Abstract
Several reports have indicated that low level of polychlorinated biphenyl (PCB) exposure can adversely affect a multitude of physiological disorders and diseases in in vitro, in vivo, and as reported in epidemiological studies. This investigation is focused on the possible contribution of two most prevalent PCB congeners in vitro in developing toxicities. We used PCB 138 and 153 at the human equivalence level as model agents to test their specificity. We chose a global approach using oligonucleotide microarray technology to investigate modulated gene expression for biological effects, upon exposure of PCBs, followed by Ingenuity Pathway Analysis (IPA), to understand the underlying consequence in developing disease and disorders. We performed in vitro studies with human peripheral blood mononuclear cells (PBMC), where PBMC cells were exposed to respective PCBs for 48 hrs. Overall, our observation on gene expression indicated that PCB produces a unique signature affecting different pathways, specific for each congener. While analyzing these data through IPA, the prominent and interesting disease and disorders were Neurological disease, Cancer, Cardiovascular disease, respiratory disease, as well as endocrine system disorders Genetic disorders, and reproductive system disease. They showed strong resemblances with in vitro, in vivo, and in the epidemiological studies. A distinct difference was observed in renal and urological diseases, organisimal injury and abnormalities, dental disease, ophthalmic disease, and psychological disorders, which are only revealed by PCB 138 exposure, but not in PCB 153. The present study emphasizes the challenges of global gene expression in vitro and was correlated with the results of exposed human population. The microarray results give a molecular mechanistic insight and functional effects, following PCB exposure. The extent of changes in genes related to several possible mode(s) of action highlights the changes in cellular functions and signaling pathways that play major roles. In addition to understanding the pathways related to mode of action for chemicals, these data could lead to the identification of genomic signatures that could be used for screening of chemicals for their potential to cause disease and developmental disorders.
Keywords: PCB 138, PCB 153, Human PBMC, Gene expression, IPA Analysis, Disease and Disorders
1. Introduction
Polychlorinated biphenyls (PCBs) make up a group of 209 individual congeners that are widespread persistent hazardous residual environmental contaminants, which have been shown to have toxic effects on various organs, including tissue of the nervous, reproductive, and in immunologic systems (Longnecker et al., 2003; Hsu et al., 2007; Hertz-Picciotto et al., 2008). Suppressed immune function can lead to increased susceptibility to infectious disease or certain types of cancers (Esseghir et al., 2007). Though PCBs were banned in the USA since 1977, persistent PCBs in our biosphere are known to cause reproductive (Loch-Caruso, 2002; Brouwer et al., 1999), neurological (Schantz et al., 2003), endocrinal (Tabb et al., 2004) and other defects. PCBs also adversely affect fetal and infant development and are immunotoxic (Wanneke et al., 2002; Lyche et al., 2004). It has been shown that congeners with chlorine substitutions in the non-ortho position are more coplanar in nature and are often associated with hepatic and reproductive toxic effects. On the other hand, congeners with chlorine substitutions at the ortho position have been shown to exert neuro- and immunotoxic effects (Kinadavanti, 2005). The most well-known mechanism related to adverse health effects such as immune suppression, hepatotoxicity, and thymic atrophy is aryl hydrocarbon (Ah) receptor-mediated pathways for dioxin-like PCBs (Safe, 1994; Van den Berg et al., 2006). Since non-coplanar PCBs (non dioxin-like) have shown a low affinity for the Ah receptor (Giesy et al., 2000), they have been regarded as potentially less toxic. However, neurotoxicity (Fisher et al., 1998), carcinogenicity (Hardell et al., 2006; Knerr et al., 2006), and changes in hormones (Cooke et al., 2001) have also been described as resulting from non-coplanar PCBs, but the mechanism is not well understood.
Several animal (Overmann et al., 1987; Allen et al., 2002) and epidemiologic studies (Rylander et al., 1996, 1998, 2000; Patandin et al., 1999; Vartiainen et al., 1998; Hertz-Picciotto et al., 2003) suggest that prenatal exposure to PCBs and related compounds result in lower birth weight. Studies using a variety of measures of exposure appear to support a reduced birth weight among infants born to women with higher fish consumption (Rylander et al., 1998a), who grew up in a fishing village or whose concentration of PCB 153 was projected to be higher at the time of the birth via kinetic modeling (Rylander et al., 1998a, b).
We have recently reported that the over expression of MT1K (Metallothionein) and CYP1A1 P450 (Cytochrome P450), can be associated with human liver disease in PCB exposures in vitro (Dutta et al., 2008). We have thus identified two most potentially significant biomarker genes, CYP1A1 (69.81 up-regulation) and MT1K (14.66 up-regulation), showing highest over-expression using PCB exposed human liver (HepG2) cells in vitro. Over expression of the CYP1A1 (Cytochrome P450) gene was specific to PCB-77 and MT1K (Metallothionein) to PCB-153. In another study, we have shown that apoptosis was the most significant cellular process pursuant to oxidative stress but each of these congeners has a unique gene expression signature, which was further validated by Taqman RT-PCR and immunoblotting studies (De et al., 2010). PCB-153 acted through TNF receptor, leading to oxidative stress through the involvement of metallothionein gene families causing apoptosis, mainly by the mitochondrial pathway. In contrast, PCB-77 acted through aryl hydrocarbon receptor, leading to oxidative stress through the involvement of cytochrome P450 (CYP1A1), and thereby causing apoptosis, by nuclear pathway. We have been able to establish that chronic exposure to PCB-153 could lead to an altered protein expression in human liver cells (HepG2) by altering several apoptotic and tumor suppressor proteins (Ghosh et al., 2007). Our current work noted some signature early disease biomarkers in PCB-exposed Slovak population (Dutta et al., 2009), and preliminary data in comparing gene expression in vitro and Slovak population indicated some similarities in their mode of actions (Ghosh et al., 2009).
In eastern Slovakia, improper disposal from the Chemko plant via the release of effluent directly into the Laborec River resulted in long-term contamination of sediment, evidenced by recent data (Kocan et al., 1994). Numerous surveys in Slovakia between 1987 and 1990 found high levels of PCBs in food (Hertzman, 1995). In the late 1980's, concentrations in breast milk in the Michalovce district averaged 4.0–4.4 mg/kg lipids (Hertzman, 1995). During 1998, the average PCB concentration (the sum of PCB-28, 52, 101, 138, 153, 156 170, 180) in human blood lipids taken from the general population living long-term in the Michalovce District was 3.5 times higher than that of the Stropkov District, and it demonstrated that PCB 153 and PCB 138 are the predominant congeners, similar to other studies (Ghosh et al., 2009; Hovander et al., 2006). Some recent epidemiological studies in this area have also shown some adverse effects in neurodevelopment, thymus size at birth (Park et al., 2008).
The gene expression profiling is considered a promising tool that may provide information more sensitive for mechanism based toxicities. Microarray is a useful method to obtain a global view of genomic changes following chemical exposures. To understand the impact of PCBs and the possible mode of action towards disease and disorder development, we have chosen a genomic approach to study the biological functions altered following a PCB-exposure on human PBMC cells in vitro. PCB 153 and PCB 138 are chemicals of our interest due to its maximum prevalence in Slovak human exposed population, and also in other PCB-exposed conditions in different studies (Gladen et al., 1999). The results were further analyzed over Ingenuity Pathway Analysis (IPA) to show a mechanistic approach of PCB-induced disease and disorder development, and compared with available epidemiological findings. The genomic results along with pathway analysis study indicate that various pathways were significantly altered by PCBs, which include, JAK/Stat signaling, Integrin signaling, fMLP Signaling in neutrophils, Oncostatin M signaling, CREB signaling on Neurons, Caveolar-midiated Endocytosis signaling, Macropinocytosis signaling, Erythropoietin signaling, Purin Metabolism and AMPK signaling. The modification in these signaling pathways leads to an alterations in endocrine, genetic, immunological, metabolic functions, and cardiovascular disease and disorders development.
2. Materials and Methods
2.1 Chemicals
Non-planar PCB-153 (2,2',4,4',5,5'-Hexachlorobiphenyl) (Product # RPC-047, CAS # 35065–27–1) and PCB-138 (2,2',3,4,4',5'- Hexachlorobiphenyl) (Product # RPC-088, CAS # 35065–28–2) with a purity >99% were used herein are products of Ultra Scientific (North Kingstown, RI). Dimethyl sulfoxide (DMSO) (Sigma, St. Louis, MO) was used for dissolving PCBs. A 2ng/µl stock solution of the PCBs was prepared in Dimethyl sulfoxide (DMSO) to the working concentrations, in the same diluents. RPMI 1640 was obtained from Invitrogen, CA, and Fetal Bovine Serum (FBS, Heat Inactivated, US Origin) was also obtained from Invitrogen, CA. Penicillin/Streptomycin was obtained from Invitrogen, California, USA. Phytohemagglutinin-M (PHA-M) was from Roche Diagonostic GmbH. Pokeweed Mitogen was from Life Technologies, USA. Trizol reagent from Invitrogen Corp. was used for RNA extraction. For microarray, GeneChip® Human Genome HU133 Plus 2.0 were obtained from Affymetrix (Santa Clara, California, USA). The RNeasy Minielute Column Kit from Qiagen(Gaithersburg, MD, USA) was used for further clean-up and concentrate RNA samples. BD Vacutainer® CPT™ (Becton Dickinson, New Jersey, USA, Cat # 362753) Cell Preparation Tube with Sodium Heparin was used for the separation of mononuclear cells from whole blood. PBS 1x sterilized solution was procured from Quality Biological Inc. (Gaithersburg, MD).
2.2 Human PBMC Culture
PBMC cells were isolated from the blood of six healthy donors with their informed consent. The study was approved by Howard University Institutional Review Board (IRB-07-GSAS-30). We did not measure the PCB concentrations of the individual, but care was taken while selecting the human volunteers. They have undergone a thorough interview about themselves and about their parents prior to collection of blood. Subjects chosen here were without any major illness and without any recent major surgical procedures. Special care was also taken about the information of any major chronic illness about their parents or they have experienced any major environmental exposures issues in their life time. If so, the subjects were not included. The subjects were also within the same age range and same sex. The technique has been adapted from Devos et al., 2004 with slight modification. Venous blood (40–50ml) was collected from each donor into the BD CPT® tube according to manufacturer’s instruction. The tubes were centrifuged at room temperature in a horizontal rotor (swing bucket) for a minimum 15 minutes 1500–1800 RCF.
The mononuclear cells (in a Buffy coat under the plasma) were collected in a fresh 15 ml tube (with cap) using a Pasture pipette. The cells were further treated by a two wash cycle with 1x PBS with centrifugation at 300 RCF and a single wash by the RPMI media supplemented with 100 units /mL penicillin G, 100 µg/mL streptomycin. The cells were re-suspended again into the culture medium (RPMI 1640) supplemented with 10% FBS (heat inactivated), 1.25 µg/ml PHA-M, 0.15 % (v/v) pokeweed mitogen, 50 µm of β-mercaptoethanol and antibiotics. PBMC was cultured at 37° C in a humidified atmosphere containing 5 % CO2 in flat bottomed cell culture plates (Nunc, USA). We prepared six plates from each donor’s blood to create 3 replicates each for each exposure with respective controls.
2.3 PCB exposure
PCB 153 and PCB 138 are of chemicals of our interest due to its maximum prevalence in Slovak human exposed population. Under this experimental study, we chose 0.87ng/ml of PCB 138 and 1.42ng/ml of PCB 153 according to the median concentration of PCBs according to the Slovak PCB-exposed Slovak population. PCB-138 and PCB-153 (dissolved in DMSO) were added to each plate individually where the final concentration of DMSO was ≤0.1%. The exposures were the same in all the experiments up to 48 hours, where we collected individual controls at 0 and 48 hours for individual donors without PCB exposures. Control cell lines were allowed to grow with DMSO only (≤0.1%of the total medium v/v) to ensure that the changes see were not due to DMSO.
2.4 RNA Extraction
RNA was extracted from the PBMC cells using an adapted Trizol Plus RNA Purification Kit (Invitrogen) according to manufacturer’s direction. Briefly, tissues were homogenized in Trizol, chloroform added, tubes mixed well and centrifuged. The aqueous layer removed to a clean, sterile tube, isopropanol was added and RNA precipitated. Samples were centrifuged to pellet RNA, washed two times with 75% ethanol and RNA re-solubilized in RNase-free H2O. Contaminating DNA was removed with the Ambion DNA-free kit. RNA concentrations were determined spectrophotometrically on a nanodrop at 230, 260 and 280 λ. RNA quality was also verified by Agilent bioanalyzer analysis using a RNA 6000 nanochip before microarray chip hybridization and RNA stored at −80 °C.
2.5 Affymetrix Chip hybridization
The RNA extracted (3 biological replicates/condition/ subject donor) was reverse transcribed to cDNA with an oligo-dT primer containing T7 RNA polymerase promoter. The cDNA was used as a template for in vitro transcription using the ENZO BioArray RNA transcript labeling kit (Affymetrix, CA). Biotin-labeled cRNA was purified, then fragmented randomly to approximately 200 bp (200mM Tris-acetate, pH 8.2, 500mM KOAc, 150mM MgOAc) prior to hybridizing to Affymetrix Human Genome Array for 16 h. The microarray was washed and stained, and fluorescent images were obtained using the Affymetrix 3000 Scanner. Quality control measures included >4-fold cRNA amplification (from total RNA/cDNA), scaling factors <2 to reach a whole-chip normalization of 800, and visual observation of hybridization patterns for chip defects for quality control. The results were cross checked with dChip software, where a model based normalization was performed (Li & Wong, 2001). The significant gene list (with all common genes) identified with Genespring and dChip with Affymetrix probe set ID at different time points were imported into the dChip and clustered based on similarity in expression. Human Genome U133 Plus 2.0 Array in our microarray gene expression analysis for PCB exposure studies includes 54,000 gene transcript, and was used during this study.
2.6 Array Quality Control
Proper quality control measures were taken throughout the procedure. Three replicates per experiment along with three controls have been performed to reduce the experimental variability. The total RNA concentration was more than 0.5µg/µl with at least four fold amplifications during labeled cRNA synthesis (Dutta et al., 2008; De et al., 2010). After scanning, first, the image was checked for alignment to grid and image contamination. Above all, the scan report generated by Gene Chip Operating Software (GCOS) had a scaling factor between 0.5 to 5, total percent of ‘P-calls’ between 30% to 50%, external controls cre>BioD>BioC>BioB and internal control GAPDH was 1 ± 0.1 (pivot table). This pivot table was then further evaluated by Hierarchical Clustering Explorer (HCE) and after this final quality control the data was analyzed with GeneSpring GX 10.0. During unsupervised clustering in Hierarchical Clustering Explorer (HCE), row by row normalization was done by mean ±SD and Euclidian distances were calculated with average linkage.
2.7 Gene Expression Data analysis
Raw data was transformed by Reduction of Invarient Probes (REDI) analysis (Expression Analysis, Durham, NC) in a two step process. First, probe hybridization intensities from all probes were used for probe normalization followed by a second step in which data from poorly performing PM probes were removed from the signal computation step. Raw data were normalized by PLIER. Differential expression was determined by Partek’s Paired t-test and filtered with p <0.01. Gene's annotations were expanded and upgraded using NCBI Entrez Gene ID, Unigene and PubMed for all significantly different genes. Transcribed sequences and expression sequence tags (ESTs) that could not be identified as to function were eliminated from the reported lists. Ingenuity Pathway software (Ingenuity®Systems, www.ingenuity.com) was employed to examine functional correlations within the different treatment groups. Data sets containing gene identifiers and corresponding expression values were uploaded into the application. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. Genes differentially expressed with p<0.01 were overlaid onto global molecular networks developed from information contained in the knowledge base. Networks were then algorithmically generated based on their connectivity. Networks were “named” on the most prevalent functional group (s) present. Canonical pathway analysis identified function specific genes significantly present within the networks.
We have used a large variety of statistical methods available in GeneSpring Gx 10.0. Briefly, probe-set analysis results derived from Microarray Suite version 5.0, and dChip (Seo et al., 2006) were used for preliminary image analysis as follows. Genes with 40% present calls (P) were selected for further analysis. Hierarchical clustering analysis was limited to probe sets with at least 40% present calls and p-value <0.05 for the Welch t-test, corrected with Benjamini and Hochberg False Discovery Rate between any two time points. A hierarchical clustering algorithm using the Pearson correlation was then used to temporally group those probe sets based on their expression patterns across the five time points. Besides GeneSpring, HCE clustering, which permits different computational algorithms and stringencies of the analysis, was employed in an interactive manner (Seo et al., 2006). Temporal clusters of genes that are specific and shared between the two disease stages, were prioritized based upon a consideration of combined support from the p-value from both dChip and MAS5.0. (GeneSpring), and visual analysis (HCE), were superimposed on gene ontology flow charts from the BayGenomics Programs in Genomic Applications (GenMAPP) (Dahlquist et al., 2002) and Ingenuity Pathway Analysis Software (IPA, CA).
“Minimum Information about a Microarray Experiment” (MIAME) compliant data has been submitted to the Gene Expression Omnibus (GEO) database. All the microarrays used in this paper can be accessed from the GEO. Individually PCB-153 microarrays can be accessed from the GEO accession number: GSE22667: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zdudfeqeaqmgsju&acc=GSE22667 and PCB-138 from GSE22632: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=xdyhjwocqwgwgtu&acc=GSE22632.
