Abstract
The C-terminal portion of adenovirus E1A suppresses ras-induced metastasis and tumorigenicity in mammalian cells; however, little is known about the mechanisms by which this occurs. In the simple eukaryote Saccharomyces cerevisiae, Ras2p, the homolog of mammalian h-ras, regulates mitogen-activated protein kinase (MAPK) and cyclic AMP-dependent protein kinase A (cAMP/PKA) signaling pathways to control differentiation from the yeast form to the pseudohyphal form. When expressed in yeast, the C-terminal region of E1A induced pseudohyphal differentiation, and this was independent of both the MAPK and cAMP/PKA signaling pathways. Using the yeast two-hybrid system, we identified an interaction between the C-terminal region of E1A and Yak1p, a yeast dual-specificity serine/threonine protein kinase that functions as a negative regulator of growth. E1A also physically interacts with Dyrk1A and Dyrk1B, two mammalian homologs of Yak1p, and stimulates their kinase activity in vitro. We further demonstrate that Yak1p is required in yeast to mediate pseudohyphal differentiation induced by Ras2p-regulated signaling pathways. However, pseudohyphal differentiation induced by the C-terminal region of E1A is largely independent of Yak1p. These data suggest that mammalian Yak1p-related kinases may be targeted by the E1A oncogene to modulate cell growth.
INTRODUCTION
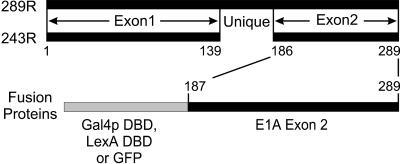
The human adenovirus early region 1A (E1A) gene encodes two major proteins of 289 and 243 residues, which differ only by the presence of an internal sequence of 46 amino acids unique to the larger protein (Figure 1). Comparison of the E1A sequences of a number of adenovirus serotypes has identified three regions of sequence conservation (Kimelman et al., 1985; van Ormondt et al., 1986), designated conserved regions 1, 2, and 3. Exon 2 of E1A encodes 104 amino acids that are not highly conserved between various adenovirus serotypes. Nevertheless, this C-terminal region of E1A is required for immortalization by E1A and transformation of rodent cells in cooperation with the products of the adenovirus E1B oncogene (Subramanian et al., 1991). In contrast to its function with the E1B proteins, the C terminus of E1A acts to repress transformation in cooperation with ras and blocks the invasive and metastatic properties of ras-transformed cells (Subramanian et al., 1989; Douglas et al., 1991; Linder et al., 1992; Boyd et al., 1993). The only known protein target of the C terminus of E1A is CtBP (Schaeper et al., 1995), a transcriptional corepressor (Criqui-Filipe et al., 1999; Furusawa et al., 1999; Meloni et al., 1999; Sewalt et al., 1999). Disruption of the interaction of E1A with CtBP enhances the ability of E1A to cooperate with activated ras to transform cells and increases the tumorigenic and metastatic capacity of these transformed cells. However, the interaction of CtBP with E1A may not completely account for these activities, because E1A mutants with deletions of other regions within the C terminus show similarly increased metastatic and tumorigenic properties, yet still retain the ability to interact with CtBP (Boyd et al., 1993; Schaeper et al., 1995).
Figure 1.
Map of the major E1A proteins and conserved regions. The two major products of E1A are 289 and 243 residues in length and differ only by the presence of an additional 46 amino acids unique to the larger. The C-terminal region of E1A corresponding to aa 187–289 was expressed as a fusion with the Gal4p DBD, LexA DBD, or GFP in this study.
Progress in elucidating the mechanisms by which the C-terminal region of E1A functions to modulate ras-induced transformation, tumorigenesis, and metastasis has been hampered by the technical difficulties associated with genetic analysis in mammalian cells. In contrast, powerful tools are available for molecular genetic analysis in the yeast Saccharomyces cerevisiae. Furthermore, expression of a foreign gene in yeast can provide an attractive means of studying gene function because many important biological processes are conserved between higher eukaryotes and yeast. Indeed, our recent studies have established that E1A interacts with conserved gene and growth regulatory pathways in yeast (Miller et al., 1995, 1996), providing a basis for more detailed investigations of the molecular mechanisms by which E1A reprograms cell growth and development.
In this study, we have examined the effects of the C-terminal domain of E1A on Ras2p-regulated growth control in S. cerevisiae. In this dimorphic yeast, the product of RAS2, the yeast homolog of mammalian h-ras, controls mitogen-activated protein kinase (MAPK) and cyclic AMP (cAMP)/protein kinase A (PKA) signaling pathways to regulate pseudohyphal/filamentous differentiation in diploid cells and invasion of the growth matrix by haploid cells (Gimeno et al., 1992; Liu et al., 1993; Mösch et al., 1996; Cook et al., 1997; Madhani et al., 1997; Robertson and Fink, 1998; Pan and Heitman, 1999). Expression of the C-terminal domain of E1A induced strong pseudohyphal growth in diploid yeast, which was independent of both the MAPK and cAMP/PKA pathways and thus functions through a novel mechanism. Using the yeast two-hybrid system, we identified an interaction between the C terminus of E1A and yeast Yak1p, a dual-specificity serine/threonine kinase (Garrett et al., 1991) that functions as a negative regulator of growth (Garrett and Broach, 1989).
MATERIALS AND METHODS
Yeast Strains, Media, and Microbiological Techniques
Yeast strains used in these experiments are listed in Table 1. All strains used for pseudohyphal growth assays are derived from the Σ1278b background (Grenson et al., 1966). Yeast culture media was prepared and yeast genetic manipulations were performed using standard techniques (Adams et al., 1998). Synthetic low-ammonia dextrose (SLAD) medium for pseudohyphal growth assay was prepared as described (Gimeno et al., 1992).
Table 1.
