Abstract
λ exonuclease degrades one strand of duplex DNA in the 5’-3’ direction to generate a 3’ overhang required for recombination. Its ability to hydrolyze thousands of nucleotides processively is attributed to its ring structure and most studies have focused on the processive phase. Here, we use single molecule FRET to reveal three phases of λ exonuclease reactions: initiation, distributive and processive phases. The distributive phase occurs at early reactions where the 3’ overhang is too short for a stable engagement with the enzyme. A mismatched base is digested five times slower than a Watson-Crick paired base and concatenating multiple mismatches has a cooperatively negative effect, highlighting the crucial role of basepairing in aligning the 5’ end toward the active site. The rate-limiting step during processive degradation appears to be the post-cleavage melting of the terminal base pair. We also found that an escape from a known pausing sequence requires enzyme backtracking.
INTRODUCTION
DNA exonucleases catalyze numerous biological processes involved in DNA replication, recombination and repair1. Bacteriophage λ encodes a 5’ to 3’ exonuclease which creates an early DNA intermediate during initiation of homologous recombination2. The λ exonuclease (λ exo) preferentially recognizes the 5’-terminal phosphate in linear duplex DNA and processively degrades the strand with the phosphate in the 5’ to 3’ direction, producing a 3’ overhang tail of the non-hydrolyzed strand3–6. Annealing factors such as Redβ further process the product of λ exo reaction during homologous recombination7, with or without the aid of host proteins such as RecA and SSB8,9.
A ring shaped structure is a recurring feature for many proteins involved with processive reactions on nucleic acid substrates10. Examples include λ-exo in DNA degradation; polynucleotide phosphorylase11,12 and exosome in RNA degradation; hexameric helicases, PCNA and β–clamp in DNA replication. Because of the central channel of the ring through which nucleic acid strands are believed to pass, the proteins do not easily fall off during reaction. λ exo indeed digests double stranded (ds) DNA in a highly processive manner (>3000 nucleotides, nt)4,5 but cuts single stranded (ss) DNA inefficiently in a distributive manner13,14. The translocation of λ-exo along DNA does not require ATP, but requires Mg2+ as a cofactor, and is fueled by the chemical energy released from the hydrolysis of the phosphodiester bond. The processive degradation rates were determined to be 10–12 nt/s by bulk biochemical studies13,15 and 13–18 nt/s by single molecule studies16–18.
Many enzymatic activities are comprised of multiple distinct stages. For instance, RNA polymerase undergoes a transition from the initiation phase to the elongation phase during transcription19. The initiation phase is characterized by a high frequency of abortive synthesis of short transcripts whereas the elongation phase shows a rapid and highly processive RNA synthesis reaction. The ribosome also undergoes several mechanistically distinct phases during translation, including initiation, elongation and termination20. The initiation steps of DNA unwinding by the RecBCD helicase/nuclease complex have also been studied in detail21–23. However, nuclease activities have not been dissected in such anatomic detail. Typically, nuclease activities have only been classified as either distributive or processive.
Recent progress in single molecule techniques has made it possible to detect rich dynamics of biological macromolecules such as enzymatic dynamics disorder18, heterogeneity24–26, transient intermediates27–29, and hidden complexity30. Here we use single molecule fluorescence resonance energy transfer (smFRET)31,32 to dissect the dynamics of λ exo and found that DNA degradation reaction cycle of this enzyme undergoes three distinct phases.
RESULTS
Single molecule fluorescence assay for λ exo
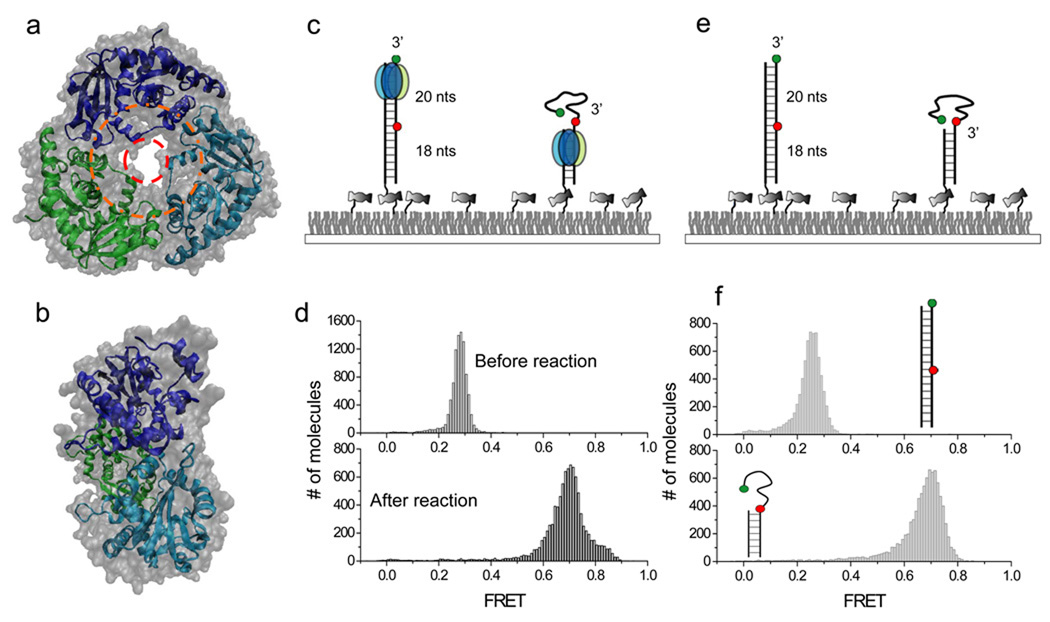
λ exo forms a homotrimeric ring-structure with a tapered central channel (Fig. 1a and 1b). The diameter at the entrance of the channel is ~30Å, wide enough to accommodate dsDNA (dotted orange ring, Fig.1a), whereas the diameter at the exit channel is ~15Å, narrow enough to sterically reject dsDNA, allowing only ssDNA to pass through (dotted red ring, Fig.1a)33. This geometrical constraint serves as an extrusion platform where dsDNA can enter but only ssDNA comes out. Here, for the sake of presentation, we will assume that the functional form of the enzyme is the trimeric ring but it should be noted that there has not been direct demonstration of this widely held assumption.
Figure 1.
Single molecule FRET assay for λ exonuclease activity. (a) Top-to-bottom view of the crystal structure of λ exo. dsDNA is thought to enter the outer ring with ~30Å diameter (orange) with ssDNA product exiting from the inner ring with ~15Å diameter. (b) Side view of the crystal structure tapered from the entrance on the right to the exit on the left. (c) Experimental schematics before (left) and after (right) degradation by λ exo. The enzyme converts dsDNA between the donor (green) and the acceptor (red) to ssDNA, causing an increase in FRET. (d) Single molecule FRET histograms before and after the degradation. (e) A partial duplex mimicking the degradation product was constructed to estimate the FRET value after the degradation. (f) Single molecule FRET histograms built from dsDNA and partial duplex in the absence of λ exo.
