Abstract
Functionally unique species contribute to the functional diversity of natural systems, often enhancing ecosystem functioning. An abundance of weakly interacting species increases stability in natural systems, suggesting that loss of weakly linked species may reduce stability. Any link between the functional uniqueness of a species and the strength of its interactions in a food web could therefore have simultaneous effects on ecosystem functioning and stability. Here, we analyse patterns in 213 real food webs and show that highly unique species consistently tend to have the weakest mean interaction strength per unit biomass in the system. This relationship is not a simple consequence of the interdependence of both measures on body size and appears to be driven by the empirical pattern of size structuring in aquatic systems and the trophic position of each species in the web. Food web resolution also has an important effect, with aggregation of species into higher taxonomic groups producing a much weaker relationship. Food webs with fewer unique and less weakly interacting species also show significantly greater variability in their levels of primary production. Thus, the loss of highly unique, weakly interacting species may eventually lead to dramatic state changes and unpredictable levels of ecosystem functioning.
Keywords: trophic interactions, predator–prey, marine, Weddell Sea, Lough Hyne, coral reef
1. Introduction
The last two decades have seen intensive research into the components of biodiversity that drive the functioning and stability of natural ecosystems. Recently, the focus of this research has shifted from studies confirming that species loss affects function and stability (e.g. [1,2]) towards understanding the mechanisms through which this process occurs (e.g. [3]). Species richness indices implicitly assume that all species contribute equally to ecosystem functioning and stability. This is an oversimplification, with numerous examples of keystone species [4], ecosystem engineers [5] and species with differential responses to changing environmental conditions [6]. Consequently, measures of functional diversity, which take into account interspecific and individual-level differences, offer the potential to identify functionally unique or redundant organisms within a system. Here, we define functional uniqueness (FU) as the originality of a species [7], which is a measure of how unique the characters of a species are relative to those of a set of other species.
A wide range of continuous trait-based measures have been developed in recent years to more accurately quantify the functional diversity of a system (e.g. [7,8]). Current research indicates that these continuous measures are more appropriate descriptors of functional diversity than categorical measures, such as functional group richness [9]. These new functional diversity measures have opened up a novel avenue of research, as it becomes clear that the functioning of an ecosystem is not governed by the phylogenetic content of its biota, rather by the functional traits of individuals [10]. Here, a functional trait is described as a component of an organism's phenotype that determines its effect on ecosystem processes and its response to environmental factors [11]. There is great promise that a trait-based assessment of natural systems will facilitate the maintenance of important ecosystem processes and services, by identifying key functional traits and functionally important species.
Studies are emerging that draw on these trait-based measures of functional diversity to identify high priority species for conservation. For example, Petchey et al. [12] have shown that trophically unique species are more susceptible to cascading extinctions, which leads to a greater than expected loss of trophic diversity after primary species loss. Continuous functional diversity measures have also been used to identify the unique impact of exotic mammalian predators on native bird populations, leading to increased probability of native species extinctions [13]. Both of these studies have attempted to identify relationships between FU and a deleterious impact on natural ecosystems, i.e. cascading extinctions and native species loss. Ascertaining further relationships between FU and attributes of naturally occurring systems may highlight potential vulnerabilities in natural communities, whose protection may help to safeguard ecosystem functioning and stability.
The distribution of interaction strengths in natural systems has repeatedly been shown to be an important determinant of stability [14–16]. Linear food chains consisting of strong interactions have been documented in many natural food webs and may drive community dynamics through cascading effects [4,17]. A prevalence of weak interactions appears to be important in dampening the destabilizing potential of these strong interactions [14]. Patterns of many weak and few strong interactions appear to be ubiquitous in nature (e.g. [18]), with loss of weak interactors leading to reduced system stability [16] and an increased mean interaction strength, which limits the coexistence of many species [19].
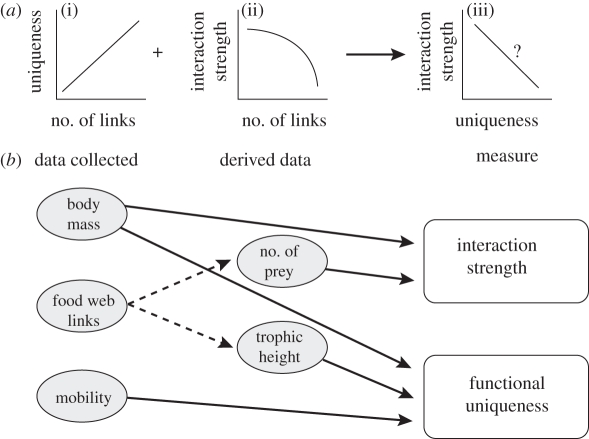
By extrapolating the results of recent studies, we have identified a potential link between the FU of a species and its mean interaction strength in a food web. Petchey et al. [12] demonstrated that often there is a positive relationship between trophic uniqueness and the number of trophic links in natural systems (figure 1a(i)). In that study, trophic uniqueness was a measure of the dissimilarity in the trophic links of one species relative to the trophic links of other species. It remains to be seen if this pattern holds for a measure of uniqueness based on functional traits. Montoya et al. [20] and O'Gorman et al. [18] have also shown that interaction strength is negatively related to number of trophic links in experimental systems (figure 1a(ii)). Combining these two relationships suggests a potential negative relationship between uniqueness and interaction strength (figure 1a(iii)). It should be noted, however, that this relationship has never been documented empirically.
Figure 1.