Yeast strains
| Strain | Genotype | Source or Reference |
|---|---|---|
| JMY38a/α | ura3-52/ura3-52 his3∷hisG/his3∷hisG leu2∷hisG/LEU2 trp1∷hisG/TRP1 | This study |
| L5986a/α | ste7∷LEU2/ste7∷LEU2 ura3-52/ura3-52 leu2∷hisG/leu2∷hisG his3∷hisG/his3∷hisG | Madhani and Fink, 1997 |
| L5987a/α | ste12∷LEU2/ste12∷LEU2 ura3-52/ura3-52 leu2∷hisG/leu2∷hisG his3∷hisG/his3∷hisG | Liu et al., 1993 |
| L6213a/α | phd1∷hisG/phd1∷hisG ura3-52/ura3-52 leu2∷hisG/leu2∷hisG his3∷hisG/his3∷hisG | Lo et al., 1997 |
| L6278a/α | kss1∷ura3∷LEU2/kss1∷ura3∷LEU2 trp1∷hisG/TRP1 ura3-52/ura3-52 leu2∷hisG/leu2∷hisG his3∷hisG/his3∷hisG | Madhani and Fink, 1997 |
| MLY61 a/α | ura3-52/ura3-52 | Pan and Heitman, 1999 |
| XPY1 a/α | bcy1∷G418/bcy1∷G418 ura3-52/ura3-52 | Pan and Heitman, 1999 |
| XPY5a/α | tpk2∷G418/tpk2∷G418 ura3-52/ura3-52 | Pan and Heitman, 1999 |
| XPY95a/α | flo8∷HygB/flo8∷HygB ura3-52/ura3-52 | Pan and Heitman, 1999 |
| XPY107a/α | flo11∷HygB/flo11∷HygB ura3-52/ura3-52 | Pan and Heitman, 1999 |
| DSY1a/α | yak1∷G418/yak1∷G418 ura3-52/ura3-52 | Pan and Heitman, 1999 |
| L40 | his3Δ200 trp1-901 leu2-3,112 ade2 LYS2∷(4lexAop-HIS3) URA3∷(8lexAop-LacZ) GAL4 | Invitrogen |
Construction of Yeast and Escherichia coli Expression Vectors
Plasmids used in this study are listed in Table 2 and the construction of those plasmids novel to this report are summarized below. Yeast expression vector pAS1U was constructed from pAS1 (Durfee et al., 1993) by subcloning the XbaI-NaeI fragment of pAS1 into the same sites of pRS426 (Christianson et al., 1992). The C-terminal domain of E1A (amino acids [aa] 187–289) was expressed as a fusion with the Gal4p DBD (aa 1–147) by subcloning an EcoRI-BamHI fragment from pMA-Ex2 (Schaeper et al., 1995) into pAS1U. The C-terminal domain of E1A was expressed as fusion with the LexA DBD from pSH2-X2, which was constructed by subcloning the same EcoRI-BamHI fragment into pSH2–1 (Hanes and Brent, 1989). The E7 protein of HPV16 was polymerase chain reaction (PCR) subcloned as an EcoRI-SalI fragment from pATH11-E7 (Carter et al., 1991) into pAS1U and pSH2–1 to make pAS1U-E7 and pSH2-E7, respectively. pRS423-LexA was constructed from pEG202 (OriGene Technologies, Rockville, MD) by subcloning the SphI-SalI fragment of pEG202 into the same sites of pRS423-ADH (Mumberg et al., 1995). pBait was constructed by moving a PvuII fragment from pRS423-LexA into the same sites of YEplac181 (Gietz and Sugino, 1988). The region encoding the C terminus of E1A was cloned as an EcoRI-BamHI fragment from pMA-Ex2 (Schaeper et al., 1995) into pBait to make pBait-X2. To construct LexA DBD fusions with E1A proteins with deletions in the C-terminal region of E1A, EcoRI-BamHI or EcoRI-SalI fragments from the previously described series of pMA-Ex2 deletion mutant plasmids (Schaeper et al., 1995) were subcloned into corresponding sites of pBait. pGFP was constructed by subcloing an NheI-HindIII fragment of pEGFP-C1 (Clontech, Palo Alto, CA) and the self-complementary oligos JMO-44 (5′-AGCTTCTGAATTCCCGGGGATCCCTGCAG-3′) and JMO-45 (5′-TCGACTCCAGGGATCCCCGGGAATTCAGA-3′) into the SpeI-SalI sites of pRS426-ADH (Mumberg et al., 1995). pGFP-X2 was constructed by subcloning an EcoRI-BamHI fragment encoding the C terminus of E1A from pMA-Ex2 into the same sites of pGFP. pRS313-STE11-4 was constructed by insertion of a BamHI-SalI fragment containing STE11-4 from plasmid pSL1509 (Stevenson et al., 1992) into pRS313 (Sikorski and Hieter, 1989). The two-hybrid prey plasmid pRS424-VP16 was constructed by subcloning a NheI-SalI fragment from pVP16 (Clontech) into the SpeI-SalI sites of pRS424-GAL1 (Mumberg et al., 1995). Drosophila CtBP was inserted into pRS424-VP16 as a BamHI fragment from plasmid h-C28 (Poortinga et al., 1998). The sequences encoding the 289R and 243R E1A proteins were PCR amplified and subcloned as EcoRI-XhoI fragments into pRS423-GAL1 to construct pRS423GAL-289R and pRS423GAL-243R, respectively (Mumberg et al., 1995). The same fragments were subcloned into pGEX-4T1 (Amersham Pharmacia Biotech, Baie d'Urfé, Québec, Canada) to construct pGST-289R and pGST-243R. pRS426-Yak1 was constructed by PCR amplification of the YAK1 coding region from yeast genomic DNA and subcloning it into BamHI and XhoI sites of pRS426-ADH (Mumberg et al., 1995).
Table 2.
Plasmids
| Gene | Plasmid | Description | Source or Reference |
|---|---|---|---|
| pAS1U | URA3 2 μm | This study | |
| pBait | LEU2 2 μm | This study | |
| pGFP | URA3 2 μm | This study | |
| pSH2-1 | HIS3 2 μm | Hanes and Brent, 1989 | |
| pMA424 | HIS3 2 μm | Ma and Ptashne, 1987 | |
| pRS313 | HIS3 CEN | Sikorski and Hieter, 1989 | |
| pRS316 | URA3 CEN | Sikorski and Hieter, 1989 | |
| pRS423 | HIS3 2 μm | Christianson et al., 1992 | |
| pRS426 | URA3 2 μm | Christianson et al., 1992 | |
| YCp50 | URA3 CEN | Rose et al., 1987 | |
| YEplac195 | URA3 μm | Gietz and Sugino, 1988 | |
| pGEX-4T-1 | Amersham Pharmacia Biotech | ||
| E1A C terminus | pAS1U-X2 | URA3 2 μm | This study |
| E1A C terminus | pGFP-X2 | URA3 2 μm | This study |
| E1A C terminus | pMA-Ex2 | HIS3 2 μm | Schaeper et al., 1995 |
| E1A C terminus | pSH2-X2 | HIS3 2 μm | This study |
| E1A C terminus | pBait-X2 | LEU2 2 μm | This study |
| E1A C terminus | pBait-X2 Δ187-221 | LEU2 2 μm | This study |
| E1A C terminus | pBait-X2 Δ227-239 | LEU2 2 μm | This study |
| E1A C terminus | pBait-X2 Δ239-254 | LEU2 2 μm | This study |
| E1A C terminus | pBait-X2 Δ255-270 | LEU2 2 μm | This study |
| E1A C terminus | pBait-X2 Δ271-284 | LEU2 2 μm | This study |
| E1A C terminus | MpBait-X2 Δ285-289 | LEU2 2 μm | This study |
| E1A 12S | pGST-243R | This study | |
| E1A 12S | pRS423GAL-243R | HIS3 2 μm | This study |
| E1A 13S | pGST-289R | This study | |
| E1A 13S | pRS423GAL-289R | HIS3 2 μm | This study |
| HPV 16 E7 | pAS1U-E7 | URA3 2 μm | This study |
| HPV 16 E7 | pSH2-E7 | HIS3 2 μm | This study |
| CtBP | pRS424-VP 16-CtBP | TRP1 2 μm | This study |
| LacZ | pIL30-URA3 | URA3 CEN | This study |
| LacZ | pIL30-HIS3 | HIS3 CEN | Laloux et al., 1994 |
| RAS2VAL19 | B1696 | URA3 CEN | Toda et al., 1985 |
| STE11-4 | pSL 1509 | URA3 CEN | Stevenson et al., 1992 |
| STE 11-4 | pRS313-STE 11 | HIS3 CEN | This study |
| STE7 | pNC318-P368 STE7 | TRP1 CEN | Trie et al., 1994 |
| STE12 | pNC252-STE12 | URA3 CEN | Errede and Ammerer, 1989 |
| TEC1 | TEC1-2mu | URA3 2 μm | Mösch and Fink, 1997 |
| PHD1 | pCG7 | URA3 2 μm | Gimeno and Fink, 1994 |
| PDE1 | YEpPDE1 | URA3 2 μm | Ma et al., 1999 |
| PDE2 | YEpPDE2 | URA3 2 μm | Ma et al., 1999 |
| BCY1 | YEplac195-BCY1 | URA3 2 μm | Pan and Heitman, 1999 |
| TPK2 | YEplac195-TPKC2 | URA3 2 μm | Pan and Heitman, 1999 |
| DYRK1 A | pGEX-2T-DYRK1A | Kentrup et al., 1996 | |
| DYRK1 B | pGEX-2T-DYRK1B | Kentrup et al., 1996 | |
| YAK1 | pJG4-5YAK1 | TRP1 2 μm | This study |
| YAK1 | pRS426-YAK1 | URA3 2 μm | This study |
Assays for Diploid Pseudohyphal/Filamentous Differentiation
Growth assays for filament formation in colonies of diploid cells were performed as described previously (Gimeno et al., 1992). Briefly, diploid yeast were transformed with expression vectors by using lithium acetate (Adams et al., 1998). Single colonies of transformed yeast were patched onto SLAD plates and scored for pseudohyphal filament formation after 2 d of growth at 30°C. Representative single colonies were directly photographed with a Leitz Diaplan light microscope equipped with a 40× long working distance objective and a Sony PowerHAD 3CCD color video camera using Northern Eclipse Imaging Analysis software (Empix Imaging, Mississauga Ontario, Canada).
