Abstract
Background
The gut is an important target organ of injury during critically ill conditions. Although Gram staining is a common and quick method for identifying bacteria, its clinical application has not been fully evaluated in critically ill conditions.
Aims
This study’s aims were to identify patterns of Gram-stained fecal flora and compare them to cultured bacterial counts and to investigate the association between the patterns and septic complications in patients with severe systemic inflammatory response syndrome (SIRS).
Methods
Fifty-two patients with SIRS were included whose Gram-stained fecal flora was classified into three patterns. In a diverse pattern, large numbers of multiple kinds of bacteria completely covered the field. In a single pattern, one specific kind of bacteria or fungi predominantly covered the field. In a depleted pattern, most bacteria were diminished in the field.
Results
In the analysis of fecal flora, the numbers of total obligate anaerobes in the depleted pattern was significantly lower than those in the diverse pattern and single pattern (p < 0.05). The concentrations of total organic acids, acetic acid, and propionic acid in the depleted pattern were significantly lower than those in diverse pattern and single pattern (p < 0.05). Mortality due to multiple organ dysfunction syndrome for the single pattern (52%) and the depleted pattern (64%) was significantly higher than that for the diverse pattern (6%) (p < 0.05).
Conclusions
Gram-stained fecal flora can be classified into three patterns and are associated with both cultured bacterial counts and clinical information. Gram-stained fecal bacteria can be used as a quick bedside diagnostic marker for severe SIRS patients.
Keywords: SIRS, Sepsis, Gram stain, Gut, Short-chain fatty acids, Flora
Introduction
The gut is considered an important target organ following severe insult such as trauma and sepsis and has an important role in promoting infectious complications and multiple organ dysfunction syndrome (MODS) from the viewpoint of deteriorated intestinal epithelia, the immune system, and commensal bacteria [1]. The gut is the “motor” of multiple organ failure, and it is now recognized that gut dysfunction is a cause for the promotion of diseases. Recently, we quantitatively evaluated microflora and environmental changes in patients with severe systemic inflammatory response syndrome (SIRS). Analysis of fecal flora confirmed that patients with severe SIRS had significantly lower total anaerobic bacterial counts (particularly Bifidobacterium and Lactobacillus) and higher Staphylococcus and Pseudomonas group counts than did healthy volunteers. Concentrations of total organic acids, and in particular beneficial short-chain fatty acids (SCFAs) such as acetic acid, propionic acid, and butyric acid in the feces, were significantly decreased in these patients, and pH was markedly increased. These results indicated that the gut flora and environment are significantly altered in patients with severe SIRS [2]. Alteration of the commensal gut flora may affect gut barrier function and systemic inflammatory responses to severe insult.
Gram staining is one of the most common and quickest methods for identifying bacteria and directing initial empiric antimicrobial therapy [3]. Gram-stained fecal flora, however, has not been fully examined under critically ill conditions. In this study, we first identified three patterns of Gram-stained fecal flora and compared them to cultured bacterial counts. Second, we evaluated their relation to septic complications and prognosis as a means of quick diagnosis of the gut environment in patients with severe SIRS.
Methods
Patients
Fifty-two severe SIRS patients admitted to the Department of Traumatology and Acute Critical Medicine, Osaka University Medical School, during the period March 2005 to April 2008 were included in our study. Severe SIRS was diagnosed in patients who fulfilled the criteria for SIRS with a serum C-reactive protein (CRP) level higher than 10 mg/dl [4], and were treated in the ICU for more than 7 days [4], and who were treated in the ICU for more than 7 days.
Enteral nutrition was initiated as quickly as possible in all patients. If infections occurred, patients were initially treated empirically for the underlying clinical syndrome and then treated according to the results of resistance testing of the isolated bacterial infection. Severe sepsis and septic shock were managed in accordance with the Surviving Sepsis Campaign guidelines [5]. The strategy for the use of antibiotics was identical throughout the study period. Fecal samples were collected serially once a week and analyzed after admission. This study was approved by the Institutional Review Board of Osaka University, and informed consent was obtained from the family of each patient.