2.8 Quantitative real-time RT-PCR analysis
The differential expression of these genes was confirmed by quantitative RT-PCR, and the expression pattern of these genes was determined by in situ hybridization. For cDNA synthesis, reverse transcriptions were done according to the Invitrogen protocol. QRT-PCR was performed with the SYBR green method by using the MyiQ Single-Color RT-PCR detection system (Bio-Rad). Primers (forward and reverse) were designed with the PRIMER3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3www.cgi) and were synthesized by Invitrogen.
We designed the following real-time PCR primers (5’–3’) for amplification of (forward/reverse): RRAD (TTTACAAGGTGCTGCTGCTGGG/ TGCCGCTGATGTCTCAATGAAC); CACNA1A (AAGGAGAGGAGGATGCGTTTC/ CAGCGTGTTGAGAGCTACCAAA); MAPK1 (CCCAAATGCTGACTCCAAAGC/ GCTCGTCACTCGGGTCGTAAT); MAP3K71P1/TAB1 (CAATCATCGCAGAGCCAGAAATC/ ACGCTCCAGAGGCGGTAAAACTC); CYP11B2 (GGCAGAGGCAGAGATGCTG/ CTTGAGTTAGTGTCTCCACCAGGA); CYP1A2 (CATCCCCCACAGCACAACAA/ TCCCACTTGGCCAGGACTTC); GDNF/GFRA1 (ACCTGGAGTTAATGTCCAACC/ GGCATATTTGAGTCACTGC); TRAP1 (TGCGAGATGTGGTAACGAAG/ CGGTGCGTCCGTCTTATAGT); and the internal control/housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ATGCCAGTGAGCTTCCCGTCAGC/ GGTATCGTGGAAGAACTCATGAC was chosen was chosen as a highly expressed constitutive gene whose expression did not change as a function of any of the experimental conditions.
Primers were synthesized by Invitrogen. PCR reactions (25 µl) were assembled in 96-well optical reaction plates with each well containing 1 µl cDNA, 1.5 µl each of 5mM forward and reverse primers, 8.5 µl water, and 12.5 µl SYBR Green Supermix with ROX internal reference dye according to manufacture’s protocol (Bio-Rad Laboratories, Hercules, CA, USA). Wells were covered with optical caps, and plates were centrifuged for 3min at 100g before loading into the instrument. Triplicate samples and no-template controls were included for each set of primers. A validation experiment was performed using serial dilutions of untreated cDNA from one entity to confirm equivalent relative efficiencies for target and housekeeping genes. Amplicon identities were supported via agarose gel electrophoresis of PCR products, which demonstrated single bands approximating the expected sizes. Replicate raw data (threshold cycle number, or Ct) for each sample were averaged and then adjusted by dividing these values by the corresponding averaged Ct for GAPDH to correct for any differences in starting quantity of material. Initial analyses were performed by using the ICYCLER system software (Bio-Rad). RT-PCR data analysis was performed using the comparative Ct method (ΔΔCt) (Livak and Schmittgen, 2001). The ΔCt value was calculated as the Cttarget –Ctreference, using GAPDH as the referenced housekeeping gene. The ΔΔCt value was calculated as the ΔCt test sample -ΔCt calibrator sample, where the mean of the control sample ΔCt values was used for the calibrator. Fold changes were calculated as 2−ΔΔCt. Statistical significance was determined by t-test (p<0.05).
2.9 Identification of Cell Processes and Pathway construction
Gene Refseq accession numbers were imported into Ingenuity Pathway Analysis (IPA) software (Ingenuity® System, Redwood City, CA, http://www.ingenuity.com) web based application. This system queries the ingenuity pathway knowledge base (IPKB) for genetic interaction. The knowledge base is derived from the scientific literature and each connection in the network is supported by previous publications. Genes which can be mapped to genetic networks are termed as “focus genes”, and are used to build networks and a score for each network is calculated fit to the user’s set of genes, which is displayed as negative log of the p-value, is an indication of the chances of the focus genes in a network being found together in random. In the current study, a score of 10 or higher was used to select highly significant biological networks. By using comparison analysis, we have also investigated the major molecular and cellular functions, physiological system developmental functions, and disease and disorder development by these two PCBs.
3. Results
3.1 Global view of changes in gene expression by PCB 153 and PCB 138
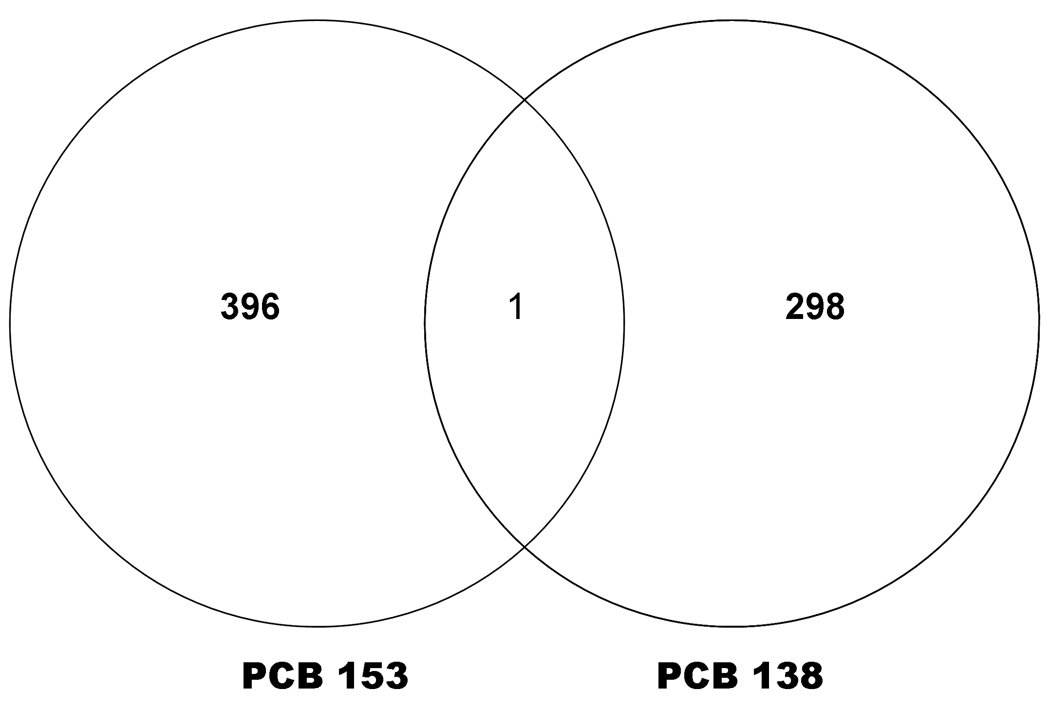
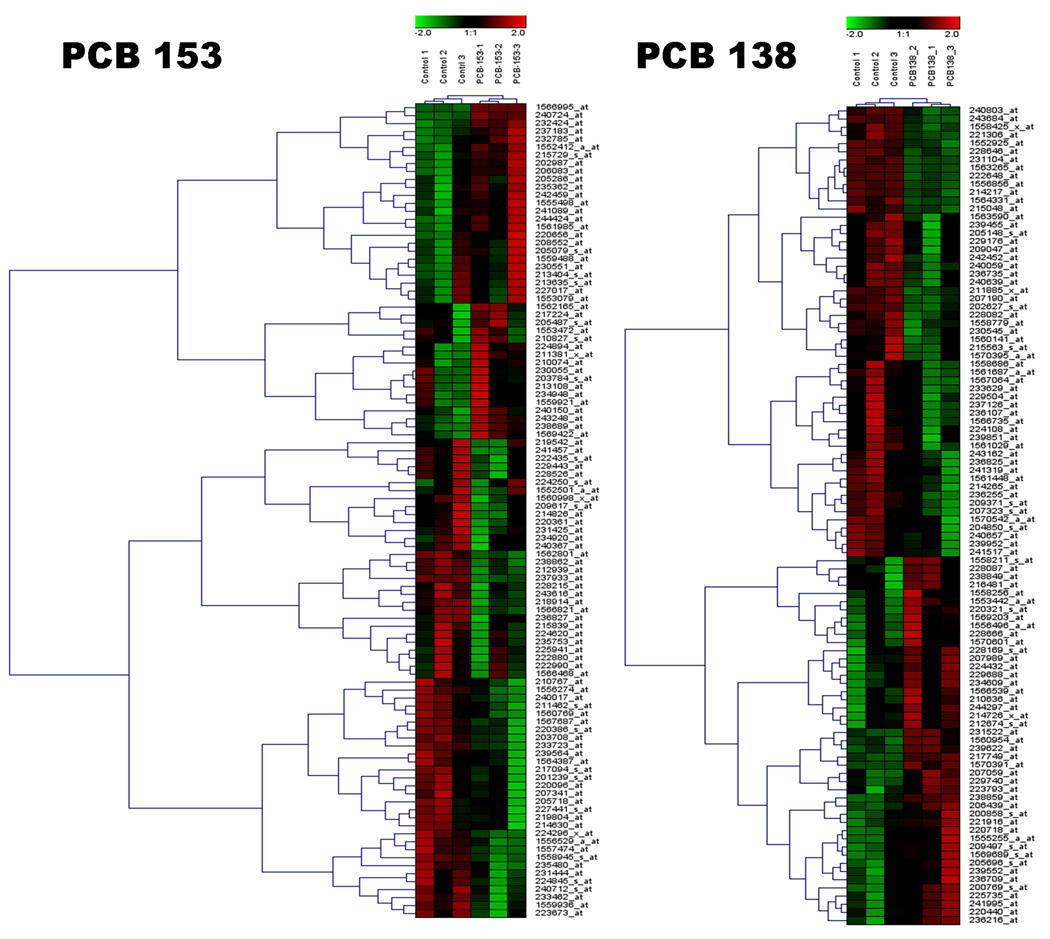
Figure 1 depicts numbers and overlap in genes differentially expressed (≥1.0 fold t-test, p<0.01) in human PBMC following exposure of PCB 153 and PCB 138. Following PCB 153 exposure, 396 transcripts were differentially expressed when compared with control, where there was no exposure of PCBs. Of that, 95 transcripts were identified by IPA analysis. Out of which, 40% (38 transcripts) were up-regulated and 60% (57 transcripts) were down regulated by PCB 153 (Table 1). Genes those that were up-regulated (with ≥1.5 fold change, t-test, p<0.01, are RRAD (6.52), VGLL1 (4.13), RMST (3.28), NAARG1L (2.65), RORB (2.44), SLC27A5 (2.28), PRDM16 (1.91), TFAP2C (1.72), GRIK4(1.68), and VWA3B (1.58); and the down-regulated genes were MGP (−3.47), MAP3K71P1 (−2.79), C4BPB (−2.73), RFX4 (−2.53), CACNA1A (−2.37), MCF2L (−2.31), SH3TC2 (−2.04), PLCZ1 (−1.90), MAPK1 (−1.82) and IQCH (−1.75), which are mostly involved in Cellular Movement, hematological system development and function, Immune Cell Trafficking, Molecular Transport, Cancer, Cellular development, Cell death, and Organ morphology (Table 3).
Figure 1. Venn diagram showing number of differentially induced by each PCB.
The similarities and differences in differential expression of genes that were expressed in human PBMC following exposure with PCB 153 and PCB 138. Numbers in non overlapping sections represent genes unique to that PCB, while numbers in overlapping area represents genes shared by both PCBs under this particular experimental condition.
Table 1.
Annotated gene transcripts differentially expressed ≥1.0 in human PBMC (t-test, p <0.01) following exposures to PCB 153. Table displays the genes that are arranged and categorized according to their Functions.
| Functions | Symbol | GenBank Accession |
Fold change | Description |
|---|---|---|---|---|
|
Molecular and Cellular Functions |
||||
| Amino Acid Metabolism | DUSP5 | NM_004419 | −1.08319 | dual specificity phosphatase 5 |
| PPP6C | NM_001123355 | −1.06838 | protein phosphatase 6, catalytic subunit |
|
| MAPK6 | NM_002748 | −1.08761 | mitogen-activated protein kinase 6 |
|
| CAMK2A | NM_015981 | 1.15705 | calcium/calmodulin- dependent |
|
| protein kinase (CaM kinase) II alpha |
||||
| Cellular Development | NRAS | NM_002524 | −1.11257 | neuroblastoma RAS viral (v- ras) oncogene homolog |
| YAP1 | NM_006106 | 1.2439 | Yes-associated protein 1, 65kDa |
|
|
Cellular Growth and Proliferation |
RRAD | BC057815 | 6.5227 | Ras-related associated with diabetes |
| SPHK1 | AK095578 | 1.03336 | sphingosine kinase 1 | |
| NRAS | NM_002524 | −1.11257 | neuroblastoma RAS viral (v- ras) oncogene homolog |
|
| CADM1 | BX641042 | 1.12279 | cell adhesion molecule 1 | |
| BAG1 | NM_004323 | 1.02958 | BCL2-associated athanogene | |
| RNA Damage and Repair | XRN2 | BC142960 | −1.14137 | 5'-3' exoribonuclease 2 |
| Small Molecule Biochemistry | DUSP5 | NM_004419 | −1.08319 | dual specificity phosphatase 5 |
| SLC23A2 | NM_203327 | 1.08323 | solute carrier family 23 (nucleobase transporters), member 2 |
|
| PPP6C | NM_001123355 | −1.06838 | protein phosphatase 6, catalytic subunit |
|
| PROKR2 | AK289995 | 1.37636 | prokineticin receptor 2 | |
| MAPK6 | NM_002748 | −1.08761 | mitogen-activated protein kinase 6 |
|
| SPHK1 | AK095578 | 1.03336 | sphingosine kinase 1 | |
| RAMP2 | BX420269 | −1.41288 | receptor (G protein-coupled) activity modifying protein 2 |
|
| CAMK2A | NM_015981 | 1.15705 | calcium/calmodulin- protein kinase (CaM kinase) II alpha dependent |
|
|
Physiological Development and Function System |
||||
|
Connective Tissue Development and Function |
TMEM123 | NM_052932 | 1.05788 | transmembrane protein 123 |
| NRAS | NM_002524 | −1.11257 | neuroblastoma RAS viral (v- ras) oncogene homolog |
|
| MAPK1 | AL157438 | −1.82015 | Mitogen-activated protein kinase 1 |
|
| Embryonic Development | TMEM123 | NM_052932 | 1.05788 | transmembrane protein 123 |
| Skeletal and Muscular | ||||
|
System Development and Function |
CACNB2 | NM_000724 | 1.27696 | calcium channel, voltage- dependent, beta 2 subunit |
| TTN | NM_133378 | −1.24884 | titin | |
| Diseases and Disorders | ||||
| Cancer | RRAD | BC057815 | 6.5227 | Ras-related associated with diabetes |
| TMEM123 | NM_052932 | 1.05788 | transmembrane protein 123 | |
| CCL27 | 1.16614 | chemokine (C-C motif) ligand 27 |
||
| WASF1 | NM_003931 | −1.15226 | WAS protein family, member 1 |
|
| CCR9 | AF145439 | 1.20783 | chemokine (C-C motif) receptor 9 |
|
| SPHK1 | AK095578 | 1.03336 | sphingosine kinase 1 | |
| PECAM1 | NM_000442 | 1.08198 | platelet/endothelial cell adhesion molecule (CD31 antigen) |
|
| IL1R1 | M27492 | −1.20114 | interleukin 1 receptor, type I | |
| NRAS | NM_002524 | −1.11257 | neuroblastoma RAS viral (v- ras) oncogene homolog |
|
| ELF3 | NM_001114309 | 1.12715 | E74-like factor 3 (ets domain transcription factor, epithelial- specific ) |
|
| ENPP2 | AK124910 | 1.07504 | ectonucleotide pyrophosphatase /phosphodiesterase 2 (autotaxin) |
|
| B3GNT6 | NM_138706 | −1.14404 | UDP-GlcNAc:betaGal beta- 1,3−N− Acetylglucosaminyl- transferase 6 (core 3 synthase) |
|
| CADM1 | BX641042 | 1.12279 | cell adhesion molecule 1 | |
| NF2 | NM_181832 | −1.12713 | neurofibromin 2 (merlin) | |
| RBL2 | BC034490 | −1.10821 | retinoblastoma-like 2 (p130) | |
| MAPK1 | AL157438 | −1.82015 | Mitogen-activated protein kinase 1 |
|
| NEK11 | AB071996 | −1.14261 | NIMA (never in mitosis gene a)- related kinase 11 |
|
| FGFR2 | NM_022970 | −1.4086 | fibroblast growth factor receptor 2 (bacteria-expressed kinase, keratinocyte gro |
|
| BAG1 | NM_004323 | 1.02958 | BCL2-associated athanogene | |
| NCAM1 | NM_181351 | −1.06549 | neural cell adhesion molecule 1 |
|
| GORASP2 | NM_015530 | −1.04165 | golgi reassembly stacking protein 2, 55kDa |
|
| Cardiovascular Disease | RNASE3 | DN998783 | 1.11144 | ribonuclease, RNase A family, 3 (eosinophil cationic protein) |
| BAG1 | NM_004323 | 1.02958 | BCL2-associated athanogene | |
| PECAM1 | NM_000442 | 1.08198 | platelet/endothelial cell adhesion molecule (CD31 antigen) |
|
| CALCA | X02330 | −1.17085 | calcitonin/calcitonin-related polypeptide, alpha |
|
| Neurological Disease | TOMM20 | NM_014765 | −1.07589 | translocase of outer mitochondrial membrane 20 homolog (yeast) |
| CCT5 | NM_012073 | −1.11151 | chaperonin containing TCP1, subunit 5 (epsilon) |
|
| ARF6 | NM_001663 | 1.00747 | ADP-ribosylation factor 6 | |
| GRIK4 | AK292726 | 1.6805 | glutamate receptor, ionotropic, kainate 4 |
|
| MGP | CR623037 | −3.47431 | matrix Gla protein | |
| GABRA4 | NM_000809 | 1.12257 | gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
|
| CACNA1A | NM_001127222 | −2.37261 | Calcium channel, voltage- dependent, P/Q type, alpha 1A subunit |
|
| PDE4B | NM_001037341 | −1.09002 | phosphodiesterase 4B, cAMP- specific (phosphodiesterase E4 dunce homolog, Drosoph |
|
| MREG | NM_018000 | 1.0484 | melanoregulin | |
| SOCS5 | NM_144949 | −1.04859 | suppressor of cytokine signaling 5 |
|
| EPHB2 | NM_004442 | 1.10964 | EPH receptor B2 | |
| CALCA | X02330 | −1.17085 | calcitonin/calcitonin-related polypeptide, alpha |
|
| DUSP5 | NM_004419 | −1.08319 | dual specificity phosphatase 5 | |
| ROBO3 | AY509035 | 1.0811 | roundabout, axon guidance receptor, homolog 3 (Drosophila) |
|
| RFX4 | AB095366 | −2.53027 | regulatory factor X, 4 (influences HLA class II expression) |
|
| CTNND2 | NM_001332 | −1.29555 | catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-r |
|
| MICALL1 | AB051455 | −1.12507 | MICAL-like 1 | |
| RTN4R | AK054602 | −1.05855 | reticulon 4 receptor | |
| SH3TC2 | NM_024577 | −2.04262 | SH3 domain and tetratricopeptide repeats 2 |
|
| RNASET2 | AK001769 | −1.22189 | ribonuclease T2 | |
| DUSP6 | 1.06845 | dual specificity phosphatase 6 | ||
| ACTB | AK125561 | −1.01689 | actin, beta | |
| TRAF3IP2 | AF136407 | 1.1812 | TRAF3 interacting protein 2 | |
| ABLIM1 | −1.12875 | actin binding LIM protein 1 | ||
| TTPA | BC041784 | −1.6207 | tocopherol (alpha) transfer protein |
|
| NF2 | NM_181832 | −1.12713 | neurofibromin 2 (merlin) | |
| ENPP2 | AK124910 | 1.07504 | ectonucleotide pyrophosphatase/phosphodi esterase 2 (autotaxin) |
|
| ITPR1 | NM_001099952 | −1.08908 | inositol 1,4,5-triphosphate receptor, type 1 |
|
| MPDZ | AB210041 | 1.53711 | multiple PDZ domain protein | |
| NCAM1 | NM_181351 | −1.06549 | neural cell adhesion molecule 1 |
|
| CORO1B | CR604899 | 1.03295 | coronin, actin binding protein, 1B |
|
| SPTLC1 | AB209757 | −1.16903 | serine palmitoyltransferase, long chain base subunit 1 |
|
| MTMR2 | AB028996 | −1.10054 | myotubularin related protein 2 |
|
| RPL31 | CR595074 | −1.01578 | ribosomal protein L31 | |
| NRAS | NM_002524 | −1.11257 | neuroblastoma RAS viral (v- ras) oncogene homolog |
|
| CADM1 | BX641042 | 1.12279 | cell adhesion molecule 1 | |
| Skeletal and Muscular Disorders | CALCA | X02330 | −1.17085 | calcitonin/calcitonin-related polypeptide, alpha |
| NEK11 | AB071996 | −1.14261 | NIMA (never in mitosis gene a)- related kinase 11 |
|
| FGFR2 | NM_022970 | −1.4086 | fibroblast growth factor receptor 2 (bacteria-expressed kinase, keratinocyte gro |
|
| BAG1 | NM_004323 | 1.02958 | BCL2-associated athanogene | |
| COL6A1 | NM_001848 | −1.57338 | collagen, type VI, alpha 1 | |
| RBL2 | BC034490 | −1.10821 | retinoblastoma-like 2 (p130) |
Table 3.