In order to dissect the λ-exo activity with high resolution at the single molecule level we designed a blunt ended dsDNA substrate in which a phosphate group is attached on the 5’ end of the hydrolyzed strand (termed the 5’ strand here). The FRET donor (Cy3) is conjugated to the 3’ end of the non-hydrolyzed strand (termed the 3’ strand) and the acceptor (Cy5) to the same strand 20 nt away from the 3’ end (Fig. 1c). FRET between Cy3 and Cy5 can report on the reaction progress as follows. As the 5’ strand is degraded, the 3’ strand becomes single stranded and then coils up34, 35 decreasing the time-averaged distance between the fluorophores, leading to increase in FRET. The other end of DNA without 5’ phosphorylation is tethered to a polymer-coated quartz surface via biotin-neutravidin interaction.
Upon adding the reaction solution containing λ exo and 2.5 mM MgCl2 to the surface-tethered DNA molecules using a flow delivery system36, digestion took place from the 5’ strand with a concomitant sliding of λ exo along the 3’ strand, resulting in a decrease in the time-averaged distance between the two fluorophores by converting a rigid dsDNA into a flexible ssDNA (Fig. 1c). Indeed, single molecule histogram of FRET efficiency E shifted from a peak at E=0.27 before the reaction to a peak at E= 0.72 after the reaction (Fig. 1d and Supplementary Results, Supplementary Fig.1). To check whether the E=0.72 state resulted from the degradation product, we constructed a partial duplex that mimics the reaction product, and also found a peak at E=0.72 from the control DNA only (Fig. 1e–f). This comparison demonstrates that single molecule FRET signal is a good measure of the DNA degradation reaction.
Protein-concentration dependent reaction phases
Fig. 2a shows representative single molecule time traces obtained from a single DNA molecule during degradation. The reaction buffer containing the protein and Mg2+ was introduced to the imaging chamber at the time indicated as “injection”. Subsequently, a protein binding event was detected as fluorescent jumps in both donor and acceptor intensities due to protein-induced fluorescence enhancement37–39 (black, upper panel). Degradation was then observed as progressive increases in FRET (blue, lower panel) caused by a decrease in donor intensity (green, middle panel) together with a concomitant increase in acceptor intensity (red, middle panel). Consistent with the histograms which showed that more than 90% of the DNA molecules are converted into ssDNA (Fig. 1d), the real time single molecule time traces showed evidence of DNA degradation for more than 90% of the DNA molecules.
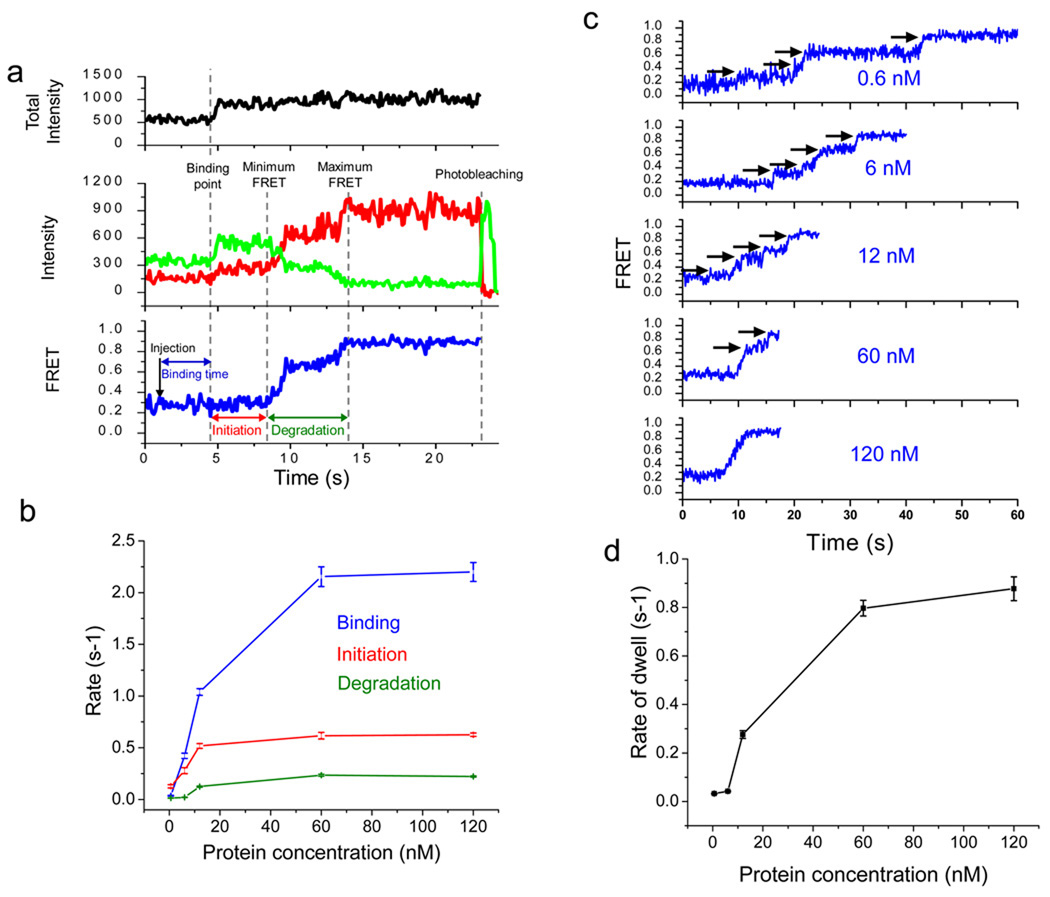
Figure 2.
λ exo carries out initiation and distributive degradation until it is stably engaged by the substrate. (a) A schematic illustration showing how binding time, initiation time and degradation time are assigned: (1) the protein binding period that begins when the reaction buffer is injected and ends right before the enhancement of both fluorescence intensities, (2) the initiation period during which fluorescence intensities increase but FRET stays at a constant low level, and (3) the degradation period during which FRET increases from the minimum to the maximum values. Total intensity is the sum of the donor and acceptor intensities (black in upper panel). Green and red curves represent the donor and acceptor intensities, respectively (middle panel), and the blue curve represents the calculated FRET efficiency (lower panel). (b) Inverse of characteristic times of binding, initiation and degradation vs. protein concentration. Each data point is an average of more than 200 molecules (see Supplementary Fig. 2–4 for their distributions). (c) Representative single molecule time traces for various protein concentrations. Below 60 nM protein, the time traces show multiple pauses, indicating frequent dissociation of λ exo from the substrate. (d) Average pause duration vs. protein concentration (see Supplementary Fig. 5 for their distributions). Error bars denote standard errors.
In order to characterize the time traces in detail, we defined three different time periods (Fig. 2a): (1) the protein binding period that begins when the reaction buffer is injected and ends when fluorescence intensities jump, (2) the initiation period during which enhanced fluorescence intensities are observed but FRET stays at a constant low level, and (3) the degradation period during which FRET increases from the minimum to the maximum values. We attributed the initiation time as the time it takes from initial protein binding until the formation of a functional DNA-protein complex capable of the nuclease reaction.
