Abstract
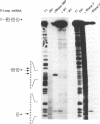
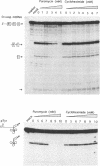
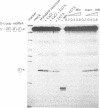
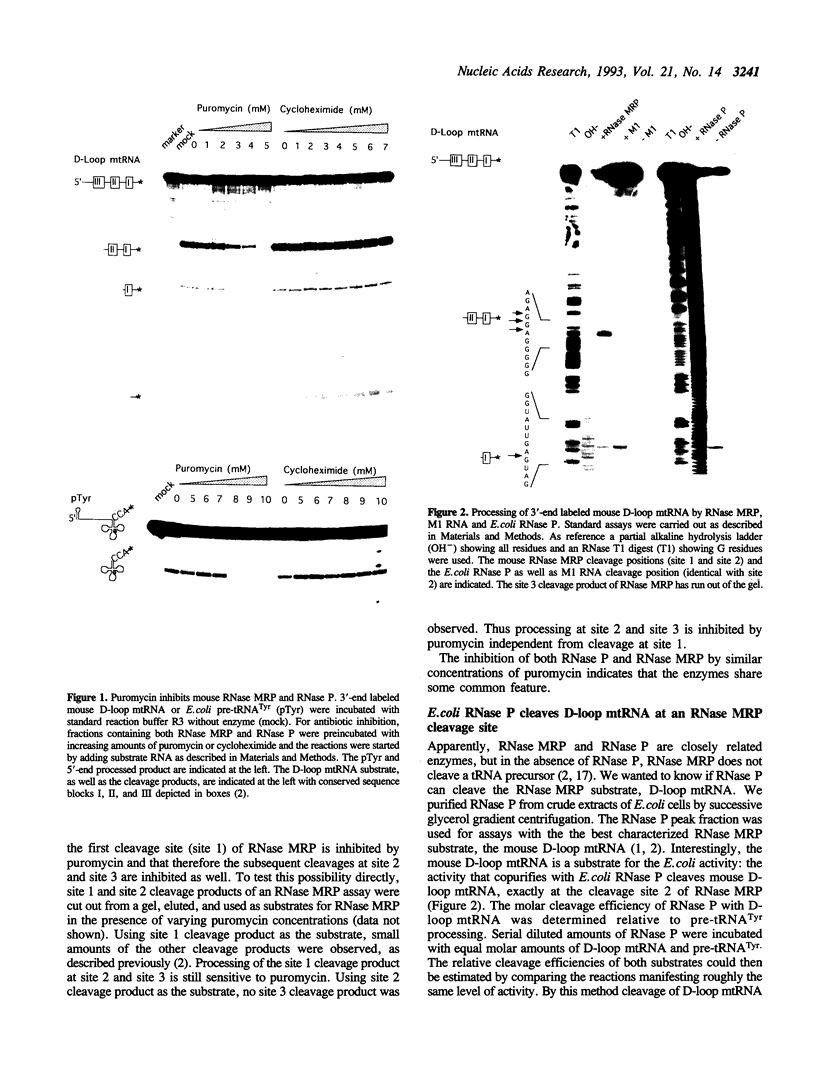
RNase MRP is a site-specific ribonucleoprotein endoribonuclease that processes RNA from the mammalian mitochondrial displacement loop containing region. RNase P is a site-specific ribonucleoprotein endoribonuclease that processes pre-tRNAs to generate their mature 5'-ends. A similar structure for the RNase P and RNase MRP RNAs and a common cleavage mechanism for RNase MRP and RNase P enzymes have been proposed. Experiments with protein synthesis antibiotics have shown that both RNase MRP and RNase P are inhibited by puromycin. We also show that E. coli RNase P cleaves the RNase MRP substrate, mouse mitochondrial primer RNA, exactly at a site that is cleaved by RNase MRP.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Kirsebom L., Talbot S. Recent studies of ribonuclease P. FASEB J. 1993 Jan;7(1):7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- Altman S. Ribonuclease P: an enzyme with a catalytic RNA subunit. Adv Enzymol Relat Areas Mol Biol. 1989;62:1–36. doi: 10.1002/9780470123089.ch1. [DOI] [PubMed] [Google Scholar]
- Bartkiewicz M., Gold H., Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989 Apr;3(4):488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- Bennett J. L., Jeong-Yu S., Clayton D. A. Characterization of a Xenopus laevis ribonucleoprotein endoribonuclease. Isolation of the RNA component and its expression during development. J Biol Chem. 1992 Oct 25;267(30):21765–21772. [PubMed] [Google Scholar]
- Boehm S. Similarities between a predicted secondary structure for the M1 RNA ribozyme and the tRNA binding center of 16 S rRNA from E. coli. FEBS Lett. 1987 Aug 17;220(2):283–287. doi: 10.1016/0014-5793(87)80830-x. [DOI] [PubMed] [Google Scholar]
- Brown G. G., Gadaleta G., Pepe G., Saccone C., Sbisà E. Structural conservation and variation in the D-loop-containing region of vertebrate mitochondrial DNA. J Mol Biol. 1986 Dec 5;192(3):503–511. doi: 10.1016/0022-2836(86)90272-x. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987 Feb;6(2):409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Hauswirth W. W., Clayton D. A. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985 Jun;4(6):1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doersen C. J., Guerrier-Takada C., Altman S., Attardi G. Characterization of an RNase P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. J Biol Chem. 1985 May 25;260(10):5942–5949. [PubMed] [Google Scholar]
- Doria M., Carrara G., Calandra P., Tocchini-Valentini G. P. An RNA molecule copurifies with RNase P activity from Xenopus laevis oocytes. Nucleic Acids Res. 1991 May 11;19(9):2315–2320. doi: 10.1093/nar/19.9.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Altman S. External guide sequences for an RNA enzyme. Science. 1990 Aug 17;249(4970):783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990 Aug 10;62(3):407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Gold H. A., Craft J., Hardin J. A., Bartkiewicz M., Altman S. Antibodies in human serum that precipitate ribonuclease P. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5483–5487. doi: 10.1073/pnas.85.15.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold H. A., Topper J. N., Clayton D. A., Craft J. The RNA processing enzyme RNase MRP is identical to the Th RNP and related to RNase P. Science. 1989 Sep 22;245(4924):1377–1380. doi: 10.1126/science.2476849. [DOI] [PubMed] [Google Scholar]
- Haas E. S., Morse D. P., Brown J. W., Schmidt F. J., Pace N. R. Long-range structure in ribonuclease P RNA. Science. 1991 Nov 8;254(5033):853–856. doi: 10.1126/science.1719634. [DOI] [PubMed] [Google Scholar]
- Karwan R., Bennett J. L., Clayton D. A. Nuclear RNase MRP processes RNA at multiple discrete sites: interaction with an upstream G box is required for subsequent downstream cleavages. Genes Dev. 1991 Jul;5(7):1264–1276. doi: 10.1101/gad.5.7.1264. [DOI] [PubMed] [Google Scholar]