High-scoring networks (Score >10) identified by Ingenuity® Pathway Analysis in PCB 153 and PCB 138 exposure to Human PBMC. Top three (3) networks (out of 5) are represented here.
| Network ID | Network | Association | Score | Focus Genes |
Functions |
|---|---|---|---|---|---|
| 1. |
RPL23A (includes EG:6147) SPAG11B, TBX19, TCR, NCAM1, NFATC1, PDE4B, Pkc(s), PRTN3, RAB11FIP1RAF1, Ras, DUSP6, F2RL1, FGF9, FN1, HGF, IFNG, IL2, IL5, ITGA4, ITGB7, MAPK1, MIR122, AIF1, AKT3, Ap1, CALCA, CCL5, CD44, CTNNB1, CTNND2, CX3CR1, CXCL10, CXCR4 |
PCB 153 | 19 | 15 | Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking |
| 2. | LGALS3, LYN, NF2, P38 MAPK, PDGFRB, PLAUR, PLG, PTK2B, PXN, SPHK1, STAT3, TNF, EFNA3, ENPP2, EPHB2, FGFR2, IgG, IL2, IL8, IL13, IL1B, IL1R1, Interferon alpha, APP, ATP1B3, AZU1, C5AR1, CBL, CCL2, CCL5, CCL27, CDKN1A, CXCR4, DUSP5, TYK2 |
PCB 153 | 13 | 12 | Cellular Movement, Hematological System Development and Function, Immune Cell Trafficking |
| 3. |
ACTB, BAG1, FSH, ITPR1, MAPK6, PDXK, TLN1 |
PCB 153 | 10 | 6 | Cancer, Cellular Movement, Molecular Transport |
| 4. | NCF1C, NCLN, PDGFRB, PPARD, PRKCA, SEPP1SOCS1SRC, STAT1, STOML2,LDL, LGALS3, LGALS8, M6PR, MAPK3, MET, MMP2, MMP3, MMP1 (includes EG:4312), Akt, AKT2, Ap1, CD44, CHN2, CHRNA3, EGFR, EPO, ERBB2, Fibrinogen, IL13, IL18, ITGB1, |
PCB 138 | 12 | 11 | Cell Cycle, Cellular Movement, Cancer |
| 5. | Vegf, ZEB1, NR0B1, PTGER2, RAB1A, RAD17, SGK1, SNAP23, STX4, STX6, TGFB1, TGFB2, TNF, TP53, DYRK3, E2F1, EZR, FAS, FOS, FOXO3, FSH, hCG, IgG, ITGB5, NFkB (complex), ACTA1, ACTA2, ATM, CCND2, CD46, CDKN1A, CHEK2, CREM, CTGF, CYP19A1 |
PCB 138 | 12 | 11 | Cell Death, Hematological Disease, Immunological Disease |
| 6. | STAT1, TGFB1, VTN MMP1 (includes EG:4312), MYC, NFkB (complex), OPRM1, PDGF BB, SERPINE1, SOCS1, HLA-DRA, IFNG, IL2, IL4, IL8, IL18, IL1B, INS, LDL, MAPK1, MAPK3, MBP, Akt, APP, C5, CASP1, CLU,CORO1C, CRP,CSH1,CXCL2,EPOR, FAS, GHRL |
PCB 138 | 11 | 10 | Antigen Presentation, Cell mediated Immune Response, Hormonal Immune Response, |
Focus Genes are italicized
In PCB 138, 298 transcripts were differentially expressed when compared with control, where there was no exposure of PCBs. Of that, 260 transcripts were identified by IPA analysis, with 56% (142 transcripts) genes were up-regulated and 44% (118 transcripts) genes were down regulated by PCB 138 (Table 2). Genes that were up-regulated (those with≥1.5 fold change, t-test, p<0.01) were TRAP1 (20.1), VWDE (2.26), CNTN5 (2,22), GFRA1 (2.03), GABRB3 (1.97), DGUOK (1.87), CMKLR1 (1.83), KLHDC1 (1.72), UNQ1887 (1.67), and GHRL (1.62); while the down-regulated genes were IQGAP3 (−3.71), DCX (−3.15), ARMC9 (−2.99), AGBL3 (−2.81), ASAM (−2.26), FUT6 (−2.22), AQP1 (−2.19), SERPINE1 (−2.17), and SIAE (1.90). These were mostly involved in Cell Cycle, Cellular Movement, Cell Death, Cancer, Hematological system development and function, Neurological diseases, Tumor morphology, Genetic disorder, and immunological diseases (Table 3).
Table 2.
Annotated gene transcripts differentially expressed ≥1.0 in human PBMC (t-test, p <0.01) following exposures to PCB 38. Table displays the genes that are arranged and categorized according to their Functions.
| Functions | Symbol | GenBank Accession |
Fold change | Description |
|---|---|---|---|---|
|
Molecular and Cellular Functions |
||||
| Cell Cycle | CETN1 | NM_004066 | −1.31524 | centrin, EF-hand protein, 1 |
| L3MBTL | AW445040 | −1.11084 | l(3)mbt-like (Drosophila) | |
| CALCR | NM–001742 | −1.28497 | calcitonin receptor | |
| MLL | AW002079 | −1.08757 | Myeloid/lymphoid or mixed- lineage leukemia (trithorax homolog, Drosophila) |
|
| CHN2 | BC038570 | 1.181 | Chimerin (chimaerin) 2 | |
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| FOXO3 | AA018818 | 1.23273 | Forkhead box O3 | |
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
| CHEK2 | BC004207 | 1.09582 | CHK2 checkpoint homolog (S. pombe) |
|
| CAMK1 | NM_003656 | 1.12275 | calcium/calmodulin- dependent protein kinase I |
|
| Cell Death | CLU | M25915 | −1.22392 | clusterin |
| MLL | AW002079 | −1.08757 | Myeloid/lymphoid or mixed- lineage leukemia (trithorax homolog, Drosophila) |
|
| PPP1R1C | AI806944 | −1.37522 | protein phosphatase 1, regulatory (inhibitor) subunit 1C |
|
| CHEK2 | BC004207 | 1.09582 | CHK2 checkpoint homolog (S. pombe) |
|
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| DYRK3 | AF186773 | 1.04097 | dual-specificity tyrosine-(Y)- phosphorylation regulated kinase 3 |
|
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
| LGALS8 | AF342815 | 1.06 | lectin, galactoside-binding, soluble, 8 (galectin 8) |
|
| CDKL3 | AI199453 | 1.28712 | cyclin-dependent kinase-like 3 | |
| SBF1 | U93181 | 1.06106 | SET binding factor 1 | |
| VDR | AA454701 | 1.04744 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
|
| AKT2 | M77198 | 1.14184 | v-akt murine thymoma viral oncogene homolog 2 |
|
| Cellular Development | EPOR | X97671 | −1.13586 | erythropoietin receptor |
| FABP7 | NM_001446 | −1.22098 | fatty acid binding protein 7, brain |
|
| MLL | AW002079 | −1.08757 | Myeloid/lymphoid or mixed- lineage leukemia (trithorax homolog, Drosophila) |
|
| EZR | AA670344 | 1.07305 | ezrin | |
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| GFRA1 | NM_005264 | 2.03021 | GDNF family receptor alpha 1 | |
| SBF1 | U93181 | 1.06106 | SET binding factor 1 | |
| DYRK3 | AF186773 | 1.04097 | dual-specificity tyrosine-(Y)- phosphorylation regulated kinase 3 |
|
| VDR | AA454701 | 1.04744 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
|
| FOXO3 | AA018818 | 1.23273 | Forkhead box O3 | |
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
|
Cellular Growth and Proliferation |
EPOR | X97671 | −1.13586 | erythropoietin receptor |
| SERPINC1 | BC022309 | −1.22562 | serpin peptidase inhibitor, clade C (antithrombin), member 1 |
|
| CLU | M25915 | −1.22392 | clusterin | |
| GRM5 | D60132 | −1.46242 | Glutamate receptor, metabotropic 5 |
|
| L3MBTL | AW445040 | −1.11084 | l(3)mbt-like (Drosophila) | |
| NEIL1 | NM_024608 | −1.29585 | nei endonuclease VIII-like 1 (E. coli) |
|
| FABP7 | NM_001446 | −1.22098 | fatty acid binding protein 7, brain |
|
| CD46 | AV649018 | −1.34411 | CD46 molecule, complement regulatory protein |
|
| MBP | NM_002385 | −1.16473 | myelin basic protein | |
| ZEB | AI743662 | −1.29392 | Zinc finger E- box binding homeobox 1 |
|
| MLL | AW002079 | −1.08757 | Myeloid/lymphoid or mixed- lineage leukemia (trithorax homolog, Drosophila) |
|
| SERPINE1 | AL574210 | −2.17396 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type |
|
| PPARD | BC002715 | 1.38438 | peroxisome proliferator- activated receptor delta |
|
| VDR | AA454701 | 1.04744 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
|
| AKT2 | M77198 | 1.14184 | v-akt murine thymoma viral oncogene homolog 2 |
|
| OPRM1 | NM_000914 | 1.15179 | opioid receptor, mu 1 | |
| DYRK3 | AF186773 | 1.04097 | dual-specificity tyrosine-(Y)- phosphorylation regulated kinase 3 |
|
| ITGB5 | NM_002213 | 1.01718 | integrin, beta 5 | |
| GHRL | AI702963 | 1.62012 | Ghrelin/obestatin preprohormone |
|
| FOXO3 | AA018818 | 1.23273 | Forkhead box O3 | |
| CHN2 | BC038570 | 1.181 | Chimerin (chimaerin) 2 | |
| CDK10 | AF153430 | 1.12072 | cyclin-dependent kinase 10 | |
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
| DKC1 | NM_001363 | 1.04563 | dyskeratosis congenita 1, dyskerin |
|
| ITGB8 | AW131039 | 1.28014 | integrin, beta 8 | |
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| GFRA1 | NM_005264 | 2.03021 | GDNF family receptor alpha 1 | |
| Cellular Movement | CCL25 | NM_005624 | −1.17796 | chemokine (C-C motif) ligand 25 |
| CLU | M25915 | −1.22392 | clusterin | |
| FABP7 | NM_001446 | −1.22098 | fatty acid binding protein 7, brain |
|
| SERPINE1 | AL574210 | −2.17396 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type |
|
| SLC22A16 | AL050350 | −1.66153 | solute carrier family 22 (organic cation transporter), member 16 |
|
| EPOR | X97671 | −1.13586 | erythropoietin receptor | |
| NES | AW028075 | −1.35515 | nestin | |
| MLL | AW002079 | −1.08757 | Myeloid/lymphoid or mixed- lineage leukemia (trithorax homolog, Drosophila) |
|
| SERPINC1 | BC022309 | −1.22562 | serpin peptidase inhibitor, clade C (antithrombin), member 1 |
|
| AKT2 | M77198 | 1.14184 | v-akt murine thymoma viral oncogene homolog 2 |
|
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
| EZR | AA670344 | 1.07305 | ezrin | |
| OPRM1 | NM_000914 | 1.15179 | opioid receptor, mu 1 | |
| GHRL | AI702963 | 1.62012 | Ghrelin/obestatin preprohormone |
|
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| ITGB8 | AW131039 | 1.28014 | integrin, beta 8 | |
| FOXO3 | AA018818 | 1.23273 | Forkhead box O3 | |
| CXCL2 ligand 2 |
BC005276 | 1.19235 | chemokine (C-X-C motif) | |
| ITGB5 | NM_002213 | 1.01718 | integrin, beta 5 | |
|
Physiological System Development and Function |
||||
|
Hair and Skin Development and Function |
CALCR | NM_001742 | −1.28497 | calcitonin receptor |
| CLU | M25915 | −1.22392 | clusterin | |
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| CHEK2 | BC004207 | 1.09582 | CHK2 checkpoint homolog (S. pombe) |
|
| AKT2 | M77198 | 1.14184 | v-akt murine thymoma viral oncogene homolog 2 |
|
| PPARD | BC002715 | 1.38438 | peroxisome proliferator- activated receptor delta |
|
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
|
Hematological System Development and Function |
CLU | M25915 | −1.22392 | clusterin |
| EPOR | X97671 | −1.13586 | erythropoietin receptor | |
| CD46 | AV649018 | −1.34411 | CD46 molecule, complement regulatory protein |
|
| SERPINC1 | BC022309 | −1.22562 | serpin peptidase inhibitor, clade C (antithrombin), member 1 |
|
| MLL | AW002079 | −1.08757 | Myeloid/lymphoid or mixed- lineage leukemia (trithorax homolog, Drosophila) |
|
| DYRK3 | AF186773 | 1.04097 | dual-specificity tyrosine-(Y)- phosphorylation regulated kinase 3 |
|
| FOXO3 | AA018818 | 1.23273 | Forkhead box O3 | |
| GFRA1 | NM_005264 | 2.03021 | GDNF family receptor alpha 1 | |
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
| Hematopoiesis | DYRK3 | AF186773 | 1.04097 | dual-specificity tyrosine-(Y)- phosphorylation regulated kinase 3 |
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
| FOXO3 | AA018818 | 1.23273 | Forkhead box O3 | |
|
Reproductive System Development and Function |
PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha |
| PPARD | BC002715 | 1.38438 | peroxisome proliferator- activated receptor delta |
|
| GHRL | AI702963 | 1.62012 | Ghrelin/obestatin preprohormone |
|
| Tissue Morphology | PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha |
| SERPINE1 | AL574210 | −2.17396 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type |
|
| Tumor Morphology | SERPINE1 | AL574210 | −2.17396 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type |
| CLU | M25915 | −1.22392 | clusterin | |
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| GFRA1 | NM_005264 | 2.03021 | GDNF family receptor alpha 1 | |
