Abstract
The Karner blue butterfly (KBB), Lycaeides melissa samuelis, is a federally protected taxon whose relationship to the Melissa blue, Lycaeides melissa, has been a point of contention during the 66 years since the KBB was first described. Using a large population-genomic dataset and a model of population divergence with migration, we investigated the relationship between the KBB and L. melissa, as well as the relationship between L. melissa and a third taxon, Lycaeides idas. We report that gene flow between the KBB and L. melissa is low, and comparable to gene flow between L. melissa and L. idas. Considering this population-genetic evidence, we conclude that the KBB is a unique evolutionary lineage that should be recognized as Lycaeides samuelis.
Keywords: conservation genetics, gene flow, Lycaeides melissa samuelis, population genomics, next-generation sequencing
1. Introduction
Estimates of gene flow among natural populations have aided in the discovery, demarcation and prioritization of units for conservation, from evolutionarily significant units (ESUs) to species [1]. Typically, efforts in conservation genetics have relied on a small number of genetic markers, and in many cases a single marker, mitochondrial DNA, which has limited the ability of conservationists to address key issues [2]. Technological advances now make it possible to sample an unprecedented number of genetic regions distributed across the genome of non-model species, though these tools have yet to be widely applied to species of conservation concern [3,4]. Here, we focus on the Karner blue butterfly (KBB), Lycaeides melissa samuelis Nabokov, and use a population-genomic dataset to address questions related to the genetic distinctness of this taxon.
Populations of the KBB occur from Minnesota to New Hampshire in association with a single larval host plant, Lupinus perennis, in habitats that include oak savannah and pine barrens [5]. Suitable KBB habitat decreased sharply in the twentieth century as a result of fire suppression and land conversion, resulting in range-wide declines in population size, ultimately leading to placement of the KBB on the US list of endangered species in 1992 [5]. Based on variation in wing pattern and genitalic morphology, the KBB was described as a subspecies of Lycaeides melissa 66 years ago by Vladimir Nabokov in 1944, who subsequently and informally revised his opinion to consider it a distinct species [6,7]. Population-genetic evidence bearing on the distinctness of the KBB was reported by Packer et al. [8], who concluded that the KBB was indistinguishable from L. melissa based on 34 allozyme loci. Subsequent molecular studies raised the possibility, contra Packer et al. [8], that the KBB might be a genetically distinct lineage, albeit with introgression of mitochondrial DNA from L. melissa [9,10].
We used a population-genomic dataset described by Gompert et al. [3] to investigate the relationship between the KBB and the closely related L. melissa. To provide a context for that comparison, we also investigated the relationship between L. melissa and a third closely related taxon, Lycaeides idas. Specifically, we asked the following questions: (i) when did the KBB and L. melissa diverge relative to L. melissa and L. idas? (ii) how do population sizes vary among the three taxa? and (iii) has there been a history of gene flow between the KBB and L. melissa? If so, how do rates of gene flow compare with those between L. melissa and L. idas? Answers to these questions will provide a more complete picture of the evolutionary history of the KBB.
2. Material and methods
The population-genomic dataset used in this study was generated using 454 pyrosequencing of individuals from 12 Lycaeides populations [3]. Here, we focus on four populations: one KBB population, two L. melissa populations (‘west’ and ‘east’) and one L. idas population (figure 1a). The ‘populations’ consist of pooled DNA of 15 individuals from three sampling localities per population. Pyrosequencing of these four populations produced 43 618 sequences, which assembled into a total of 9955 contiguous alignments (contigs), with an average of 329 base pairs per alignment. The KBB population consisted of individuals from Fort McCoy (Wisconsin), Necedah (Wisconsin) and Saratoga (New York). Further locality information, and details of data generation are given in the electronic supplementary material.