We performed dwell time analysis of all three periods as a function of protein concentration ranging from 0.6 nM to 120 nM (monomer concentration) (Supplementary Fig. 2–5). The protein binding period showed a dependence on protein concentration as expected (Fig. 2b). Interestingly, the initiation period also showed protein concentration dependence. Presently, there is no demonstration that the active form is the trimeric ring and our method cannot directly probe the assembly state of the protein. Nevertheless, the data demonstrate that a single binding event of protein(s) is not enough to catalyze reactions that are detectable via FRET change. Both the rate of binding and the rate of initiation initially increase linearly with protein concentration, not sigmoidally, suggesting that these steps do not represent the assembly of a trimer from monomers in solution. The initiation time reached a plateau of about 1.6 s at saturating protein concentrations, significantly longer than the estimated time scale of protein delivery via flow (< 0.4 s). Therefore, the initiation period likely involves a conformational change of the protein complex as well as protein binding.
Unexpectedly, the degradation period was also shortened with increasing protein concentration (Fig. 2b). Close examination of individual traces showed several pauses during the reaction (indicated by black arrows in Fig.2c). In addition, the average pause dwell times were longer at lower protein concentrations (Fig. 2d and supplementary Table1). Therefore, the concentration-dependent overall speed of degrading the first 20 bp is attributed to the tendency of the functional enzyme to dissociate so that the reaction pauses until rebinding of a functional enzyme unit. At present, we cannot determine whether the enzyme complex dissociates as a whole or in part. The apparent saturation in the rate of re-initiating degradation at high protein concentrations (Fig. 2d) hints that an additional step (other than protein binding) is required for the rescue to occur. This concentration-dependent behavior is characteristic of distributive enzymes and therefore is surprising for the highly processive λ exo. We postulated that λ exo may perform distributive degradation events during initial degradation until the 3’ ssDNA overhang generated is of sufficient length to engage the enzyme stably. If so, our FRET-based assay should show processive degradation once the 3’ overhang becomes long enough. We tested this prediction next.
Protein-concentration independent reaction phase
We lengthened the upstream of the original substrate by 23 bp, longer than the enzyme footprint (~13–14nts)15, in order to monitor degradation after λ exo is stably engaged by the 3’ strand. The reference FRET values measured in the absence of protein from the extended construct and its partial duplex mimicking the degradation product were ~0.27 and ~0.55, respectively (Fig. 3a). FRET values of the substrate before the reaction (+λ-exo, −Mg2+) and after the reaction (+λ-exo, +Mg2+) (Fig. 3b) agreed with the reference FRET values, demonstrating that the extended construct is efficiently degraded by λ-exo under our experimental conditions. We also carried out a gel-based degradation assay to see whether fluorophore labeling of the 3’ strand negatively impacts the degradation of the 5’ strand. The gel assay showed that the 3’ strand was efficiently degraded with a minimal effect on the reaction (Supplementary Fig. 6).
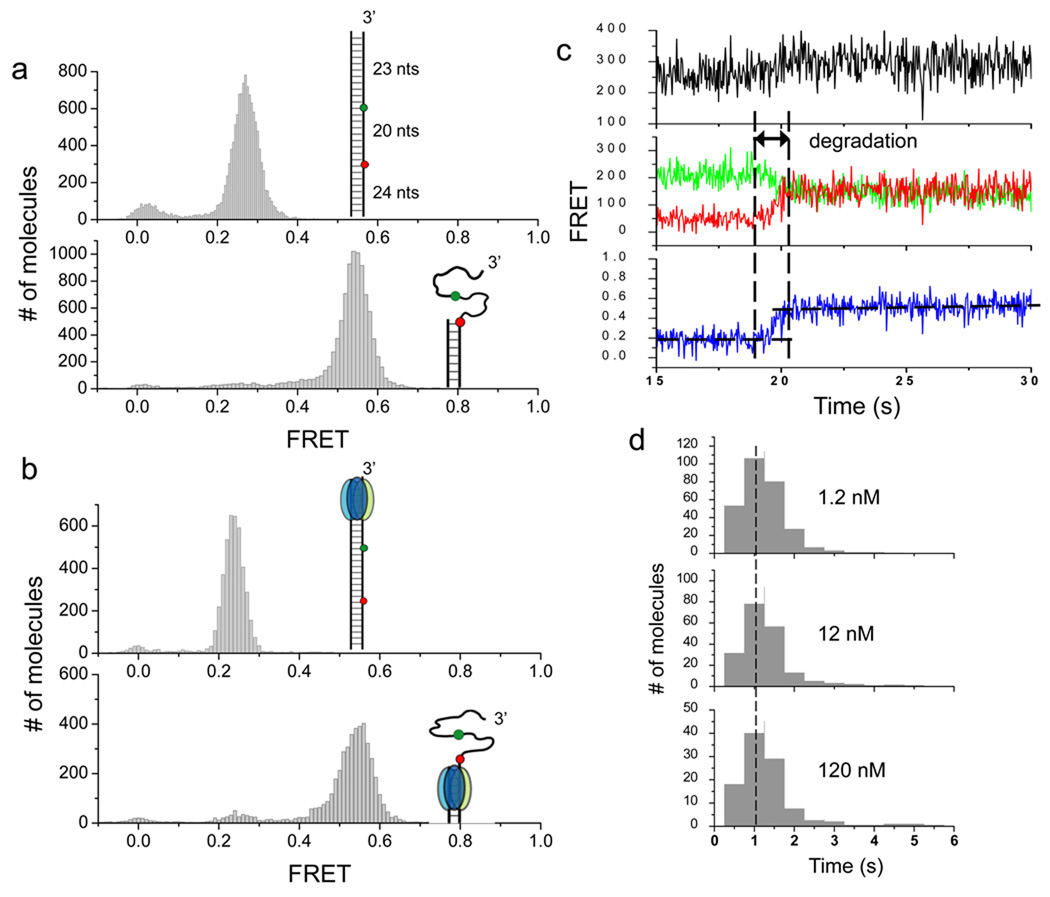
Figure 3.
After stable engagement with the 3’ overhang, λ exo processively degrades the substrate without dissociating. (a) In order to detect DNA degradation after the protein has been stabled engaged with the substrate, the original DNA construct used in Figures 1 and 2 are extended by 23 bp to its upstream (inset of the upper panel). The extension of 23 bp is longer than the footprint of the enzyme (13–14 nt), ensuring that the substrate can be fully engaged when the enzyme passes the region between two fluorophores (inset, upper panel). A partial duplex mimicking the reaction product was also constructed (inset, lower panel). Single molecule FRET histograms of the two constructs in the absence of the protein are shown. (b) Single molecule FRET histograms obtained after λ exo was injected into reaction chambers without Mg2+ (upper panel) and with Mg2+ (lower panel). Comparison of histograms between a and b verified that DNA degradation indeed occurred. (c) A representative set of time traces from a single molecule of the extended construct during degradation (120 nM protein). The degradation period measured by FRET is marked. (d) Histograms of degradation times at different protein concentrations are identical to each other, showing that the FRET increase from the extended construct reports on the processive phase.
The real time single molecule FRET time traces of the extended construct showed a gradual FRET increase that begins 1–200 seconds after protein injection (Fig. 3c and Supplementary Fig. 7). The delay time between protein injection and commencement of degradation beyond the first 23 bp was protein concentration dependent (Supplementary Fig. 7), consistent with the proposed distributive degradation activity on the first 20 bp observed from the original construct. Therefore, the distributive activity on the original construct cannot be due to the terminal labeling of the 3’ strand with a fluorophore.