| AKT2 | M77198 | 1.14184 | v-akt murine thymoma viral oncogene homolog 2 |
|
| Diseases and Disorders | ||||
| Cancer | AQP1 | AL518391 | −2.19529 | aquaporin 1 (Colton blood group) |
| SERPINE1 | AL574210 | −2.17396 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type |
|
| SERPINC1 | BC022309 | −1.22562 | serpin peptidase inhibitor, clade C (antithrombin), member 1 |
|
| EPOR | X97671 | −1.13586 | erythropoietin receptor | |
| PRIM2 | AL121975 | −1.24788 | primase, DNA, polypeptide 2 (58kDa) |
|
| DCX | NM_000555 | −3.15137 | doublecortex; lissencephaly, X-linked (doublecortin) |
|
| L3MBTL | AW445040 | −1.11084 | l(3)mbt-like (Drosophila) | |
| NEIL1 | NM_024608 | −1.29585 | nei endonuclease VIII-like 1 (E. coli) |
|
| CD46 | AV649018 | −1.34411 | CD46 molecule, complement regulatory protein |
|
| MLL | AW002079 | −1.08757 | Myeloid/lymphoid or mixed- lineage leukemia (trithorax homolog, Drosophila) |
|
| CCL25 | NM_005624 | −1.17796 | chemokine (C-C motif) ligand 25 |
|
| CLU | M25915 | −1.22392 | clusterin | |
| FABP7 | NM_001446 | −1.22098 | fatty acid binding protein 7, brain |
|
| SGK2 | NM_016276 | −1.10332 | serum/glucocorticoid regulated kinase 2 |
|
| ZEB1 | AI743662 | −1.29392 | Zinc finger E-box binding homeobox 1 |
|
| NES | AW028075 | −1.35515 | nestin | |
| CEP70 | NM_024491 | −1.23922 | centrosomal protein 70kDa | |
| PPP1R1C | AI806944 | −1.37522 | protein phosphatase 1, regulatory (inhibitor) subunit 1C |
|
| CHRNA3 | BC000513 | −1.27941 | cholinergic receptor, nicotinic, alpha 3 |
|
| PPARD | BC002715 | 1.38438 | peroxisome proliferator- activated receptor delta |
|
| CHN2 | BC038570 | 1.181 | Chimerin (chimaerin) 2 | |
| ITGB5 | NM_002213 | 1.01718 | integrin, beta 5 | |
| DYRK3 | AF186773 | 1.04097 | dual-specificity tyrosine-(Y)- phosphorylation regulated kinase 3 |
|
| TRAP1 | AA720770 | 20.0997 protein 1 | TNF receptor-associated | |
| CHEK2 | BC004207 | 1.09582 | CHK2 checkpoint homolog (S. pombe) |
|
| OPRM1 | NM_000914 | 1.15179 | opioid receptor, mu 1 | |
| GHRL | AI702963 | 1.62012 | Ghrelin/obestatin preprohormone |
|
| AK2 | NM_013411 | 1.07258 | adenylate kinase 2 | |
| ITGB8 | AW131039 | 1.28014 | integrin, beta 8 | |
| SBF1 | U93181 | 1.06106 | SET binding factor 1 | |
| HLA-DRA | M60333 | 1.09279 | major histocompatibility complex, class II, DR alpha |
|
| CDKL3 | AI199453 | 1.28712 | cyclin-dependent kinase-like 3 | |
| CDK10 | AF153430 | 1.12072 | cyclin-dependent kinase 10 | |
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
| ACY1 | AA176362 | 1.52601 | Aminoacylase 1 | |
| DRD5 | NM_000798 | 1.08916 | dopamine receptor D5 | |
| CAMK1 | NM_003656 | 1.12275 | calcium/calmodulin- dependent protein kinase I |
|
| RNF139 | AF064801 | 1.10115 | ring finger protein 139 | |
| AKT2 | M77198 | 1.14184 | v-akt murine thymoma viral oncogene homolog 2 |
|
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| MAT2A | NM_005911 | 1.07712 | methionine adenosyltransferase II, alpha |
|
| EZR | AA670344 | 1.07305 | ezrin | |
| FOXO3 | AA018818 | 1.23273 | Forkhead box O3 | |
| GFRA1 | NM_005264 | 2.03021 | GDNF family receptor alpha 1 | |
| VDR | AA454701 | 1.04744 | vitamin D (1,25-dihydroxyvitamin D3) receptor |
|
| LGALS8 | AF342815 | 1.06 | lectin, galactoside-binding, soluble, 8 (galectin 8) |
|
|
Endocrine System Development and Function |
NR0B1 | NM_000475 | −1.77275 | nuclear receptor subfamily 0, group B, member 1 |
| CYP1A2 | AF182274 | −1.14416 | cytochrome P450, family 1, subfamily A, polypeptide 2 |
|
| ITGB5 | NM_002213 | 1.01718 | integrin, beta 5 | |
| Genetic Disorder | AQP1 | AL518391 | −2.19529 | aquaporin 1 (Colton blood group) |
| SERPINE1 | AL574210 | −2.17396 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type |
|
| ANKH | AI672354 | −1.34784 | ankylosis, progressive homolog (mouse) |
|
| CHRNA3 | BC000513 | −1.27941 | cholinergic receptor, nicotinic, alpha 3 |
|
| MLL | AW002079 | −1.08757 | Myeloid/lymphoid or mixed- lineage leukemia (trithorax homolog, Drosophila) |
|
| MBP | NM_002385 | −1.16473 | myelin basic protein | |
| SERPINC1 | BC022309 | −1.22562 | serpin peptidase inhibitor, clade C (antithrombin), member 1 |
|
| CLU | M25915 | −1.22392 | clusterin | |
| DCX | NM_000555 | −3.15137 | doublecortex; lissencephaly, X-linked (doublecortin) |
|
| FABP7 | NM_001446 | −1.22098 | fatty acid binding protein 7, Brain |
|
| LDB3 | AA211481 | −1.1146 | LIM domain binding 3 | |
| CD46 | AV649018 | −1.34411 | CD46 molecule, complement regulatory protein |
|
| PRIM2 | AL121975 | −1.24788 | primase, DNA, polypeptide 2 (58kDa) |
|
| SGK2 | NM_016276 | −1.10332 | serum/glucocorticoid regulated kinase 2 |
|
| CEP70 | NM_024491 | −1.23922 | centrosomal protein 70kDa | |
| CTNS | AK001327 | −1.31839 | cystinosis, nephropathic | |
| GRM5 | D60132 | −1.46242 | Glutamate receptor, metabotropic 5 |
|
| ZEB1 | AI743662 | −1.29392 | Zinc finger E-box binding homeobox 1 |
|
| RIMS1 | AF263310 | −1.18856 | regulating synaptic membrane exocytosis |
|
| EPOR | X97671 | −1.13586 | erythropoietin receptor | |
| SLC16A2 | NM_006517 | −1.22395 | solute carrier family 16, member 2 (monocarboxylic acid transporter 8) |
|
| SH3BP2 | AB000462 | −1.35407 | SH3-domain binding protein 2 |
|
| GLIS3 | AI277316 | −1.0471 | GLIS family zinc finger 3 | |
| CALCR | NM_001742 | −1.28497 | calcitonin receptor | |
| NR0B1 | NM_000475 | −1.77275 | nuclear receptor subfamily 0, group B, member 1 |
|
| NES | AW028075 | −1.35515 | nestin | |
| HSPA6 | NM_002155 | −1.08626 | heat shock 70kDa protein 6 (HSP70B’) |
|
| GFRA1 | NM_005264 | 2.03021 | GDNF family receptor alpha 1 | |
| CXCL2 | BC005276 | 1.19235 | chemokine (C-X-C motif) ligand 2 |
|
| ADD1 | AL556041 | 1.17831 | adducin 1 (alpha) | |
| ACY1 | AA176362 | 1.52601 | Aminoacylase 1 | |
| CHEK2 | BC004207 | 1.09582 | CHK2 checkpoint homolog (S. pombe) |
|
| ADAR | NM_001111 | 1.04582 | adenosine deaminase, RNA- specific |
|
| ITGB5 | NM_002213 | 1.01718 | integrin, beta 5 | |
| NEFL | BF055311 | 1.22744 | neurofilament, light polypeptide 68kDa |
|
| CDK10 | AF153430 | 1.12072 | cyclin-dependent kinase 10 | |
| PAX9 | NM_006194 | 1.58502 | paired box 9 | |
| ZNF81 | AI434443 | 1.29507 | Zinc finger protein 81 | |
| `RNF139 | AF064801 | 1.10115 | ring finger protein 139 | |
| HLA-DRA | M60333 | 1.09279 | major histocompatibility complex, class II, DR alpha |
|
| LGALS8 | AF342815 | 1.06 | lectin, galactoside-binding, soluble, 8 (galectin 8) |
|
| GHRL | AI702963 | 1.62012 | Ghrelin/obestatin preprohormone |
|
| DRD5 | NM_000798 | 1.08916 | dopamine receptor D5 | |
| UNC93B1 | NM_030930 | 1.0883 | unc-93 homolog B1 (C. elegans) |
|
| SEPP1 | AV653290 | 1.23209 | Selenoprotein P, plasma, 1 | |
| OPRM1 | NM_000914 | 1.15179 | opioid receptor, mu 1 | |
| DKC1 | NM_001363 | 1.04563 | dyskeratosis congenita 1, dyskerin |
|
| CHN2 | BC038570 | 1.181 | Chimerin (chimaerin) 2 | |
| VDR | AA454701 | 1.04744 | vitamin D (1,25-dihydroxyvitamin D3) receptor |
|
| PPARD | BC002715 | 1.38438 | peroxisome proliferator- activated receptor delta |
|
| AKT2 | M77198 | 1.14184 | v-akt murine thymoma viral oncogene homolog 2 |
|
| ITGB8 | AW131039 | 1.28014 | integrin, beta 8 | |
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
| TRAP1 | AA720770 | 20.0997 | TNF receptor-associated protein 1 |
|
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| Hematological Disease | EPOR | X97671 | −1.13586 | erythropoietin receptor |
| MLL | AW002079 | −1.08757 | Myeloid/lymphoid or mixed- lineage leukemia (trithorax homolog, Drosophila) |
|
| CHRNA3 | BC000513 | −1.27941 | cholinergic receptor, nicotinic, alpha 3 |
|
| SERPINC1 | BC022309 | −1.22562 | serpin peptidase inhibitor, clade C (antithrombin), |
|
| SERPINE1 | AL574210 | −2.17396 | member 1 serpin peptidsase inhibitor, clade E (nexin, plasminogen activator inhibitor type |
|
| PRIM2 | AL121975 | −1.24788 | primase, DNA, polypeptide 2 (58kDa) |
|
| CD46 | AV649018 | −1.34411 | CD46 molecule, complement regulatory protein |
|
| CALCR | NM_001742 | −1.28497 | calcitonin receptor | |
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| OPRM1 | NM_000914 | 1.15179 | opioid receptor, mu 1 | |
| VDR | AA454701 | 1.04744 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
|
| CHEK2 | BC004207 | 1.09582 | CHK2 checkpoint homolog (S. pombe) |
|
| CDKL3 | AI199453 | 1.28712 | cyclin-dependent kinase-like 3 | |
| CDK10 | AF153430 | 1.12072 | cyclin-dependent kinase 10 | |
| GFRA1 | NM_005264 | 2.03021 | GDNF family receptor alpha 1 | |
| EXOC4 | AI964022 | 1.12501 | exocyst complex component 4 | |
| AKT2 | M77198 | 1.14184 | v-akt murine thymoma viral oncogene homolog 2 |
|
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
| ITGB5 | NM_002213 | 1.01718 | integrin, beta 5 | |
| Reproductive System | ||||
| Disease | SERPINC1 | BC022309 | −1.22562 | serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| CLU | M25915 | −1.22392 | clusterin | |
| CD46 | AV649018 | −1.34411 | CD46 molecule, complement regulatory protein |
|
| EPOR | X97671 | −1.13586 | erythropoietin receptor | |
| NES | AW028075 | −1.35515 | nestin | |
| ZEB1 | AI743662 | −1.29392 | Zinc finger E-box binding homeobox 1 |
|
| FABP7 | NM_001446 | −1.22098 | fatty acid binding protein 7, brain |
|
| PPP1R1C | AI806944 | −1.37522 | protein phosphatase 1, regulatory (inhibitor) subunit 1C |
|
| SERPINE1 | AL574210 | −2.17396 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type |
|
| PRKCA | NM_002737 | 1.06631 | protein kinase C, alpha | |
| ITGB5 | NM_002213 | 1.01718 | integrin, beta 5 | |
| CHN2 | BC038570 | 1.181 | Chimerin (chimaerin) 2 | |
| DRD5 | NM_000798 | 1.08916 | dopamine receptor D5 | |
| GHRL | AI702963 | 1.62012 | Ghrelin/obestatin preprohormone |
|
| SBF1 | U93181 | 1.06106 | SET binding factor 1 | |
| CAMK1 | NM_003656 | 1.12275 | calcium/calmodulin- dependent protein kinase I |
|
| CHEK2 | BC004207 | 1.09582 | CHK2 checkpoint homolog (S. pombe) |
|
| PPARD | BC002715 | 1.38438 | peroxisome proliferator- activated receptor delta |
|
| EZR | AA670344 | 1.07305 | ezrin | |
| LGALS8 | AF342815 | 1.06 | lectin, galactoside-binding, soluble, 8 (galectin 8) |
|
| CDK10 | AF153430 | 1.12072 | cyclin-dependent kinase 10 | |
| DYRK3 | AF186773 | 1.04097 | dual-specificity tyrosine-(Y)- phosphorylation regulated kinase 3 |
|
| SRC | BG767702 | 1.48672 | v-src sarcoma (Schmidt- Ruppin A-2) viral oncogene homolog (avian) |
|
| AKT2 | M77198 | 1.14184 | v-akt murine thymoma viral oncogene homolog 2 |
|
| FOXO3 | AA018818 | 1.23273 | Forkhead box O3 | |
| OPRM1 | NM_000914 | 1.15179 | opioid receptor, mu 1 | |
| CDKL3 | AI199453 | 1.28712 | cyclin-dependent kinase-like 3 | |
| VDR | AA454701 | 1.04744 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
Out of those 95 and 260 differentially expressed gene transcripts by PCB 153 and PCB 138 respectively, only one gene (PRIM2, 215709_at) was found to be common, which is a DNA primase, p58 subunit. This gene plays a key role in both the initiation of DNA replication and synthesis of Okazaki fragments, and was down-regulated −1.18 and −1.28 in PCB 153 and PCB 138, respectively.
