Abstract
Two homodimeric proteins, alpha 2 and MCM1, are required to repress transcription of a-cell type specific genes in haploid yeast alpha-cells. In vitro studies by others of the interactions of these proteins with operator DNA have suggested that MCM1 binds to the middle and alpha 2 to the ends of the 31 bp operator. We have previously shown that alpha 2 organizes chromatin structure adjacent to the operator; in the presence of alpha 2 repressor, a precisely positioned nucleosome abuts the operator in both minichromosomes and the genome. We present in vivo footprinting evidence consistent with occupancy of the operator by MCM1 in both a- and alpha-cells and by alpha 2 repressor in alpha-cells. Interestingly, our in vivo results differ from previous in vitro work in detail. In contrast to the broad block of reagent accessibility to DNA by the factors seen in vitro, we find a pattern of strand-specific protection or augmented reactivity in vivo. The in vivo results are consistent with genetic data concerning transcriptional regulation of a-cell specific genes and corroborate the crystallographic data of others.
Full text
PDF

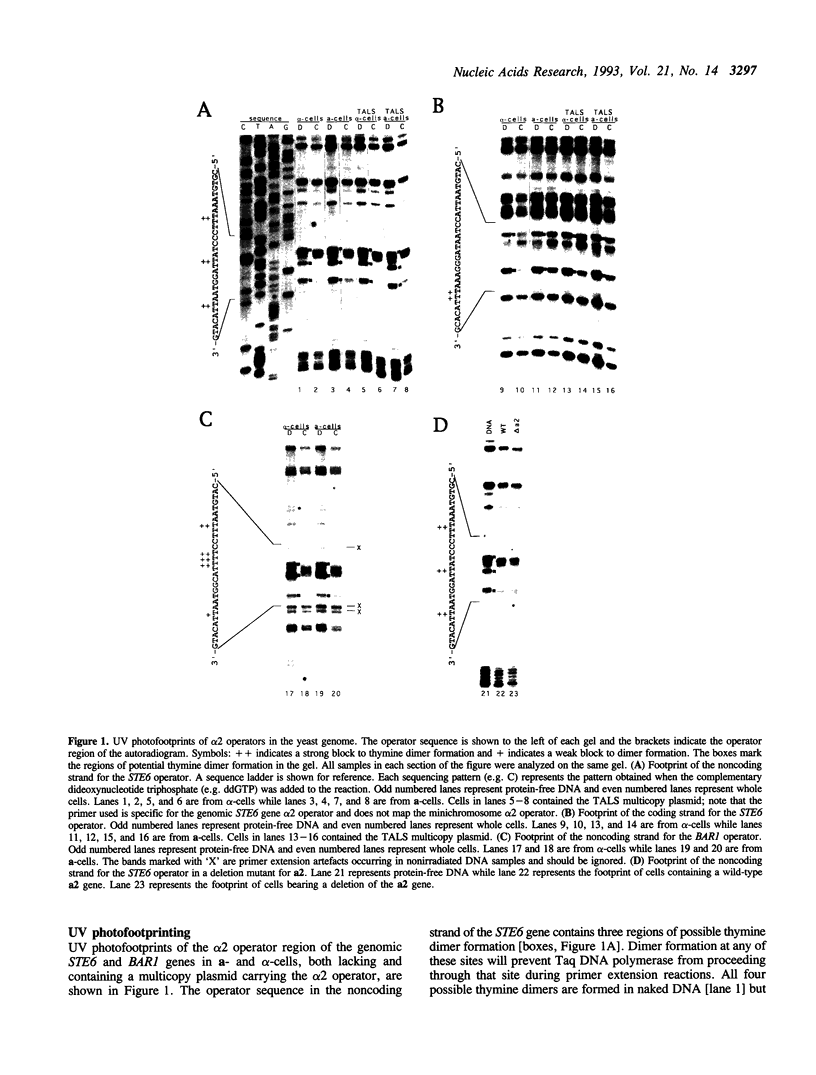
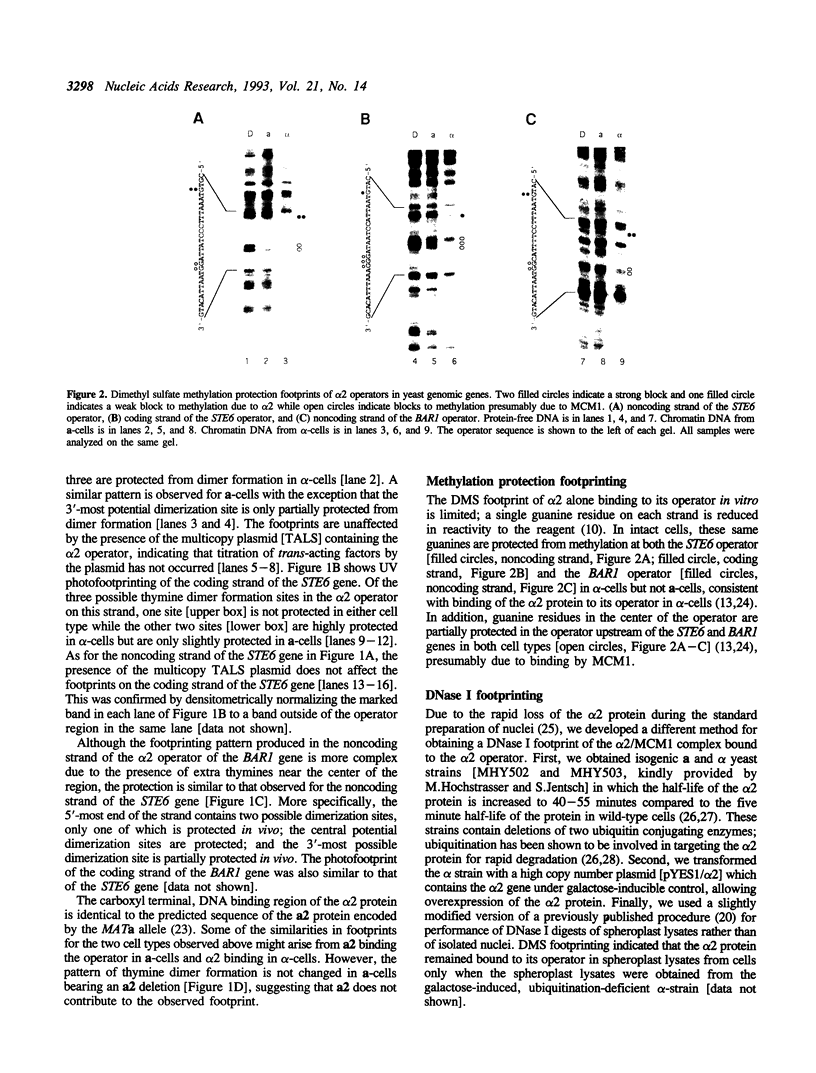
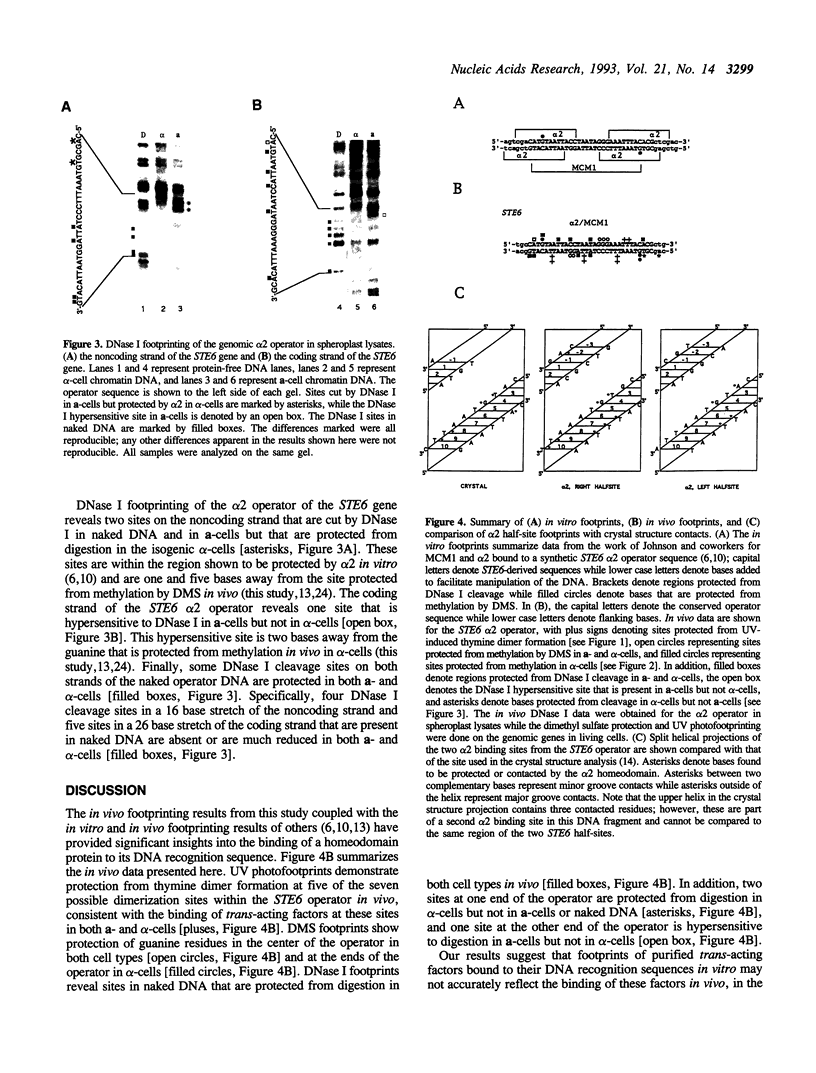

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axelrod J. D., Majors J. An improved method for photofootprinting yeast genes in vivo using Taq polymerase. Nucleic Acids Res. 1989 Jan 11;17(1):171–183. doi: 10.1093/nar/17.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M. M., Wang J. C. Use of light for footprinting DNA in vivo. Nature. 1984 Jun 21;309(5970):682–687. doi: 10.1038/309682a0. [DOI] [PubMed] [Google Scholar]
- Bender A., Sprague G. F., Jr MAT alpha 1 protein, a yeast transcription activator, binds synergistically with a second protein to a set of cell-type-specific genes. Cell. 1987 Aug 28;50(5):681–691. doi: 10.1016/0092-8674(87)90326-6. [DOI] [PubMed] [Google Scholar]
