Abstract
KSHV LANA1, a latent protein expressed during chronic infection to maintain viral genome, inhibits major histocompatibility complex class I (MHC I) peptide presentation in cis as a means of immune evasion. Through deletional cloning, we localized this function to the LANA1 central repeat 1 (CR1) subregion. Other CR subregions retard LANA1 translation and proteasomal processing but do not markedly inhibit LANA1 peptide processing by MHC I. Inhibition of proteasomal processing ablates LANA1 peptide presentation. Direct expression of LANA1 within the endoplasmic reticulum (ER) overcomes CR1 inhibition suggesting that CR1 acts prior to translocation of cytoplasmic peptides into the ER. By physically separating CR1 from other subdomains, we show that LANA1 evades MHC I peptide processing by a mechanism distinct from other herpesviruses including Epstein-Barr virus (EBV). Although LANA1 and EBV EBNA1 are functionally similar, they appear to use different mechanisms to evade host cytotoxic T lymphocyte surveillance.
Introduction
Viral latency is a mechanism for reducing expression of proteins susceptible to immune recognition. During latency, the large human DNA tumor viruses, Epstein-Barr virus (EBV or HHV4) and Kaposi's sarcoma-associated herpesvirus (KSHV or HHV8) exist as episomes that replicate in tandem with the cell genome using host replication proteins (Collins and Medveczky, 2002). To be retained in cells, however, the expression of several viral proteins from the latent viral genome must be maintained. The Epstein-Barr nuclear antigen 1 (EBNA1) and latency-associated nuclear antigen 1 (LANA1) are analogous to each other and act to tether the viral episomes to the host genome for EBV and KSHV respectively, allowing proper segregation of virus during cell division (Ballestas et al., 1999; Barbera et al., 2004; Grogan et al., 1983; Harris et al., 1985). Both viral proteins have DNA binding domains at each end with long intervening central repeat sequences.
EBNA1 has been extensively studied for its intrinsic properties that retard peptide antigen presentation. The central EBNA1 Gly-Ala repeat region (GAr) was first shown by Masucci and colleagues to inhibit its own proteasomal degradation in cis, thus reducing the pool of EBNA1 peptides available for antigen processing (Levitskaya et al., 1995). Proteasomes are directly coupled to the peptide translocation machinery that pumps viral peptides into the lumen of the endoplasmic reticulum (ER), allowing efficient loading of viral peptides onto MHC I prior to transport to the plasma membrane (Yewdell, 2007). More recently it has been reported that misfolded proteins which are rapidly shuttled into proteasomal pathways comprise a major source for MHC-presented peptides (Schubert et al., 2000). Up to 20% of all newly synthesized cellular proteins are misfolded, so-called defective ribosomal products (DRiPs), and processed in this manner thus allowing robust and rapid immune surveillance sampling of foreign proteins (Yewdell and Nicchitta, 2006). EBNA1 GAr appears to avoid DRiP formation by retarding its own ribosomal translation (Tellam et al., 2007; Yin et al., 2003). While slower EBNA1 translation generates fewer misfolded EBNA1 molecules, sufficient amounts of this viral protein accumulate because of decreased proteasomal protein turnover (Tellam et al., 2007; Yin et al., 2003). Since the EBNA1 GAr domain is responsible for both inhibiting protein degradation inhibition and retarding translation, distinguishing the relative significance of these two effects for EBNA1 immune evasion has been difficult.
KSHV is a gammaherpesvirus relative of EBV that causes Kaposi's sarcoma (KS), primary effusion lymphoma and a subset of multicentric Castleman's disease (Cesarman et al., 1995; Chang et al., 1994; Soulier et al., 1995). With the onset of the African AIDS pandemic, KS is now the most commonly reported neoplasm for adults in parts of sub-Saharan Africa (Parkin et al., 2008). Although KS is a major unmet public health burden in Africa, this problem will likely best be addressed through development of inexpensive vaccines or immunotherapy. Identifying mechanisms used by KSHV to avoid immune recognition will assist in the identification of effective KSHV vaccine antigens.
Similar to EBV EBNA1, KSHV LANA1 also has a central repeat (CR) domain (321-937 aa). The LANA1 CR is comprised of repeats rich in glutamine (Q), glutamate (E), and aspartate (D). Unlike EBNA1 GAr repeats, the LANA1 repeats are imperfect and may be further divided into three subdomains: CR1, CR2, and CR3 (Kwun et al., 2007). Using the entire central repeat region, Zaldumbide et al. reported that the CR functions as a cis-acting inhibitor of MHC-antigen processing, analogous to EBNA1 GAr (Zaldumbide et al., 2007). More detailed analyses have been hampered by the difficulty in cloning these extensively repeated domains. We previously showed that a junctional domain between CR2 and CR3 participates in retardation of LANA1 translation, and similar to the EBNA1 GAr, this effect is likely to be caused by peptide rather than mRNA structures (Kwun et al., 2007). We also found that some LANA1 isoforms have abnormally prolonged half-lives, suggesting that the CR may avoid CTL processing through the same mechanisms previously described for EBNA1 GAr (Kwun et al., 2007). Here, we have mapped the individual CR domains involved in surface presentation of ovalbumin peptides fused to various LANA1 constructs. We find that neither CR2 nor CR3 are primarily responsible for peptide presentation evasion. Instead, CR1 appears to block peptide presentation prior to translocation of peptides into the ER. These data show for the first time that that LANA1 and EBNA1 use different mechanisms to avoid antiviral CTL recognition and indicate that gammaherpesvirus immune evasion is more diverse than previously assumed, providing important implications for design of T cell based immunotherapeutic approaches.