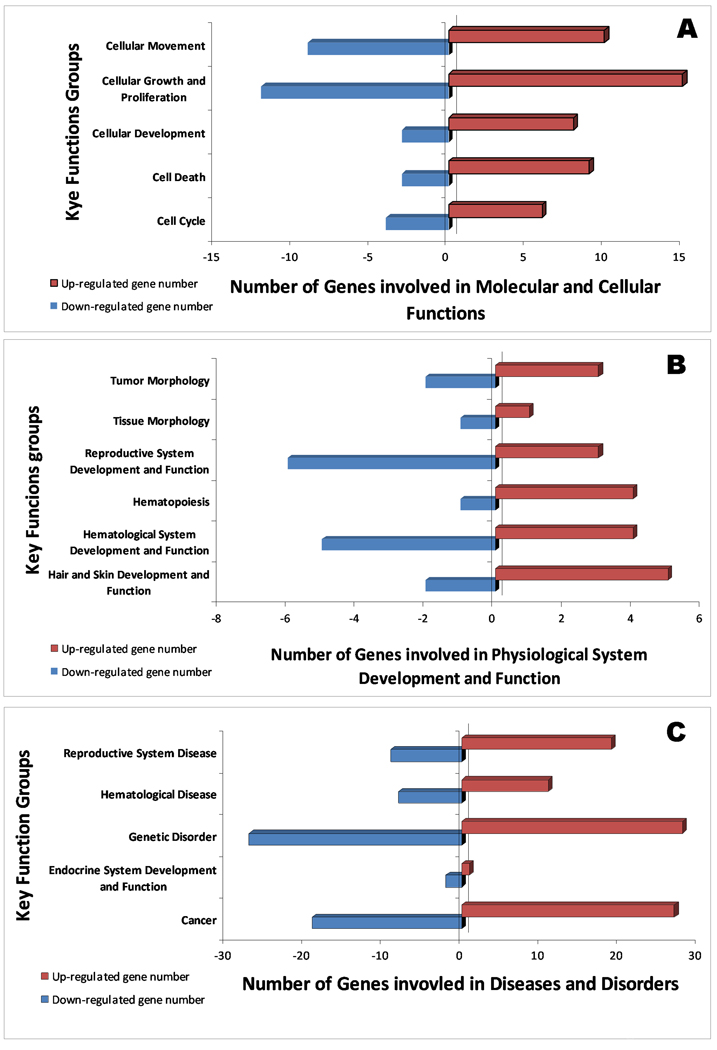
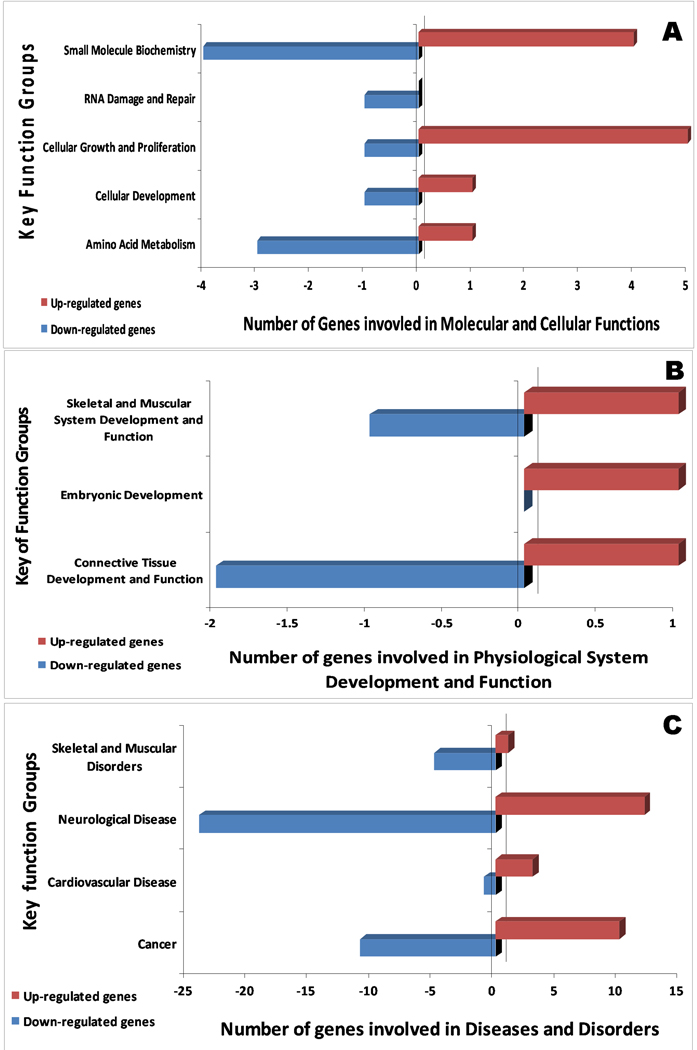
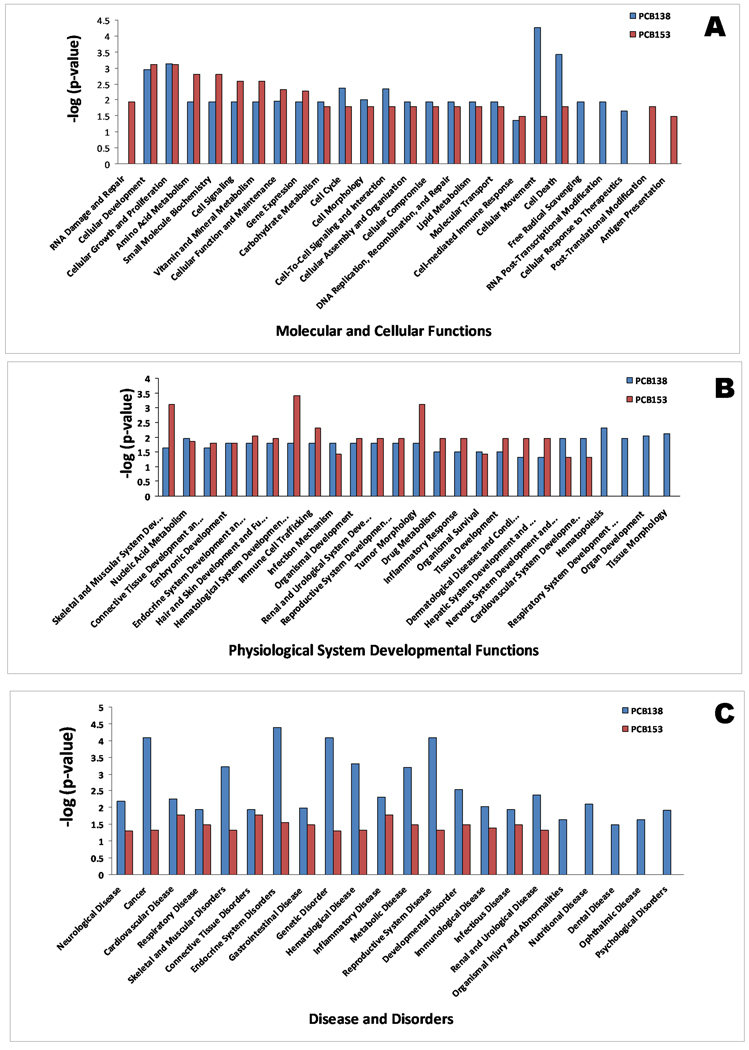
Figure 2 and 3 summarize the major functional categories and direction of changes of genes that were up/down-regulated in human PBMC following PCB 138 and 153 exposures, respectively. In PCB 138, cellular growth, cell death, and cellular movement were highly impacted, with most of the genes up-regulated in their molecular and cellular functions (Figure 2A). This has also been reflected by mostly up-regulated genes in reproductive disease, hematological disease, genetic disorder, and cancer (Figure 2 A–C). In contrast, a more down regulation trend was observed in PCB 153 (Figure 3A–C). Together, these data suggest that key functional groups are different in those two PCBs, which are playing the critical roles in developing diseases in respective PCBs. In the current work, the biological effects caused by PCB 153 and PCB 138 can be found in three levels; gene function level (Table 1 & 2), network level (Table 3), and integrated level (Figure 7).
Figure 2.
Key functional group and their number of genes involvement in PCB 138 exposures by IPA analysis in molecular and cellular functions (A), physiological system development and function (B), and disease and disorders (C), which are affected by PCB 138 exposure in human PBMC. Bar shows the number of genes in each ontological group up (right) or down (left) regulated.
Figure 3.
Key functional group and their number of genes involvement in PCB 153 exposures by IPA analysis. Depiction of the key functional groups in molecular and cellular functions (A), physiological system development and function (B), and disease and disorders (C) which are affected by PCB 153 exposure in human PBMC. Bar shows the number of genes in each ontological group up (right) or down (left) regulated.
Figure 7.
Key functional differences in developing toxicity by PCB 153 and 138 in human PBMC in molecular and cellular functions (A), physiological system development and function (B), and disease and disorders (C) in human PBMC cells following PCB 153 and PCB 138 exposures.
3.2 Identification of biological network functions by PCB 153 and PCB 138
We investigated the biological interaction among the genes associated with PCB 153 and PCB 138 exposures using Ingenuity Pathway Analysis tool. Analysis of top genes with greatest magnitude of differential expression associated with PCB 153 and PCB 138 exposure, with a p-value cut-off 0.001 and fold change ≥ 1.0 (up- or down-regulated) showed three significant networks each (score ≥ 10.0). Networks are listed in Table 3. While networks consider all possible interactions, canonical pathway analysis queries genes in pre-defined and well characterized biological pathways. It was clear from the analysis that two PCBs acted differently on the gene expression following the different networks and downstream pathway activation.
3.3 Biological networks induced by PCB 153 exposures
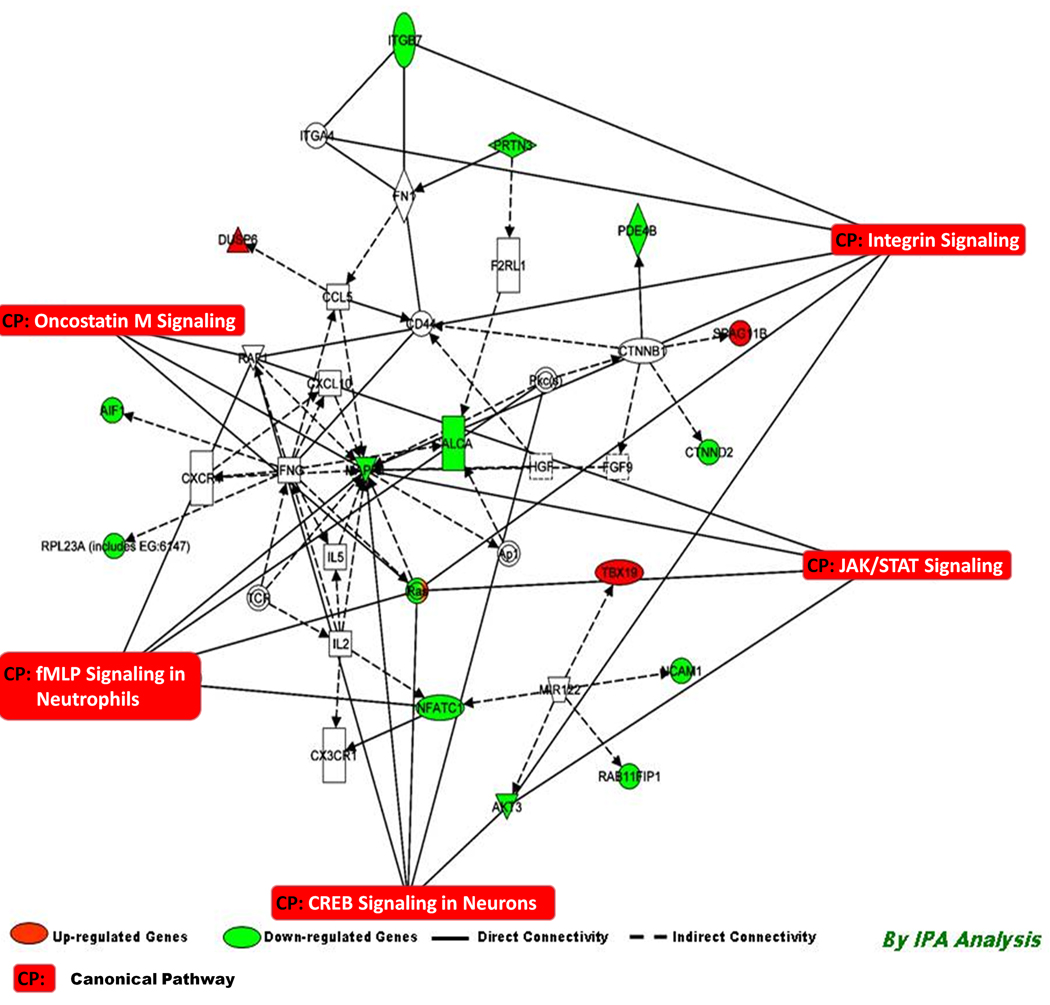
The top scoring networks (Network 1–2, Score = 19–13) identified for PCB 153 exposures include genes involved in cellular movement, hematological system development and function, as well as immune cell trafficking (Table 1). Network 3 (Score = 10, Table 3) associated with PCB 153 exposure includes genes involved in cancer, Cellular movement, and molecular transport. Details of focus genes are shown in Table 3. With the canonical pathway analysis, five top pathways were identified to be significantly associated with PCB 153 exposure, viz. JAK/Stat Signaling, Integrin signaling, fMLP signaling in neutrophils, Onctostatin M signaling, and CREB signaling in neurons (Figure 4).
Figure 4.
Connectivity of differentially expressed genes in the important signaling pathway following exposure of PCB 153 in human PBMC depicting the connectivity between genes expressed (≥1.0, t-test, p<0.001) and the important signaling pathways in human PBMC in vitro following exposure of PCB 153. Geometric figures in red denote up-regulated genes and those are green indicate down-regulation. The network shows genes mainly involved in cell cycle and cell. Solid interconnecting lines show the genes that are directly connected and the dotted lines signify the indirect connection between the genes and cellular functions. Canonical functions that are highly represented are shown within the box signaling (JAK/Stat, Integrin, Oncostatim M signaling, and fMLP signaling in Neutrophils). Genes in uncolored notes were not identified as differentially expressed in our experiment and were integrated into computational generated networks based on evidence stored in the IPA (Ingenuity Software) knowledge memory indicating relevance of this network.
3.4 Biological networks induced by PCB 138 exposures
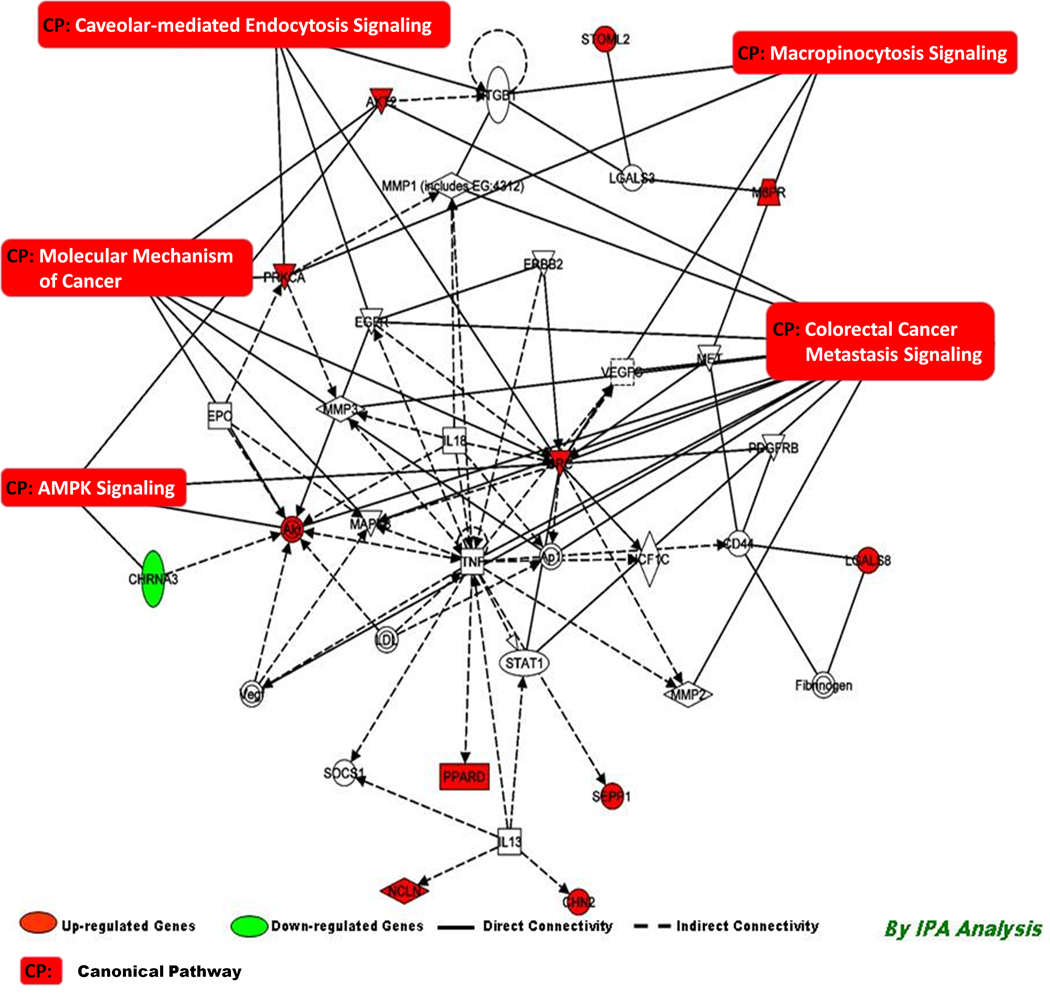
The top scoring networks (Network 4–5, Score = 12) associated with PCB 138 exposures include genes involved in cancer, cell cycle, cellular movement, cell death, hematological disease, and immunological disease (Table 3). Network 6 (Score = 11, Table 3) associated with PCB 138 exposure includes genes involved in Antigen presentation, and cell mediated immune response. Details of focus genes are shown in Table 3. With the canonical pathway analysis, five top pathways were significantly associated with PCB 138 exposure. These were Molecular mechanism of cancer, Colorectal cancer metastasis signaling, AMPL signaling, Caveolar-mediated endocystosis signaling, and Macropinocytosis signaling (Figure 5).
Figure 5.
Connectivity of differentially expressed genes in the important signaling pathway following exposure of PCB 138 in human PBMC depicting the connectivity between genes expressed (≥1.0, t-test, p<0.001) and the important signaling pathways in human PBMC in vitro following exposure of PCB 138. Geometric figures in red denote up-regulated genes and those are green indicate down-regulation. The network shows genes mainly involved in cell cycle and cell signaling. Solid interconnecting lines shows the genes that are directly connected and the dotted lines signify indirect connection between the genes and cellular functions. Canonical functions that are highly represented are shown within the box (AMPK Signaling, Molecular mechanism of Cancer, Caveloar-mediated Endocytosis, Macropinocytosis, and Colorectal cancer metastasis signaling). Genes in uncolored notes were not identified as differentially expressed in our experiment and were integrated into computational generated networks based on evidence stored in the IPA (Ingenuity Software) knowledge memory indicating relevance of this network.