- Dranginis A. M. Binding of yeast a1 and alpha 2 as a heterodimer to the operator DNA of a haploid-specific gene. Nature. 1990 Oct 18;347(6294):682–685. doi: 10.1038/347682a0. [DOI] [PubMed] [Google Scholar]
- Dranginis A. M. Regulation of STA1 gene expression by MAT during the life cycle of Saccharomyces cerevisiae. Mol Cell Biol. 1989 Sep;9(9):3992–3998. doi: 10.1128/mcb.9.9.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Goutte C., Johnson A. D. a1 protein alters the DNA binding specificity of alpha 2 repressor. Cell. 1988 Mar 25;52(6):875–882. doi: 10.1016/0092-8674(88)90429-1. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989 Dec 14;342(6251):749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M., Varshavsky A. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell. 1990 May 18;61(4):697–708. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- Huibregtse J. M., Evans C. F., Engelke D. R. Comparison of tRNA gene transcription complexes formed in vitro and in nuclei. Mol Cell Biol. 1987 Sep;7(9):3212–3220. doi: 10.1128/mcb.7.9.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Keleher C. A., Goutte C., Johnson A. D. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988 Jun 17;53(6):927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992 Feb 21;68(4):709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- MacKay V. L., Welch S. K., Insley M. Y., Manney T. R., Holly J., Saari G. C., Parker M. L. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc Natl Acad Sci U S A. 1988 Jan;85(1):55–59. doi: 10.1073/pnas.85.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Dean A., Simpson R. T. Yeast alpha 2 repressor positions nucleosomes in TRP1/ARS1 chromatin. Mol Cell Biol. 1990 May;10(5):2247–2260. doi: 10.1128/mcb.10.5.