Results
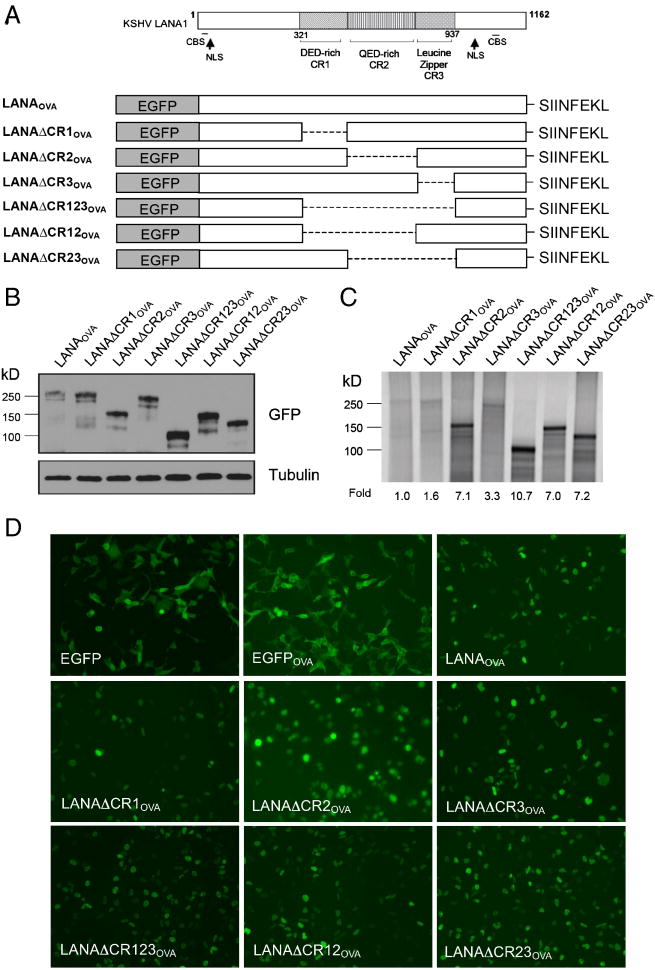
To identify subregions responsible for self-inhibition of LANA1 protein degradation, we generated LANA1 deletion constructs of CR domains (LANAΔCR123) and each CR subdomain (LANAΔCR1, LANAΔCR2, LANAΔCR3), in the context of full-length LANA1 (Kwun et al., 2007). A PEST-sequence destabilized EGFP (d1EGFP) protein (Clontech) was cloned into the C-terminus (Fig. 1A) or N-terminus (Fig. S1) and protein turnover rates were assessed by measuring fluorescence. LANA1 full-length and deletion mutant constructs were transfected into 293 cells and allowed to recover for 24 hours. Cells were then treated with 100 μg/ml cycloheximide (CHX) for 12 hours to inhibit new protein synthesis and residual EGFP-tagged protein was determined by fluorimetry.
Fig. 1.
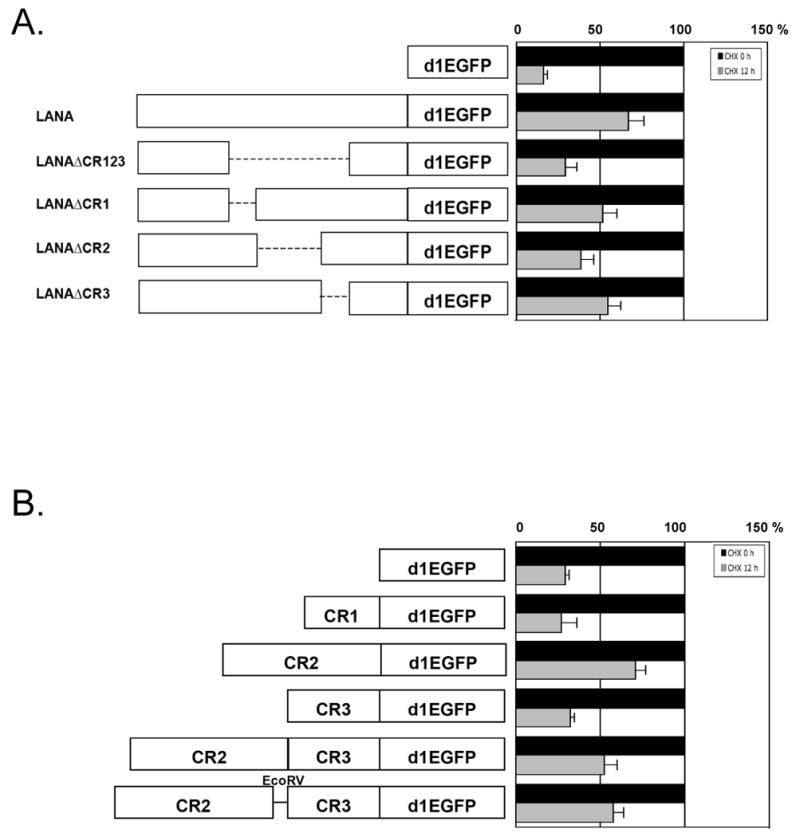
The central repeat (CR) 2 domain of LANA1 inhibits LANA1 turnover. (A) LANA1 repeat regions (CR1, CR2, CR3) were individually and in combination deleted from full-length LANA1, and cloned as fusion proteins with a destabilized enhanced green fluorescent protein (d1EGFP) (left panel) to ensure rapid proteasomal turnover. P value=0.007 (d1EGFP and LANAd1EGFP), 0.065 (LANAd1EGFP and LANAΔCR123d1EGFPd), 0.0225 (LANAd1EGFP and LANAΔCR2d1EGFP). (B) Turnover of individual LANA1 CR regions and subregions cloned into d1EGFP were also determined. P value=0.0441 (d1EGFP and CR2-d1EGFP), 0.022 (CR2-d1EGFP and CR1-d1EGFP), 0.0106 (CR2-d1EGFP and CR3-d1EGFP), 0.0325 (CR2-d1EGFP and CR2CR3-d1EGFP). Normalized EGFP fluorescence at time 0 (A and B right panel, black bars) was used as a reference to compare EGFP fluorescence 12 hours after CHX treatment (grey bars) to assess protein turnover. Results are average values performed in triplicate from independent experiments. P values between different groups, were obtained by two-tailed Student's t test using Prism software (GraphPad).