In contrast to the protein-concentration dependent delay time, the average reaction duration of degrading the DNA segment from bp 23 to 43 was not dependent on protein concentration (Fig. 3d), indicating that FRET increase is now reporting on the processive phase of the reaction. The degradation rate determined by the number of nt divided by degradation time (20 nt/1.1 s) is ~18 nt/s, and stayed constant over a wide range of protein concentration from 1.2 nM to 120 nM. This is at least 4 fold faster than the concentration-dependent overall rates (~4.4 nt/s at maximum) during the distributive phase. We further tested whether the rate of the processive degradation can be recovered from the time trajectories of the distributive phase by removing the pauses (Supplementary Fig. 8). The overall reaction time showed very similar distributions once the pauses have been removed. Therefore, the time spent in the paused states accounts for most of the differences in the apparent reaction rate between processive and distributive phases. Our observations provide experimental support for the proposal that the encirclement of the trimeric ring around the ssDNA generated is the reason for rapid and highly processive activity of λ exo because the processive phase does not begin until up to 23 nt of 3’ ssDNA overhang is generated.
Next we site-specifically labeled λ exo at the N-terminus with CoA-547 fluorophore, which is spectrally equivalent to Cy3, using Sfp phosphopantetheinyl transferase40,41 (Supplementary Methods) with a labeling efficiency of 17%. We used this fluorescently labeled protein on the same extended construct used in Fig 3a, with Cy3 and Cy5 labels, designed to probe the processive phase. This combination allowed us to monitor protein binding and dissociation via signal changes in the Cy3 channel while at the same time observing the processive degradation via changes of FRET between the fluorophores on the DNA. Since this measurement does not require the determination of absolute FRET efficiencies, we calculated the apparent FRET efficiencies based on the acceptor intensity divided by the sum of donor and acceptor intensities, regardless of whether there are more than one donor present or not.
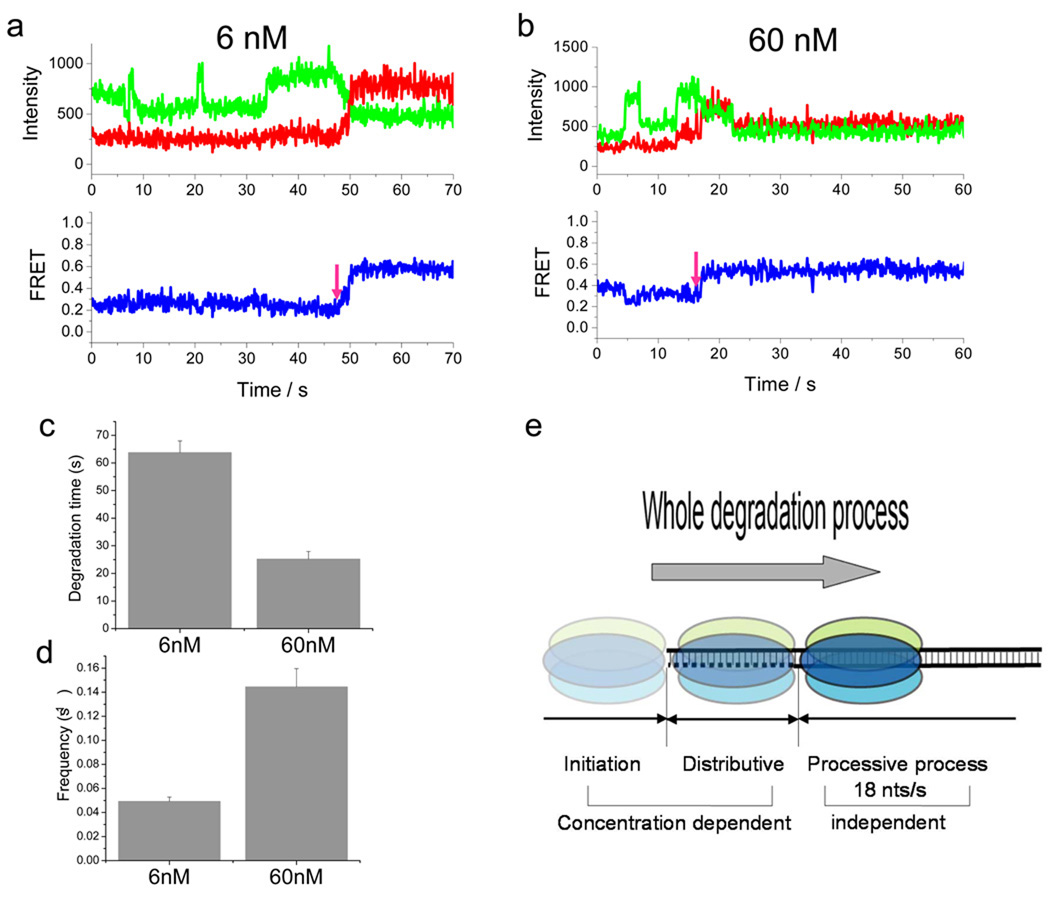
We tested two different concentrations of the enzyme, 6 nM and 60 nM. We observed discrete intensity fluctuations of Cy3 channel between high and low values, likely due to binding and dissociation of labeled proteins, until the processive degradation started (Fig. 4a and b). We measured the degradation time of the first 23 bp of the DNA by determining the time it takes until FRET starts to increase (see the magenta arrows). The degradation time, which reflects the distributive phase, decreased with higher protein concentration (Fig. 4c). The frequency of binding events, in contrast, was higher at the higher protein concentration (Fig. 4d). We also note that the binding/unbinding events do not occur once the processive phase starts (Fig. 4a and b), showing that the events observed are specific to the distributive phase. In summary, the experiments using the labeled enzyme further support our model that multiple binding and dissociation events occur in the distributive phase. Whether the binding and dissociating unit is a monomer or a trimer could not be determined due to the low labeling efficiency.
Figure 4.
λ exo performs distributive degradation before complete engagement to DNA. (a–b) Representative single molecule time traces of the acceptor and donor channel signals obtained from using the extended construct (used in Fig. 3) but in the presence of 6 nM (a) and 60 nM (b) CoA-547-labeled protein. Multiple events of increase and decrease in donor channel signal are observed before FRET starts to increase upon degradation beyond the first 23 bp. Note that the binding/unbinding events do not occur once the processive phase starts, showing that the events observed are specific to the distributive phase. (c) Average degradation time of the first 23 bp of the extended construct determined from the time between protein addition and the moment that FRET starts to increase (see the magenta arrows). (d) Average frequency of donor intensity increase/decrease events before FRET starts to increase. (e) Model of DNA degradation of λ exo. The degradation activity of λ exo is divided into three phases: initiation, distributive, and processive degradation.