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Shimizu M., Johnson L., Grunstein M., Simpson R. T. Stable nucleosome positioning and complete repression by the yeast alpha 2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992 Mar;6(3):411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Smith D. L., Johnson A. D. Flexibility of the yeast alpha 2 repressor enables it to occupy the ends of its operator, leaving the center free. Genes Dev. 1988 Jul;2(7):807–816. doi: 10.1101/gad.2.7.807. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Roth S. Y., Szent-Gyorgyi C., Simpson R. T. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991 Oct;10(10):3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. L., Johnson A. D. A molecular mechanism for combinatorial control in yeast: MCM1 protein sets the spacing and orientation of the homeodomains of an alpha 2 dimer. Cell. 1992 Jan 10;68(1):133–142. doi: 10.1016/0092-8674(92)90212-u. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Jensen R., Herskowitz I. Control of yeast cell type by the mating type locus: positive regulation of the alpha-specific STE3 gene by the MAT alpha 1 product. Cell. 1983 Feb;32(2):409–415. doi: 10.1016/0092-8674(83)90460-9. [DOI] [PubMed] [Google Scholar]
- Tan S., Ammerer G., Richmond T. J. Interactions of purified transcription factors: binding of yeast MAT alpha 1 and PRTF to cell type-specific, upstream activating sequences. EMBO J. 1988 Dec 20;7(13):4255–4264. doi: 10.1002/j.1460-2075.1988.tb03323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. L., Herskowitz I. Sequences upstream of the STE6 gene required for its expression and regulation by the mating type locus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2536–2540. doi: 10.1073/pnas.83.8.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]