As seen in Fig. 1A, full-length LANA1 protein markedly stabilizes the turnover of PEST-EGFP sequences: 67.5% of LANA1-d1EGFP protein remains 12 hours after CHX inhibition of new protein synthesis while nearly all unfused vector (d1EGFP) protein has undergone degradation during the same period. To exclude the possible effect of the position of GFP tag artificially affecting the stabilization of the fusion proteins, both C- (Fig. 1A) and N-terminally (Fig. S1) fused PEST-EGFP constructs were examined and show similar patterns. When each repeat domain is expressed as a PEST-EGFP expression construct individually or in combination with other repeat domains, only LANA1 CR2-containing peptides have significantly diminished protein turnover (Fig. 1B). Taken together, these data suggest that CR2 primarily inhibits cis protein turnover in the context of the larger LANA1 structure and also that the primary structure of CR2 alone is sufficient to retard proteasomal degradation.
To determine whether, like EBNA1, LANA1 domains retarding cis LANA1 translation also inhibit proteasomal degradation, we examined a CR2-EcoRV-CR3 fragment containing an inserted EcoRV DNA sequence that disrupts the junction between the CR2 and CR3 domains. We previously found that this insertion diminishes LANA1 CR translation retardation mediated by the CR2CR3 junction (Kwun et al., 2007). Turnover of CR2-EcoRV-CR3, however, was unchanged from the parental fragment having an intact CR2-CR3 junction suggesting that this region is not active in preventing protein turnover. Unlike EBNA1, the translation retardation of KSHV LANA1 (CR2-CR3 junction) can be physically separated from its proteasomal inhibition function (CR2 domain).
We took advantage of an ability to dissociate these two processing functions to determine their importance in MHC class I presentation. LANA1 full-length and deletion constructs were fused with EGFP in the N-terminus and the ovalbumin (OVA) peptide SIINFEKL in the C-terminus (Fig. 2A). Surface presentation of SIINFEKL by the murine MHC I allele H2Kb is detected by flow cytometry using the 25-D1.16 antibody (eBioscience) directed against the peptide bound to the H2Kb cleft (Porgador et al., 1997). We expressed each construct in human 293KbC2 cells engineered for stable expression of murine H2Kb (a kind gift of Jonathan Yewdell) (Tscharke et al., 2005). Expression of each deletion construct was confirmed by immunoblotting (Fig. 2B), in vitro translation (Fig. 2C), as well as fluorescence microscopy (Fig. 2D).
Fig. 2.
Construction and expression of EGFP-LANAOVA. (A) Schematic diagram of EGFP-LANA1OVA constructs used to measure SIINFEKL antigen presentation. Protein expression was determined by immunoblotting (B), in vitro translation labeled with [35S]-Met (C) and fluorescence of the EGFP-LANA1 fusion proteins (D).
As expected, the constructs with deleted CR2 or CR3 domains had higher translation efficiencies compared to full-length LANA1 since this disrupts the CR2-CR3 junction (Fig. 2C). Moreover, expression of the CR1 deletion construct had similar translation efficiency as full-length LANA1, consistent with the site of translation retardation lying between CR2 and CR3. These proteins thus accumulated in the nucleus similar to parental LANA1 due to retention of nuclear localization signals present in the N and C-termini of LANA1 (Fig. 2D).
Two color flow cytometry for allophycocyanin (APC)-conjugated 25-D1.16 antibody (eBioscience) and EGFP were next used to measure SIINFEKL surface expression in cells expressing the various LANA1 deletion constructs. EGFP vector and EGFPOVA constructs were used as negative and positive controls, respectively. Consistent with the results of Zaldumbide et al. (Zaldumbide et al., 2007), the LANA1-SIINFEKL fusion protein shows marked reduction of SIINFEKL presentation compared to the EGFPOVA control. We also confirm that LANA1 inhibition of antigen presentation is dependent on the central repeat region since the fusion protein lacking this segment (LANAΔCR123OVA) is presented to a similar extent as the EGFPOVA control protein (Fig. 3A and 3B). Surprisingly, we find that neither the CR2-CR3 junction, which impedes LANA1 translation (Fig. 2C), nor the CR2 domain, which inhibits proteasomal processing (Fig. 1A and 1B) are predominantly responsible for SIINFEKL immune evasion (Fig. 3A and 3B). Instead, all LANA1 constructs lacking CR1, including deletion of CR1 alone (LANAΔCR1OVA), deletion of the entire CR region (LANAΔCR123OVA) and deletion of CR1CR2 (LANAΔCR12OVA), show markedly increased SIINFEKL peptide presentation compared to LANAOVA. In constrast, LANA1 proteins with the CR2 region deleted (LANAΔCR2OVA and LANAΔCR23OVA) inhibit MHC I peptide presentation similar to the parental LANA1 molecule. Transfection efficiency for each of the constucts was similar (Fig. 2D), as monitored through the expression of EGFP protein by flow cytometry analysis (50 to 70% of 25,000 cells were positive for EGFP expression, Fig. 3A), regardless of translation efficiency (Fig. 2C). The amount of total MHC class I level on the surface determined by flow cytometry using anti-mouse MHC class I (H-2Kb) (clone AF6-88.5.5.3, eBioscience) was unchanged despite LANA1 expression (data not shown) indicating that inhibition of SIINFEKL peptide presentation is not due to a general downregulation of MHC I surface expression by LANA1. In addition, cotransfection of CR1 and EGFPOVA from separate plasmids does not inhibit SIINFEKL presentation (data not shown), indicating that CR1 acts in cis to prevent MHC presentation of SIINFEKL rather than in trans. Since surface expression level of MHC I is unchanged by LANA1, it is unlikely that total MHC I is targeted for trans downregulation as is the case with the KSHV K3 and K5 proteins (Coscoy, 2007).