λ exo undergoes three phases of degradation
Based on our data, we can classify the degradation activity of λ exo into three phases: the initiation phase, the distributive degradation phase, and the processive degradation phase (Fig. 4e). Protein concentration is a key determinant governing both the rate of initiation and the rate of distributive degradation whereas the rate of processive degradation is independent of protein concentration. The initiation phase is a process forming a catalytically functional DNA-protein complex whereas the distributive degradation phase is an intermediate process through which the initiation phase transits to the processive phase, presumably by enzyme encircling of the 3’ ssDNA overhang generated. Finally, processive degradation is a highly efficient process during which thousands of nt are digested without enzyme dissociation.
Base pair mismatches slow down degradation reaction
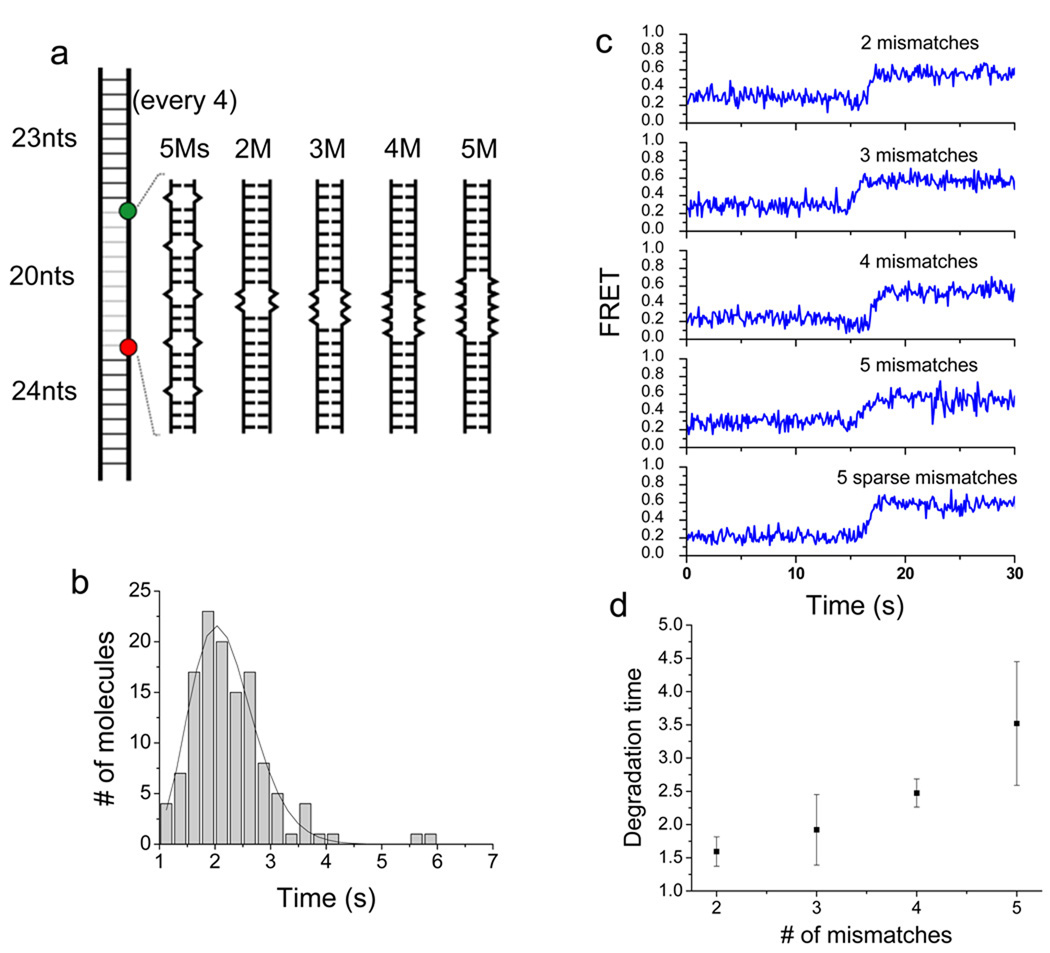
Each catalytic cycle of λ exo is composed of (1) hydrolytic scission of a phosphodiester-bond, (2) 5’ to 3’ translocation along DNA by 1 nt, and (3) melting of the next base pair at the junction between the 3’ ssDNA tail and the duplex. Presently, it is unclear how these events are coordinated and temporally ordered. A previous study found that the enzyme digests DNA regions that are rich in AT basepairs faster than GC-rich regions, suggesting that melting is rate-limiting in the overall reaction18. This finding suggests that the melting of duplex, presumably at the terminal basepair, is prerequisite for the next round of cleavage reaction. If so, introducing single basepair mismatches to the duplex DNA may allow the enzyme to digest the DNA faster because the enzyme does not need to break base-pairing interactions. We examined the role of DNA base pairing on degradation. Five mismatches are placed, one every 4 nt over the 20 nt distance between two fluorophores in the extended construct that reports on the processive phase (5Ms in Fig. 5a). Contrary to our expectation, we found that the degradation of this sparsely mismatched construct is two times slower than the fully basepaired construct (~ 2 sec vs. ~1 sec overall reaction time) (Fig. 5b, Fig. 3d and Supplementary Fig.9). Since the presence of five isolated mismatches slowed down the reaction of 20 bp region by a factor of two, we can also deduce that the reaction at a single mismatch site is ~5 times slower than that at an intact one (Supplementary Fig. 10). We further examined the effect of mismatches by introducing from 2 to 5 consecutive mismatches (2M to 5M in Fig. 5a). Example single molecule FRET time traces are shown in Fig. 5c. The average degradation time increased with increasing number of mismatches (Fig. 5d). Interestingly, the increase was nonlinear (i.e. faster than a linear increase) showing that the effect of sequential mismatches is synergistic. We suggest that base-pairing interaction plays a stimulatory role during the DNA degradation by λ exo, possibly by aligning the 5’ end of the degradation strand toward the active site of the protein. In addition, five sparse mismatches over 20 basepair region slowed down the reaction by a factor of 2 whereas five consecutive mismatches slowed down the reaction by a factor of 3.5. These results suggest that the helicity of duplex is important for processive degradation by λ exo.
Figure 5.
DNA mismatches impede the degradation activity of λ exo in a synergistic manner. (a) Schematics of DNA constructs with periodically sparse (5Ms) and consecutive (2M to 5M) mismatches. (b) Histogram of degradation time obtained from periodically mismatched DNA (5Ms). (c) Representative single molecule FRET time traces taken from DNA constructs with various mismatch configurations outlined in (a). (d) Average degradation time as a function of the number of consecutive mismatches shows that the effective mismatches is non-additive if they are concatenated.