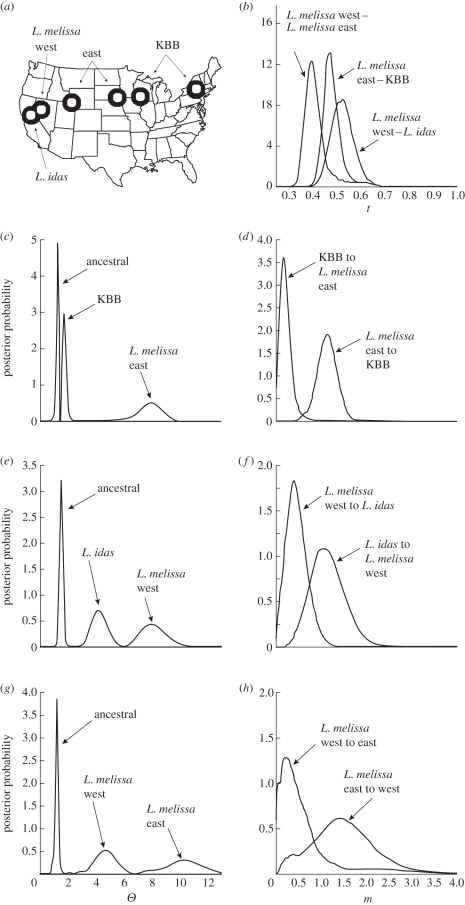
Figure 1.
(a) Map of sampled locations and taxa in North America and posterior probability distributions for parameters as follows: (b) splitting times, (c,e and g) populations sizes and (d,f and h) migration rates. Height of curves corresponds to the estimated probability that a given parameter value is true, given the data (95% confidence intervals are reported in table 1).
To investigate the population-genetic history of the KBB and related taxa, we used the isolation with migration model (IM) of Nielsen & Wakeley [11] and Hey & Nielsen [12], as implemented in the software IMa [13]. IMa uses Markov chain Monte Carlo (MCMC) sampling to obtain maximum-likelihood estimates of six parameters: ancestral population size (ΘA), current population sizes (Θ1 and Θ2), rates of migration between two populations subsequent to an initial split (m1 and m2) and the timing of divergence (t). We used separate runs of IMa to investigate the histories of the following pairs of populations: KBB and L. melissa east; L. melissa east and L. melissa west; and L. melissa west and L. idas (figure 1a). Note that the comparisons involving L. melissa are made by geographical proximity, with L. melissa east being closest to the KBB, and L. melissa west closest to L. idas. The number of genetic regions (contigs) involved in the comparisons are as follows: 317 regions for KBB and L. melissa, 256 for the comparison within L. melissa and 188 for L. melissa and L. idas (a total of 440 unique regions across all comparisons). These analyses included contigs with a minimum of three sequences per population; the maximum number per population was 198, and the mean was 9.16 sequences per contig per population. For each pair of populations, 15 independent MCMC simulations were run for 500 000 generations, with a burn in of 100 000 generations; results were combined prior to maximum-likelihood parameter estimation. Additional details of the IM model are discussed in the electronic supplementary material.
The parameters estimated by IMa are in units that incorporate mutation rate (see electronic supplemental material). Given mutation rates (per gene), these parameters can be converted into years or individuals. We do not have an estimate of genome-wide mutation rate for Lycaeides. Instead, we use the parameters directly to draw comparative conclusions about the histories of our taxa. We note also that our taxa have differing numbers of generations per year (KBB has two, L. melissa has two to three and L. idas has one), which would complicate a conversion of the splitting parameter (t) into years. Instead, t can be thought of as the evolutionarily effective amount of time that has passed since divergence.