Assay for FG(TyA)::LacZ Activity
The plasmids pIL30-URA3 or pIL30-HIS3 (Laloux et al., 1994), which contain the LacZ gene under the transcriptional control of a filamentation response element (Mösch et al., 1996), were used to monitor MAPK-mediated activation of transcription during pseudohyphal growth. To examine the effect of the C-terminal domain of E1A on MAPK signaling, diploid yeast of the Σ1278 background were transformed with a reporter plasmid together with plasmids expressing the E1A fusion protein. β-Galactosidase assays were prepared as previously described (Mösch et al., 1996). Protein concentrations from the clarified extracts were determined using the Bradford assay (Bio-Rad Laboratories, Hercules, CA), with bovine serum albumin as a standard. β-Galactosidase activity (nmol/min/mg protein) was calculated using the following equation (OD420 × 1.7)/(0.0045 × protein concentration [mg/ml] × extract volume [ml] × time [min]) (Adams et al., 1998).
Yeast Two-Hybrid Screen
Yeast strain L40 (Invitrogen, Carlsbad, CA) was transformed as described by Gietz et al. (1997) with pBait-X2 and a yeast genomic library in plasmid pJG4–5 (OriGene Technologies) kindly provided by Dr. M. Christman (University of Virginia, Charlottesville, VA). About 2 × 107 yeast transformants were screened for the ability to grow in the absence of histidine.
Expression of Recombinant Glutathione S-transferase (GST) Fusion Proteins in E. coli
Vectors expressing GST fusions of either E1A, DYRK1A, or DYRK1B (Kentrup et al., 1996) were introduced into BL21 E. coli cells and recombinant fusion proteins were prepared and purified using glutathione-Sepharose as described by the manufacturer (Amersham Pharmacia Biotech, Baie d'Urfé, Québec, Canada).
Preparation of Yeast Cell Extracts and Coprecipitation Testing
Yeast cells transformed with pRS423GAL-289R and pRS423GAL-243R, which express the larger or smaller major E1A protein under the control of the GAL1 promoter, were grown in synthetic complete selection medium containing glucose until the cell density reached 0.8. Cells were pelleted, washed with water, and resuspended in selection medium containing galactose for 6 h. Cells were collected by centrifugation, resuspended in 100 mM Tris-HCl buffer (pH7.5) containing 1 mM dithiothreitol and 20% glycerol, and disrupted with glass beads by vortexing. Glass beads and cell debris were removed by centrifugation at 4°C. The supernatant was used for in vitro interaction assays with E. coli-produced GST-DYRK fusions. Purified GST-DYRK fusion proteins were incubated with yeast extracts containing E1A overnight at 4°C in phosphate-buffered saline buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) for protein complex formation. GST-DYRK protein complexes were purified by affinity absorption to glutathione-Sepharose by using standard procedures, separated by SDS-PAGE, and E1A was detected by Western blotting with the E1A monoclonal antibody M73 (Harlow et al., 1985).
Assays of Protein Kinase Activity
To detect the effect of E1A on DYRK phosphorylation activity, GST-DYRK proteins were incubated with GST-E1A fusion protein in phosphorylation buffer (33 mM HEPES, 6.6 mM manganese chloride, 6.6 mM magnesium chloride, 0.7 mM dithiothreitol) containing 0.66 μg/μl histone H3 (Sigma-Aldrich Canada, Oakville, Ontario, Canada) for 2 h at 4°C. Reactions were started by introduction of 2 μCi of [32P]ATP (Amersham Pharmacia Biotech) and were carried out at 30°C for 30 min. Reactions were terminated by the addition of 2× sample loading buffer and boiling. Samples were resolved on 15% SDS-PAGE gels, which were then dried and subjected to analysis using a Molecular Dynamics phosphorImager (Sunnyvale, CA).
RESULTS
C-Terminal Domain of E1A Enhances Yeast Pseudohyphal Differentiation
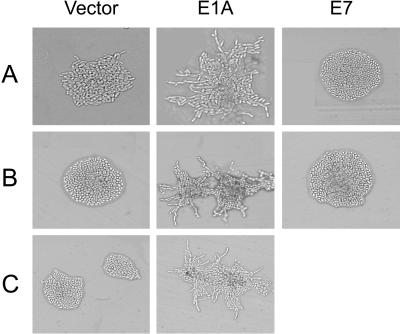
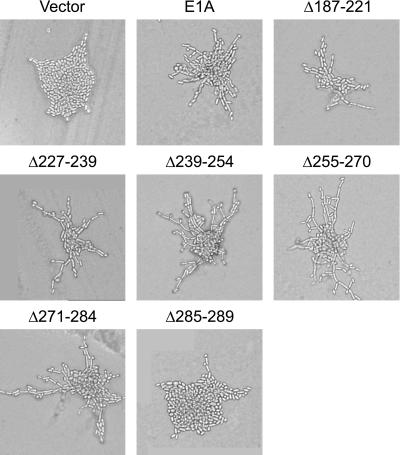
Under conditions of nitrogen starvation, diploid yeast of the Σ1278b background elongate, begin to bud in a unipolar manner, and form chains of cells in a process that has been referred to as pseudohyphal differentiation (Gimeno et al., 1992). Pseudohyphal differentiation is regulated by the RAS2 product via MAPK and cAMP/PKA signaling pathways (see INTRODUCTION). We reasoned that the C terminus of E1A might affect Ras2p function in yeast cells as it does ras function in mammalian cells, and examined whether expression of E1A in yeast affected diploid pseudohyphal growth. Expression of the C terminus (residues 187–289) of E1A fused to the yeast Gal4p DNA binding domain (DBD), the prokaryotic LexA DBD, or green fluorescent protein (GFP) (Figure 1) strongly enhanced yeast pseudohyphal growth compared with yeast transformed with the parent vectors (Figure 2). This effect appeared specific to E1A, as otherwise identical vectors expressing the comparably sized human papilloma virus (HPV) 16 E7 protein fused to the Gal4p or LexA DBD had no effect on pseudohyphal growth (Figure 2). Enhancement of pseudohyphal growth by E1A was observed on low-nitrogen medium but not on rich medium (our unpublished results). We also assessed the ability of a series of deletion mutants within the C-terminal domain of E1A to induce pseudohyphal growth (Figure 3). The region of E1A required for induction of pseudohyphal growth was mapped to aa 284–289, the last five residues of E1A.