A DNA bubble can cause a degradation arrest
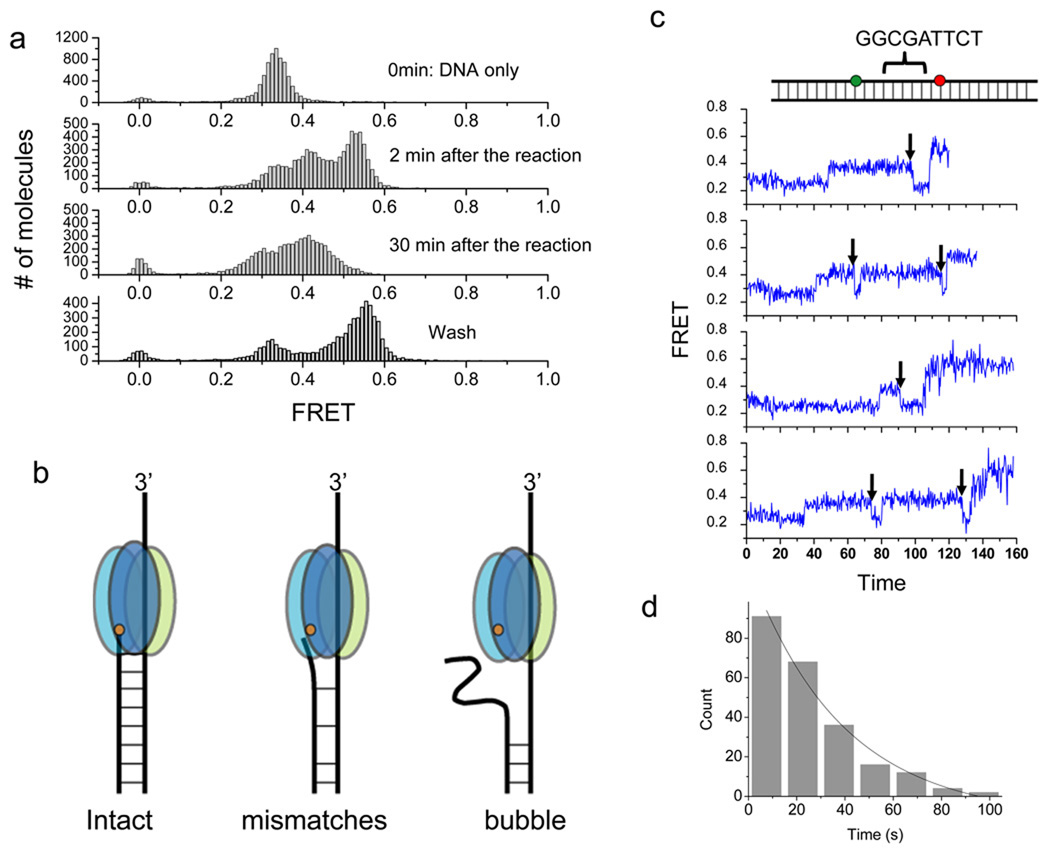
As a further test, we designed a bubble construct with a stretch of 18 mismatches between the two fluorophores to see if degradation further slows down (bottom panel inset in Supplementary Fig. 11). Single molecule FRET efficiency histograms at different reaction time are shown in Fig. 6a. The histogram for the undigested bubble DNA shows a FRET peak at E=0.33, which is higher than ~0.27 FRET measured from the fully basepaired DNA, likely because of the collapse of the ssDNA segment between the two fluorophores. The histogram obtained 2 min into the reaction shows a broad FRET distribution, ranging from 0.2 to 0.6, instead of the narrow FRET peak at 0.55 observed from the standard substrate, suggesting that the bubble structure significantly delays the degradation reaction by λ exo (Supplementary Fig. 11). The histogram in the third panel of Fig. 6a shows that λ exo is captured or stalled in the bubble region even after 30 min reaction. This is in contrast to the histogram obtained from 2 min reaction of the full intact duplex substrate which showed a narrow peak, indicating that the λ exo passed through the labeled region and reached the downstream of the substrate (Fig 3b, lower panel). When the proteins in solution were washed out by flushing the chamber with additional reaction buffer after 30 min, the major FRET peak shifted to high FRET due to ssDNA collapse after λ exo dissociation (Fig. 6a, fourth panel). These data overall suggest that the enzyme is unable to degrade the DNA through a large bubble.
Figure 6.
Degradation arrest on a bubble DNA and escape from a known pausing sequence. (a) Single molecule FRET histograms of a bubble DNA construct with 18 nt mismatches between the donor and the acceptor. DNA only (upper panel); 2 min incubation with 120 nM protein (second panel); 30 min incubation (third panel); and washed after 30 min incubation (bottom panel) (b) Model of λ exo degradation intact, mismatched and bubble DNA. The bubble causes degradation arrest. The orange dot represents the active site of λ exo. (c) Representative single molecule time traces of FRET efficiency during degradation of a DNA construct containing the known pausing sequence (cartoon on the top) at 120 nM protein. After it pauses, λ exo appears to backtrack (FRET decrease, black arrows) before continuing to degrade the DNA. (d) Histogram of pause duration and an exponential fit. The average pause duration is 24.2s ± 1.3 (229 molecules).
When λ exo encounters a mismatch or a bubble, the 5’ end of the degradation strand would become unconstrained (Fig. 6b), which may result in a misalignment of the 5’ end from the active site of the enzyme due to poor helicity caused by mismatches. In addition, the degradation end may now fluctuate away from the active site so that λ exo may not grab the 5’ end readily whereas the 5’ end on the intact B-form helix is maintained in the correct orientation, guiding it to the active site. In the case of the bubble, a fork or “Y” branch shape is created upon cleavage at the beginning of bubble and the 5’ end might completely deviate out of the tapered engaging ring (Fig. 6b). The enzyme is thus trapped on the 3’ strand for a long period of time, resulting in the arrest of degradation (third panel of Fig. 6a). We carried out a gel assay to confirm that a large bubble (20 nt) causes the arrest of degradation (Supplementary Fig. 12), consistent with the single molecule data.
Escape mechanism from sequence-dependent pausing
We constructed a DNA containing the GGCGATTCT sequence in order to reproduce the pausing behavior of the enzyme originally reported in 200317. We inserted the sequence in the middle of our duplex construct so that if there is a pause, it will show up during the processive phase of the reaction. Indeed, we observed a reproducible pause at ~0.4 FRET (Fig. 6c), and the pause time was 24.2 ± 1.3 s on average (229 molecules: Fig. 6d). This experiment shed new light on how the enzyme can be rescued from the paused state. Before it progresses past the pause site, it often moves backwards, indicated by a FRET decrease (marked by black arrows in Fig. 6c). Over 90% of observed molecules (230 of 248 molecules) showed backtracking before continued degradation. This backtracking, reminiscent of RNA polymerase backtracking34, may be explained as follows.
When λ exo digests DNA one nucleotide at a time, its precise registration may help the enzyme recognize a particular sequence that provides a strong interaction between the enzyme and DNA sequences17. In contrast, when λ exo moves back, or backtracks from the pause site, such a motion occurs presumably by diffusion because there is no DNA hydrolysis involved in the backward movement. During the diffusion, the enzyme may not register every single nucleotide allowing the bypass of the pausing sequence site. The average lifetime of the paused state was about a factor of two shorter in optical tweezers experiments17. One possibility is that application of ~1 pN of force may aid backtracking on ssDNA, required for escaping from the paused state. Backward movements were very infrequently observed in the processive mode of our standard sequence (less than 0.2%, an example in Supplementary Fig. 13).
