. J. Cell Biol. 1995;129:1251–1262. doi: 10.1083/jcb.129.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen IS, Taphouse C, Halfant KA, Gomer RH. Regulation and processing of a secreted protein that mediates sensing of cell density in Dictyostelium. Development. 1991;113:1375–1385. doi: 10.1242/dev.113.4.1375. [DOI] [PubMed] [Google Scholar]
- Zouwail S, Pettitt TR, Dove SK, et al. Phospholipase D activity is essential for actin localization and actin-based motility in Dictyostelium. Biochem J. 2005;389:207–214. doi: 10.1042/BJ20050085. [DOI] [PMC free article] [PubMed] [Google Scholar]