Abstract
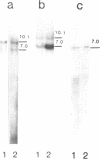
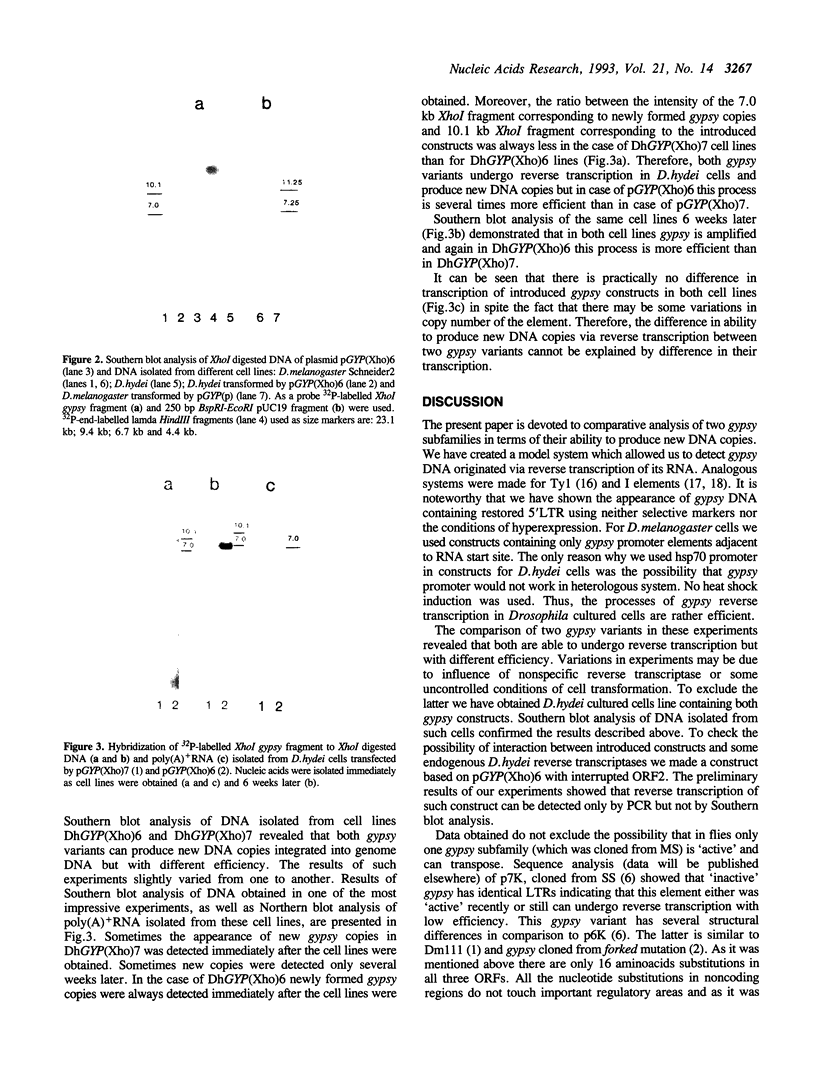
Plasmid DNA constructs containing 5' end truncated retrotransposon gypsy were introduced into Drosophila cultured cells. Appearance of new complete DNA copies with reconstructed via reverse transcription 5'LTR were detected by PCR after transient expression and by Southern blot analysis of genome DNA of stably transformed cells. Two gypsy subfamilies supposed to be different in transpositional activity were analyzed in terms of their ability to produce new DNA copies via reverse transcription in D. hydei cultured cells. It was demonstrated that both gypsy variants undergo retrotransposition but with different efficiency.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arkhipova I. R., Mazo A. M., Cherkasova V. A., Gorelova T. V., Schuppe N. G., Llyin Y. V. The steps of reverse transcription of Drosophila mobile dispersed genetic elements and U3-R-U5 structure of their LTRs. Cell. 1986 Feb 28;44(4):555–563. doi: 10.1016/0092-8674(86)90265-5. [DOI] [PubMed] [Google Scholar]
- Bayev A. A., Jr, Lyubomirskaya N. V., Dzhumagaliev E. B., Ananiev E. V., Amiantova I. G., Ilyin Y. V. Structural organization of transposable element mdg4 from Drosophila melanogaster and a nucleotide sequence of its long terminal repeats. Nucleic Acids Res. 1984 Apr 25;12(8):3707–3723. doi: 10.1093/nar/12.8.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Di Nocera P. P., Dawid I. B. Transient expression of genes introduced into cultured cells of Drosophila. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7095–7098. doi: 10.1073/pnas.80.23.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S., Heidmann T. An indicator gene for detection of germline retrotransposition in transgenic Drosophila demonstrates RNA-mediated transposition of the LINE I element. EMBO J. 1991 Jul;10(7):1927–1937. doi: 10.1002/j.1460-2075.1991.tb07719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. I., Belyaeva E. S., Aslanian M. M. Autonomous transposition of gypsy mobile elements and genetic instability in Drosophila melanogaster. Mol Gen Genet. 1990 Nov;224(2):303–308. doi: 10.1007/BF00271566. [DOI] [PubMed] [Google Scholar]
- Kim A. I., Belyaeva E. S. Transposition of mobile elements gypsy (mdg4) and hobo in germ-line and somatic cells of a genetically unstable mutator strain of Drosophila melanogaster. Mol Gen Genet. 1991 Oct;229(3):437–444. doi: 10.1007/BF00267467. [DOI] [PubMed] [Google Scholar]
- Liubomirskaia N. V., Arkhipova I. R., Il'in Iu V. Transcriptsiia mobil'nogo élementa Drozofily MDG4 v usloviiakh gipertermii. Genetika. 1990 Oct;26(10):1720–1728. [PubMed] [Google Scholar]
- Lyubomirskaya N. V., Arkhipova I. R., Ilyin Y. V., Kim A. I. Molecular analysis of the gypsy (mdg4) retrotransposon in two Drosophila melanogaster strains differing by genetic instability. Mol Gen Genet. 1990 Sep;223(2):305–309. doi: 10.1007/BF00265067. [DOI] [PubMed] [Google Scholar]
- Marlor R. L., Parkhurst S. M., Corces V. G. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol. 1986 Apr;6(4):1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrokhi L. J., Obolenkova L. A., Priimägi A. F., Ilyin Y. V., Gerasimova T. I., Georgiev G. P. The nature of unstable insertion mutations and reversions in the locus cut of Drosophila melanogaster: molecular mechanism of transposition memory. EMBO J. 1985 Dec 30;4(13B):3781–3787. doi: 10.1002/j.1460-2075.1985.tb04148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolell J., Bender W., Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Bender W. Sequences of the gypsy transposon of Drosophila necessary for its effects on adjacent genes. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9650–9654. doi: 10.1073/pnas.85.24.9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Bender W. The anterobithorax and bithorax mutations of the bithorax complex. EMBO J. 1986 Sep;5(9):2293–2303. doi: 10.1002/j.1460-2075.1986.tb04497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélisson A., Finnegan D. J., Bucheton A. Evidence for retrotransposition of the I factor, a LINE element of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4907–4910. doi: 10.1073/pnas.88.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H., Pirrotta V. A transposable P vector that confers selectable G418 resistance to Drosophila larvae. EMBO J. 1985 Jan;4(1):167–171. doi: 10.1002/j.1460-2075.1985.tb02332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]