DISCUSSION
We have developed a single molecule FRET assay that allows us to monitor the real time enzymatic action of individual functional units of λ exonuclease. Different DNA substrates were designed to have high sensitivity in different stages of reaction. We find that DNA degradation cycle of λ exo undergoes multiple phases of reaction, namely, an initiation phase and both distributive and processive degradation phases. Our findings here open a new way of conceptualizing the molecular basis for processive enzymes. For example, the mechanism by which proteins form catalytically competent complexes with DNA 22,37 has long been a topic of interest but ensemble averaging in bulk studies can mask the detailed steps (see Supplementary Fig. 14). Our single molecule assay bypasses the limitation of ensemble averaging and shows that the initiation phase that begins upon first protein binding event is also dependent on protein concentration (Fig. 2b). With the direct readout of the initiation process in hand, future studies may be able to reveal how enzyme mutations for example at the trimeric interfaces and putative DNA binding sites affect the initiation process.
This study also found an additional distributive phase between initiation and processive degradation. The distributive process was characterized by a high tendency of dissociation from the substrate. λ exo carries out multiple rounds of short DNA degradation events punctuated by enzyme dissociation and re-association. The distributive phase might be biologically important because it can act as a lag phase that allows the processive enzyme to interact with other viral factors, e.g., Redβ9 and Redγ, and host proteins such as RecA and SSB, to regulate the degradation reaction. A similar perspective on the abortive transcription of RNA polymerase has also been proposed as a potential point of regulation42,43.
Mismatches and bubbles are lesions that frequently occur in the cell and here we investigated how they affect the processive phase of degradation using the high spatiotemporal resolution of our assay. We found that multiple mismatches slow down the processive degradation rate of λ exo in a non-additive manner. Fluctuations of the 5’ end liberated from the base pairing of helix serves as a barrier so that λ exo takes a longer time to form the functional DNA-protein complex acting on the 5’-P terminal end, again supporting the importance of the 5’ end recognition for efficient resection.
How can our observation that a pre-melted duplex caused by mismatches delays the degradation be reconciled with the previous finding that the GC-rich region of the DNA substrate is degraded more slowly than the AT-rich region18? The terminal basepair may remain intact until the hydrolytic scission, and then melt to allow the release of the single nucleotide product. If the melting of the terminal basepair is slower for GC regions even after the cleavage reaction, the two results can be reconciled into the following order of reaction cycle (scission → base-pair melting → nucleotide release → 5’ to 3’ translocation). Although the duplex is pre-melted via mismatches ahead of the reaction cycle, the completion of the degradation cycle will slow down if the scission is retarded due to the deviation of the 5’ end from a catalytically competent complex. For the intact duplex degradation, our data and the results from a previous report 18 in combination suggest that the rate-limiting step is the post-cleavage melting of the terminal basepair.
METHODS
Protein and DNA
λ exonuclease was purchased from New England Biolabs (Catalog # M0262S) for DNA-labeled experiments. The protein used for the protein-labeled experiment in Fig. 4 was expressed and purified as described in Supplementary Methods. All DNA oligonucleotides were purchased from Integrated DNA Technologies (IDT). Biotin was conjugated at either the 5’ end of the 3’ strand (original construct used in Figs. 1 and 2) or the 3’ end of the 5’ degradation strand (extended construct used in Fig. 3). Both Cy3 and Cy5 were incorporated into the 3’ DNA strand by an internal modification during DNA synthesis by IDT (iCy3 and iCy5). Sequences of DNA and positions of biotin and fluorophores are provided in Supplementary Methods.
Experimental conditions
DNA constructs were immobilized on a polyethylene glycol (PEG, Laysan Bio Inc.)-coated quartz surface as shown in Fig. 1 and 3 to minimize nonspecific surface adsorption of proteins44,45. Immobilization was achieved by a specific molecular interaction between neutravidin (Pierce) and biotin moieties at the ends of DNA and biotin-modified PEG on the surface. 50–100 pM concentrations of DNA molecules were added to the imaging chamber to obtain an appropriate density for single molecule imaging. A glucose and glucose oxidase were added to remove oxygen that causes rapid photobleaching of fluorescent dyes. The reaction buffer contained 67 mM Glycine-KOH (pH 9.4), 2.5 mM MgCl2, 50 µg/ml BSA, 1 mg/ml Trolox46 (Sigma Aldrich) and an oxygen scavenging system of 1mg/ml glucose oxidase (Sigma Aldrich) and 0.4% (w/v)D-glucose (Sigma Aldrich). A flow chamber was constructed by assembling a microscope slide and a coverslip together with 3M™ double-sided tape and sealing with epoxy36, 47. The holes on the slide were used for the inlet and outlet of solution exchange36. A 1 mL syringe was connected to the flow chamber through tubing, and a pipette tip that contains a reaction solution was tightly plugged into an inlet hole preventing a buffer leakage. When the syringe was pulled, the solution was introduced into the chamber at the rate of ~10 µl/s. Degradation reaction was started by injecting the reaction buffer containing λ exo to an imaging chamber at room temperature.
Single-Molecule Data Acquisition
Total internal reflection fluorescence microscopy was employed to excite a FRET donor (Cy3) on the DNA using an Nd:YAG laser (532 nm, 75 mW, Crystal Laser). The fluorescence emission light from Cy3 and Cy5 was collected by a water immersion objective lens (UPlanApo 60×, Olympus) and then sent through a 550 nm long-pass filter48. The fluorescence emission light was further separated into donor and acceptor signals with a 630 nm dichroic mirror (Chroma) and was detected by back-illuminated electron-multiplying charge-coupled device (EMCCD, Andor) with a time resolution of 30 or 100 ms. Fluorescence signals of donor and acceptor were amplified before camera readout. Thus, both recorded fluorescence intensities of Cy3 and Cy5 are in an arbitrary unit (a.u.). The data were recorded in a video file format by an in-house software written in Visual C++. Single molecule intensities were extracted from the recorded video file by IDL software and FRET efficiency is calculated as the ratio of intensities, Intensityacceptor/(Intensitydonor +Intensityacceptor) after correcting for cross-talk between the donor and acceptor channels. All data were analyzed with programs written in MATLAB and plotted in Origin.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Raul Roy and Xinghua Shi for experimental help and Jeehae Park for helpful discussions. G.L. was supported by the Jane Coffin Childs Medical Institute. Funds were provided by grants from National Science Foundation (0646550, 0822613) and National Institutes of Health (GM065367). T.H. is an investigator with Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
G.L. performed single molecule and ensemble fluorescent experiments. J.Y. performed single molecule experiments. G.L. and B.L. expressed and purified λ exo, and B.L. carried out the protein labeling. G.L., J.Y., B.L. and T.H. designed the experiments, analyzed the data, and wrote the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Ceska TA, Sayers JR. Structure-specific DNA cleavage by 5 ' nucleases. Trends Biochem.Sci. 1998;23:331–336. doi: 10.1016/s0968-0004(98)01259-6. [DOI] [PubMed] [Google Scholar]