(a) Observed (i,ii) and predicted (iii) relationships involving interaction strength, FU and number of trophic links. (b) Flow diagram highlighting the data employed to calculate FU and interaction strength. The FU of species i, FUi, is calculated as FUi = di•/d••, where, di• = Σjdij, d•• = Σjdi• and dij is the measure of functional trait dissimilarity between species i and species j used by Pavoine et al. [7]. Interaction strength is calculated as aij = −b × (Mj−0.25/sj), where Mj is the body mass of predator j, sj is the number of prey species consumed by predator j and the exponent of −0.25 is based on allometric scaling relationships to approximate basal metabolic rate per unit biomass. Note the interdependence of the two measures on body size, which also suggests a probable relationship. See electronic supplementary material for a more detailed description of these two measures.
Body size is an important determinant of interaction strength in nature. Large-bodied species typically eat smaller-bodied ones and predator–prey body size allometry has been successfully used to predict the strength of interactions between species [21]. Body size is also an important trait, contributing to many aspects of a species' function in an ecological system, e.g. metabolic rate, nutrient turnover, home range, ingestion rate and secondary production [22]. As such, it is necessary to consider body size in the estimation of both interaction strength and FU (figure 1b). This interdependency on body size may also lead to a relationship between these two measures. A major aim of the present study is to assess whether factors other than this interdependence on body size may be driving any potential relationship.
Here, we use data from natural and experimental food webs to test whether a relationship exists between FU and interaction strength. Our main hypothesis is that there is a negative relationship between these two measures, based on trends identified in the aforementioned studies. If such an association exists, we will also explore the possible drivers of this relationship. As outlined above, highly unique species are more vulnerable to cascading extinctions [12] and if they are also likely to be weak interactors, this will have important consequences on the stability of real food webs. Loss of weak interactors is unlikely to produce effects that are immediately apparent in an ecosystem, such as trophic cascades or widespread changes in ecosystem functioning. However, increased variability of ecosystem processes and reduced resistance may gradually undermine the system in the long term [16]. If a link between FU and interaction strength is identified, this may reveal an important mechanism driving the relationship between biodiversity and ecosystem functioning or stability.
2. Material and methods
(a). Sources of empirical data
To investigate the mechanisms that might link FU and interaction strength (mean direct effect, or MDE), we examined published data from 210 experimentally assembled food webs and supplemented this with an investigation of data from three natural food webs. The experimental food webs originated from studies carried out at Lough Hyne, southwest Ireland [16,18]. Here, large subtidal exclusion cages were used to manipulate the diversity of 10 species of decapods, echinoderms and small fish. These manipulated species acted as top predators in the mesocosms, with lower trophic levels recruiting naturally through the mesh of the cages over experimental time periods ranging from 35 to 425 days. After community assembly had taken place, the species richness of these experimental food webs ranged from 42 to 77 taxa. This variation in taxon diversity was similar to the variation observed in many documented natural ecosystems (e.g. [23–25]), resulting in a spectrum of complex food webs that differed in predator richness, community diversity and time taken for community assembly (see electronic supplementary material, figure S1).
Exclusion cages have the potential to reduce water flow and light penetration, as well as altering the behaviour of organisms [26]; thus it is unclear whether these factors may influence potential patterns in the experimental communities. To investigate if a relationship between FU and MDE exists in communities uninfluenced by cages, we examined the existing information on three natural ecosystems: the Weddell Sea (WS) [27], the entire Lough Hyne (LH) food web (Jacob unpublished) and a Caribbean coral reef (CCR) [28]. These food webs consist of 490, 345 and 247 taxa, respectively, with a high proportion of taxa resolved to species level (see electronic supplementary material, table S1).
For all the food webs described above, we were able to obtain information on three important animal species traits that contribute to the functioning of these complex systems: body mass, mobility and trophic height. These three traits are directly related to a wide range of aspects of the phenotype of a species. As previously mentioned, body mass is linked to metabolic rate, nutrient turnover, home range, ingestion rate and secondary production [22]. Mobility is associated with feeding type, foraging methods and response to predation risk or prey defences [29,30]. Trophic height summarizes diet composition and susceptibility to predation. As such, a large amount of functional information can be summarized in these three easily measurable traits [27].
We also acquired data on all the predator–prey links in each system. Sources for these food web links are detailed in the publications outlined above and include gut content analysis, site-specific field observations and literature research. Some of the links described are likely to occur only at certain times of the year or may never be realized at the site in question. This is a limitation facing any study that employs composite webs (see [31]), but it is currently the best alternative to intensive sampling and gut content analysis over a long period of time, which has currently been achieved for only a limited number of food web studies, e.g. the Ythan Estuary [23], Tuesday Lake [24] and Broadstone Stream [25]. These are the only data we require to calculate FU and MDE in the chosen communities (figure 1b; derivations of these two measures can be found in the electronic supplementary material).
For each community, we performed a linear regression between log(FU) and log(MDE) and recorded the slopes, r2 values and intercepts. All data were log-transformed to meet the assumptions of normality and homogeneity of variance (with exploration of residuals versus fitted values and normal Q–Q plots). In order to understand the importance of individual traits, such as body size, and the potential for joint effects of individual traits on FU and MDE to cause the observed relationships, we repeated the analyses described above three times, once with each of the three traits excluded.