Pattern Classification from Fecal Gram Staining
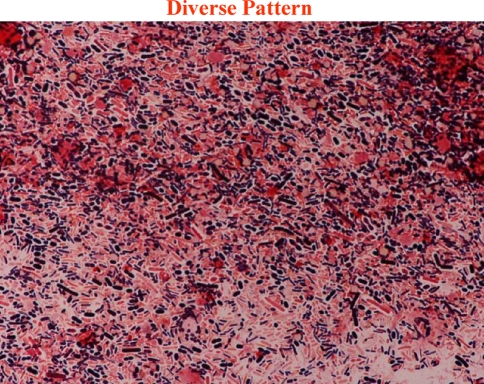
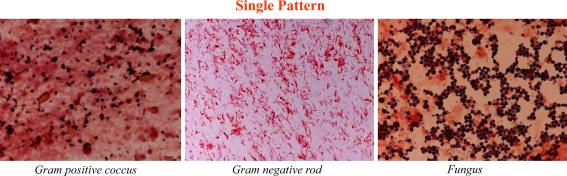
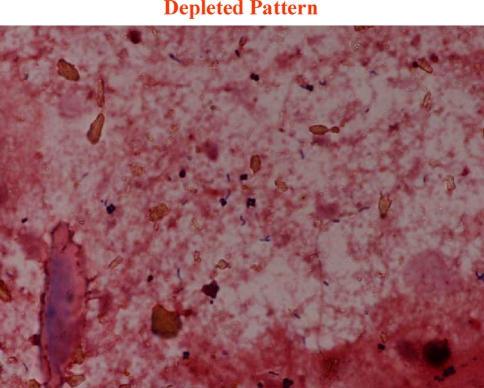
We classified Gram-stained fecal flora into three patterns in accordance with the patterns of bacteria we could identify (Fig. 1). The classification of each sample was certified at the Division of Infection Control and Prevention in our hospital by an infectious disease physician who had no knowledge of the clinical course of each patient. In the diverse pattern, large numbers of multiple kinds of bacteria completely covered the field. In the single pattern, one specific kind of bacteria or fungi predominantly covered the field. Figure 2 shows that Gram-positive cocci (GPC), Gram-negative rods (GNR), and fungi dominated the field. In the depleted pattern, most bacteria were diminished in the field (Fig. 3). Alterations in gut flora usually progress from a diverse pattern to a single pattern and then onwards to a depleted pattern.
Fig. 1.
Pattern classification from fecal Gram stain. In the diverse pattern, numerous kinds of bacteria totally covered the field
Fig. 2.
Pattern classification from fecal Gram stain. In the single pattern, specified bacteria (only one or two kinds of bacteria) dominantly covered the field. These pictures show that Gram-positive coccus (left), Gram-negative rod (middle) and fungus (right) dominantly cover the field
Fig. 3.
Pattern classification from fecal Gram stain. In the depleted pattern, most bacteria were diminished in the field. The deterioration of gut flora usually occurs from a diverse pattern to a single pattern, and from a single pattern to a depleted pattern
Culture of Fecal Bacteria
Feces were collected in a test tube, which was maintained under anaerobic conditions in an atmosphere of 7% H2 and 5% CO2 in N2. The test tube was cooled in an icebox before culture. VL-G roll tube agar [6] supplemented with 0.2% cellobiose and 0.2% maltose (modified VL-G roll tube agar) was used to determine total anaerobe counts. Different media were used for selective isolation of different microorganisms: modified VL-G roll tube agar to which 80 μg/ml vancomycin and 1 μg/ml kanamycin were added for Bacteroidaceae; CW agar (Nikken Bio Medical Laboratory Inc., Kyoto, Japan) for lecithinase-positive Clostridium; MPN agar [7] for Bifidobacterium; COBA agar [8] for Enterococcus; LBS agar (Becton Dickinson and Company, Cockeysville, MD) supplemented with 0.8% Laboratory Lemco powder (Oxoid Co. Ltd, Basingstoke, UK) for Lactobacillus; Staphylococcus medium no. 110 agar (Nissui Pharmaceutical Co., Ltd, Tokyo, Japan) for Staphylococcus; DHL agar (Nissui Pharmaceutical Co., Ltd) for Enterobacteriaceae; NAC agar (Nissui Pharmaceutical Co., Ltd) for Pseudomonas; and GS agar (Nissui Pharmaceutical Co., Ltd) for Candida. CW agar and LBS agar were cultured anaerobically at 37°C for 72 h. After incubation, colonies on plates were counted and Gram stained. Numbers of viable bacteria per Gram of feces (wet weight) were calculated. All bacterial counts (colony-forming units (CFU)/g of wet feces) were transformed to a logarithmic scale (log10CFU) for ease of statistical analysis. The lower limit of bacterial detection with this procedure was 1000 CFU/g of feces for the obligate anaerobes, Bacteroidaceae and Bifidobacterium, and 100 CFU/g of feces for other bacteria. The reproducibility and stability of these measurements were shown previously [9].