Fig. 3.
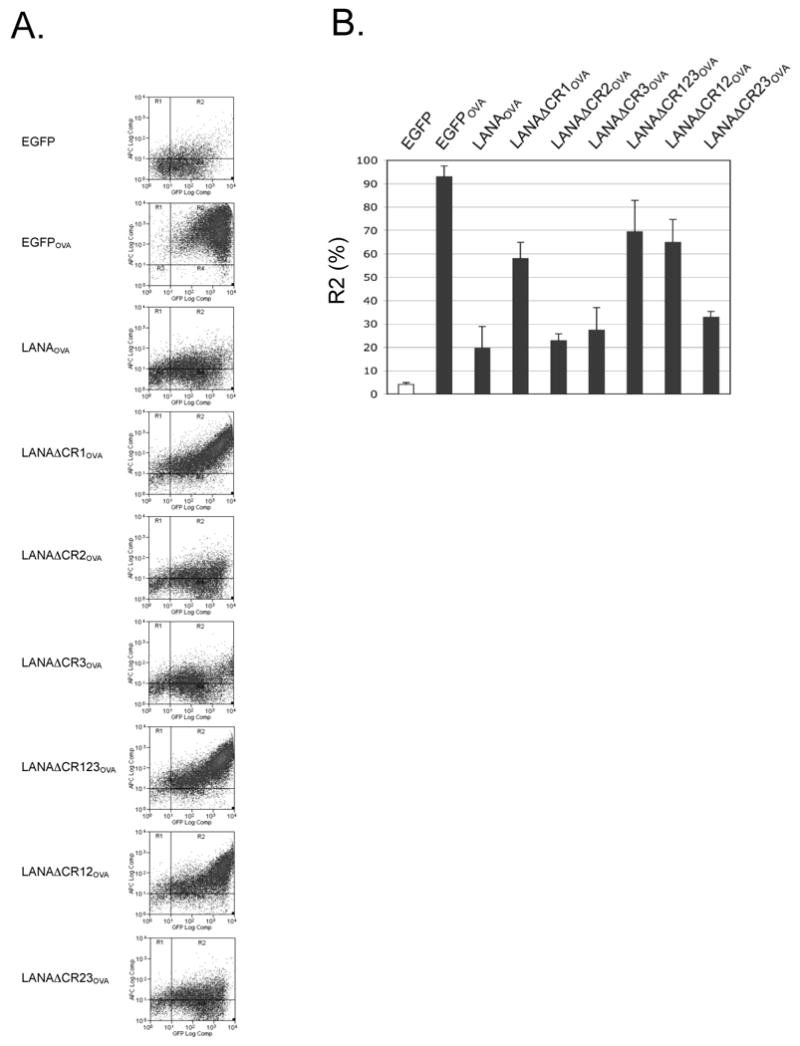
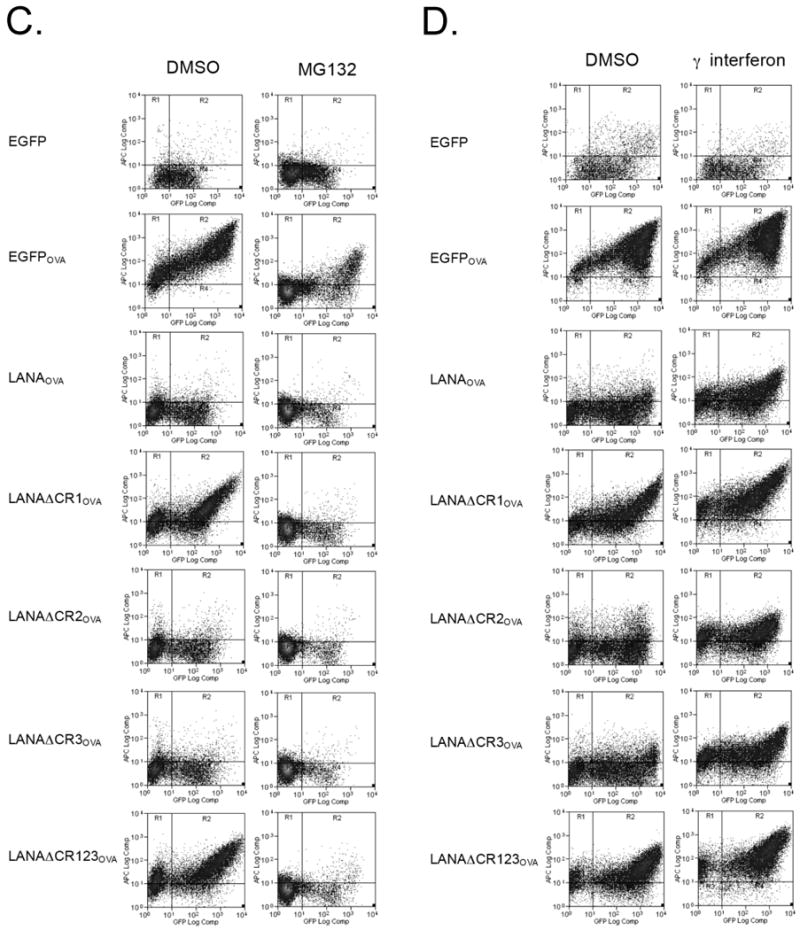
The CR1 region of LANA1 inhibits MHC I peptide antigen presentation in cis. (A) Representative results from flow cytometry analysis for SIINFEKL presentation on 293KbC2 cells expressing EGFP-LANA1OVA constructs (Fig. 2A). Cells were stained with APC anti-mouse MHC class I Kb-SIINFEKL (25-D1.16) 24 h after transfection. Samples were gated for EGFP positivity to ensure construct expression and then the percentage SIINFEKL presenting cells were determined by APC positivity. All experiments were repeated at least three times using either APC or Phycoerythrin (PE) 25-D1.16 (eBioscience) for detection and show similar results. (B) Quantitation of SIINFEKL presentation from three independent experiments (mean±S.D.). P=0.0001 (EGFP and EGFPOVA), 0.0023 (EGFPOVA and LANAOVA), 0.0049 (LANAOVA and LANAΔCR1OVA), 0.0135 (LANAOVA and LANAΔCR123OVA), 0.0311 (LANAOVA and LANAΔCR12OVA). P values between different groups, were obtained by two-tailed Student's t test using Prism software (GraphPad). (C, D) The effect of different pretreatment conditions on antigen presentation. Cells were pretreated with MG132 (proteasome inhibitor) (C), interferon gamma (immunoproteasome inducer) (D) and then SIINFEKL peptide presentation was determined by flow cytometry analysis.