(b). Examination of stability in the mesocosm communities
The experimental LH communities contained treatments where large predators were manipulated on a richness gradient from 0 to 10 (more precisely, the predator richness levels were 0, 1, 4, 6, 7, 8 and 10). The manipulated predators were typically among the most unique species in the experimental food webs that developed. To investigate whether the loss of functionally unique (and potentially weakly interacting) species had an effect on the stability of the mesocosm communities, we compared both the temporal and spatial coefficients of variation (CV) of primary production in highly simplified food webs, i.e. containing 0–1 manipulated consumers (66 food webs), to more complex webs, i.e. containing 4–10 manipulated consumers (144 food webs). Primary production was measured as the square root of chlorophyll a, obtained from glass slides within the mesocosms (see [16] for details).
The temporal CV of primary production was calculated as the CV of a given experimental food web through consecutive sampling sessions (similar to Steiner [32]). The spatial CV of primary production was calculated as the CV across replicate food webs within each sampling period. By replicate food webs, we mean mesocosm communities from the same sampling period for which manipulated consumer identities (and not just richness) were equivalent, i.e. the composition of the core manipulated community had to be identical between replicates. This equated to three or four replicates of each food web treatment (with treatments described in more detail in [16,18]). This spatial CV measure has previously been used to assess the consistency in ecosystem process rates of replicate communities (e.g. [33]).
(c). Randomizations to test importance of functional traits
To determine which functional traits are driving any potential relationship between FU and MDE, we carried out a randomization test on the data from the 210 experimental LH food webs. For each of the 210 communities, we created three sets of randomizations, each containing 1000 randomized communities. These three sets randomized body mass, mobility and food web links (and thus trophic height), respectively. For each randomization, we estimated the slope, r2 value and intercept as described above. If the empirical value of a statistic was outside the 2.5 or 97.5 per cent quantiles of the randomized distribution, we concluded that they could not have occurred by chance and thus were significantly different from the randomized distributions at p < 0.05.
To randomize body mass and mobility, we sampled without replacement from the existing empirical data for the selected food web. Complete randomization of the links in a food web would lead to unrealistic food web structures, isolated species and ranges of trophic height that are not representative of the experimental communities. Consequently, we used the niche model [34] to maintain a realistic food web structure during randomization of the existing links. This preserved the fractions of top, intermediate and basal species, the means and variabilities of generality, vulnerability and food chain length, and the degree of cannibalism, omnivory, looping and trophic similarity in the randomized food webs.
3. Results
(a). Empirical patterns—experimental communities
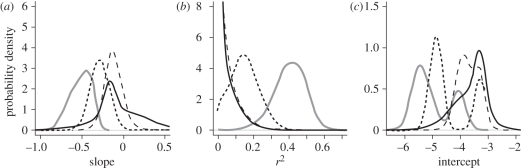
The analysis of the relationship between FU and interaction strength for the 210 experimental food webs reveals a number of consistent trends. First, there is a significant negative relationship (linear regression: p < 0.0001) between log(FU) and log(MDE), so that increasing FU is associated with a reduction in MDE for each of the 210 food webs (solid grey line in figure 2a; note the negative values for all empirical slopes on the x-axis). Over 90 per cent of these relationships have an r2 value greater than 0.3 (solid grey line in figure 2b). The distribution of intercepts for the 210 relationships appears bimodal (solid grey line in figure 2c). Further investigation of the data shows that experimental food webs with manipulated predator richness of 0 or 1 are distributed around the right-hand peak of the distribution (less-negative intercepts), while webs with a higher manipulated predator richness are distributed around the left-hand peak of the distribution (more negative intercepts). This separation of simplified webs (0–1 manipulated species) and more complex webs (4–10 species) acts as a further justification for the groupings we use to analyse stability effects in the webs (see §3d).
Figure 2.
The distribution of (a) slopes, (b) r2 values and (c) intercepts from the linear regressions of log(FU) against log(MDE) for the 210 experimental food webs. The solid grey lines depict the distributions for the empirical data. The black lines illustrate how these distributions shift after randomizing mobility (dotted black line), body mass (dashed black line) and food web links (solid black line). Probability densities were calculated using a Gaussian kernel density estimator.
Note that repeating this analysis with each of the three traits (body mass, mobility and trophic height) excluded in turn from the calculation of FU had no qualitative effect on this relationship, i.e. significant negative relationships remained for all 210 webs (see electronic supplementary material, figure S2).
(b). Empirical patterns—natural ecosystems
Investigation of the WS, entire LH and CCR food webs appears to confirm many of the patterns outlined in the experimental communities above for naturally occurring ecosystems (see electronic supplementary material, figure S3). We carried out a multiple linear regression between log(FU) and log(MDE) on all the natural food web data, with food web identity as an additional factor. There is a significant negative relationship (p < 0.001) between log(FU) and log(MDE) for all three systems, with slopes (mean ± s.e.) of WS = −0.37 ± 0.029, LH = −0.50 ± 0.043 and CCR = −0.17 ± 0.047. There is a large amount of unexplained variation in these relationships, however, with r2 values of WS = 0.26, LH = 0.28 and CCR = 0.05. The intercepts for all three systems (WS = −6.13 ± 0.113, LH = −6.24 ± 0.153, CCR = −5.59 ± 0.148) fall within the range of the higher predator richness treatments in the experimental communities (left-hand peak of the solid grey line in figure 2c). While the intercepts in the WS and LH webs are not significantly different from one another (p = 0.53), the CCR is significantly different from both the WS (p = 0.014) and LH (p = 0.006).