Determination of Fecal Organic Acid Concentrations
A portion of the feces was isolated, weighed, mixed with 0.15 M perchloric acid at a fourfold volume, and stored at 4°C for 12 h. The mixture was centrifuged at 4°C at 15,000 rpm for 10 min, and the supernatant was filtrated with a 0.45-μm membrane filter (Millipore Japan Ltd, Tokyo, Japan), and sterilized. The sample was analyzed for organic acids by high-performance liquid chromatography, which was performed with a Waters system (Waters 432 Conductivity Detector; Waters Co., Milford, MA) equipped with two columns (Shodex RSpack KC-811; Showa Denko Co., Ltd, Tokyo, Japan). The concentrations of organic acids were calculated with the use of external standards, and the reproducibility and stability of these measurements were shown previously [10].
Surveillance and Definition of Infections
We defined infectious complications as infections that occurred after the diagnosis of severe SIRS during the ICU stay. Body temperature was measured continuously. Surveillance cultures from urine, blood, and sputum were performed routinely for each patient. In cases of suspected infection, laboratory tests, chest X-rays, and computed tomography scanning were performed when necessary. The definition of enteritis had to meet criteria including at least an acute onset of diarrhea (liquid stools for more than 12 h) with or without vomiting or fever (>38°C), no likely noninfectious cause, and enteric pathogens detected by laboratory testing. Bacterial infection was diagnosed in accordance with the Centers for Disease Control Definitions [11]. Bacteremia was defined as one positive blood culture.
Statistical Analysis
We evaluated fecal flora and gut environmental factors of the most advanced Gram-stain pattern in each patient. The incidences of enteritis, bacteremia, and mortality were also evaluated for each pattern. Results are expressed as mean ± SD values for each pattern. Statistical analysis was performed with the Mann–Whitney test where appropriate. Statistical significance was determined at p < 0.05.
Results
Patient characteristics are listed in Table 1. The principal origin of SIRS was sepsis detected in a total of 45 out of 52 patients. There were 17 patients with diverse pattern, 21 patients with single pattern, and 14 patients with depleted pattern. The three groups did not differ in terms of age, sex, or Acute Physiology and Chronic Health Evaluation (APACHE) II score on admission. There was no significant difference in the number of antibiotic types administered among the three patterns. The duration of use of antibiotics was 14.6 ± 12.1 days in the diverse pattern, 19.7 ± 12.6 days in the single pattern, and 19.1 ± 9.9 days in the depleted pattern, and there was no significant difference among the three patterns. In the single pattern, GPC was dominant in six patients, GNR in nine patients, and fungus in six patients.
Table 1.