We next examined whether LANA1 antigen is processed in a proteasomal-dependent or -independent fashion (e.g., autophagy) by pretreating 293KbC2 cells transfected with various LANA1OVA constructs with MG132 (Fig. 3C). As seen in Figure 3C, presentation is largely abolished by MG132 pretreatment, indicating that LANA1 is processed for MHC I presentation through canonical proteasomal processing. Preliminary experiments suggest that neither rapamycin nor chloroquine pretreatment modifies LANA1OVA presentation, consistent with autophagy playing a small role, if any, in LANA1 immune processing (unpublished data). To determine if immunoproteasomal processing is critical to LANA1 presentation, we pretreated cells with interferon gamma (Fig. 3D). Relatively higher antigen presentation of LANA1 suggests that immunoproteasomal processing enhances LANA1 presentation.
That LANA1 CR1 inhibits presentation in cis but does not either retard synthesis of LANA1 or its proteasomal degradation suggests that it might reduce translocation of LANA1 peptides from the cytosol into the ER for loading onto MHC I. We therefore bypassed this step by introducing endoplasmic reticulum (ER) signal peptide (SP) sequences (Persson et al., 1980) into the N-terminal of EGFP-LANA1OVA constructs (SP-LANAOVA, SP-LANADΔCR1OVA) and then examined expression and antigen presentation. SP construct-expressing cells (green) were also loaded with ER-tracker (red), a marker for the ER, to determine the localization of LANA1 (Fig. 4A). In contrast to LANAOVA without SP, SP-LANAOVA is efficiently processed for MHC I presentation (Fig. 4B). Both LANADΔCR1OVA proteins, with or without the SP, undergo similar levels of antigen presentation suggesting that CR1 inhibits pre-ER steps in peptide presentation (Fig. 4B). Pretreatment of SP-LANAOVA with MG132 diminished peptide presentation and so we cannot exclude that this protein undergoes retro-translocation and proteasomal degradation.
Fig. 4.
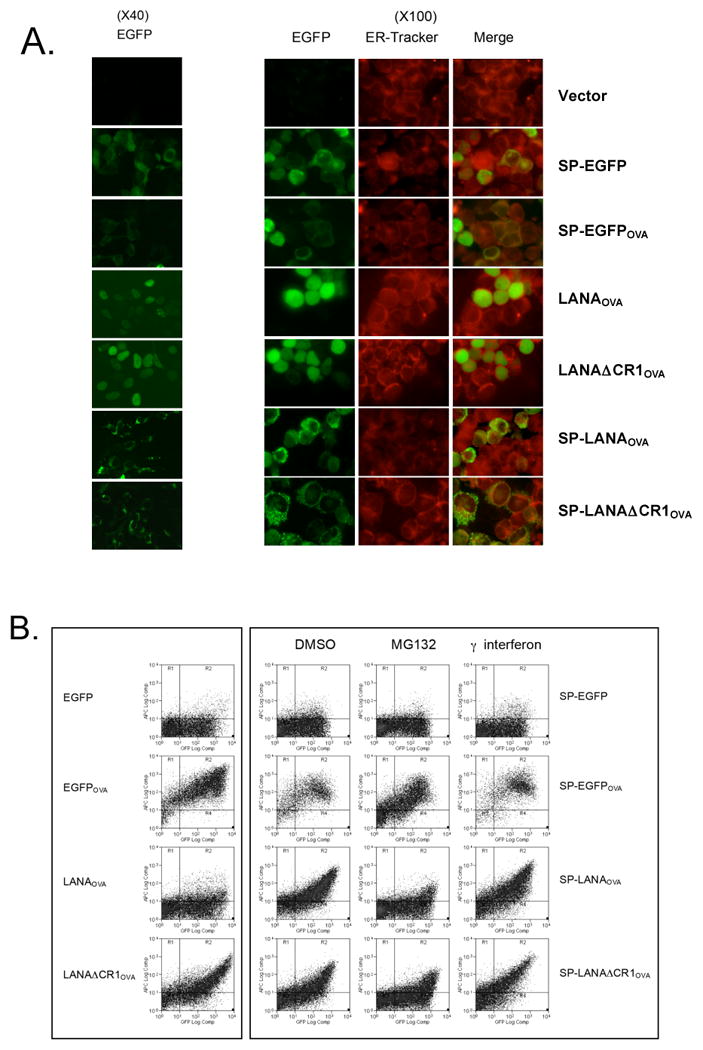
The effect of endoplasmic reticulum (ER) targeting signal peptide (SP) on LANA1 antigen presentation. (A) LANA1 expression was determined with EGFP (green) and ER-Tracker (red) fluorescence in living cells. The EGFP signal of SP constructs precisely overlapped with that of the ER-Tracker. (B) Flow cytometry analysis for SIINFEKL presentation on 293KbC2 cells expressing either EGFP-LANA1OVA or SP-EGFP-LANA1OVA constructs with pretreatment of MG132 and interferon gamma.