Figure 2.
Effect of the C-terminal domain of E1A on yeast pseudohyphal growth. Wild-type diploid yeast of the Σ1278b background (JMY38 a/α) were transformed with a control vector; with vectors expressing the C-terminal region of E1A fused to the Gal4p DBD (A), LexA DBD (B), or GFP (C); or with vectors expressing the HPV 16 E7 protein similarly fused to the Gal4p (A) or LexA DBD (B). Transformed yeast were grown on SLAD medium for 2 d at 30°C and photographed.
Figure 3.
Induction of pseudohyphal growth by E1A mutants containing small deletions in the C-terminal region. Wild-type diploid yeast of the Σ1278b background (JMY38 a/α) were transformed with a control vector, a vector expressing the C-terminal region of E1A fused to the Gal4p DBD, or similar vectors expressing mutant forms of the C-terminal region of E1A containing the indicated amino acid deletions. Transformed yeast were grown on SLAD medium for 2 d at 30°C and photographed.
Independence of the E1A Effect from the MAPK Signal Transduction Pathway
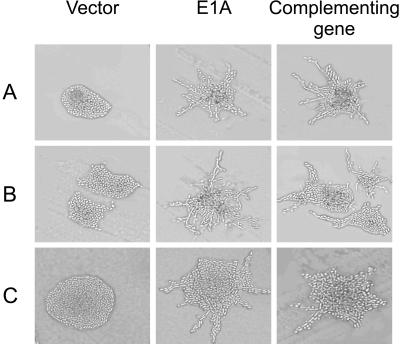
The MAP kinase signaling cascade consists of Ste20p, Ste11p, Ste7p, Kss1p, and the transcription factor Ste12p, which functions together with another transcription factor, Tec1p, to activate expression of genes required for pseudohyphal growth (Madhani and Fink, 1998). To test whether the C-terminal domain of E1A enhanced pseudohyphal differentiation through this MAPK cascade, we expressed the C-terminal region of E1A in diploid strains that are homozygous for the disruption of genes encoding components of this pathway. Disruption of STE7, KSS1, or STE12 abolished pseudohyphal growth in yeast that were not expressing E1A (Figure 4). However, expression of the C-terminal region of E1A strongly induced pseudohyphal growth in these mutant strains (Figure 4), indicating that this domain of E1A enhanced pseudohyphal growth independently of this yeast MAPK signal transduction pathway.
Figure 4.
Ability of the C-terminal domain of E1A to induce pseudohyphal growth in yeast mutated for components of the MAPK signal transduction pathway. Diploid yeast strains with homozygous disruptions of STE7 (L5986) (A), KSS1 (L6278) (B), or STE12 (L5987) (C) were transformed with either a control vector without E1A (left column) or a vector expressing the C-terminal region of E1A (middle column). Transformants were grown on SLAD medium for 2 d at 30°C and photographed. Diploid yeast of the same strains transformed only with complementing vectors expressing STE7 (A), KSS1 (B), or STE12 (C) are shown in right column.
To test this conclusion further, we used an FG(TyA)::LacZ reporter construct, which is activated by Ste12p and Tec1p in response to MAPK activation under nitrogen starvation conditions (Madhani and Fink, 1997). In diploid wild-type yeast, this reporter was strongly stimulated by expression of the constitutively active STE11-4 allele. However, transcription from this reporter was not induced by expression of the C-terminal region of E1A (Table 3). Moreover, expression of the C-terminal region of E1A did not induce reporter gene transcription in homozygous ste12Δ diploids (Table 3), although it enhanced pseudohyphal growth in this strain (Figure 4C). Thus, the biochemical analyses support the conclusion from the genetic studies that the C-terminal domain of E1A stimulates pseudohyphal growth independently of the MAPK cascade.
Table 3.
Effect of the C-terminal portion of E1A on expression of the FG(TyA)∷LacZ reporter
| Strain | Relevant genotype | Plasmid | β-Galactosidase activity* (nmol/(min × mg)) |
|---|---|---|---|
| JMY38a/α | WT | Vector | 15 ± 4.6 |
| C terminus of E1A | 19 ± 10 | ||
| STE11-4 | 110 ± 54 | ||
| L5987a/α | ste12Ε/ste12Ε | Vector | 4.5 ± 1.6 |
| C terminus of E1A | 1.9 ± 1.0 | ||
| STE12 | 32 ± 6.3 |
Average of three independent experiments with standard deviation.
Independence of the E1A Effect from the cAMP/PKA Signal Transduction Pathway
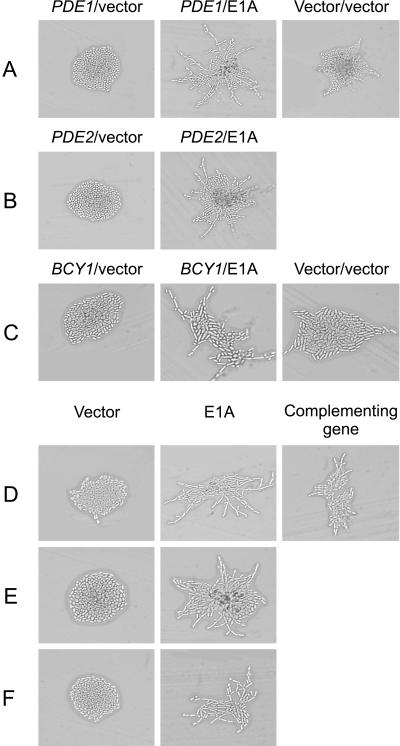
Ras2p also stimulates pseudohyphal growth via the cAMP/PKA pathway. In particular, Ras2p stimulates adenylate cyclase activity, and the resultant accumulation of cAMP activates Tpk2p, one of the three yeast isoforms of the catalytic subunit of PKA (Robertson and Fink, 1998). Tpk2p phosphorylates the transcription factor Flo8p, which is involved in the activation of transcription from genes required for pseudohyphal growth, including FLO11 (Pan and Heitman, 1999). To test if the C-terminal domain of E1A enhanced pseudohyphal differentiation through the cAMP/PKA signal transduction pathway, we first expressed the C-terminal region of E1A in diploid strains overexpressing genes that negatively regulate this pathway. To determine whether induction of pseudohyphal growth by the C-terminal region of E1A is dependent on increased levels of cAMP, we tested the effect of overexpressing PDE1 or PDE2, which encode low- and high-affinity phosphodiesterases, respectively (Ma et al., 1999). Although overexpression of either PDE1 or PDE2 inhibited pseudohyphal growth in yeast that were not expressing E1A, this had no effect on the ability of E1A to induce pseudohyphal growth (Figure 5, A and B). Similarly, overexpression of BCY1, which encodes the negative regulatory subunit of PKA (Toda et al., 1987), abolished pseudohyphal growth in yeast that were not expressing E1A, but did not affect enhancement of pseudohyphal growth by the C-terminal domain of E1A (Figure 5C). Thus, pseudohyphal growth by E1A does not require increased levels of cAMP or enhancement of PKA signaling.