Patient characteristics
| Diverse pattern | Single pattern | Depleted pattern | |
|---|---|---|---|
| Number | 17 | 21 | 14 |
| Agea | 58.1 ± 13.7 | 67.0 ± 16.7 | 63.8 ± 13.9 |
| Sex (male/female) | 14/3 | 14/7 | 8/6 |
| APACHE IIa | 15.4 ± 6.5 | 17.5 ± 8.6 | 17.7 ± 7.6 |
| Number of antibiotic types | 4.1 ± 1.8 | 5.9 ± 2.8 | 5.5 ± 3.7 |
| Sampling day from admission | 16.6 ± 13.9 | 21.3 ± 13.6 | 19.4 ± 9.4 |
| Origins of SIRS | |||
| Sepsis | 13 | 20 | 12 |
| Pneumonia | 2 | 6 | 1 |
| Abdominal infection | 3 | 8 | 9 |
| Meningitis | 2 | 0 | 1 |
| Encephalitis | 1 | 0 | 0 |
| Necrotizing fasciitis | 4 | 4 | 0 |
| Others | 1 | 2 | 1 |
| Trauma | 4 | 0 | 2 |
| Burn | 0 | 1 | 0 |
| Infectious complications | |||
| Enteritis | 1 | 4 | 6 |
| Bacteremia | 6 | 15 | 9 |
| Mortality | 1 | 11 | 9 |
APACHE Acute Physiology and Chronic Health Evaluation, SIRS systemic inflammatory response syndrome
amean ± SD
In the analysis of fecal flora, the number of total obligate anaerobes including Bacteroidaceae and Bifidobacterium in the depleted pattern were significantly lower than those in the diverse pattern and the single pattern (p < 0.05) (Table 2). The number of total facultative anaerobes including Lactobacillus, Enterobacteriaceae, and Enterococcus in the depleted pattern were significantly lower than those in the diverse pattern and the single pattern (p < 0.05).
Table 2.
Fecal flora of each pattern
| Diverse pattern | Single pattern | Depleted pattern | Normal | |
|---|---|---|---|---|
| Total obligate anaerobes | 9.7 ± 0.7 | 8.8 ± 2.6 | 4.4 ± 2.9*# | 10.5 ± 0.5 |
| Bacteroidaceae | 8.9 ± 2.0 | 7.7 ± 3.2 | 3.9 ± 2.9*# | 10.1 ± 0.4 |
| Bifidobacterium | 7.8 ± 2.4 | 5.8 ± 3.1* | 2.2 ± 1.1*# | 9.6 ± 0.7 |
| Lecitinase-positive Clostridium | 2.5 ± 1.5 | 2.6 ± 2.2 | 2.1 ± 0.7 | 2.1 ± 0.7 |
| Total facultative anaerobes | 8.2 ± 1.5 | 8.4 ± 1.2 | 6.6 ± 1.6*# | 7.5 ± 0.4 |
| Lactobacillus | 5.5 ± 2.3 | 5.2 ± 2.3 | 3.5 ± 1.9*# | 5.0 ± 1.0 |
| Bnterobacteriaceae | 5.3 ± 2.6 | 4.3 ± 2.5 | 2.2 ± 0.8*# | 7.4 ± 0.8 |
| Enterococcus | 8.0 ± 1.6 | 7.7 ± 2.0 | 5.5 ± 2.1*# | 7.0 ± 0.9 |
| Staphylococcus | 5.2 ± 1.6 | 4.8 ± 2.1 | 5.1 ± 1.8 | 2.7 ± 0.8 |
| Pseudomonas | 2.0 ± 0.7 | 3.4 ± 2.0* | 1.8 ± 0.0# | ND |
| Candida | 3.4 ± 1.4 | 3.3 ± 2.4 | 3.3 ± 1.9 | 2.0 ± 0.5 |
ND not detected. Values are mean ± SD (log10 colony-forming units/g feces)
* p < 0.05 versus diverse pattern, # p < 0.05 versus single pattern
In the analysis of organic acids, the concentrations of total organic acids, acetic acid, and propionic acid in the depleted pattern were significantly lower than those in the diverse pattern and the single pattern (p < 0.05) (Table 3).
Table 3.