Discussion
Cytotoxic T lymphocyte (CTL) recognition of infected host cells is critical to immune control of KSHV infection (Wilkinson et al., 2002). The response is initiated when intracellular viral proteins are degraded by the proteasomal machinery, allowing short peptide sequences to be processed and presented on the infected cell surface bound to major histocompatibility class I (MHC I) antigens (Schubert et al., 2000). Using ova-fusion constructs, we examined LANA1 cis-inhibition of antigen presentation. While our studies provide general insights into LANA1 antigen presentation, caution is needed in interpreting them and our results require confirmation under more natural settings.
To understand the mechanisms of LANA1 CTL immune evasion, we previously examined translational retardation and turnover inhibition mediated by LANA1 (Kwun et al., 2007). We found that a LANA1 peptide sequence at the junction of the CR2 and CR3 subdomains was required to retard translation of LANA1 mRNA, an effect similar to the EBNA1 GAr. We also found that LANA1 proteins containing the entire CR domain inhibit proteasomal turnover as has been previously described (Kwun et al., 2007). Although these findings could suggest the possibility for common mechanisms to evade MHC I presentation for latent viral protein peptides by the large DNA tumor viruses, this is not always the case as the herpesvirus saimiri (HVS) reduces mRNA levels of the open reading frame (ORF73) protein homologous to KSHV LANA1 to avoid MHC class I antigen presentation (Gao et al., 2009). Careful analysis of the individual LANA1 CR domains also indicates that KSHV and EBV use different mechanisms to evade CTL immune responses against their major latency proteins.
Here we show that the LANA1 CR1 domain is primarily responsible for in cis prevention of MHC I presentation of LANA1 peptides. Unlike EBV EBNA1 (Apcher et al., 2009), this effect can be physically separated from the translation retardation and proteasome inhibition domains, suggesting that neither is critical for LANA1 evasion of antigen presentation. Surprisingly, CR1's immune evasion mechanism appears to be more complex than simple inhibition of proteasomal processing. The CR2 domain expressed as an isolated fragment markedly retards cis proteasomal processing yet its contribution to inhibition of MHC I peptide presentation is minimal compared to CR1.
Since MHC I presentation inhibition occurs in cis and yet proteasomal processing is not markedly inhibited by CR1, one possibility is that CR1 domain decouples proteasomal processing of LANA1 from the ER translocation machinery. Immunoproteasomal processing is tightly linked to surface of the ER membrane so that peptides generated by processing are efficiently translocated by transporter associated with antigen presentation 1 (TAP1) (Brooks et al., 2000). Herpes simplex virus ICP47 binds to TAP1 and blocks transport of viral peptides into the ER (Fruh et al., 1995; Hill et al., 1995; York et al., 1994). Cytomegalovirus also blocks peptide transport by producing a protein, US6, that blocks TAP1 (Hengel et al., 1997; Lehner et al., 1997). When we bypass this machinery using a ER signal peptide to translocate LANA1 directly into the ER, both LANA1 and LANA1 lacking a CR1 domain are efficiently presented to MHC I. LANA1 expressed into the lumen of the ER might be processed by ER aminopeptidase associated with antigen processing (ERAAP) (Blanchard et al., 2010) or by retrotranslocation and proteasomal processing of LANA1 protein, either of which may be TAP-dependent. This raises the possibility that CR1 delocalizes the active immunoproteasome from the TAP machinery so that peptides generated in cis are not efficiently translocated into the ER. LANA1 does not affect expression of TAP1 or immunoproteasome components, LMP2 or LMP7 (data not shown), consistent with CR1 not disrupting the processing and translocation machinery itself and suggesting a potentially novel mechanism of viral immune evasion from CTL recognition. Despite their functional similarities, EBV EBNA1 and KSHV LANA1 avoid CTL recogntion through different mechanisms and LANA1 serves as a unique model for interrogating this system.
KSHV proteins have been shown to possess a remarkable array of immune evasion activities (Coscoy, 2007). Understanding how KSHV avoids effective immune responses during latency is critical for development of effective vaccines to prevent and treat tumors caused by this virus. Our findings suggest that a LANA1 therapeutic vaccine candidate lacking the CR1 domain may be more effective than the parental molecule for inducing a functional CTL response against virus infected cells. Regardless of whether or not CR1 deletion candidates ultimately are useful for therapeutic vaccines, there is a striking need for KSHV vaccine candidates that can be employed in sub-Saharan Africa where KSHV is ubiquitous and where KS is now one of the most commonly reported tumors (Chang et al., 1996; Chokunonga et al., 1999; Wabinga et al., 1993).
Materials and methods
Plasmids
LANA1 full-length/deletion and the central repeat subdomain (CR1, CR2, CR3 or CR2/CR3) containing constructs were generated by PCR with a BC-1 DNA template and fused to EcoRI/HindIII sites of pd1EGFP-N1 encoding a destabilized EGFP in the C-terminus as described previously (Kwun et al., 2007) to examine protein turnover. A destabilized EGFP was also amplified by PCR using primers (pd1EGFP-sense, GGA TCC GCC ACC ATG GTG AGC AAG GGC GAG GAG CTG; pd1EGFP-antisense, GAA TTC CAC ATT GAT CCT AGC AGA AGC ACA) and inserted into BamHI/EcoRI sites in the N-terminus of LANA1 constructs (Fig. S1). For flow cytometry studies of antigen presentation, LANA1 full length or deletion mutants of CR region generated with various PCR reactions using primers (Kwun et al., 2007) inserted into EcoRI/HindIII sites. The EGFPOVA expression construct, enhanced GFP (EGFP) was amplified from the pEGFP-C1 vector (Clontech) by PCR and inserted into BamHI/EcoRI sites (Kwun et al., 2007). The chicken ovalabumin epitope SIINFEKL peptide sequence was introduced into HindIII/XhoI sites using direct ligation of following primers: SIIN-Forward 5′-AAG CTT AGC ATA ATT AAT TTC GAA AAG CTC TAA GCG GCC GCG CTC GAG-3′; SIIN-Reverse 5′-CTC GAG CGC GGC CGC TTA GAG CTT TTC GAA ATT AAT TAT GCT AAG CTT-3′. For SP constructs, N-terminal ER signal peptide sequence was introduced in front of EGFP using following primers: SP-sense 5′-GGA TCC GCC ACC ATG AGG TAC ATG ATT TTA GGC TTG CTC GCC CTT GCG GCA GTC TGC AGC GCT ATG GTG AGC AAG GGC GAG GAG; EGFP-antisense 5′-CTT GAA TTC CTT GTA CAG CTC GTC CAT GC-3′. Correct insert sequences were determined by DNA sequencing for all plasmids.