3. Results
MCMC simulations produced posterior probability distributions for most of our focal parameters that were sharp, suggesting high confidence in our estimates (figure 1; point estimates and 95% confidence intervals are given in table 1). Estimated splitting times for the three comparisons were clustered (figure 1b). Estimates of population sizes found small ancestral populations in all cases, larger contemporary populations for L. melissa and L. idas, and a small contemporary effective population size for the KBB (figure 1c,e and g). Estimated rates of migration were asymmetrical in all cases (figure 1d,f and h). For example, gene flow from L. melissa east to KBB was higher than the reverse direction (figure 1d). The highest rates of gene flow were observed in the intraspecific comparison of the two L. melissa populations: note the point estimates as well as the higher upper bounds of the confidence intervals relative to the comparisons involving the KBB and L. idas (table 1).
Table 1.
Estimates of divergence times (t), migration rates (m), Θ (four times the effective population size times μ), and 95% confidence intervals (see supplementary text for more details). Lowercase letters after population names refer to panels in figure 1.
| comparison | t |
| KBB and L. melissa east (b) | 0.475 (0.425–0.655) |
| L. melissa west and L. idas (b) | 0.525 (0.445–0.625) |
| L. melissa west and L. melissa east (b) | 0.395 (0.355–0.605) |
| direction of migration | m |
| KBB to L. melissa east (d) | 0.165 (0.025–0.925) |
| L. melissa east to KBB (d) | 1.125 (0.665–1.535) |
| L. melissa west to L. idas (f) | 0.405 (0.075–0.975) |
| L. idas to L. melissa west (f) | 1.085 (0.495–1.945) |
| L. melissa west to L. melissa east (h) | 0.205 (0.035–6.285) |
| L. melissa east to L. melissa west (h) | 1.395 (0.245–3.565) |
| population | Θ |
| KBB (c) | 1.715 (1.502–1.997) |
| L. melissa east (c) | 7.936 (5.533–9.527) |
| L. melissa west (e) | 7.924 (6.404–10.052) |
| L. idas (e) | 4.123 (3.211–5.491) |
| L. melissa west (g) | 4.648 (2.981–6.277) |
| L. melissa east (g) | 10.388 (7.203–13.906) |
4. Discussion
The ecological and morphological distinctness of the KBB have been well established. Differences in wing pattern and genitalic variation distinguish the KBB from other Lycaeides [6,14]. Ecologically, the KBB uses a single larval host, whereas the closely related L. melissa uses a large number of legumes [15]. Previous genetic work has suggested varying levels of hybridization with L. melissa, or very recent derivation from a common ancestor [3,9]. We find evidence for limited gene flow between L. melissa and the KBB. In particular, we find rates of gene flow between those taxa that are lower than rates of gene flow between populations of L. melissa, and comparable to rates of gene flow between L. melissa and L. idas. It is difficult to make direct comparisons between these results and previous work in this system, given the data involved here which is different in both quantity and quality. However, times of divergence and population size estimates agree with previous work in this system: the split between L. idas and L. melissa, for example, encompasses a greater amount of evolutionary time than the split between L. melissa and the KBB, and the estimated population size for the KBB is smaller than for L. melissa.
Although questions of taxonomy are of secondary interest to the investigation of evolutionary processes, we conclude by asking if subspecific status for the KBB is appropriate given what has been learned in the last 66 years. There are no thresholds for multi-locus genetic distinctness or levels of gene flow that are widely agreed upon as being consistent with taxonomic species status [2,16]. Given the ubiquity and diversity of introgression that has been discovered in recent years for many taxa [17], it is hard to imagine how a universal, taxonomic-genetic threshold would be possible or even desirable. Instead, we suggest that sufficiently powerful genetic data can be used in a comparative sense to address localized taxonomic problems. We report levels of gene flow between the KBB and L. melissa that are comparable to levels of gene flow between L. melissa and L. idas, which are recognized as distinct taxa. Rates of gene flow between the KBB and L. melissa are also lower than rates detected between conspecific populations of L. melissa. In light of the comparative picture of gene flow presented here based on hundreds of loci, coupled with what we know about the distinctiveness of the KBB in terms of host plant association, morphology and life-history variation, we conclude that the current subspecific designation for the KBB is inappropriate, and that the KBB should be elevated to Lycaeides samuelis Nabokov.