- 2.Haber JE. In-Vivo Biochemistry - Physical Monitoring of Recombination Induced by Site-Specific Endonucleases. Bioessays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 3.Little JW, Lehman IR, Kaiser AD. An Exonuclease Induced by Bacteriophage Lambda .I. Preparation of Crystalline Enzyme. Journal of Biological Chemistry. 1967;242 672-&. [PubMed] [Google Scholar]
- 4.Little JW. An Exonuclease Induced by Bacteriophage Lambda .2. Nature of Enzymatic Reaction. Journal of Biological Chemistry. 1967;242 679-&. [PubMed] [Google Scholar]
- 5.Carter DM, Radding CM. Role of Exonuclease and Beta Protein of Phage Lambda in Genetic Recombination .2. Substrate Specificity and Mode of Action of Lambda Exonuclease. Journal of Biological Chemistry. 1971;246 2502-&. [PubMed] [Google Scholar]
- 6.Thomas KR, Olivera BM. Processivity of DNA Exo-Nucleases. Journal of Biological Chemistry. 1978;253:424–429. [PubMed] [Google Scholar]
- 7.Radding CM. Regulation of Lambda Exonuclease .I. Properties of Lambda Exonuclease Purified from Lysogens of Lambdat11 and Wild Type. Journal of Molecular Biology. 1966;18 doi: 10.1016/s0022-2836(66)80243-7. 235-&. [DOI] [PubMed] [Google Scholar]
- 8.Black LW. DNA Packaging in Dsdna Bacteriophages. Annual Review of Microbiology. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 9.Muyrers JPP, Zhang YM, Buchholz F, Stewart AF. RecE/RecT and Red alpha/Red beta initiate double-stranded break repair by specifically interacting with their respective partners. Genes & Development. 2000;14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 10.Hingorani MM, O'Donnell M. Toroidal proteins: Running rings around DNA. Current Biology. 1998;8:R83–R86. doi: 10.1016/s0960-9822(98)70052-1. [DOI] [PubMed] [Google Scholar]
- 11.Guissani A. Processive And Synchronous Mechanism Of Polynucleotide Phosphorylase Phosphorolysis - Comparison Between Experimental Results And Those Calculated From A Theoretical-Study. European Journal Of Biochemistry. 1977;79:233–243. doi: 10.1111/j.1432-1033.1977.tb11801.x. [DOI] [PubMed] [Google Scholar]
- 12.Klee CB, Singer MF. Processive Degradation Of Individual Polyribonucleotide Chains .2. Micrococcus Lysodeikticus Polynucleotide Phosphorylase. Journal Of Biological Chemistry. 1968;243 923-&. [PubMed] [Google Scholar]
- 13.Subramanian K, Rutvisuttinunt W, Scott W, Myers RS. The enzymatic basis of processivity in lambda exonuclease. Nucleic Acids Research. 2003;31:1585–1596. doi: 10.1093/nar/gkg266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sriprakash KS, Lundh N, Mooonhuh M, Radding CM. Specificity of Lambda-Exonuclease Interactions with Single-Stranded-DNA. Journal of Biological Chemistry. 1975;250:5438–5445. [PubMed] [Google Scholar]
- 15.Mitsis PG, Kwagh JG. Characterization of the interaction of lambda exonuclease with the ends of DNA. Nucleic Acids Research. 1999;27:3057–3063. doi: 10.1093/nar/27.15.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dapprich J. Single-molecule DNA digestion by lambda-exonuclease. Cytometry. 1999;36:163–168. doi: 10.1002/(sici)1097-0320(19990701)36:3<163::aid-cyto3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 17.Perkins TT, Dalal RV, Mitsis PG, Block SM. Sequence-dependent pausing of single lambda exonuclease molecules. Science. 2003;301:1914–1918. doi: 10.1126/science.1088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Oijen AM, et al. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 19.Young BA, Gruber TM, Gross CA. Views of transcription initiation. Cell. 2002;109:417–420. doi: 10.1016/s0092-8674(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 20.Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Translation at the single-molecule level. Annual Review of Biochemistry. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- 21.Lucius AL, Wong CJ, Lohman TM. Fluorescence stopped-flow studies of single turnover kinetics of E-coli RecBCD helicase-catalyzed DNA unwinding. Journal of Molecular Biology. 2004;339:731–750. doi: 10.1016/j.jmb.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Wong CJ, Lucius AL, Lohman TM. Energetics of DNA end binding by E-coli RecBC and RecBCD helicases indicate loop formation in the 3'-single-stranded DNA tail. Journal of Molecular Biology. 2005;352:765–782. doi: 10.1016/j.jmb.2005.07.056. [DOI] [PubMed] [Google Scholar]
- 23.Wong CJ, Rice RL, Baker NA, Ju T, Lohman TM. Probing 3 '-ssDNA loop formation in E-coli RecBCD/RecBC-DNA complexes using non-natural DNA: A model for "Chi" recognition complexes. Journal of Molecular Biology. 2006;362:26–43. doi: 10.1016/j.jmb.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Zhuang XW, et al. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 25.Zhuang XW, et al. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 26.Rothwell PJ, et al. Multiparameter single-molecule fluorescence spectroscopy reveals heterogeneity of HIV-1 reverse transcriptase: primer/template complexes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1655–1660. doi: 10.1073/pnas.0434003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hohng S, et al. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 29.Woodside MT, et al. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi J, Dertouzos J, Gafni A, Steel D, Palfey BA. Single-molecule kinetics reveals signatures of half-sites reactivity in dihydroorotate dehydrogenase A catalysis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5775–5780. doi: 10.1073/pnas.0510482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stryer L, Haugland RP. Energy Transfer - a Spectroscopic Ruler. Proceedings of the National Academy of Sciences of the United States of America. 1967;58 doi: 10.1073/pnas.58.2.719. 719-&. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha T, et al. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovall R, Matthews BW. Toroidal structure of lambda-exonuclease. Science. 1997;277:1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- 34.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha TJ. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophysical Journal. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selvin P, Ha T. Single-Molecule Techniques: A Laboratory Manual. 1 ed. Cold Spring Harbor Laboratory Press; 2008. pp. 3–36. [Google Scholar]
- 37.Maluf NK, Fischer CJ, Lohman TM. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. Journal of Molecular Biology. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 38.Luo G, Wang M, Konigsberg WH, Xie XS. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2007;104:12610–12615. doi: 10.1073/pnas.0700920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myong S, et al. Cytosolic Viral Sensor RIG-I Is a 5 '-Triphosphate-Dependent Translocase on Double-Stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin J, et al. Single-cell FRET imaging of transferrin receptor trafficking dynamics by Sfp-catalyzed, site-specific protein labeling. Chemistry & Biology. 2005;12:999–1006. doi: 10.1016/j.chembiol.2005.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nature Protocols. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 42.Hsu LM. Promoter clearance and escape in prokaryotes. Biochimica Et Biophysica Acta-Gene Structure And Expression. 2002;1577:191–207. doi: 10.1016/s0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 43.Roberts JW. Biochemistry - RNA polymerase, a scrunching machine. Science. 2006;314:1097–1098. doi: 10.1126/science.1135746. [DOI] [PubMed] [Google Scholar]
- 44.Ha T, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 45.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasnik I, McKinney SA, Ha T. Nonblinking and longlasting single-molecule fluorescence imaging. Nature Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 47.Grossman D, van Hoof A. RNase II structure completes group portrait of 3 ' exoribonucleases. Nat. Struct. Mol. Biol. 2006;13:760–761. doi: 10.1038/nsmb0906-760. [DOI] [PubMed] [Google Scholar]
- 48.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.