In vitro transcription/translation and immunoblotting
LANA1 constructs (Fig. 2A) were digested and linearized with XhoI enzyme. DNA templates were in vitro transcribed using T3 RNA polymerase and Riboprobe in vitro transcription system (Promega). RNA concentrations were determined by UV spectroscopic measurement (NanoVue, GE Healthcare) at 260 nm, and 0.5 molar equivalents of RNA were calculated for use in uncoupled in vitro translation with a rabbit reticulocyte lysate system (Promega) and [35S]-methionine (GE Healthcare). Reaction products were resolved on SDS-PAGE gel, dried, exposed overnight to screens, and read using a Typhoon imager (GE Healthcare). All the expression levels for LANA1 construct were also confirmed by immunoblotting with anti-GFP (Santa Cruz) or anti-alpha-tubulin (Sigma).
Analysis of GFP fluorescence (protein turnover)
HEK293 cells were grown in Dulbeccos' Modified Eagle Medium (DMEM) supplemented with 10% FBS and transfected with Lipofectamine-2000 (Invitrogen), with either d1EGFP or LANA-d1EGFP constructs (Fig. 1). To examine inhibition of GFP turnover, pd1EGFP-N1 (Kwun et al., 2007) encoding a destabilized EGFP with an estimated half-life (t1/2) of 1 h was used for constructing fusions to CR subdomains or LANA full-length/deletion mutant constructs at the N terminus of EGFP. Twenty-four hours after transfection, cells placed in DMEM with 100 μg/ml CHX (Sigma) to inhibit new protein synthesis or in an equal volume of DMSO diluent as control. Samples were harvested 12 h after CHX or control treatment and lysed in buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% SDS, 3 mM EDTA, 1% Triton X-100, 1 mM NaF, and 1 mM Na orthovanadate) supplemented with proteinase inhibitors. Fluorescent protein determination was measured using 50 μg of protein in 100 μl of PBS using a Synergy 2 microplate reader (BioTek).
Flow cytometry analysis for antigen presentation
HEK293 cells stably expressing the mouse class I allele H-2Kb (referred to as 293KbC2 kindly provided from Dr. Yewdell (Tscharke et al., 2005) were grown in DMEM with 10% FBS supplemented with 0.5 mg/ml G418 (Hyclone). For flow cytometry analysis, 293KbC2 cells were transfected with 1 μg of EGFP-LANA1OVA constructs (Fig. 3) or SP-EGFP-LANA1OVA constructs (Fig. 4) using Fugene 6 (Roche) and harvested and washed with PBS 24 hrs after transfection and stained with allophycocyanin (APC) or phycoerytherin (PE) anti-mouse MHC class I Kb-SIINFEKL (25-D1.16) (eBioscience) for 30 min at 4 degree. Cells were washed twice with PBS and analyzed using a DAKO CyAn high speed analyzer. For each experiment, gating was performed for positive EGFP fluorescence, and 25,000 events were collected and analyzed. The cells were treated with MG132 (Sigma, 20 μM for 12 h) or interferon gamma (eBioscience, 1000 U/mL for 24 h) before harvest. All experiments were repeated at least three times for reproducibility, with representative experiments shown.
Fluorescence microscopy
For ER labeling, the cells were incubated at 37°C for 30 min with ER-Tracker Red (Molecular Probes) and processed by manufacturers method. Cells were analyzed using Nikon TS100 with Spot insight digital camera or an Olympus AX70 epifluorescence microscope equipped with a Spot RT digital camera.
Statistic analysis
Data were compared by analysis of variance with paired student's t-test using Prism software (GraphPad). Values were considered significant at p< 0.05.