The relationship between log(FU) and log(MDE) is particularly weak in the CCR web (r2 = 0.05). To understand why this relationship is significantly weaker than the other two natural webs, we investigated the taxonomic resolution of the three food webs, i.e. whether organisms were identified to species level or into broader taxonomic categories. We expected the taxonomic resolution to be lower in the CCR web because it is noticeably skewed towards intermediate species (see electronic supplementary material, table S1 and figure S1). There is no significant difference in taxonomic resolution between the WS and LH webs (χ2 = 3.58, d.f. = 1, p = 0.06), but significantly fewer taxa were resolved to species level in the CCR than in either the WS (χ2 = 75.4, d.f. = 1, p < 0.0001) or the LH (χ2 = 39.1, d.f. = 1, p < 0.0001) webs. To investigate whether lower resolution data would alter the observed relationships in the WS and LH webs, we grouped subsets of species by their taxonomic class (similar to the CCR), such that the percentage of taxa resolved to species level was reduced to approximately 90 per cent (WS: 182 species, 26 trophic species; LH: 151 species, 16 trophic species). This led to a weaker relationship between log(FU) and log(MDE) in the two food webs, with shallower slopes (WS = −0.30 and LH = −0.20) and greatly reduced r2 values (WS = 0.06 and LH = 0.04).
(c). Randomizations to test importance of functional traits
The randomizations shed some light on the functional traits that might be driving the relationship between log(FU) and log(MDE). All three traits appear to make an important contribution to the relationship, because randomizing the data for each trait often produces a weaker linear relationship (table 1). One notable exception is the distribution of slopes after randomizing food web links. Here, only 18 per cent of empirical slopes differed from the slopes of the randomized food webs. This is a consequence of the large range of slopes produced by randomizing food web links (with the empirical pattern regularly falling within this broad random distribution).
Table 1.
A comparison of the empirical, linear relationships between log(FU) and log(MDE) with the relationships after randomizing the data for either mobility, body mass or food web links. Shown are the percentages of empirical relationships that were more extreme than 95 per cent of the relationships from the corresponding randomized data. Mobility, body mass and food web links were randomized 1000 times for each of the 210 empirical webs.
| slope | r2 value | intercept | |
|---|---|---|---|
| mobility | 69.05 | 79.52 | 79.05 |
| body mass | 99.05 | 99.05 | 100 |
| links | 17.62 | 100 | 81.90 |
The weakest disruption of the relationship occurs after randomizing mobility, with the overall pattern remaining qualitatively the same. Similar to the empirical patterns, the slope of the relationship between log(FU) and log(MDE) is always negative (dotted black line in figure 2a) and highly significant (linear regression: p < 0.0001). After randomization, over 71 per cent of r2 values are greater than 0.1 (dotted black line in figure 2b). The distribution of intercepts still appears to be bimodal (dotted black line in figure 2c). Randomizing mobility does not significantly alter the range of intercepts from that of the empirical webs (χ2 = 2.27, d.f. = 1, p = 0.13).
Randomizing body mass leads to a greater deterioration of the relationship between log(FU) and log(MDE). While the slope of the relationship is typically still negative (dashed black line in figure 2a), 99 per cent of the relationships are no longer significant (linear regression: p > 0.05) and 86 per cent have r2 values less than 0.1 (dashed black line in figure 2b). While the intercepts still appear to be bimodal (dashed black line in figure 2c), the range of intercepts is significantly different from the empirical webs (χ2 = 76.0, d.f. = 1, p < 0.0001).
Randomizing food web links qualitatively disrupts the relationship between log(FU) and log(MDE) even further. Here, 34 per cent of the webs show a positive relationship between log(FU) and log(MDE) after randomizing food web links (solid black line in figure 2a), none of these relationships are significant and 87 per cent have r2 values less than 0.1 (solid black line in figure 2b). The bimodal distribution of intercepts breaks down (solid black line in figure 2c), and the range of intercepts is significantly different from that of the empirical webs (χ2 = 126.9, d.f. = 1, p < 0.0001).
(d). Comparison of simplified and complex experimental communities
The manipulated predators were consistently among the most unique species in the experiment. Therefore, simplified food webs with manipulated predator richness of 0 or 1 are more likely to contain fewer highly unique species than complex food webs with manipulated predator richness of 4–10. For the relationships between log(FU) and log(MDE), simplified food webs have significantly weaker slopes (t = 11.41; d.f. = 199; p < 0.0001) and less-negative intercepts (t = 43.83; d.f. = 194; p < 0.0001) than the complex food webs. Both the temporal (t = 2.77; d.f. = 41; p = 0.0085) and spatial (t = 4.33; d.f. = 27; p = 0.0002) CV of simplified food webs were also significantly higher than for complex food webs (figure 3).
Figure 3.
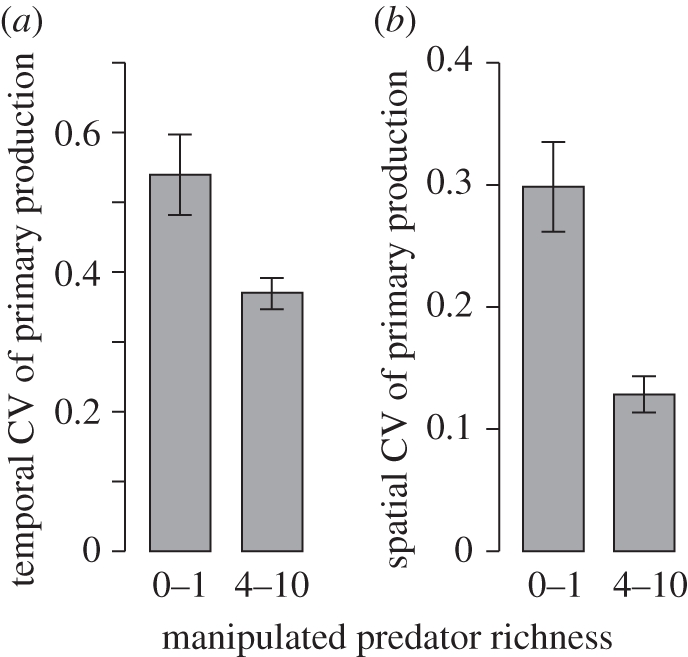
Stability effects in the experimental communities. Stability was measured as the (a) temporal and (b) spatial coefficient of variation (CV) of primary production. In both cases, a higher CV equates to lower stability. Effects were partitioned out to compare the stability of diverse communities (4–10 manipulated predators) to simplified communities (0–1 manipulated predators).