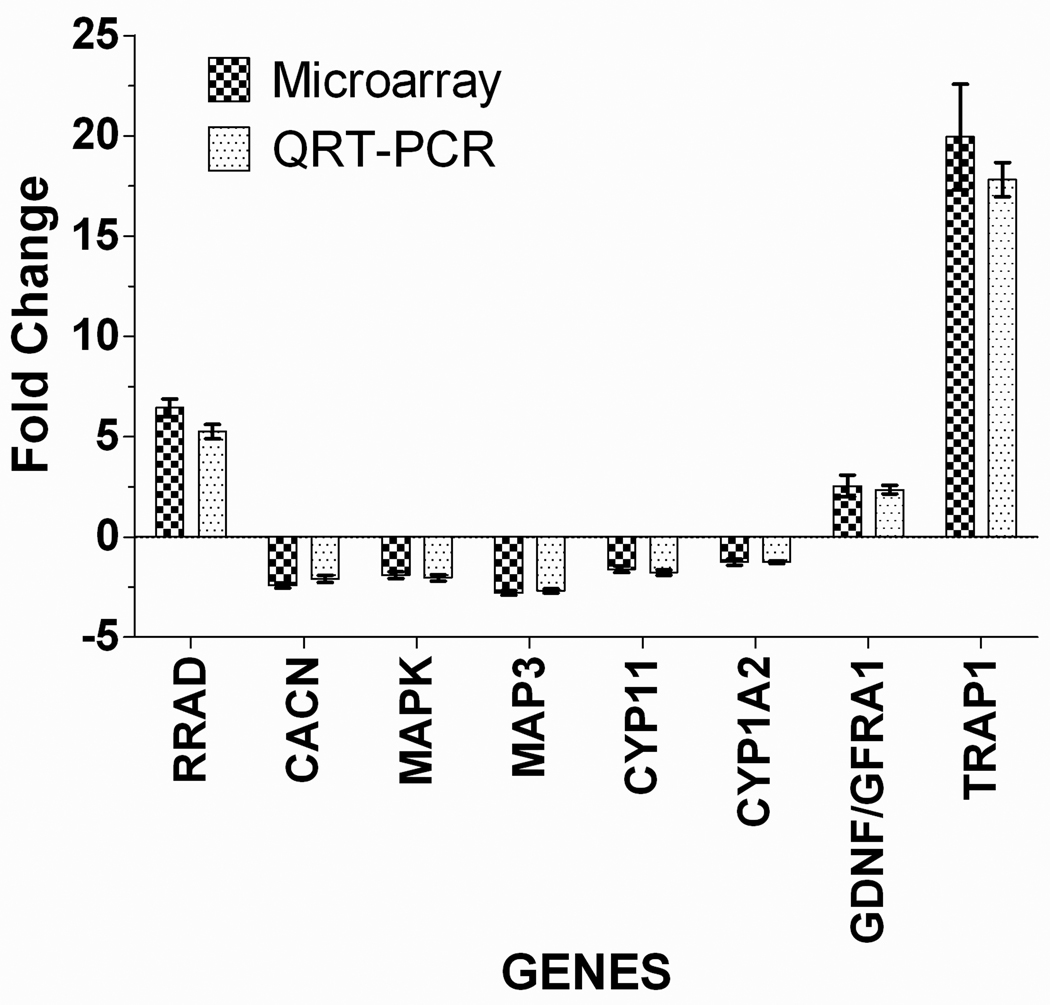
3.5 RT-PCR confirmation of selected genes from Arrays
Gene expression level of selected genes for validation of array data was determined by using quantitative Real Time Polymerase Chain Reaction (qRT-PCR) (Figure 6). Genes selected that were significantly changed (1.5 fold), fell within two experimental exposure conditions and/or were known to be impacted by PCB exposures. Ras-related associated with diabetes (RRAD) and TRP (Transient receptor potential) channel family −1 (TRAP1), a mitochondrial heat shock protein (HSP), were the genes found in the study that significantly changed (up-regulated) following PCB 153 (6.52 fold) and PCB 138 (20.09 fold) exposure on human PBMC cells respectively. They are known to play an important role in developing disease and disorders.
Figure 6.
The Comparison of microarray and RT-PCR data in human PBMC for the select genes following PCB 153 and PCB 138 exposure. Data are expressed as fold change. Error bars represent standard errors of means (±SEM).
The mechanism of action suggested that TRAP1 acts as an antagonist of ROS and protects cells from GzmM-mediated apoptosis. GzmM cleaves TRAP1 and abolishes its antagonistic function to ROS, resulting in ROS accumulation. Silencing TRAP1 through RNA interference increases ROS accumulation, whereas TRAP1 over-expression attenuates ROS production. ROS accumulation is in accordance with the release of cytochrome c from mitochondria and enhances GzmM-mediated apoptosis (Hua et al., 2007).
4. Discussion
The present study demonstrates that gene expression in PBMC can be significantly modulated in vitro by exposure to PCBs. Our research is focused on the possible contribution of PCBs towards disease and disorder development through global gene expression studies. We used oligonucleotide microarray technology to resolve these issues. The hypothesis is based on several epidemiological findings that PCB causes a deficit in neurodevelopment, cognitive impairment and growth (Hertz-Picciotto et al., 2008; Kodavanti, 2005; Sonneborn et al., 2008; Takahashi et al., 2009). PCB 153 and 138 were chosen due to their relevance and relative abundance in the actual human epidemiological situation (Hertz-Picciotto et al., 2008). The scope of this research was also to examine a cellular (in vitro) model that can best mimic the human in vivo situation. This cellular model (PBMC in vitro) of gene expression pretense future applications in biomonitoring studies of environmental toxic exposures (PCBs here) for its susceptibility in developing disease and disorders in an epidemiological situation. Here we report the induction of differential gene expression in this model, where our approach was to identify the possible mode of action and risk towards disease and disorder development, based on Ingenuity Pathway Analysis.
In our previous findings, we have shown that several PCB induced altered gene expression in the human genome have been associated with specific diseases and CYP1A1 and MT1K are the congener specific biomarker genes that are responsible for liver diseases induced by PCBs (Dutta et al., 2008). The most common PCB congener, 2,2',4,4',5,5'-hexachlorobiphenyl (PCB 153), is considered a useful marker of body burden of PCB (Ayotte et al., 2003; Hauser et al., 2003a, b; Axmon et al., 2004; Rignell-Hydborn et al., 2004; Rignell-Hydborn et al., 2005), because it correlates very well with the overall total PCB concentration (Grimval et al., 1997; Glynn et al., 2003). Moreover, among 44 American Vietnam veterans, the PCB 153 concentration has been shown to be well correlated with the total PCB-derived TEQ (Gladen et al., 1999).
In the present study, our main target was to correlate the possible modes of action(s) towards its toxicity in developing disease and disorders through differential gene expression and pathway analysis. In our study, PBMC in vitro study platform was chosen, as it was being widely used by many researchers to determine the susceptibility of any drug and chemical toxicant, and their toxic mechanism induced by transcriptional expression that would reflect those produced in in vivo. (Japour et al., 1993; Devos et al., 2004; van Leeuwen et al., 2005; Bethell et al., 2007). Most of these methods have been developed to determine early biological effects due to exposure to xenobiotics. In general, these effects are detectable long before clinical symptoms appear (Bonassi and Au, 2002; van Delft et al., 1998). It would be of significant benefit to molecular epidemiology that would give (mechanism-based) information on several health effects simultaneously, and that would be more suitable for monitoring effects at low exposure levels. Our discussions here are mainly focused on the disease and disorder development by these two congeners, eliciting the underlying mechanism of action, the differences and similarities in between them, and the possibilities of future use of these data. This validates our experimental design and selection of PBMC in our study.
The microarray analysis data from this study clearly indicate that there was the difference in differential expression of genes by these two PCB congeners, with only one gene in common (Figure 1). When the data were compared by Ingenuity Pathway Analysis, it also reveals their probable mode of action in a different way, with a few similarities in disease and disorder development. However, their magnitudes were different (Figure 7).
In PCB 153, RRAD, RROB, SLC27A5, CACNA1A, LILRA5, MAP3K71P, CYP11B2 has shown notable expression among which up-regulation of RRAD, RROB, SLC27A5 is important in conjecture that RRAD over-expression is associated with insulin resistance in Type II (non-insulin-dependent) diabetes mellitus (Reynet and Khan, 1993). Rad (Ras associated with diabetes) GTPase is the prototypic member of a subfamily of Ras-related small G proteins. Rad is a low molecular weight GTPase that is over-expressed in skeletal muscle of some patients with type II diabetes mellitus and/or obesity. A review of literature suggest that RARD over-expression is associated with insulin resistance in Type II (non-insulin-dependent) diabetes mellitus (Reynet and Khan, 1993) and also over-expression of Rad inhibits glucose uptake in cultured muscle and fat cells (Moyers et al., 1996). An increased expression of this mitochondrial HSP could be part of a pro-survival signaling pathway aimed to evade toxic effects and protects the cells from oxidative stress and apoptosis (Gesuladi et al., 2007). Studies have indicated the protective properties of over-expression of the cytosolic inducible member of the HSP70 family, Hsp72, few studies have investigated the protective potential of Hsp75 against ischemic injury (Voloboueva et al., 2008). Over-expression of Rad in adipocytes and muscle cells in culture, results in diminished insulin-stimulated glucose uptake (Ilany et al., 2006). Rad is the prototypic member of a family of novel Ras-related GTPases that is normally expressed in heart, skeletal muscle, and lung and that has been shown to exhibit a novel form of bi-directional interaction with the nm23 metastasis suppressor. Rad may also act as an oncogenic protein in breast tissues and demonstrate a potential mechanism by which interaction between Rad and nm23 may regulate growth and tumorigenicity of breast cancer (Tseng et al., 2001). RROB, a RAR-related orphan receptor B may affect the regulation of estrogen target genes in uterus and adipose tissue of adult off springs. These effects may result from interactions with developmental processes, reproductive abnormalities, adult functions, or a combination of them (Hsu et al., 2007; Ceccatelli et al., 2006). SLC27A5 (FATP5) is a solute carrier family 27 (fatty acid transporter), member 5, is a liver-specific member of the FATP/SlC27 family, which has been shown to exhibit both fatty acid transport and bile acid-CoA ligase activity in vitro. The over-expression of FATP5 mediates the uptake of long-chain fatty acids (LCFAs) in cultured mammalian cells, which have a functional alteration in bile acid metabolism, lipid metabolism, and body weight regulation and is also linked to obesity (Ceccatelli et al., 2006; Hubbard et al., 2006). Results of the studies on prenatal PCB exposure in Eastern Slovakian population also indicate similar observation, such as a reduced birth weight in male infants, less postnatal growth, impaired development, impaired immune-response, and lower thyroid hormone level (Hertz-Picciotto et al., 2008; Park et al., Sonneborn et al., 2008). These also support our observation associated with the endocrine system disorder, genetic disorder, Immunological diseases, metabolic diseases, and cardiovascular diseases, the top five bio-functions. The metabolic impairments to PCB 153 exposure are demonstrated in the above diseased states.
In the pathway analysis, three IPA network associated with PCB 153 exposure includes genes involved in cellular movement, hematological system development and function, immune cell trafficking, molecular transport, and cancer (Table 3). A key network gene, MAPK1, is a member of the MAP kinase family. MAP kinases act as an integration point for multiple biochemical signals, and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation, and development, which can be activated by pro-inflammatory cytokines and environmental stress (Voong et al., 2008). Down-regulation of MAPK1 in our study could represent dysregulation of one of the networks that regulates cell growth through cell-cycle control, apoptosis, or can act as a potential tumor suppressor gene (Huang et al., 2008). However, although some recent studies have strongly indicated the incidences of cancers in PCBs–exposed population in Eastern Slovakia (Pavuk et al., 2004) as well as in Central Europe (Benko et al., 2009), but is however, controversial (Golden and Kimbrough, 2009). PCB experimental studies provide data that are used to regulate and control human exposure, although the epidemiologic evidence fails to establish PCBs as human carcinogens. Thus, what is used for population risk assessment may not be appropriate for individual-risk assessment or concluding that a causal relationship exists between PCB exposure and cancer risk (Shields, 2006). This is because the cancer incidences was significantly lower in the exposed areas than in the general population of Slovak and The Czech Republic, which are recognized as the most heavily PCB-contaminated sites in the world (Benko et al., 2009).
In PCB 138, TRAP1, CNTN5, GFRA1, VWDE, and CYP1A2 has shown notable expression, among which, up-regulation of TRAP1, CNTN5, GFRA1 are important in conjecture of Tumor necrosis factor-associated protein 1 (TRAP-1), identical to heat shock rprotein75 (HSP75), which is a member of the HSP family of molecular chaperones that interact with the retinoblastoma protein during mitosis and after heat shock (Chen et al., 1996). It is substantially homologous to members of the 90-kDa family of heat-shock proteins (HSP90), is an important molecular chaperone for proteins that are involved in numerous cellular processes. A number of cell signaling molecules, such as steroid hormone receptors and protein kinases, require HSP90 for maintenance in an active state within the cell (Masuda et al., 2004). The over-expression of TRAP1 may act as an antagonist of ROS and protects cells from GzmM-mediated apoptosis. GzmM cleaves TRAP1 and abolishes its antagonistic function to ROS, resulting in ROS accumulation. Silencing TRAP1 through RNA interference increases ROS accumulation, whereas TRAP1 over-expression attenuates ROS production. ROS accumulation is in accordance with the release of cytochrome c from mitochondria and enhances GzmM-mediated apoptosis. (Hua et al., 2007). The human NB-2 gene (CNTN5), is a glycosylphosphatidylinositol (GPI)-anchored neuronal membrane protein that functions as a cell adhesion molecule. It may play a role in the formation of axon connections in the developing nervous system, and may contribute to human neurological disorders (Kamei et al., 2000).
GFRA1 (GDNF receptor family A-1) are over-expressed in a subset of estrogen receptor–positive tumors. Germ line–activating oncogenic mutations in RET allow this receptor to signal independently of GFRA-1and its ligand glial cell–derived neurotrophic factor (GDNF) to promote a spectrum of endocrine neoplasias (Esseghir et al., 2007). To date, the majority of in vitro studies on wild-type RET/GFRa1 signaling have focused either on neuronal lines that have the endogenous expression of these receptors or on epithelial and fibroblast lines in which the receptors are expressed ectopically. Studies in these systems have corroborated the phenotype of RET-, GFRA1-, and GDNF-deficient mice in demonstrating a role for GDNF signaling in promoting cell proliferation, neuroprotection, and migration. Less well studied is the role of GDNF signaling in cancer, although it has been shown that GDNF can promote cell migration/chemotaxis and invasion of a RET+/GFRa1+ pancreatic cell line and that these activities are dependent on the activation of the MAPK and PI3K pathways (Viet et al., 2004). Our Ingenuity Pathway analysis towards key functional aspects in molecular and cellular functions, physiological system development and function due to PCB 138 exposures also substantiate the findings (Figure 7A–C). On the contrary, while fewer studies reported an association between PCB body burden and a higher incidence of breast cancer (Aronson et al., 2000), the majority of the epidemiological studies did not find an association between PCB exposure and an increased incidence of breast cancer (laden et al., 2002). Laboratory studies have demonstrated that PCBs may induce a broad range of estrogenic and anti-estrogenic or androgenic effects but a variety of factors, including the dose and duration of exposure, and more importantly, the degree of chlorination will influence the nature and severity of effect. It is unclear if other environmental factors have contributed to the lower incidence of breast or prostate cancer in the Michalovce district of Slovakia. The known anti-estrogenic properties of OH and methylsulfonyl PCB metabolites may help to explain the lower incidence of breast cancer in the Michalovce District as well as the observed lower incidence of bladder malignancies in Uherske Hradiste District (Benko et al., 2009).
Over-expression of glial cell line-derived neurotrophic factor (GDNF), the ligand for GFRA-1, shows an increase in the self-renewal of spermatogonial stem cells in the testes (Meng et al., 2000). Therefore, GFRA-1 is considered the most specific marker for spermatogonial stem cells (Lucas et al., 2009). In epidemiological setting, it has been reported on a study of two hundred adolescents in Flanders (Belgium), who had accidental moderate exposure to PCBs, had shown that pubertal development of boys and girls is delayed. In one such suburb near two waste incinerators, fewer boys reached the adult stages of genital development and pubic hair growth. In addition, in the same suburb, fewer girls had reached the adult stage of breast development. Left plus right testicular volume was lower in both polluted areas than in the control area but was not related to the current exposure of the adolescents to polychlorinated aromatic hydrocarbons (PCAHs). Through endocrine disruption, environmental exposure to PCAHs may interfere with sexual maturation and in the end adversely affect human reproduction (Den Hond et al., 2002). Our disease and disorder analysis study by IPA analysis also indicated such abnormalities (Endocrine System Disorder, Reproductive System Disorder) which were also reflected in the physiological system development and function (Figure 7C).