Fecal organic acids of each pattern
| Diverse pattern | Single pattern | Depleted pattern | Normal | |
|---|---|---|---|---|
| Total organic acids | 76.5 ± 32.1 | 70.4 ± 55.2 | 20.3 ± 12.5*# | 88.4 ± 21.2 |
| Succinic acid | 6.8 ± 9.6 | 7.9 ± 16.9 | 0.6 ± 1.3 | 0.9 ± 1.2 |
| Lactic acid | 4.8 ± 9.8 | 9.9 ± 16.4* | 12.4 ± 9.9* | 0.5 ± 0.3 |
| Formic acid | 1.6 ± 3.2 | 2.0 ± 3.4 | 1.0 ± 1.7 | 0.4 ± 0.3 |
| Acetic acid | 9.0 ± 6.9 | 8.5 ± 10.4 | 0.0 ± 0.0*# | 50.8 ± 13.1 |
| Propionic acid | 49.1 ± 21.6 | 38.3 ± 30.5 | 6.3 ± 7.2*# | 18.7 ± 6.8 |
| Butyric acid | 5.4 ± 6.4 | 3.2 ± 5.4 | 0.0 ± 0.0* | 16.6 ± 6.7 |
| Isovaleric acid | 0.8 ± 1.6 | 0.6 ± 1.3 | 0.0 ± 0.0 | 1.4 ± 0.7 |
| Valeric acid | 0.1 ± 0.3 | 0.3 ± 0.9 | 0.0 ± 0.0 | 0.6 ± 0.4 |
| pH | 6.7 ± 1.1 | 6.6 ± 1.1 | 7.3 ± 0.7# | 6.6 ± 0.3 |
Values are mean ± SD (μmol/g of feces)
* p < 0.05 versus diverse pattern, # p < 0.05 versus single pattern
In the analysis of septic complications, the incidence of enteritis for the depleted pattern (43%) was significantly higher than that for the diverse pattern (6%) and the single pattern (19%) (p < 0.05). There was no significant difference in the incidence of pneumonia between the three patterns (depleted pattern, 29%; single pattern, 57%; diverse pattern, 57%).The incidence of bacteremia for single pattern (71%) was significantly higher than that for the diverse pattern (35%) (p < 0.05). There was no significant difference in the incidence of bacteremia between the depleted pattern and the diverse pattern or single pattern. Analysis of the blood cultures detected Staphylococcus species in 18 patients, Enterococcus faecalis in six patients, Escherichia coli in five patients, Pseudomonas in four patients, and other bacteria as shown in Table 1. The mortality rates due to MODS for the single pattern (52%) and depleted pattern (64%) were significantly higher than that for the diverse pattern (6%) (p < 0.05). In the patients who died, the mean time to death was 6 days after admission in the diverse pattern, 40.9 ± 17.8 days in the single pattern, and 45.2 ± 38.3 days in the depleted pattern. In the patients who survived, the mean time to hospital discharge was 41.8 ± 26.0 days after admission in the diverse pattern, 43.7 ± 22.6 days in the single pattern, and 51.2 ± 33.9 days in the depleted pattern. There was no significant difference in time to discharge among the three patterns.
Discussion
The gut is a target organ of injury after severe insults such as sepsis, trauma, burns, shock, bleeding, infection, and all kinds of stress [12]. The gut under severe insult is considered to have an important role in promoting infectious complications and MODS in regard to intestinal epithelia, the immune system, and commensal bacteria [1]. Proper treatment for the digestive tract can thus be of great value in SIRS patients. The characterization of alternations in the gut flora can be an indication of patient status and a possible guide for appropriate treatment. Validated markers for quick diagnosis are needed for timely and appropriate treatment of SIRS patients. To our knowledge, this is the first report in which the pattern of Gram-stained fecal flora has been evaluated for potential use as a quick diagnostic marker of the gut environment in patients with severe SIRS.
Gram staining is one of the most common and simplest methods for identifying bacteria in sputum. According to the guidelines of the American Thoracic Society and the Infectious Diseases Society of America, the results of Gram staining can be used to direct initial empiric antimicrobial therapy and may increase the diagnostic value [3]. Fecal Gram staining is not usually used clinically because each bacterial species cannot be identified specifically because feces contain many kinds of obligate anaerobes. However, in critically ill conditions, severe SIRS patients had 100–10,000 fewer total anaerobes, including Bifidobacterium and Lactobacillus, as compared to healthy volunteers [2]. The decrease in altered gut flora was so great that we could observe different patterns of Gram-stained fecal bacteria under the microscope. Therefore, the recognition of such patterns can be of diagnostic value.
In the diverse pattern, numerous and different kinds of bacteria covered the field. In the analysis of fecal cultures, the number of obligate anaerobes was almost within the normal range. In the single pattern, specific bacteria dominated the field. Fecal Gram staining could not identify each bacterial species, but such a simplified pattern of Gram-stained fecal flora could indicate a significant alteration in gut flora. In fact, the number of total obligate anaerobes in the single pattern was lower than that in the diverse pattern. The dominant bacteria in the single pattern differed in each case, but the number of Pseudomonas was significantly higher than in the diverse pattern. In the depleted pattern, most bacteria were diminished in the field. In the analysis of fecal flora, the number of both total obligate anaerobes and total facultative anaerobes in depleted pattern were significantly lower than those in the diverse pattern and single pattern (p < 0.05). These results indicate that the pattern of Gram staining could be associated with a decrease in fecal bacterial numbers and could be a diagnostic marker for altered gut flora.