4. Discussion
Our analysis of 213 real food webs indicates that there is a consistent negative relationship between FU and interaction strength. This relationship can sometimes be quite weak, but for all 213 webs there is a tendency for highly unique species to have the weakest mean interaction strength in the system. This pattern is consistent with previously observed relationships [12,18,20] and has several important implications. Highly unique species have been shown to be more vulnerable to cascading extinctions in natural and model food webs [12]. This suggests that the extinction of any species in a system is likely to trigger the further loss of functionally unique species, i.e. increasing the probability of more unique species becoming extinct. Current research shows that the loss of any species should reduce the stability of a system [16,35], whether it is a strong or weak interactor. Loss of weak interactors rarely leads to significant cascading effects on ecosystem processes in the short term, such as those seen after the loss of keystone species [4] or strong interactors [16]. However, the loss of weak interactors has been shown to be associated with an erosion of stabilizing structures, increasing variability of ecosystem processes and reducing the resistance of the community to invasions and extinctions [16], which may lead to deterioration of the system in the long term [14,15]. Given the increased probability of highly unique species becoming extinct [12] and the likelihood that these species will also be weak interactors (figure 2a), this may lead to subtle, but detrimental effects on system stability (such as those seen in figure 3), which may slowly undermine the integrity of the system.
The randomizations reveal that, of the three functional traits considered in this study, body mass and trophic height are the two most important drivers of the relationship between FU and MDE (figure 2). While randomizing mobility led to quantitative differences, the pattern remained qualitatively the same, as increasing uniqueness was still always associated with weaker interaction strengths. It is important to note that the inherent interdependence of both FU and MDE on body mass applies to the randomizations as well as the empirical analysis. If this relationship were solely driven using body size in both these measures, we should still find a relationship between FU and MDE after randomization (note that the same randomized body masses are used to calculate FU and MDE, it is only the empirical pattern that is not maintained). The relationship between FU and MDE breaks down after randomizing body mass, however, with low r2 values and even some positive relationships observed (see dashed black line in figure 2a,b). Further evidence that the observed relationship between FU and MDE is not driven by joint effects of body size is the persistence of significant negative relationships even when body size is excluded from the analyses (see electronic supplementary material, figure S2).
This suggests that there is some arrangement of body masses in nature driving the relationship between FU and MDE and not just the joint dependence of these two measures on body mass. For example, in aquatic systems, communities are highly size-structured, with many small organisms and few larger ones. Large organisms also typically only eat smaller ones. As such, large organisms are more likely to be functionally unique (fewer species of similar physical structure) and also likely to spread their interactions over a greater number of prey, possibly leading to weaker interaction strengths. For the webs analysed, the most unique species and the weakest interactors often tended to be the largest organisms. It should also be noted that large organisms are more susceptible to extinctions, as predicted by increased environmental warming [36] and commercial harvesting trends [37]. This also increases the likelihood of highly unique, weak interactors being lost from natural ecosystems.
Traits such as body mass and trophic height are inextricably linked owing to the inherent size structuring of most natural food webs [22,38] and this may explain their similar influence in driving the patterns we observe. Furthermore, both traits are likely to play an important role in determining FU, as well as MDE. Body mass has been shown to scale with metabolic rate, nutrient turnover, home range, ingestion rate and secondary production [22], all of which are important factors regulating interaction strength. Trophic height tends to be positively related to generality, i.e. number of prey (e.g. [24]), which may lead to a dilution of mean interaction strength by dissipating interactions among multiple prey species. This may contribute to an observed negative relationship between interaction strength and number of trophic links [18,20], which is an important driver of the association with FU. Trophic height also tends to be negatively related to vulnerability, i.e. number of predators (e.g. [24]) and this trait has been shown to directly relate to interaction strength [39]. As such, body mass and trophic height appear to be dominant traits, whose functional roles influence many important properties of natural systems, including the strength of trophic interactions.
In the experimental food webs, we observed a bimodal distribution of intercepts in the 210 relationships between FU and MDE. This bimodal distribution appears to be a result of the number of large manipulated predators present in the experimental treatments. High manipulated predator richness (4–10 species) led to intercepts that were comparable to the natural ecosystems under investigation, i.e. the WS, LH and CCR webs. Simplified communities with 0 or 1 manipulated predators had significantly greater intercepts. The simplified communities also had significantly shallower slopes. This suggests that the loss of large or top predators from a community will lead to a significant change in the link between FU and MDE. This is most likely driven by a reduction in the functional diversity of these simplified communities, with fewer unique species present (several of the most unique species throughout the experimental communities were manipulated predators). Loss of functional diversity can have serious implications for resource dynamics and ecosystem stability, as the value and range of functional traits present in a system strongly influence short-term fluxes of matter and energy [40]. There is also a possibility that a change in the distribution of interactions may be causing a weakening of the relationship with FU. With fewer weak interactions to dampen the destabilizing potential of strong interactions, system stability is likely to be greatly reduced [14]. It is difficult to say from this study whether these changes are caused directly, through the loss of the large predators, or indirectly, through secondary extinctions after the top predators are removed. Further investigation is required to shed light on these issues.