Cytochrome P-450 1A2 (CYP1A2) is an enzyme involved in the metabolic activation of some carcinogens and is believed to be induced by xenobiotics. This gene encodes a member of the cytochrome P450 superfamily of enzymes. The cytochrome P450 proteins are monooxygenases which catalyze many reactions involved in drug metabolism and synthesis of cholesterol, steroids and other lipids. The protein encoded by this gene localizes to the endoplasmic reticulum, and its expression is induced by some polycyclic aromatic hydrocarbons (PAHs). Many PAHs not only induce CYP1 family enzymes but are also bioactivated by these enzymes to a wide range of metabolites, some of which are highly carcinogenic. CYP1A2 is one of the major enzymes that bioactivate a number of procarcinogens and thus induction of CYP1A2 may increase the carcinogenicity of these compounds. They diminish the potency of the PAH-mediated up-regulation of CYP1 and, therefore, may decrease the carcinogenic risks associated with bioactivated PAHs (Vakharia et al., 2001). Very few studies, however, have investigated the association between environmental exposures and in vivo CYP1A2 activity in humans. In our study CYP1A2 is down-regulated by PCB 138 exposure in human PBMC, which fails to protect and may lead to organismal injury and abnormalities that have also been shown by Ingenuity analysis. In agreement with our results, it was also shown that maternal mouse hepatic CYP1A2, by sequestering dioxin and thus altering the pharmacokinetics, protects the embryos from toxicity and birth defects (Dragin et al., 2006). These results support the notion that CYP1A2 activity may be a marker of an early adverse biological effect of exposure to PCBs in humans (Fitzgerald et al., 2005).
In conjunction with the individual gene analysis described above, we used Ingenuity Pathway Analysis (IPA) software to further evaluate the microarray data from human PMBC following exposure to PCB 153 and 138. With Ingenuity, we identified several distinct different pathways, by which PCBs may exert its toxicity in relation to the development of different disease and disorder. Networks with gene/protein interaction in cellular movement and cell signaling were highly represented, with majority of genes down-regulated (Figure 4), with key signaling process in Jak/stat, Integrin, Oncoststin M signaling, CREB signaling in neurons. These data are in agreement with the majority which shows effects related to environmental neurotoxicity (Coasta, 1998), and related to cognitive functioning in adolescents (Newman et al., 2009). In PCB 138, network shows genes mainly involved in cell cycle, cell death, and caner, majority of genes up-regulated (Figure 5), with key canonical functions, Endocytosis signaling, Macropinocytosis signaling, AMPK signaling, Colorectal cancer metastasis signaling, and Molecular mechanism of cancer. Recent studies indicate that regulation of mGluR surface expression plays a critical role in neuronal function and is critical to neuronal signaling and activity-dependent synaptic plasticity. Caveolin-1, an adaptor protein that associates with lipid rafts and the main coat protein of caveolae, binds to and colocalizes with metabotropic glutamate receptors 1/5 (mGluR1/5). As the downstream effectors, it may contribute to regulation of synaptic efficacy. The impact of caveolin-1 on neuronal signaling may become prominent in pathological situations in which its expression is dysregulated, such as neuronal injury, ischemia, and Alzheimer’s disease (Francesconi et al., 2009). In our case, the PCB 138 exposure to human PBMC cells in vitro led to a constant elevation of AMPK activity, indicating an additional response in gene expression. 5'-AMP-activated protein kinase (AMPK) serves as an energy sensor and is at the center of control for a large number of metabolic reactions, thereby playing a crucial role in Type 2 diabetes and other human diseases. Environmental stress regulates the intracellular localization of AMPK, and upon recovery from heat shock or oxidant exposure, AMPK accumulates in the nuclei (Kodiha et al., 2007). Apoptosis is the best characterized form of programmed cell death. However, non-apoptotic forms of cell death are now recognized as playing significant roles during embryonic development, neurodegeneration, and cancer regression (Lockshin and Zakeri, 2004). Moreover, the majority of degenerating cells do not exhibit chromatin condensation typical of apoptosis. These observations provide evidence for a necrosis-like form of cell death initiated by dysregulation of Macropinocytosis. This raises the possibility that the explanation for the differential sensitivity to methuosis could reside not only at the level of induction of Macropinocytosis but also at the level of intracellular trafficking or membrane channel function (Overmeyer et al., 2008), where genes connected to this signaling pathway in our study are mostly up-regulated (Figure 5).
In the comparison of results through Gene expression and IPA analysis between PCB 153 and PCB 138, it is found that two molecules act differently in terms of differential gene expression, and towards disease and developmental processes in significantly different pathways. It substantiates the fact of congener specificity, and substitution pattern of chlorine atoms of these PCBs towards body burden and toxicity. Typically, in human studies, PCB congeners are grouped, either as total PCBs or the sum of several commonly found congeners, or are indexed by a single congener, usually PCB 153 (Longnecker et al., 2003). However, studies using a summative PCB measure do not reveal which types of congeners are influential, and what features of these congeners are linked to affect (Newman et al., 2009). Despite similarities in molecular and cellular functions between these two congeners, marked differences were observed in cellular movement, cell death, by these two PCB congeners. Among distinct differences, the free radical scavenging, RNA post-translational modification, and cellular response to therapeutics were observed in PCB 153 exposure, and RNA damage and repair, and antigen presentation was observed in PCB 138 exposures only (Figure 7A). In the physiological system and developmental function, the marked differences were observed in hematopeiesis, respiratory system development and function, organ development, and tissue morphology, which were only triggered by PCB 138 (Figure 7B). While investigating the disease and disorder development through IPA, most prominent and interesting disease and disorder were Neurological disease, Cancer, Cardiovascular disease, respiratory disease, endocrine system disorders, genetic disorders, and reproductive system disease, which has strong resemblances with in vitro, in vivo, and in epidemiological situations. A distinct difference was observed in renal and urological diseases, organisimal injury and abnormalities, dental disease, ophthalmic disease, and psychological disorders, which are only revealed by PCB 138 exposure, but not by PCB 153. More importantly, the intensities of disease and disorders are higher in all cases for PCB 138 (Figure 7C), showing its higher potencies in toxic effect.
5. Conclusions
The present study demonstrates that gene expression in PBMC can be significantly modulated in vitro by exposure to genotoxic chemicals. By exposing human PBMC, we identified genes that were differentially expressed for each agent individually (differentially expressed at the human equivalence level (see Figure 8). Thus, hypothetically monitoring the expression levels for these (groups of) genes in a biomonitoring study may discriminate persons exposed to the specific agents from those who are not exposed. The present study emphasizes the challenges of global gene expression in vitro and correlated with the results human exposed population. The microarray results from the present study give a molecular mechanistic insight, and functional effects, following PCB exposure. The extent of changes in genes related to several possible mode(s) of action highlights the changes in cellular functions and signaling pathways to play major roles. In addition to understanding the pathways related to mode of action for chemicals, these genomic data could lead to identification of genomic signatures that could be used for screening of chemicals for their potential to cause disease and disorder development.
Figure 8.
Heat maps of relative expression changes in human PBMC following PCB 153 and PCB 138 exposures showing relative expression changes in human PBMC following PCB 153 (top 102 genes out of 397, A) and PCB 138 (top 100 genes out of 297, B) exposures. Red denotes up-regulation, black no difference, and green down-regulation; where brighter green is increasingly repressed and red increasingly induced by PCBs. The hierarchical clustering showed that control (column 1–3) and the treated (column 4–6) are grouped together and was based on average linkage with a Pearson correlation.
Overall, our results do show promise that large scale evaluation of changes in gene expression using microarrays, combined with a cell-based assay system, may become a useful tool for toxicity evaluation. A recent work (Blomme et al., 2009) towards the use of toxicogenomics (or the use of gene expression profiling in toxicology) represents an attractive approach to predict toxicity and to gain a mechanistic understanding of toxic changes is also in accordance with our current work. Even with the limited compound set evaluated in this study, it is apparent that each compound may produce its own unique expression profile (signature gene expression), and that similarities in profiles between compounds may indicate similarities in toxic mechanism. However, a much larger database comprised of many (perhaps hundreds) of compounds (mixture of PCBs equivalent to an epidemiological situation) will be needed in order to produce accurate determinations of toxicity based on similarity of expression profiles. Though the effects of the congeners are not identical, the synergistic and/or antagonistic effects of individual gene/genes in the pathway cannot be ruled out in actual human exposure scenario.
Acknowledgements
These studies are supported by the 1UO1ES016127-01 grant from the National Institute of Environmental Health Sciences (NIEHS/NIH). Thanks are also due to General Clinical Research Center (GCRC), and Dr. Annapurni V. Jaym Trouth of Neuroscience Department at Howard University in supporting us towards blood collection from healthy donors as per approved HU IRB # IRB-07-GSAS-30. We are indebted to Prof. Prabir K. Chakraborty, Professor Emeritus of Uniformed Services University of the Health Sciences, Bethesda for his careful editing while revising the manuscript. Its contents are solely the responsibility of the authors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
There is no conflict of interest among the authors in the present work.
References
- Allen JR, Barsotti DA, Carstens LA. Residual effects of polychlorinated biphenyls on adult nonhuman primates and their offspring. J Toxicol Environ Health. 2002;6:55–66. doi: 10.1080/15287398009529830. [DOI] [PubMed] [Google Scholar]
- Aronson KJ, Miller AB, Woolcott CG, Sterns EE, McCready DR, Lickley LA, et al. Breast adipose tissue concentrations of polychlorinated biphenyls and other organochlorines and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2000;9:55–63. [PubMed] [Google Scholar]
- Axmon A, Rylander L, Stromberg U, Jönsson B, Nilsson-Ehle P, Hagmar L. Polychlorinated biphenyls in serum and time to pregnancy. Environ Res. 2004;96:186–195. doi: 10.1016/j.envres.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Ayotte P, Muckle G, Jacobson JL, Jacobson SW, Dewailly E. Inuit Cohort Study. Assessment of pre- and postnatal exposure to polychlorinated biphenyls: lessons from the Inuit Cohort Study. Environ Health Perspect. 2003;111:1253–1258. doi: 10.1289/ehp.6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencko V, Rames J, Ondrusova M, Plesko I, Jurickova L, Trnovec T. Human exposure to polyhalogenated hydrocarbons and incidence of selected malignancies -central European experience. Neoplasma. 2009;56:353–357. doi: 10.4149/neo_2009_04_353. [DOI] [PubMed] [Google Scholar]
- Bethell R, De Muys J, Lippens J, Richard A, Hamelin B, Ren C, Collins P. In vitro interactions between apricitabine and other deoxycytidine analogues. Antimicrob Agents Chemother. 2007;51:248–253. doi: 10.1128/AAC.01204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme EA, Yang Y, Waring JF. Use of toxicogenomics to understand mechanisms of drug-induced hepatotoxicity during drug discovery and development. Toxicol Lett. 2009;186:22–31. doi: 10.1016/j.toxlet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Bonassi S, Au WW. Biomarkers in molecular epidemiology studies for health risk prediction. Mutation Research. 2002;511:73–86. doi: 10.1016/s1383-5742(02)00003-0. [DOI] [PubMed] [Google Scholar]
- Brouwer A, Longnecker MP, Birnbaum LS, Cogliano J, Kostyniak P, Moore J, et al. Characterization of potential endocrine-related health effects at low-dose levels of exposure to PCBs. Environ Health Perspect. 1999;107:639–649. doi: 10.1289/ehp.99107s4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccatelli R, Faass O, Schlumpf M, Lichtensteiger W. Gene expression and estrogen sensitivity in rat uterus after developmental exposure to the polybrominated diphenylether PBDE 99 and PCB. Toxicology. 2006;220:104–116. doi: 10.1016/j.tox.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Chen CF, Chen Y, Dai K, Chen P-L, Riley DJ, Lee WH. A new member of the hsp90 family of molecular chaperones interacts with the retinoblastoma protein during mitosis and after heat shock. Mol Cell Biol. 1996;16:4691–4699. doi: 10.1128/mcb.16.9.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke PS, Robertson LW, Hansen LG. Disruption of steroid hormone signaling by PCBs. Lexington: The University Press of Kentucky; 2001. [Google Scholar]
- Costa LG. Neurotoxicity testing: a discussion of in vitro alternatives. Environ Health Perspect. 1998;106 Suppl 2:505–510. doi: 10.1289/ehp.98106505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- De S, Ghosh S, Chatterjee R, Chen YQ, Moses L, Kesari A, et al. PCB congener specific oxidative stress response by microarray analysis using human liver cell line. Environ Int. 2010;36:907–917. doi: 10.1016/j.envint.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hond E, Roels HA, Hoppenbrouwers K, Nawrot T, Thijs L, Vandermeulen C, et al. Sexual maturation in relation to polychlorinated aromatic hydrocarbons: Sharpe and Skakkebaek's hypothesis revisited. Environ Health Perspect. 2002;110:771–776. doi: 10.1289/ehp.02110771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos S, Van Den Heuvel R, Hooghe R, Hooghe-Peters EL. Limited effect of selected organic pollutants on cytokine production by peripheral blood leukocytes. Eur Cytokine Netw. 2004;15:145–151. [PubMed] [Google Scholar]
- Devos S, Van Den Heuvel R, Hooghe R, Hooghe-Peters EL. Limited effect of selected organic pollutants on cytokine production by peripheral blood leukocytes. Eur Cytokine Netw. 2004;5:145–151. [PubMed] [Google Scholar]
- Dragin N, Dalton TP, Miller ML, Shertzer HG, Nebert DW. For dioxin-induced birth defects, mouse or human CYP1A2 in maternal liver protects whereas mouse CYP1A1 and CYP1B1 are inconsequential. J Biol Chem. 2006;281:18591–18600. doi: 10.1074/jbc.M601159200. [DOI] [PubMed] [Google Scholar]
- Dutta SK, Ghosh S, De S, Hoffman EP. CYP1A1 and MT1K are Congener Specific Biomarker Genes for Liver Diseases Induced by PCBs. Environ Toxicol Pharmacol. 2008;25:218–221. doi: 10.1016/j.etap.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Dutta SK, Ghosh S, Zang S, Trnovec T, Palkovicova L, Sovcikova E, et al. Identification of Early Disease Biomarkers in 45 Months PCB-Exposed Slovak Population [Abstract] Epidemiology. 2009;20:S131. [Google Scholar]
- Esseghir S, Todd SK, Hunt T, Poulsom R, Plaza-Menacho I, Reis-Filho JS, Isacke CM. A role for glial cell derived neurotrophic factor induced expression by inflammatory cytokines and RET/GFR alpha 1 receptor up-regulation in breast cancer. Cancer Res. 2007;67:11732–11741. doi: 10.1158/0008-5472.CAN-07-2343. [DOI] [PubMed] [Google Scholar]
- Fischer LJ, Seegal RF, Ganey PE, Pessah IN, Kodavanti PR. Symposium overview: Toxicity of non-coplanar PCBs. Toxicol Sci. 1998;41:49–61. doi: 10.1006/toxs.1997.2386. [DOI] [PubMed] [Google Scholar]
- Fitzgerald EF, Hwang SA, Lambert G, Gomez M, Tarbell A. PCB exposure and in vivo CYP1A2 activity among Native Americans. Environ Health Perspect. 2005;113:272–277. doi: 10.1289/ehp.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi A, Kumari R, Zukin RS. Regulation of group I metabotropic glutamate receptor trafficking and signaling by the caveolar/lipid raft pathway. J Neurosci. 2009;29:3590–3602. doi: 10.1523/JNEUROSCI.5824-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesualdi NM, Chirico G, Pirozzi G, Costantino E, Landriscina M, Esposito F. Tumor necrosis factor-associated protein 1 (TRAP-1) protects cells from oxidative stress and apoptosis. Stress. 2007;10:342–350. doi: 10.1080/10253890701314863. [DOI] [PubMed] [Google Scholar]