Figure 5.
Ability of the C-terminal domain of E1A to induce pseudohyphal growth in yeast with alterations in the cAMP/PKA signal transduction pathway. The indicated transformants were grown on SLAD medium for 2 d at 30°C and photographed (A–E). Wild-type diploid yeast (JMY38 a/α) were transformed with vectors overexpressing PDE1 (A), PDE2 (B), or BCY1 (C) and either a control vector without E1A (left column) or a vector expressing the C-terminal region of E1A (middle column). Yeast of the same strain transformed with two empty control vectors are shown in the right column. (D–F) Diploid yeast strains with homozygous disruptions of TPK2 (XPY5) (D), FLO8 (XPY95) (E), or FLO11 (XPY107) (F) were transformed with either a control vector without E1A (left column) or a vector expressing the C-terminal region of E1A (middle column). (D) Cells of strain XPY95 transformed with a vector expressing TPK2 are shown in the right column.
We further tested whether the C-terminal region of E1A functioned via the cAMP/PKA signal transduction pathway by disrupting components of this cascade and examining whether this would block the enhancement of pseudohyphal growth conferred by E1A. We expressed the C-terminal domain of E1A in diploid strains homozygous for disruptions in TPK2, FLO8, or FLO11. Each of these disruptions abolished pseudohyphal growth in yeast that were not expressing E1A, but had no effect on the ability of E1A to induce pseudohyphal growth (Figure 5, D–F), strongly supporting the conclusion that the C-terminal domain of E1A enhances pseudohyphal growth independently of the cAMP/PKA signaling pathway.
E1A Requires Phd1p for Function
PHD1 encodes a putative transcription factor that enhances pseudohyphal growth independently of the MAPK and cAMP pathways (Chandarlapaty and Errede, 1998; Pan and Heitman, 2000). Because the C-terminal region of E1A also functions independently of the MAPK and cAMP pathways, we asked whether it depends on Phd1p for function. Consistent with previous reports, the phd1Δ strain formed pseudohyphae poorly (Lo et al., 1997) and this was fully complemented by introduction of PHD1 (Figure 6). Expression of the C-terminal domain of E1A failed to enhance pseudohyphal growth in this strain, indicating that it requires Phd1p for function.
Figure 6.
Dependence of the E1A effect on Phd1p. A diploid yeast strain homozygously disrupted for PHD1 (L6213) was transformed with a control vector without E1A, a vector expressing the C-terminal region of E1A, or a vector expressing PHD1. Transformants were grown on SLAD medium for 2 d at 30°C and photographed.
Interaction of Yeast Yak1p with the C-Terminal Domain of E1A
To attempt to identify proteins with which the C-terminal domain of E1A interacts to stimulate pseudohyphal growth, we used the yeast two-hybrid screen. L40 yeast cells were transformed with a bait plasmid expressing the C-terminal region of E1A fused to the DNA binding domain of LexA and a library of yeast genomic DNA fragments fused to a transcriptional activation domain. We isolated one positive clone encoding a C-terminal fragment (aa 163–807) of the yeast dual specificity serine/threonine protein kinase Yak1p. Yak1p interacted specifically with the C-terminal domain of E1A and not with conserved region 2 of E1A or with comparably sized fragments of mouse CBP or human SUG1 fused to LexA (our unpublished results). Interestingly, Yak1p was originally identified as a negative regulator of cell growth that functions in opposition to the RAS-regulated cAMP/PKA pathway (Garrett and Broach, 1989; Ward and Garrett, 1994), but little is known about Yak1p function in yeast pseudohyphal signaling.
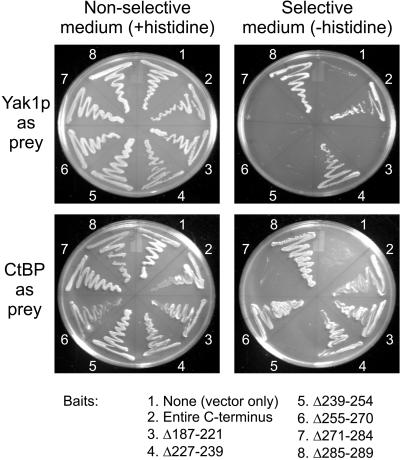
To identify the regions within the C-terminal domain of E1A that are required for interaction with Yak1p, two-hybrid interaction studies were performed with plasmids expressing a series of deletion mutants within the C-terminal region of E1A fused to the LexA binding domain. In addition to Yak1p, we used CtBP, the only other cellular protein known to interact with the C-terminal domain of E1A, as a control. The region of the C-terminal domain required for interaction with Drosophila CtBP was mapped to aa 271–284, which contain the CtBP binding motif (PLDLS), and aa 239–254 (Figure 6). This is consistent with a previous report (Boyd et al., 1993), although the mutant Δ239–254 retained a measurable, but weaker interaction with human CtBP. This difference may be related to species differences between human and Drosophila CtBP or differences in the fusion constructs. Using the same deletion mutants, we determined that aa 187–221 and aa 239–284 of the C-terminal domain of E1A are required for interaction with Yak1p (Figure 7).
Figure 7.
Interaction of yeast Yak1p with the E1A C-terminal deletion mutants in the yeast two-hybrid assay. Yeast strain L40 was transformed with expression vectors for the indicated LexA-E1A fusions and expression vectors for YAK1 or CtBP fused to a transcriptional activation domain. Transformed yeast were streaked on nonselective plates (+histidine) and selection plates (−histidine) and allowed to grow at 30°C for 3 d. For each E1A mutant used as “bait,” the indicated numbers are inclusive and refer to the amino acid residues deleted with respect to the 289R E1A protein.
Interaction of E1A with Mammalian Homologs of Yak1p
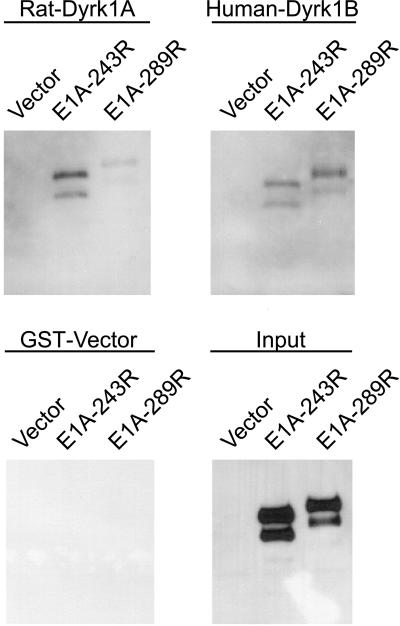
Yak1p-related proteins represent a novel subfamily of protein kinases with unique structural and enzymatic features, which have been categorized as the dual-specificity, Yak-related kinases (Dyrks) (Becker and Joost, 1999). The catalytic domain of yeast Yak1p shares the highest homology with the mammalian Dyrk1 proteins (Kentrup et al., 1996). To ask whether E1A might physically interact with the Dyrks, we conducted in vitro protein binding assays by using the rat DYRK1A and human DYRK1B products expressed as GST fusion proteins in E. coli (Figure 8). Both the 243R and 289R E1A proteins were efficiently coprecipitated with the recombinant GST-Dyrk1A and GST-Dyrk1B, but not with GST alone, suggesting that mammalian Dyrk1A and Dyrk1B proteins physically interact with E1A.