Evidence for the destabilizing effect of unique species loss can be found from the comparison of temporal and spatial CV of primary production in simplified communities (0 or 1 manipulated predators) to more realistic assemblages (4–10 manipulated predators). Note again that the manipulated predators are typically among the most unique species in these food webs. Here, a higher level of temporal and spatial variability in ecosystem process rates equates to a lower level of stability [32,33]. The simplified food webs are clearly much less stable, with a significantly higher CV of primary production (both temporal and spatial) than food webs with high manipulated predator richness (figure 3). Higher temporal CV in the simplified communities is a result of greater fluctuations in ecosystem process rates through time. The increased spatial CV in the simplified food webs suggests that there is little consistency in the ecosystem process rates of these communities, with largely unpredictable levels of primary production in replicate food webs. Such an outcome highlights the potentially detrimental effects of the relationship between FU and MDE outlined above. As highly unique, weakly interacting species are lost, the system continues to function at a sustainable level in the short term. However, ecosystem process rates become unpredictable and may be more vulnerable to environmental fluctuations or further perturbations [35], increasing the possibility of a shift in community composition and structure.
The weakest negative relationship between FU and MDE was found to occur in the CCR web. Unlike the WS and LH food webs, the CCR is heavily skewed towards intermediate level species, with only 2 per cent of species described as basal resources or top predators (see electronic supplementary material, table S1). This appears to be largely owing to the low level of taxonomic resolution in the basal species, where algal and phytoplankton species are grouped together. Both the WS and LH webs contain detailed identification of algal and phytoplankton species and the links to their consumers. Additionally, there is a much lower level of taxonomic resolution throughout the CCR web compared with both the WS and LH webs (see electronic supplementary material, table S1), with many species grouped into classes such as Bivalvia, Gastropoda and Polychaeta. The importance of highly resolved food web data was emphasized by the grouping of some species into classes in the WS and LH webs and re-analysis of the data. Even with 90 per cent of taxa resolved to species level, the relationship between FU and MDE is greatly weakened in both webs, with shallower slopes and r2 values less than 0.1 (similar to the CCR web). Without highly resolved species information across all levels of a food web, important patterns and relationships may be missed (see also [41]).
The deleterious impact of the observed relationship between FU and interaction strength should not be underestimated, because of its potential to be the foundation of a hidden unravelling of ecosystem structure and dynamics. The rivet hypothesis [42] suggests that food webs can tolerate a certain level of perturbation under which they are able to function normally in a steady state. While this may have no immediate discernible effect, over time small perturbations erode away the resistance of the community to change, a feature shown to be associated with the loss of weak interactors [16]. A relatively minor perturbation can then cause the system to switch to a new state, where ecosystem process rates and the delivery of ecosystem services are largely unpredictable, and from which it cannot easily return. Even if conditions revert to those that were originally present, the system would not return to its original state immediately or entirely, a situation known as hysteresis [43]. This will make restoration of ecosystem services, whether for economic or aesthetic purposes, time-consuming and expensive. Given the observed negative relationship between FU and interaction strength, as well as the susceptibility of highly unique species to secondary extinctions [12] and the increased variability of primary production in simplified communities, it is likely that the loss of biodiversity from natural systems will lead to debilitating effects that are largely undetectable until it is too late to prevent a catastrophic state change.
Acknowledgements
The work was funded by the SIZEMIC research network (supported by the European Science Foundation) and the Irish Research Council for Science Engineering and Technology. E.J.O'G. is an EMPOWER Postdoctoral Fellow. O.L.P. is a Royal Society University Research Fellow.