- Ghosh S, De S. Dutta SK: Altered Protein Expressions in Chronic PCB-153 Induced Human Liver (HepG2) cells. Int J Toxicol. 2007;26:203–212. doi: 10.1080/10915810701352648. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Dutta SK, Zang S, Trnovec T, Palkovicova L, Sovcikova E, et al. PCB Exposure In Vitro (PBMC): Differential Gene Expression, Pathway Analysis for Possible Mode(s) of Actions, and Disease Development in Comparison with PCB-Exposed Slovak Population [Abstract] Epidemiology. 2009;20:S130. [Google Scholar]
- Giesy JP, Kannan K, Blankenship AL, Jones PD, Hilscherova K. Dioxin-like and non-dioxin-like toxic effects of polychlorinated biphenyls (PCBs): implications for risk assessment. Cent Eur J Public Health. 2000;8 Suppl:43–55. [PubMed] [Google Scholar]
- Gladen BC, Longnecker MP, Schecter AJ. Correlations among polychlorinated biphenyls, dioxins, and furans in humans. Am J Ind Med. 1999;35:15–20. doi: 10.1002/(sici)1097-0274(199901)35:1<15::aid-ajim3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Glynn AW, Wolk A, Aune M, Atuma S, Zettermark S, Maehle-Schmid M, et al. Serum concentrations of organochlorines in men: a search for markers of exposure. Sci Total Environ. 2000;263:197–208. doi: 10.1016/s0048-9697(00)00703-8. [DOI] [PubMed] [Google Scholar]
- Golden R, Kimbrough R. Weight of evidence evaluation of potential human cancer risks from exposure to polychlorinated biphenyls: an update based on studies published since 2003. CritRev Toxicol. 2009;39:299–331. doi: 10.1080/10408440802291521. [DOI] [PubMed] [Google Scholar]
- Grimvall E, Rylander L, Nilsson-Ehle P, Nilsson U, Strömberg U, Hagmar L, Ostman C. Monitoring of polychlorinated biphenyls in human blood plasma: methodological developments and influence of age, lactation, and fish consumption. Arch Environ Contam Toxicol. 1997;32:329–336. doi: 10.1007/s002449900193. [DOI] [PubMed] [Google Scholar]
- Hardell L, Andersson SO, Carlberg M, Bohr L, van Bavel B, Lindström G, et al. Adipose tissue concentrations of persistent organic pollutants and the risk of prostate cancer. J Occup Environ Med. 2006;48:700–707. doi: 10.1097/01.jom.0000205989.46603.43. [DOI] [PubMed] [Google Scholar]
- Hauser R, Chen Z, Pothier L, Ryan L, Altshul L. The relationship between human semen parameters and environmental exposure to polychlorinated biphenyls and p,p'-DDE. Environ Health Perspect. 2003a;111:1505–1511. doi: 10.1289/ehp.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser R, Singh NP, Chen Z, Pothier L, Altshul L. Lack of an association between environmental exposure to polychlorinated biphenyls and p,p'-DDE and DNA damage in human sperm measured using the neutral comet assay. Hum Reprod. 2003b;18:2525–2533. doi: 10.1093/humrep/deg508. [DOI] [PubMed] [Google Scholar]
- Hertzman C. A report for the Environmental Action Programme for Central and Eastern Europe. World Bank; 1995. Environment and Health in Eastern Europe. [Google Scholar]
- Hertz-Picciotto I, Park Hye-Y, Dostal M, Kocan A, Trnovec T, Sram R. Prenatal Exposures to Persistent and Non-Persistent Organic Compounds and Effects on Immune System Development. Basic & Clin Pharmacol & Toxicol. 2008;102:146–154. doi: 10.1111/j.1742-7843.2007.00190.x. [DOI] [PubMed] [Google Scholar]
- Hertz-Piccotto I, Trnovec T, Kocan A, Charles MJ, Ciznar P, Langer P, Sovcikova E, James R. PCB and Early Childhood Development in Slovakia: Study Design and Background. Fresenius Environ Bull. 2003;12:208–214. [Google Scholar]
- Hovander L, Linderholm L, Athanasiadou M, Athanassiadis I, Bignert A, Fängström B, et al. Levels of PCBs and their metabolites in the serum of residents of a highly contaminated area in eastern Slovakia. Environ Sci Technol. 2006;40:3696–3703. doi: 10.1021/es0525657. [DOI] [PubMed] [Google Scholar]
- Hsu PC, Pan MH, Li LA, Chen CJ, Tsai SS, Guo YL. Exposure in utero to 2,2',3,3',4,6'-hexachlorobiphenyl (PCB 132) impairs sperm function and alters testicular apoptosis-related gene expression in rat offspring. Toxicol Appl Pharmacol. 2007;221:68–75. doi: 10.1016/j.taap.2007.01.027. [DOI] [PubMed] [Google Scholar]
- Hua G, Zhang Q, Fan Z. Heat shock protein 75 (TRAP1) antagonizes reactive oxygen species generation and protects cells from granzyme M-mediated apoptosis. J Biol Chem. 2007;282:20553–20560. doi: 10.1074/jbc.M703196200. [DOI] [PubMed] [Google Scholar]
- Huang C, Liu LY, Li ZF, Wang P, Ni L, Song LP, Xu DH, Song TS. Effects of small interfering RNAs targeting MAPK1 on gene expression profile in HeLa cells as revealed by microarray analysis. Cell Biol Int. 2008;32:1081–1090. doi: 10.1016/j.cellbi.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Hubbard B, Doege H, Punreddy S, Wu H, Huang X, Kaushik VK, Mozell RL, et al. Mice deleted for fatty acid transport protein 5 have defective bile acid conjugation and are protected from obesity. Gastroenterology. 2006;130:1259–1269. doi: 10.1053/j.gastro.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Ilany J, Bilan PJ, Kapur S, Caldwell JS, Patti ME, Marette A, Kahn CR. Over-expression of Rad in muscle worsens diet-induced insulin resistance and glucose intolerance and lowers plasma triglyceride level. Proc Natl Acad Sci USA. 2006;103:4481–4486. doi: 10.1073/pnas.0511246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japour AJ, Mayers DL, Johnson VA, Kuritzkes DR, Beckett LA, Arduino JM, et al. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Takeda Y, Teramoto K, Tsutsumi O, Taketani Y, Watanabe K. Human NB-2 of the contactin subgroup molecules: chromosomal localization of the gene (CNTN5) and distinct expression pattern from other subgroup members. Genomics. 2000;69:113–119. doi: 10.1006/geno.2000.6310. [DOI] [PubMed] [Google Scholar]
- Knerr S, Schrenk D. Carcinogenicity of “non-dioxinlike” polychlorinated biphenyls. Crit Rev Toxicol. 2006;36:663–694. doi: 10.1080/10408440600845304. [DOI] [PubMed] [Google Scholar]
- Kocan A, Petrik J, Drobna B, Chovancova J. Levels of PCBs and some organochlorine pesticides in the human population of selected areas of the Slovak Republic. I. Blood. Chemosphere. 1994;29:2315–2325. doi: 10.1016/0045-6535(94)90400-6. [DOI] [PubMed] [Google Scholar]
- Kodavanti PR. Neurotoxicity of persistent organic pollutants: possible modes(s) of action and further considerations. Dose-Response. 2005;3:273–305. doi: 10.2203/dose-response.003.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodiha M, Rassi JG, Brown CM, Stochaj U. Localization of AMP kinase is regulated by stress, cell density, and signaling through the MEK-->ERK1/2 pathway. Am J Physiol Cell Physiol. 2007;293:C1427–C1436. doi: 10.1152/ajpcell.00176.2007. [DOI] [PubMed] [Google Scholar]
- Laden F, Ishibe N, Hankinson SE, Wolff MS, Gertig DM, Hunter DJ, Kelsey KT. Polychlorinated biphenyls, cytochrome 450 1A1 and breast cancer risk in the Nurses’ Health Study. Cancer Epidemiol Biomarkers Prev. 2002;11:1560–1565. [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression indexcomputation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loch-Caruso R. Uterine muscle as a potential target of polychlorinated biphenyls during pregnancy. Int J Hyg Environ Health. 2002;205:121–130. doi: 10.1078/1438-4639-00137. [DOI] [PubMed] [Google Scholar]
- Lockshin RA, Zakeri Z. Apoptosis, autophagy, and more. Int J Biochem Cell Biol. 2004;36:2405–2419. doi: 10.1016/j.biocel.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Longnecker MP, Wolff MS, Gladen BC, Brock JW, Grandjean P, Jacobson JL, Korrick SA, et al. Comparison of polychlorinated biphenyl levels across studies of human neurodevelopment. Environ Health Perspect. 2003;111:65–70. doi: 10.1289/ehp.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas B, Fields C, Hofmann MC. Signaling Pathways in Spermatogonial Stem Cells and Their Disruption by Toxicants. Birth Defects Research (Part C) 2009;87:35–42. doi: 10.1002/bdrc.20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyche J, Larsen H, Skaare JU, Tverdal A, Dahl E, Johansen G, Ropstad E. Effects of perinatal exposure to low doses of PCB 153 and PCB 126 on lymphocyte proliferation and hematology in goat kids. J Toxicol Environ Health A. 2004;67:889–904. doi: 10.1080/15287390490443740. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Shima G, Aiuchi T, Horie M, Hori K, Nakajo S, et al. Involvement of tumor necrosis factor receptor-associated protein 1 (TRAP1) in apoptosis induced by beta-hydroxyisovalerylshikonin. J Biol Chem. 2004;279:503–515. doi: 10.1074/jbc.M404256200. [DOI] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvönen ME, Parvinen M, de Rooij DG, Hess MW, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- Moyers JS, Bilan PJ, Reynet C, Kahn CR. Over-expression of Rad inhibits glucose uptake in cultured muscle and fat cells. J Biol Chem. 1996;271:23111–23116. doi: 10.1074/jbc.271.38.23111. [DOI] [PubMed] [Google Scholar]
- Newman J, Gallo MV, Schell LM, DeCapriom AP, Denhamm M, Deane GD. Akwesasne Task Force on Environment. Analysis of PCB congeners related to cognitive functioning in adolescents. Neurotoxicology. 2009;30:686–696. doi: 10.1016/j.neuro.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmann SR, Kostas J, Wilson LR, Shain W, Bush B. Neurobehavioral and somatic effects of perinatal PCB exposure in rats. Environ Res. 1987;44:56–70. doi: 10.1016/s0013-9351(87)80086-5. [DOI] [PubMed] [Google Scholar]
- Overmeyer JH, Kaul A, Johnson EE, Maltese WA. Active ras triggers death in glioblastoma cells through hyperstimulation of macropinocytosis. Mol Cancer Res. 2008;6:965–977. doi: 10.1158/1541-7786.MCR-07-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Hertz-Picciotto I, Petrik J, Palkovicova L, Kocan A, Trnovec T. Prenatal PCB exposure and thymus size at birth in neonates in Eastern Slovakia. Environ Health Perspect. 2008;116:104–109. doi: 10.1289/ehp.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patandin S, Dagnelie PC, Mulder PG, Op de Coul E, van der Veen JE, Weisglas-Kuperus N, et al. Dietary exposure to polychlorinated biphenyls and dioxins from infancy until adulthood: A comparison between breast-feeding, toddler and long-term exposure. Environ Health Perspect. 1999;107:45–51. doi: 10.1289/ehp.9910745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavuk M, Cerhan JR, Lynchm CF, Schecterm A, Petrikm J, Chovancovam J, Kocanm A. Environmental exposure to PCBs and cancer incidence in eastern Slovakia. Chemosphere. 2004;54:1509–1520. doi: 10.1016/j.chemosphere.2003.10.038. [DOI] [PubMed] [Google Scholar]
- Reynet C, Kahn CR. Rad: a member of the Ras family over-expressed in muscle of type II diabetic humans. Science. 1993;262:1441–1444. doi: 10.1126/science.8248782. [DOI] [PubMed] [Google Scholar]
- Rignell-Hydbom A, Rylander L, Giwercman A, Jönsson BAG, Lindh L, Eleuteri P, et al. Exposure to PCB and p,p'-DDE and human sperm chromatin integrity. Environ Health Perspect. 2005;113:175–179. doi: 10.1289/ehp.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rignell-Hydbom A, Rylander L, Giwercman A, Jönsson BAG, Nilsson-Ehle P, Hagmar L. Exposure to CB-153 and p,p'-DDE and male reproductive function. Hum Reprod. 2004;19:2066–2075. doi: 10.1093/humrep/deh362. [DOI] [PubMed] [Google Scholar]
- Rylander L, Stromberg U, Dyremark E, Nilsson-Ehle P, Ostman C, Hagmar L. Polychlorinated biphenyls in blood plasma among Swedish female fish consumers in relation to low birth weight. Am J Epidemiol. 1998a;147:493–502. doi: 10.1093/oxfordjournals.aje.a009476. [DOI] [PubMed] [Google Scholar]
- Rylander L, Stromberg U, Hagmar L. Agreement between reported fish consumption obtained by two interviews and its impact on the results in a reproduction study. Eur J Epidemiol. 1998b;14:93–97. doi: 10.1023/a:1007488117066. [DOI] [PubMed] [Google Scholar]
- Rylander L, Stromberg U, Hagmar L. Dietary intake of fish contaminated with persistent organochlorine compounds in relation to low birth weight. Scand J Work Environ Health. 1996;22:260–266. doi: 10.5271/sjweh.140. [DOI] [PubMed] [Google Scholar]
- Rylander L, Strömberg U, Hagmar L. Lowered birth weight among infants born to women with a high intake of fish contaminated with persistent organochlorine compounds. Chemosphere. 2000;40:1255–1262. doi: 10.1016/s0045-6535(99)00377-x. [DOI] [PubMed] [Google Scholar]
- Safe SH. Polychlorinated biphenyls (PCBs): environmental impact, biochemical and toxic responses, and implications for risk assessment. Crit Rev Toxicol. 1994;24:87–149. doi: 10.3109/10408449409049308. [DOI] [PubMed] [Google Scholar]
- Schantz SL, Widholm JH, Rice DC. Effects of PCB exposure on neuropsychological function in children. Environ Health Perspect. 2003;111:357–376. doi: 10.1289/ehp.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Gordish-Dressman H, Hoffman EP. An interactive power analysis tool for microarray hypothesis testing and generation. Bioinformatics. 2006;22:808–814. doi: 10.1093/bioinformatics/btk052. [DOI] [PubMed] [Google Scholar]
- Shields PG. Understanding population and individual risk assessment: the case of polychlorinated biphenyls. Cancer Epidemiol Biomarkers Prev. 2006;15:830–839. doi: 10.1158/1055-9965.EPI-06-0222. [DOI] [PubMed] [Google Scholar]
- Sonneborn D, Park HY, Petrik J, Kocan A, Palkovicova L, Trnovec T, et al. Prenatal polychlorinated biphenyl exposures in eastern Slovakia modify effects of social factors on birth weight. Paediatric and Perinatal Epidemiology. 2008;22:202–213. doi: 10.1111/j.1365-3016.2008.00929.x. [DOI] [PubMed] [Google Scholar]
- Tabb MM, Kholodovych V, Grun F, Zhou C, Welsh WJ, Blumberg B. Highly Chlorinated PCBs Inhibit the Human Xenobiotic Response Mediated by the Steroid and Xenobiotic Receptor (SXR) Environ Health Perspect. 2004;112:163–169. doi: 10.1289/ehp.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Negishi T, Imamura M, Sawano E, Kuroda Y, Yoshikawa Y, Tashiro T. Alterations in gene expression of glutamate receptors and exocytosis-related factors by a hydroxylated-polychlorinated biphenyl in the developing rat brain. Toxicology. 2009;257:17–24. doi: 10.1016/j.tox.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Tseng YH, Vicent D, Zhu J, Niu Y, Adeyinka A, Moyers JS, et al. Regulation of growth and tumorigenicity of breast cancer cells by the low molecular weight GTPase Rad and nm23. Cancer Res. 2001;61:2071–2079. [PubMed] [Google Scholar]
- Vakharia DD, Liu N, Pause R, Fasco M, Bessette E, Zhang QY, Kaminsky LS. Effect of metals on polycyclic aromatic hydrocarbon induction of CYP1A1 and CYP1A2 in human hepatocyte cultures. Toxicol Appl Pharmacol. 2001;170:93–103. doi: 10.1006/taap.2000.9087. [DOI] [PubMed] [Google Scholar]
- van Delft JH, Baan RA, Roza L. Biological effect markers for exposure to carcinogenic compounds and their relevance for risk assessment. Critical Review Toxicology. 1998;28:477–510. doi: 10.1080/10408449891344254. [DOI] [PubMed] [Google Scholar]
- Van den Berg M, Birnbaum LS, Denison M, De Vito M, Farland W, Feeley M, et al. The 2005 World Health Organization reevaluation of human and Mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicol Sci. 2006;93:223–241. doi: 10.1093/toxsci/kfl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen DM, Gottschalk RW, van Herwijnen MH, Moonen EJ, Kleinjans JC, van Delft JH. Differential gene expression in human peripheral blood mononuclear cells induced by cigarette smoke and its constituents. Toxicol Sci. 2005;86:200–210. doi: 10.1093/toxsci/kfi168. [DOI] [PubMed] [Google Scholar]
- Vartiainen T, Jaakkola JJ, Saarikoski S, Tuomisto J. Birth weight and sex of children and the correlation to the body burden of PCDDs/PCDFs and PCBs of the mother. Environ Health Perspect. 1998;106:61–66. doi: 10.1289/ehp.9810661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit C, Genze F, Menke A, Hoeffert S, Gress TM, Gierschik P, Giehl K. Activation of phosphatidylinositol 3-kinase and extracellular signalregulated kinase is required for glial cell line-derived neurotrophic factor-induced migration and invasion of pancreatic carcinoma cells. Cancer Res. 2004;64:5291–5300. doi: 10.1158/0008-5472.CAN-04-1112. [DOI] [PubMed] [Google Scholar]
- Voloboueva LA, Duan M, Ouyang Y, Emery JF, Stoy C, Giffard RG. Over expression of mitochondrial Hsp70/Hsp75 protects astrocytes against ischemic injury in vitro. J Cereb Blood Flow Metab. 2008;28:1009–1016. doi: 10.1038/sj.jcbfm.9600600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voong LN, Slater AR, Kratovac S, Cressman DE. Mitogen-activated protein kinase ERK1/2 regulates the class II transactivator. J Biol Chem. 2008;283:9031–9039. doi: 10.1074/jbc.M706487200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winneke G, Walkowiak J, Lilienthal H. PCB-induced neurodevelopmental toxicity in human infants and its potential mediation by endocrine dysfunction. Toxicology. 2002;181–182:161–165. doi: 10.1016/s0300-483x(02)00274-3. [DOI] [PubMed] [Google Scholar]