Figure 8.
Coprecipitation of E1A with mammalian homologs of Yak1p. Purified recombinant rat GST-Dyrk1A or human GST-Dyrk1B was incubated with yeast extracts containing either the 243R or 289R E1A proteins (see MATERIALS AND METHODS). The GST-Dyrk protein complexes were then pulled down with glutathione-Sepharose beads and the proteins were analyzed by SDS-PAGE and imunoblotted with a monoclonal antibody against E1A. The empty GST vector was used as a negative control, and the input levels of E1A were as shown.
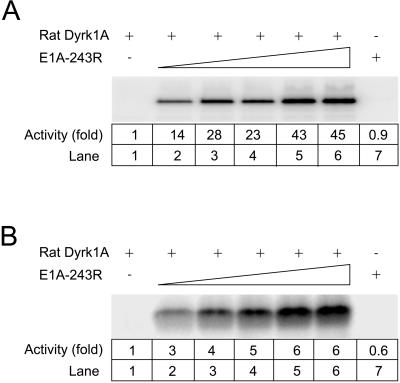
An unusual enzymatic property of Yak1p-related kinases is their ability to catalyze tyrosine-directed autophosphorylation, as well as the phosphorylation of serine/threonine residues in exogenous substrates (Becker and Joost, 1999). To define the biochemical consequences of the interaction of E1A with mammalian Dyrks, we examined the effect of E1A on recombinant rat Dyrk1A kinase activity in vitro. Recombinant GST-E1A significantly enhanced the ability of rat GST-Dyrk1A to phosphorylate itself (Figure 9A) and histone H3 (Figure 9B) as substrate in a dose-responsive manner. No effect on Dyrk1A activity was observed upon addition of GST alone (our unpublished results). Thus, the interaction between Dyrk1A and E1A stimulated its kinase activity.
Figure 9.
Effect of E1A on kinase activity of recombinant rat Dyrk1A. (A) Effect of E1A on autophosphorylation. GST-Dyrk1A was incubated with GST-E1A 243R in phosphorylation buffer for 2 h on ice. Following the addition of [γ-32P]ATP, samples were incubated for 30 min at 30°C, resolved on 15% SDS-PAGE, and analyzed using a Molecular Dynamics phosphoImager. Lane 1 contains 12 μg of GST-Dyrk1A. Lanes 2–6 contain 12 μg of GST-Dyrk1A with 2, 4, 6, 8, or 12 μg of GST-E1A 243R. Lane 7 contains 12 μg of GST-E1A 243R. (B) Effect of E1A on phosphorylation of histone H3. The assay was performed as described in A except for the presence of histone H3. Fold changes in kinase activity are indicated below each lane.
Regulation of Pseudohyphal Growth by Yak1p
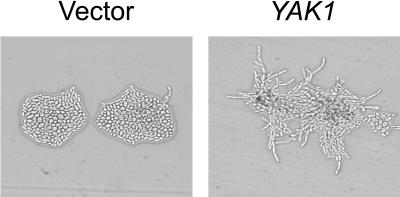
Overexpression of YAK1 in wild-type diploid yeast induced strong pseudohyphal growth compared with yeast transformed with a control vector (Figure 10). However, a previous study had reported that YAK1 is not essential for pseudohyphal differentiation, because diploid yeast with homozygous deletions in YAK1 were still able to undergo pseudohyphal growth (Pan and Heitman, 1999). We also tested the ability of various regulators of pseudohyphal growth to induce filamentation in diploid yak1Δ yeast. Expression of the constitutively active RAS2VAL19, of components of the MAPK cascade (STE11–4, STE12, or TEC1), or of TPK2 (the central component of the cAMP/PKA pathway) all induced strong pseudohyphal growth in a wild-type strain, but not in the yak1Δ mutant (Figure 11), indicating that Yak1p is essential for Ras2p regulation of pseudohyphal growth through both the MAPK and cAMP/PKA pathways. In contrast, overexpression of PHD1 induced strong pseudohyphal growth in both the wild-type and yak1Δ strain. Similarly, expression of the C terminus of E1A induced pseudohyphal growth in both the wild-type and yak1Δ mutant strains, although to a somewhat lesser extent in the yak1Δ mutant strain. These results are consistent with a role for Yak1p in modulating both the MAPK and cAMP/PKA signaling pathways that control pseudohyphal differentiation.
Figure 10.
Effect of Yak1p overexpression on yeast pseudohyphal growth. Wild-type diploid yeast (JMY38 a/α) were transformed with a control vector or a vector expressing full-length YAK1 under the transcriptional control of the ADH1 promoter. Transformed yeast were grown on SLAD medium for 2 d at 30°C and photographed.
Figure 11.
Genetic analysis of the role of Yak1p in regulating pseudohyphal growth. Wild-type diploid yeast (MLY61 a/α) or yeast disrupted for YAK1 (DSY1 a/α) were transformed with a control vector (A), or with vectors expressing RAS2VAL19 (B), STE11-4 (C), STE12 (D), TEC1 (E), TPK2 (F), PHD1 (G), or the C terminus of E1A (H). Transformed cells were then transferred to SLAD plates, allowed to grow for 2 d at 30°C, and photographed.
DISCUSSION
The E1A proteins of adenovirus have opposing effects on the functions of the mammalian ras oncogene product (Mymryk, 1996). Although E1A cooperates with activated ras to oncogenically transform cells (Ruley, 1983), it also suppresses ras-induced metastasis and tumorigenicity (Subramanian et al., 1989; Douglas et al., 1991; Linder et al., 1992; Boyd et al., 1993). Little is known about the mechanisms by which E1A modulates ras function in mammalian cells. The development of a simple model system in which the interactions between E1A and ras could be analyzed genetically would facilitate the elucidation of these interactions and their attendant regulatory pathways. Because extensive experimentation has shown that many regulatory mechanisms are conserved between the simple eukaryote S. cerevisiae and higher eukaryotic cells, we decided to investigate the effects of E1A on Ras2p function in this budding yeast.
In this study, we demonstrated that the C-terminal domain of adenovirus E1A strongly enhanced yeast pseudohyphal growth (Figure 2) and this requires the last five residues of E1A (Figure 3). E1A functions independently of the Ras2p-regulated MAPK and cAMP/PKA pathways to enhance pseudohyphal growth (Figures 4 and 5), suggesting that it functions either downstream of these pathways or via a third parallel regulatory pathway. This is further supported by our observation that the induction of pseudohyphal growth by the C-terminal region of E1A requires Phd1p (Figure 6), an enhancer of pseudohyphal growth that can function independently of the MAPK and cAMP/PKA pathways (Chandarlapaty and Errede, 1998; Pan and Heitman, 2000).