References
- 1.Naeem S., Thompson L. J., Lawler S. P., Lawton J. H., Woodfin R. M. 1994. Declining biodiversity can alter the performance of ecosystems. Nature 368, 734–737 10.1038/368734a0 (doi:10.1038/368734a0) [DOI] [Google Scholar]
- 2.Tilman D., Reich P. B., Knops J. M. H. 2006. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 441, 629–632 10.1038/nature04742 (doi:10.1038/nature04742) [DOI] [PubMed] [Google Scholar]
- 3.Yachi S., Loreau M. 2007. Does complementary resource use enhance ecosystem functioning? A model of light competition in plant communities. Ecol. Lett. 10, 54–62 10.1111/j.1461-0248.2006.00994.x (doi:10.1111/j.1461-0248.2006.00994.x) [DOI] [PubMed] [Google Scholar]
- 4.Estes J. A., Duggins D. O. 1995. Sea otters and kelp forests in Alaska: generality and variation in a community ecological paradigm. Ecol. Monogr. 65, 75–100 10.2307/2937159 (doi:10.2307/2937159) [DOI] [Google Scholar]
- 5.Coleman F. C., Williams S. L. 2002. Overexploiting marine ecosystem engineers: potential consequences for biodiversity. Trends Ecol. Evol. 17, 40–44 10.1016/S0169-5347(01)02330-8 (doi:10.1016/S0169-5347(01)02330-8) [DOI] [Google Scholar]
- 6.Brown J. H., Whitham T. G., Ernest S. K. M., Gehring C. A. 2001. Complex species interactions and the dynamics of ecological systems: long-term experiments. Science 293, 643–650 10.1126/science.293.5530.643 (doi:10.1126/science.293.5530.643) [DOI] [PubMed] [Google Scholar]
- 7.Pavoine S., Ollier S., Dufour A. B. 2005. Is the originality of a species measurable? Ecol. Lett. 8, 579–586 10.1111/j.1461-0248.2005.00752.x (doi:10.1111/j.1461-0248.2005.00752.x) [DOI] [Google Scholar]
- 8.Petchey O. L., Gaston K. J. 2002. Functional diversity (FD), species richness and community composition. Ecol. Lett. 5, 402–411 10.1046/j.1461-0248.2002.00339.x (doi:10.1046/j.1461-0248.2002.00339.x) [DOI] [Google Scholar]
- 9.Petchey O. L., Hector A., Gaston K. J. 2004. How do different measures of functional diversity perform? Ecology 85, 847–857 10.1890/03-0226 (doi:10.1890/03-0226) [DOI] [Google Scholar]
- 10.Naeem S., Wright J. P. 2003. Disentangling biodiversity effects on ecosystem functioning: deriving solutions to a seemingly insurmountable problem. Ecol. Lett. 6, 567–579 10.1046/j.1461-0248.2003.00471.x (doi:10.1046/j.1461-0248.2003.00471.x) [DOI] [Google Scholar]
- 11.Reiss J., Bridle J. R., Montoya J. M., Woodward G. 2009. Emerging horizons in biodiversity and ecosystem functioning research. Trends Ecol. Evol. 24, 505–514 10.1016/j.tree.2009.03.018 (doi:10.1016/j.tree.2009.03.018) [DOI] [PubMed] [Google Scholar]
- 12.Petchey O. L., Eklof A., Borrvall C., Ebenman B. 2008. Trophically unique species are vulnerable to cascading extinction. Am. Nat. 171, 568–579 [DOI] [PubMed] [Google Scholar]
- 13.Blackburn T. M., Petchey O. L., Cassey P., Gaston K. J. 2005. Functional diversity of mammalian predators and extinction in island birds. Ecology 86, 2916–2923 10.1890/04-1847 (doi:10.1890/04-1847) [DOI] [Google Scholar]
- 14.McCann K., Hastings A., Huxel G. R. 1998. Weak trophic interactions and the balance of nature. Nature 395, 794–798 10.1038/27427 (doi:10.1038/27427) [DOI] [Google Scholar]
- 15.Neutel A. M., Heesterbeek J. A. P., de Ruiter P. C. 2002. Stability in real food webs: weak links in long loops. Science 296, 1120–1123 10.1126/science.1068326 (doi:10.1126/science.1068326) [DOI] [PubMed] [Google Scholar]
- 16.O'Gorman E. J., Emmerson M. C. 2009. Perturbations to trophic interactions and the stability of complex food webs. Proc. Natl Acad. Sci. USA 106, 13 393–13 398 10.1073/pnas.0903682106 (doi:10.1073/pnas.0903682106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bascompte J., Melian C. J., Sala E. 2005. Interaction strength combinations and the overfishing of a marine food web. Proc. Natl Acad. Sci. USA 102, 5443–5447 10.1073/pnas.0501562102 (doi:10.1073/pnas.0501562102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Gorman E. J., Jacob U., Jonsson T., Emmerson M. C. 2010. Interaction strength, food web topology and the relative importance of species in food webs. J. Anim. Ecol. 79, 682–692 [DOI] [PubMed] [Google Scholar]
- 19.Kokkoris G. D., Jansen V. A. A., Loreau M., Troumbis A. Y. 2002. Variability in interaction strength and implications for biodiversity. J. Anim. Ecol. 71, 362–371 10.1046/j.1365-2656.2002.00604.x (doi:10.1046/j.1365-2656.2002.00604.x) [DOI] [Google Scholar]
- 20.Montoya J. M., Emmerson M. C., Woodward G. 2005. Perturbations and indirect effects in complex food webs. In Dynamic food webs: multispecies assemblages, ecosystem development, and environmental change (eds De Ruiter P. C., Wolters V., Moore J. C.), pp. 369–380 Amsterdam, The Netherlands: Academic Press [Google Scholar]
- 21.Emmerson M. C., Raffaelli D. 2004. Predator–prey body size, interaction strength and the stability of a real food web. J. Anim. Ecol. 73, 399–409 10.1111/j.0021-8790.2004.00818.x (doi:10.1111/j.0021-8790.2004.00818.x) [DOI] [Google Scholar]