The analysis of fecal organic acid concentrations in the present study showed that the concentrations of SCFAs such as acetate, propionate, and butyrate associated with depleted pattern were significantly lower than those of the other two groups. SCFAs are the metabolic end products of the colonic microflora fermentation of carbohydrates and glycoproteins [13]. The reduction in fecal concentrations of organic acids in the depleted pattern can be explained by the significantly reduced number of total obligate anaerobes in the gut. SCFAs are utilized mainly by intestinal epithelial cells as energy substrates, and some are absorbed into the portal flow to the liver and utilized as systemic energy sources [14]. Butyrate plays an important role in gene expression [15] and possesses anti-inflammatory activity (e.g., inhibition of NF-κB, IL-12, and TNF-α, and an increase in IL-10) [16]. The significantly decreased organic acid concentrations including SCFAs observed with the single pattern and depleted pattern in this study may reflect the altered immune response to pathogens. Further studies are required to clarify the mechanism and influence of decreased organic acids in the gut.
In the present study, mortality due to MODS associated with the single pattern (52%) and depleted pattern (64%) was significantly higher than that in the diverse pattern (6%) (p < 0.05). The diverse pattern, single pattern, and depleted pattern appear to represent a continuum of abnormality depending on the severity of the patient’s condition. The depleted pattern gradually changed to diverse pattern as the patient’s condition improved. Commensal gut flora has important and specific metabolic, trophic, and protective functions. Equilibrium between species of resident bacteria provides stability of the microbial population within an individual under normal conditions [17]. The presence of complexes of intestinal microflora provides protection against colonization by many pathogenic infectious agents and is a phenomenon referred to as “colonization resistance” [18]. These reports suggest that the commensal gut flora, especially obligate anaerobes, play an essential role in homeostasis and protection from injury. Our previous results showed that the decrease of total obligate anaerobes was significantly associated with septic mortality in patients with severe SIRS [19]. The altered gut flora including decreased counts of obligate total anaerobic bacteria may lead to decreased intestinal resistance to pathogens in critically ill patients.
In the present study, the incidences of bacteremia associated with the single pattern (71%) were significantly higher than those for the diverse pattern (35%) (p < 0.05). With the single pattern, the GPC identified by fecal Gram staining mainly consisted of Staphylococcus and Enterococcus species. GNR consist of Bacteroidaceae species, Enterobacteriaceae, Pseudomonas aeruginosa, and fungi in fecal Gram stains consist of Candida species. In this analysis of gut flora, each dominant fecal GPC, GNR, and fungus was not always cultured from blood when bacteremia was detected. One reason was that the bacteria from blood culture could come from various organs. Another reason was that alteration of gut flora can influence systemic immune function. Steffen reported that in patients who showed bacterial translocation, Gram-negative facultative anaerobes, particularly E. coli, were the most common organisms identified in mesenteric lymph nodes and serosal scrapings under laparotomy; obligate anaerobes were cultured less frequently, despite their dominance in the gut [20]. These results showed that translocated bacteria are not always from dominant fecal bacteria. At the same time, various types of strong antibiotics change the dominant bacteria in only a few days. In our study, broad-spectrum antibiotics having anti-anaerobic activity were used in all patients. Further evaluation of the clinical impact of bacterial translocation on severe SIRS is needed.
A quick evaluation of gut flora can be useful for the development of a novel digestive tract treatment to intervene directly in the composition of gut flora. It takes several days to obtain results from anaerobic cultures, whereas the Gram staining method can be done immediately at the bedside. In the present study, we demonstrated that it is possible to use fecal Gram staining to classify fecal bacteria into several patterns in SIRS patients. In addition, these patterns appeared to represent a different status of the gut flora and environment and were associated with septic complications as well as mortality in these patients. In this way, the pattern of fecal Gram-stained bacteria can be used as a quick diagnostic marker for gut flora and septic complications prior to treatment. Recently, we reported that synbiotics maintained the gut flora and environment and decreased the incidence of septic complications in patients with severe SIRS. Thus, further study is needed to investigate the mechanism and interaction between altered gut flora and septic complications as well as effective early treatment for the gut.