Using a yeast two-hybrid interaction screen to identify proteins that interact with the C terminus of E1A, we isolated a clone encoding aa 163–807 of the yeast Yak1p protein. Yak1p is a protein kinase of 807 amino acids that functions as a negative regulator of the Ras2p-regulated cAMP/PKA signal transduction pathway (Garrett and Broach, 1989). The cAMP/PKA signal transduction pathway is essential for the progression of yeast cells through the G0/G1 transition of the cell cycle (Garrett and Broach, 1989; Ward and Garrett, 1994). Yeast Yak1p was originally identified in a screen for mutants that suppress the growth defect in RAS mutant strains (Garrett and Broach, 1989). YAK1 mutation also restores growth in a strain lacking the three redundant PKA catalytic subunit genes (Ward and Garrett, 1994). Thus, Yak1p appears to act as an antagonist of the Ras2p-cAMP/PKA pathway and as a negative regulator of growth. However, Yak1p arrests growth only in yeast strains that are attenuated in the Ras2p-cAMP/PKA pathway and overexpression has no apparent effects on otherwise wild-type yeast cells (Garrett et al., 1991).
We performed a number of tests to determine whether Yak1p plays a role in regulating the signal transduction pathways that control pseudohyphal growth. Overexpression of YAK1 induced strong pseudohyphal growth in diploid yeast cells (Figure 10). In addition, disruption of both copies of YAK1 in diploid yeast had profound effects on the ability of Ras2p, MAPK or cAMP/PKA components to stimulate pseudohyphal differentiation (Figure 11). A role for Yak1p in pseudohyphal growth is consistent with previous work showing that Yak1p kinase activity is stimulated by nitrogen starvation (Garrett et al., 1991), the same signal used to stimulate pseudohyphal growth in these studies. Although our studies provide strong evidence that the Yak1p kinase modulates both the RAS-dependent MAPK and cAMP/PKA pathways, the exact effect on RAS-regulated signal transduction remains to be addressed. The recent observation that Yak1p interacts with the Hrt1p component of the Skp1p-Cdc53p-F-box complex (Uetz et al., 2000) may provide insight into the mechanism by which Yak1p affects pseudohyphal growth. The Skp1p-Cdc53p-F-box complex normally ubiquitinates the G1 cyclins Cln1p and Cln2p, signaling their proteolytic destruction (Skowyra et al., 1999). Interference with this process by Yak1p might stabilize Cln1p or Cln2p, both of which are essential for pseudohyphal growth, and each of which can promote cell elongation when overexpressed (Loeb et al., 1999). Alternatively, overexpression of Yak1p has also been shown to suppress the growth defects in late mitotic mutants, which characteristically exhibit increased levels of the G2/M cyclin Clb2p (Jaspersen et al., 1998). Interestingly, disruption of CLB2 induces constitutive pseudohyphal growth and overexpression of CLB2 can inhibit pseudohyphal growth (Ahn et al., 1999), suggesting that Yak1p could enhance pseudohyphal growth by reducing Clb2p expression or antagonizing Clb2p function.
Disruption of YAK1 had no effect on the ability of Phd1p and little effect on the ability of the C terminus of E1A to stimulate pseudohyphal growth (Figure 11). These results are consistent with previous observations that Phd1p functions independently of the MAPK and cAMP/PKA pathways to induce pseudohyphal growth (Chandarlapaty and Errede, 1998). Importantly, although the C-terminal region of E1A binds to Yak1p, this appears to mediate only a small portion of the ability of E1A to induce pseudohyphal differentiation. This is consistent with our genetic (Figures 4 and 5) and biochemical data (Table 3) demonstrating that the C-terminal region of E1A induces pseudohyphal growth independently of the MAPK and cAMP/PKA pathways, but requires Phd1p (Figure 6). This is also in agreement with our observation that the regions of E1A that interact with Yak1p are not essential for induction of pseudohyphal growth (Figures 3 and 7).
Homologs of yeast YAK1 have been recently cloned and characterized. These include Drosophila MNB (Tejedor et al., 1995); Dictyostelium YAKA (Souza et al., 1998); and mammalian DYRK1A, DYRK1B, DYRK1C, DYRK2, DYRK3, DYRK4, and DYRK4B (Kentrup et al., 1996; Becker et al., 1998). Although the precise function of the Dyrks has yet to be defined, they probably play an important role in regulating cell cycle and differentiation. Dictyostelium YAKA is required for the initiation of development, and overexpression of YAKA causes cell cycle arrest in nutrition-rich medium, promoting developmental events (Souza et al., 1998). In Drosophila, mutation of MNB results in specific defects in the development of the central nervous system (Tejedor et al., 1995). In humans, DRYK1A is located in the “Down syndrome critical region” of chromosome 21 (Chen and Antonarakis, 1997), suggesting that it too may be involved in development.
In addition to binding to Yak1p, we demonstrated that E1A interacts with rat Dyrk1A and human Dyrk1B in vitro (Figure 8), suggesting that the C-terminal domain of E1A targets a conserved sequence present in both yeast Yak1p and mammalian homologs. Importantly, the interaction of E1A with rat Dyrk1A enhanced the ability of Dyrk1A to phosphorylate itself and histone H3 in vitro (Figure 9), indicating that E1A could potentially activate Dyrk function at inappropriate times.
We determined that the interaction of E1A with Yak1p requires two separate regions spanning aa 187–221 and 241–284 of E1A (Figure 7). This region encompasses that required for the interaction of E1A with CtBP, but is more extensive. Mutants within the region spanning aa 241–284 are impaired for the ability to immortalize primary rodent cells, and fail to block ras-induced tumorigenesis and metastasis in rodent systems (Schaeper et al., 1995). Unfortunately, mutants in the region spanning aa 187–221 were not tested in that study. However, the existing data suggest a possible connection between these activities in mammalian cells and the interaction of E1A with Dyrks.
In conclusion, we have identified yeast Yak1p and the mammalian Dyrk1 proteins as a new family of cellular regulatory proteins targeted by the C-terminal region of E1A. Our data suggest that Yak1p modulates Ras2p signaling to regulate yeast pseudohyphal differentiation. By analogy, the Dyrk proteins may function similarly in mammalian cells to modulate ras function. Interestingly, the targeting of mammalian Dyrks by E1A may contribute to the ability of E1A to negatively modulate ras-induced tumorigenicity and metastasis.
ACKNOWLEDGMENTS
We thank Dr. G. Hammond and Dr. J. Torchia for critical reading of the manuscript and Nik Avvakomov, Michael Shuen, and Jay Loftus for technical assistance. We thank Drs. G. Fink, J. Heitman, J. Thevelein, A. Dranginis, M. Christman, G. Chinnadurai, S. Parkhurst, D. Galloway, W. Becker, J. Torchia, and J. Broach for generous gifts of plasmids and strains. We also thank Dr. J. Pringle and anonymous reviewers for their helpful comments. This work was supported by grants from the Medical Research Council of Canada, The London Health Sciences Center, and The University of Western Ontario Academic Development Fund awarded to J.S.M., and National Institutes of Health Grant GM-28920 awarded to M.M.S. J.S.M. is supported by a Scholarship from the Canadian Institutes of Health Research.