Supplementary Material
Acknowledgments
This work was funded through NIH NCI121930. Suzane Ramos da Silva was supported in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior grant BEX1049/05-4. Rusung Tan is a Senior Scholar of the Michael Smith Foundation for Health Research. Patrick S. Moore and Yuan Chang were supported by American Cancer Society Research Professorships. The authors thank Ernest M. Meyer for flow cytometric assistance, Philip Glass and Terry Riley for useful insights on large viral repeat region sequences, and William H. Chambers for suggestions on experimental design and preliminary analysis of MHC I processing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apcher S, Komarova A, Daskalogianni C, Yin Y, Malbert-Colas L, Fahraeus R. mRNA translation regulation by the Gly-Ala repeat of Epstein-Barr virus nuclear antigen 1. J Virol. 2009;83:1289–1298. doi: 10.1128/JVI.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency- associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- Barbera AJ, Ballestas ME, Kaye KM. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J Virol. 2004;78:294–301. doi: 10.1128/JVI.78.1.294-301.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard N, Kanaseki T, Escobar H, Delebecque F, Nagarajan NA, Reyes-Vargas E, Crockett DK, Raulet DH, Delgado JC, Shastri N. Endoplasmic reticulum aminopeptidase associated with antigen processing defines the composition and structure of MHC class I peptide repertoire in normal and virus-infected cells. J Immunol. 2010;184:3033–3042. doi: 10.4049/jimmunol.0903712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks P, Murray RZ, Mason GG, Hendil KB, Rivett AJ. Association of immunoproteasomes with the endoplasmic reticulum. Biochem J. 2000;352(Pt 3):611–615. [PMC free article] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;265:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Chang Y, Ziegler J, Wabinga H, Katangole-Mbidde E, Boshoff C, Schulz T, Whitby D, Maddalena D, Jaffe HW, Weiss RA, Moore PS. Kaposi's sarcoma-associated herpesvirus and Kaposi's sarcoma in Africa. Uganda Kaposi's Sarcoma Study Group. Arch Intern Med. 1996;156:202–204. [PubMed] [Google Scholar]
- Chokunonga E, Levy LM, Bassett MT, Borok MZ, Mauchaza BG, Chirenje MZ, Parkin DM. Aids and cancer in Africa: the evolving epidemic in Zimbabwe. AIDS. 1999;13:2583–2588. doi: 10.1097/00002030-199912240-00012. [DOI] [PubMed] [Google Scholar]
- Collins CM, Medveczky PG. Genetic requirements for the episomal maintenance of oncogenic herpesvirus genomes. Adv Cancer Res. 2002;84:155–174. doi: 10.1016/s0065-230x(02)84005-2. [DOI] [PubMed] [Google Scholar]
- Coscoy L. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat Rev Immunol. 2007;7:391–401. doi: 10.1038/nri2076. [DOI] [PubMed] [Google Scholar]
- Fruh K, Ahn K, Djaballah H, Sempe P, van Endert PM, Tampe R, Peterson PA, Yang Y. A viral inhibitor of peptide transporters for antigen presentation. Nature. 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- Gao J, Coulson JM, Whitehouse A, Blake N. Reduction in RNA levels rather than retardation of translation is responsible for the inhibition of major histocompatibility complex class I antigen presentation by the glutamic acid-rich repeat of herpesvirus saimiri open reading frame 73. J Virol. 2009;83:273–282. doi: 10.1128/JVI.01532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan EA, Summers WP, Dowling S, Shedd D, Gradoville L, Miller G. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology, and chromosome binding. Proc Natl Acad Sci USA. 1983;80:7650–7653. doi: 10.1073/pnas.80.24.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Young BD, Griffin BE. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt's lymphoma-derived cell lines. J Virol. 1985;56:328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengel H, Koopmann JO, Flohr T, Muranyi W, Goulmy E, Hammerling GJ, Koszinowski UH, Momburg F. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity. 1997;6:623–632. doi: 10.1016/s1074-7613(00)80350-7. [DOI] [PubMed] [Google Scholar]
- Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- Kwun HJ, da Silva SR, Shah IM, Blake N, Moore PS, Chang Y. Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen 1 Mimics Epstein-Barr Virus EBNA1 Immune Evasion through Central Repeat Domain Effects on Protein Processing. J Virol. 2007;81:8225–8235. doi: 10.1128/JVI.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner PJ, Karttunen JT, Wilkinson GW, Cresswell P. The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc Natl Acad Sci USA. 1997;94:6904–6909. doi: 10.1073/pnas.94.13.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitskaya J, Coram M, Levitsky V, Imreh S, Stigerwald-Mullen PM, Klein G, Kurilla MG, Masucci MG. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Sitas F, Chirenje M, Stein L, Abratt R, Wabinga H. Part I: Cancer in Indigenous Africans--burden, distribution, and trends. Lancet Oncol. 2008;9:683–692. doi: 10.1016/S1470-2045(08)70175-X. [DOI] [PubMed] [Google Scholar]
- Persson H, Jornvall H, Zabielski J. Multiple mRNA species for the precursor to an adenovirus-encoded glycoprotein: identification and structure of the signal sequence. Proc Natl Acad Sci USA. 1980;77:6349–6353. doi: 10.1073/pnas.77.11.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- Tellam J, Fogg MH, Rist M, Connolly G, Tscharke D, Webb N, Heslop L, Wang F, Khanna R. Influence of translation efficiency of homologous viral proteins on the endogenous presentation of CD8+ T cell epitopes. J Exp Med. 2007;204:525–532. doi: 10.1084/jem.20062508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscharke DC, Karupiah G, Zhou J, Palmore T, Irvine KR, Haeryfar SM, Williams S, Sidney J, Sette A, Bennink JR, Yewdell JW. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J Exp Med. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wabinga HR, Parkin DM, Wabwire-Mangen F, Mugerwa J. Cancer in Kampala, Uganda, in 1989-91: changes in incidence in the era of AIDS. Int J Cancer. 1993;54:5–22. doi: 10.1002/ijc.2910540106. [DOI] [PubMed] [Google Scholar]
- Wilkinson J, Cope A, Gill J, Bourboulia D, Hayes P, Imami N, Kubo T, Marcelin A, Calvez V, Weiss R, Gazzard B, Boshoff C, Gotch F. Identification of Kaposi's sarcoma-associated herpesvirus (KSHV)-specific cytotoxic T-lymphocyte epitopes and evaluation of reconstitution of KSHV-specific responses in human immunodeficiency virus type 1-Infected patients receiving highly active antiretroviral therapy. J Virol. 2002;76:2634–2640. doi: 10.1128/JVI.76.6.2634-2640.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW. Plumbing the sources of endogenous MHC class I peptide ligands. Curr Opin Immunol. 2007;19:79–86. doi: 10.1016/j.coi.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006;27:368–373. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Yin Y, Manoury B, Fahraeus R. Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science. 2003;301:1371–1374. doi: 10.1126/science.1088902. [DOI] [PubMed] [Google Scholar]
- York IA, Roop C, Andrews DW, Riddell SR, Graham FL, Johnson DC. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- Zaldumbide A, Ossevoort M, Wiertz EJ, Hoeben RC. In cis inhibition of antigen processing by the latency-associated nuclear antigen I of Kaposi sarcoma herpes virus. Mol Immunol. 2007;44:1352–1360. doi: 10.1016/j.molimm.2006.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.