- Karwan R. RNase MRP/RNase P: a structure-function relation conserved in evolution? FEBS Lett. 1993 Mar 15;319(1-2):1–4. doi: 10.1016/0014-5793(93)80025-p. [DOI] [PubMed] [Google Scholar]
- Kiss T., Filipowicz W. Evidence against a mitochondrial location of the 7-2/MRP RNA in mammalian cells. Cell. 1992 Jul 10;70(1):11–16. doi: 10.1016/0092-8674(92)90528-k. [DOI] [PubMed] [Google Scholar]
- Kiss T., Marshallsay C., Filipowicz W. 7-2/MRP RNAs in plant and mammalian cells: association with higher order structures in the nucleolus. EMBO J. 1992 Oct;11(10):3737–3746. doi: 10.1002/j.1460-2075.1992.tb05459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Nishimura S. Enzymatic RNA sequencing. Methods Enzymol. 1989;180:154–163. doi: 10.1016/0076-6879(89)80099-0. [DOI] [PubMed] [Google Scholar]
- Manam S., Van Tuyle G. C. Separation and characterization of 5'- and 3'-tRNA processing nucleases from rat liver mitochondria. J Biol Chem. 1987 Jul 25;262(21):10272–10279. [PubMed] [Google Scholar]
- Ojala D., Merkel C., Gelfand R., Attardi G. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell. 1980 Nov;22(2 Pt 2):393–403. doi: 10.1016/0092-8674(80)90350-5. [DOI] [PubMed] [Google Scholar]
- Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981 Apr 9;290(5806):470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Reed R. E., Baer M. F., Guerrier-Takada C., Donis-Keller H., Altman S. Nucleotide sequence of the gene encoding the RNA subunit (M1 RNA) of ribonuclease P from Escherichia coli. Cell. 1982 Sep;30(2):627–636. doi: 10.1016/0092-8674(82)90259-8. [DOI] [PubMed] [Google Scholar]
- Reimer G., Raska I., Scheer U., Tan E. M. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res. 1988 May;176(1):117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Robertson H. D., Altman S., Smith J. D. Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid presursor. J Biol Chem. 1972 Aug 25;247(16):5243–5251. [PubMed] [Google Scholar]
- Saccone C., Pesole G., Sbisá E. The main regulatory region of mammalian mitochondrial DNA: structure-function model and evolutionary pattern. J Mol Evol. 1991 Jul;33(1):83–91. doi: 10.1007/BF02100199. [DOI] [PubMed] [Google Scholar]
- Sbisà E., Nardelli M., Tanzariello F., Tullo A., Saccone C. The complete and symmetric transcription of the main non coding region of rat mitochondrial genome: in vivo mapping of heavy and light transcripts. Curr Genet. 1990 Mar;17(3):247–253. doi: 10.1007/BF00312616. [DOI] [PubMed] [Google Scholar]
- Sbisà E., Tullo A., Nardelli M., Tanzariello F., Saccone C. Transcription mapping of the Ori L region reveals novel precursors of mature RNA species and antisense RNAs in rat mitochondrial genome. FEBS Lett. 1992 Jan 27;296(3):311–316. doi: 10.1016/0014-5793(92)80311-4. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Bennett J. L., Dairaghi D. J., Clayton D. A. Secondary structure of RNase MRP RNA as predicted by phylogenetic comparison. FASEB J. 1993 Jan;7(1):208–213. doi: 10.1096/fasebj.7.1.7678563. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Clayton D. A. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev. 1992 Oct;6(10):1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- Topper J. N., Bennett J. L., Clayton D. A. A role for RNAase MRP in mitochondrial RNA processing. Cell. 1992 Jul 10;70(1):16–20. doi: 10.1016/0092-8674(92)90529-l. [DOI] [PubMed] [Google Scholar]
- Vioque A. Protein synthesis inhibitors and catalytic RNA. Effect of puromycin on tRNA precursor processing by the RNA component of Escherichia coli RNase P. FEBS Lett. 1989 Mar 27;246(1-2):137–139. doi: 10.1016/0014-5793(89)80269-8. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Singh R., Reddy R. Rat nucleolar 7-2 RNA is homologous to mouse mitochondrial RNase mitochondrial RNA-processing RNA. J Biol Chem. 1989 Sep 5;264(25):14835–14839. [PubMed] [Google Scholar]