REFERENCES
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- Ahn S-H, Acurio A, Kron SJ. Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol Biol Cell. 1999;10:3301–3316. doi: 10.1091/mbc.10.10.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker W, Joost H-G. Structural and functional characteristics of Dyrk, a novel subfamily of protein kinases with dual specificity. Prog Nucleic Acid Res Mol Biol. 1999;62:1–17. doi: 10.1016/s0079-6603(08)60503-6. [DOI] [PubMed] [Google Scholar]
- Becker W, Weber Y, Wetzel K, Eirmbter K, Tejedor FJ, Joost H-G. Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J Biol Chem. 1998;273:25893–25902. doi: 10.1074/jbc.273.40.25893. [DOI] [PubMed] [Google Scholar]
- Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JJ, Yaegashi N, Jenison SA, Galloway DA. Expression of human papillomavirus proteins in yeast Saccharomyces cerevisiae. Virology. 1991;182:513–521. doi: 10.1016/0042-6822(91)90592-y. [DOI] [PubMed] [Google Scholar]
- Chandarlapaty S, Errede B. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:2884–2891. doi: 10.1128/mcb.18.5.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Antonarakis SE. Localization of a human homologue of the Drosophila mnb and rat Dyrk genes to chromosome 21q22.2. Hum Genet. 1997;99:262–265. doi: 10.1007/s004390050350. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cook JG, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signaling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- Criqui-Filipe P, Ducret C, Maira SM, Wasylyk B. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 1999;18:3392–3403. doi: 10.1093/emboj/18.12.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Gopalakrishnan S, Quinlan MP. Modulation of transformation of primary epithelial cells by the second exon of the Ad5 E1A12S gene. Oncogene. 1991;6:2093–2103. [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn AE, Lee W-H, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Errede B, Ammerer G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 1989;3:1349–1361. doi: 10.1101/gad.3.9.1349. [DOI] [PubMed] [Google Scholar]
- Furusawa T, Moribe H, Kondoh H, Higashi Y. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor δEF1. Mol Cell Biol. 1999;19:8581–8590. doi: 10.1128/mcb.19.12.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett S, Broach J. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 1989;3:1336–1348. doi: 10.1101/gad.3.9.1336. [DOI] [PubMed] [Google Scholar]
- Garrett S, Menold MM, Broach JR. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Mol Cell Biol. 1991;11:4045–4052. doi: 10.1128/mcb.11.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Triggs-Raine B, Robbins A, Graham KC, Woods RA. Identification of proteins that interact with a protein of interest: applications of the yeast two-hybrid system. Mol Cell Biochem. 1997;172:67–79. [PubMed] [Google Scholar]
- Gimeno CJ, Fink GR. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Grenson M, Mousset M, Wiame JM, Bechet J. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim Biophys Acta. 1966;127:325–338. doi: 10.1016/0304-4165(66)90387-4. [DOI] [PubMed] [Google Scholar]
- Hanes SD, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- Harlow E, Franza BR, Jr, Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Gotoh Y, Yashar BM, Errede B, Nishida E, Matsumoto K. Stimulatory effects of yeast and mammalian 14–3-3 proteins on the Raf protein kinase. Science. 1994;265:1716–1719. doi: 10.1126/science.8085159. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentrup H, Becker W, Heukelbach J, Wilmes A, Schürmann A, Huppertz C, Kainulainen H, Joost H-G. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. J Biol Chem. 1996;271:3488–3495. doi: 10.1074/jbc.271.7.3488. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Miller JS, Porter D, Roberts BE. E1a regions of the human adenoviruses and of the highly oncogenic simian adenovirus 7 are closely related. J Virol. 1985;53:399–409. doi: 10.1128/jvi.53.2.399-409.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloux I, Jacobs E, Dubois E. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 1994;22:999–1005. doi: 10.1093/nar/22.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S, Popowicz P, Svensson C, Marshall H, Bondesson M, Akusjärvi G. Enhanced invasive properties of rat embryo fibroblasts transformed by adenovirus E1A mutants with deletions in the carboxy-terminal exon. Oncogene. 1992;7:439–443. [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Lo H-J, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Loeb JD, Kerentseva TA, Pan T, Sepulveda-Becerra M, Liu H. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics. 1999;153:1535–1546. doi: 10.1093/genetics/153.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Ma P, Wera S, Van Dijck P, Thevelein JM. The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling. Mol Biol Cell. 1999;10:91–104. doi: 10.1091/mbc.10.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. The riddle of MAP kinase signaling specificity. Trends Genet. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- Meloni AR, Smith EJ, Nevins JR. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc Natl Acad Sci USA. 1999;96:9574–9579. doi: 10.1073/pnas.96.17.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ME, Cairns BR, Levinson RS, Yamamoto KR, Engel DA, Smith MM. Adenovirus E1A specifically blocks SWI/SNF-dependent transcriptional activation. Mol Cell Biol. 1996;16:5737–5743. doi: 10.1128/mcb.16.10.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ME, Engel DA, Smith MM. Cyclic AMP signaling is required for function of the N-terminal and CR1 domains of adenovirus E1A in Saccharomyces cerevisiae. Oncogene. 1995;11:1623–1630. [PubMed] [Google Scholar]
- Mösch H-U, Fink GR. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch H-U, Roberts RL, Fink GR. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Mymryk JS. Tumor suppressive properties of the adenovirus 5 E1A oncogene. Oncogene. 1996;13:1581–1589. [PubMed] [Google Scholar]
- Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Heitman J. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol Cell Biol. 2000;20:8364–8372. doi: 10.1128/mcb.20.22.8364-8372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson LS, Fink GR. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Ruley HE. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt RGAB, Gunster MJ, van der Vlag J, Satijn DPE, Otte AP. C-Terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol Cell Biol. 1999;19:777–787. doi: 10.1128/mcb.19.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Souza GM, Lu S, Kuspa A. YakA, a protein kinase required for the transition from growth to development in Dictyostelium. Development. 1998;125:2291–2302. doi: 10.1242/dev.125.12.2291. [DOI] [PubMed] [Google Scholar]
- Stevenson BJ, Rhodes N, Errede B, Sprague GF., Jr Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- Subramanian T, La Regina M, Chinnadurai G. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1a protein. Oncogene. 1989;4:415–420. [PubMed] [Google Scholar]
- Subramanian T, Malstrom SE, Chinnadurai G. Requirement of the C-terminal region of adenovirus E1a for cell transformation in cooperation with E1b. Oncogene. 1991;6:1171–1173. [PubMed] [Google Scholar]
- Tejedor F, Zhu XR, Kaltenbach E, Ackermann A, Baumann A, Canal I, Heisenberg M, Fischbach KF, Pongs O. Minibrain: a new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron. 1995;14:287–301. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Scott JD, McMullen B, Hurwitz M, Krebs EG, Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- van Ormondt H, Maat J, Dijkema R. Comparison of nucleotide sequences of the early E1a regions for subgroups A, B, and C of human adenoviruses. Gene. 1986;12:63–76. doi: 10.1016/0378-1119(80)90016-5. [DOI] [PubMed] [Google Scholar]
- Ward MP, Garrett S. Suppression of a yeast cyclic AMP-dependent protein kinase defect by overexpression of SOK1, a yeast gene exhibiting sequence similarity to a developmentally regulated mouse gene. Mol Cell Biol. 1994;14:5619–5627. doi: 10.1128/mcb.14.9.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]