Acknowledgments
This study was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan and Yakult Bio-Science Foundation.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Clark JA, Coopersmith CM. Intestinal crosstalk: a new paradigm for understanding the gut as the “motor” of critical illness. Shock. 2007;28:384–393. doi: 10.1097/shk.0b013e31805569df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimizu K, Ogura H, Goto M, et al. Altered gut flora and environment in patients with severe SIRS. J Trauma. 2006;60:126–133. doi: 10.1097/01.ta.0000197374.99755.fe. [DOI] [PubMed] [Google Scholar]
- 3.American Thoracic Society; Infectious Diseases Society of America Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005;171:388–416. doi: 10.1164/rccm.200405-644ST. [DOI] [PubMed] [Google Scholar]
- 4.Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. 1992. Chest. 2009;136(5 Suppl):e28. [DOI] [PubMed]
- 5.Dellinger RP, Levy MM, Carlet JM, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit Care Med. 2008;36:296–327. doi: 10.1097/01.CCM.0000298158.12101.41. [DOI] [PubMed] [Google Scholar]
- 6.Azuma R, Suto T. Validity of transfer of the taxonomical position of Corynebacterium pseudopyogenes from genus Corynebacterium to genus Actinomyces. In: Izuka H, Hasegawa T, eds. Proceeding of the 1st international conference on culture collection. Tokyo: University of Tokyo Press; 1970:493–505.
- 7.Tanaka R, Mutai M. Improved medium for selective isolation and enumeration of Bifidobacterium. Appl Environ Microbiol. 1980;40:866–869. doi: 10.1128/aem.40.5.866-869.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petts DN. Colistin-oxolinic acid-blood agar: a new selective medium for streptococci. J Clin Microbiol. 1984;19:4–7. doi: 10.1128/jcm.19.1.4-7.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugawara G, Nagino M, Nishio H, et al. Perioperative synbiotic treatment to prevent postoperative infectious complications in biliary cancer surgery: a randomized controlled trial. Ann Surg. 2006;244:706–714. doi: 10.1097/01.sla.0000219039.20924.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi H, Yajima T. Correlation between water-holding capacity of different types of cellulose in vitro and gastrointestinal retention time in vivo of rats. J Sci Food Agric. 1992;60:139–146. doi: 10.1002/jsfa.2740600202. [DOI] [Google Scholar]
- 11.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36:309–332. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Moore FA, Moore EE, Poggetti R, et al. Gut bacterial translocation via the portal vein: a clinical perspective with major torso trauma. J Trauma. 1991;31:629–636. doi: 10.1097/00005373-199105000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 14.Rémésy C, Demigné C, Morand C. Metabolism of short chain fatty acids in the liver. In: Cummings JH, Rombeau JL, Sakata T, editors. Physiological and clinical aspects of short-chain fatty acids. Cambridge: Cambridge University Press; 1995. pp. 171–190. [Google Scholar]
- 15.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 16.Saemann MD, Bohmig GA, Osterreicher CH, et al. Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000;14:2380–2382. doi: 10.1096/fj.00-0359fje. [DOI] [PubMed] [Google Scholar]
- 17.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 18.Vollaard EJ, Clasener HA. Colonization resistance. Antimicrob Agents Chemother. 1994;38:409–414. doi: 10.1128/aac.38.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu K, Ogura H, Goto M, et al. The persistent decrease of total obligate anaerobes in gut is related to the high mortality in patients with severe SIRS. Crit Care Med. 2006;34(Suppl):A1. [Google Scholar]
- 20.Steffen EK, Berg RD, Deitch EA. Comparison of translocation rates of various indigenous bacteria from the gastrointestinal tract to the mesenteric lymph node. J Infect Dis. 1988;157:1032–1038. doi: 10.1093/infdis/157.5.1032. [DOI] [PubMed] [Google Scholar]