- 22.Woodward G., Ebenman B., Emmerson M. C., Montoya J. M., Olesen J. M., Valido A., Warren P. H. 2005. Body size in ecological networks. Trends Ecol. Evol. 20, 402–409 10.1016/j.tree.2005.04.005 (doi:10.1016/j.tree.2005.04.005) [DOI] [PubMed] [Google Scholar]
- 23.Hall S. J., Raffaelli D. G. 1991. Food web patterns: lessons from a species-rich food web. J. Anim. Ecol. 60, 823–842 [Google Scholar]
- 24.Jonsson T., Cohen J. E., Carpenter S. R. 2005. Food webs, body size, and species abundance in ecological community description. Adv. Ecol. Res. 36, 1–84 10.1016/S0065-2504(05)36001-6 (doi:10.1016/S0065-2504(05)36001-6) [DOI] [Google Scholar]
- 25.Woodward G., Speirs D. C., Hildrew A. G. 2005. Quantification and resolution of a complex, size-structured food web. Adv. Ecol. Res. 36, 85–135 10.1016/S0065-2504(05)36002-8 (doi:10.1016/S0065-2504(05)36002-8) [DOI] [Google Scholar]
- 26.Miller L. P., Gaylord B. 2007. Barriers to flow: the effects of experimental cage structures on water velocities in high-energy subtidal and intertidal environments. J. Exp. Mar. Biol. Ecol. 344, 215–228 10.1016/j.jembe.2007.01.005 (doi:10.1016/j.jembe.2007.01.005) [DOI] [Google Scholar]
- 27.Jacob U. 2005. Trophic dynamics of Antarctic Shelf ecosystems—food webs and energy flow budgets. PhD thesis (online at http://nbn-resolving.de/urn:nbn:de:gbv:46-diss000118684), University of Bremen, Germany [Google Scholar]
- 28.Opitz S. 1993. A quantitative model of the trophic interactions in a Caribbean coral reef ecosystem. In Trophic models of aquatic ecosystems. ICLARM Conference Proceedings 26, Metro Manila, Philippines (eds Christensen V., Pauly D.), pp. 259–267 Metro Manila, Philippines: ICLARM; Copenhagen, Denmark: International Council for the Exploration of the Sea; Copenhagen, Denmark: DANIDA. [Google Scholar]
- 29.Duffy J. E., Hay M. E. 1994. Herbivore resistance to seaweed chemical defense: the roles of mobility and predation risk. Ecology 75, 1304–1319 10.2307/1937456 (doi:10.2307/1937456) [DOI] [Google Scholar]
- 30.Stachowicz J. J., Hay M. 1999. Reduced mobility is associated with compensatory feeding and increased diet breadth of marine crabs. Mar. Ecol.-Prog. Ser. 188, 169–178 10.3354/meps188169 (doi:10.3354/meps188169) [DOI] [Google Scholar]
- 31.Raffaelli D. G., Hall S. J. 1996. Assessing the relative importance of trophic links in food webs. Food webs: integration of patterns and dynamics (eds Polis G. A., Winemiller K. O.), pp. 185–191 New York, NY: Chapman & Hall [Google Scholar]
- 32.Steiner C. F. 2005. Temporal stability of pond zooplankton assemblages. Freshw. Biol. 50, 105–112 10.1111/j.1365-2427.2004.01310.x (doi:10.1111/j.1365-2427.2004.01310.x) [DOI] [Google Scholar]
- 33.Fukami T., Naeem S., Wardle D. A. 2001. On similarity among local communities in biodiversity experiments. Oikos 95, 340–348 10.1034/j.1600-0706.2001.950216.x (doi:10.1034/j.1600-0706.2001.950216.x) [DOI] [Google Scholar]
- 34.Williams R. J., Martinez N. D. 2000. Simple rules yield complex food webs. Nature 404, 180–183 10.1038/35004572 (doi:10.1038/35004572) [DOI] [PubMed] [Google Scholar]
- 35.Yachi S., Loreau M. 1999. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl Acad. Sci. USA 96, 1463–1468 10.1073/pnas.96.4.1463 (doi:10.1073/pnas.96.4.1463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petchey O. L., McPhearson P. T., Casey T. M., Morin P. J. 1999. Environmental warming alters food-web structure and ecosystem function. Nature 402, 69–72 10.1038/47023 (doi:10.1038/47023) [DOI] [Google Scholar]
- 37.Pauly D., Christensen V., Dalsgaard J., Froese R., Torres F. 1998. Fishing down marine food webs. Science 279, 860–863 10.1126/science.279.5352.860 (doi:10.1126/science.279.5352.860) [DOI] [PubMed] [Google Scholar]
- 38.Jennings S., Pinnegar J. K., Polunin N. V. C., Boon T. W. 2001. Weak cross-species relationships between body size and trophic level belie powerful size-based trophic structuring in fish communities. J. Anim. Ecol. 70, 934–944 10.1046/j.0021-8790.2001.00552.x (doi:10.1046/j.0021-8790.2001.00552.x) [DOI] [Google Scholar]
- 39.Rossberg A. G., Brannstrom A., Dieckmann U. 2010. How trophic interaction strength depends on traits. Theoret. Ecol. 3, 13–24 10.1007/s12080-009-0049-1 (doi:10.1007/s12080-009-0049-1) [DOI] [Google Scholar]
- 40.Diaz S., Cabido M. 2001. Vive la difference: plant functional diversity matters to ecosystem processes. Trends Ecol. Evol. 16, 646–655 10.1016/S0169-5347(01)02283-2 (doi:10.1016/S0169-5347(01)02283-2) [DOI] [Google Scholar]
- 41.Martinez N. D. 1991. Artifacts or attributes: effects of resolution on the little-rock lake food web. Ecol. Monogr. 61, 367–392 10.2307/2937047 (doi:10.2307/2937047) [DOI] [Google Scholar]
- 42.Ehrlich P. A., Ehrlich A. H. 1981. Extinction: the causes and consequences of the disappearance of species. New York, NY: Random House [Google Scholar]
- 43.Beisner B. E., Haydon D. T., Cuddington K. 2003. Alternative stable states in ecology. Front. Ecol. Environ. 1, 376–382 10.1890/1540-9295(2003)001[0376:ASSIE]2.0.CO;2 (doi:10.1890/1540-9295(2003)001[0376:ASSIE]2.0.CO;2) [DOI] [Google Scholar]