References
- 1.DeSalle R., Amato G. 2004. The expansion of conservation genetics. Nat. Rev. Genet. 5, 702–712 10.1038/nrg1425 (doi:10.1038/nrg1425) [DOI] [PubMed] [Google Scholar]
- 2.Forister M. L., Nice C. C., Fordyce J. A., Gompert Z., Shapiro A. M. 2008. Considering evolutionary processes in the use of single-locus genetic data for conservation, with examples from the Lepidoptera. J. Insect Conserv. 12, 37–51 10.1007/s10841-006-9061-6 (doi:10.1007/s10841-006-9061-6) [DOI] [Google Scholar]
- 3.Gompert Z., Forister M. L., Fordyce J. A., Nice C. C., Williamson R. J., Buerkle C. A. 2010. Bayesian analysis of molecular variance in pyrosequences quantifies population genetic structure across the genome of Lycaeides butterflies. Mol. Ecol. 19, 2455–2473 10.1111/j.1365-294X.2010.04727.x (doi:10.1111/j.1365-294X.2010.04727.x) [DOI] [PubMed] [Google Scholar]
- 4.Shaffer H. B., Thomson R. C. 2007. Delimiting species in recent radiations. Syst. Biol. 56, 896–906 10.1080/10635150701772563 (doi:10.1080/10635150701772563) [DOI] [PubMed] [Google Scholar]
- 5.Andow D. A., Baker R. J., Lane C. 1994. Karner blue butterfly: a symbol of a vanishing landscape. Minnesota Agricultural Experiment Station, University of Minnesota: Miscellaneous publication/Minnesota Agricultural Experiment Station, University of Minnesota. St Paul, Minnesota [Google Scholar]
- 6.Nabokov V. 1949. The nearctic members of the genus Lycaeides Hubner (Lycaenidae, Lepidoptera). Bull. Mus. Comp. Zool. 101, 479–541 [Google Scholar]
- 7.Nabokov V. 1975. From letter to Robert Dirig. In Nabokov's butterflies (eds Boyd B., Pyle R. M.), pp. 713–714 Boston, MA: Beacon Press [Google Scholar]
- 8.Packer L., Taylor J. S., Savignano D. A., Bleser C. A., Lane C. P., Sommers L. A. 1998. Population biology of an endangered butterfly, Lycaeides melissa samuelis (Lepidoptera; Lycaenidae): genetic variation, gene flow, and taxonomic status. Can. J. Zool.-Rev. Can. Zool. 76, 320–329 10.1139/cjz-76-2-320 (doi:10.1139/cjz-76-2-320) [DOI] [Google Scholar]
- 9.Gompert Z., Nice C. C., Fordyce J. A., Forister M. L., Shapiro A. M. 2006. Identifying units for conservation using molecular systematics: the cautionary tale of the Karner blue butterfly. Mol. Ecol. 15, 1759–1768 10.1111/j.1365-294X.2006.02905.x (doi:10.1111/j.1365-294X.2006.02905.x) [DOI] [PubMed] [Google Scholar]
- 10.Nice C. C., Anthony N., Gelembiuk G., Raterman D., Ffrench-Constant R. 2005. The history and geography of diversification within the butterfly genus Lycaeides in North America. Mol. Ecol. 14, 1741–1754 10.1111/j.1365-294X.2005.02527.x (doi:10.1111/j.1365-294X.2005.02527.x) [DOI] [PubMed] [Google Scholar]
- 11.Nielsen R., Wakeley J. 2001. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158, 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hey J., Nielsen R. 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167, 747–760 10.1534/genetics.103.024182 (doi:10.1534/genetics.103.024182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hey J., Nielsen R. 2007. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc. Natl Acad. Sci. USA 104, 2785–2790 10.1073/pnas.0611164104 (doi:10.1073/pnas.0611164104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucas L. K., Fordyce J. A., Nice C. C. 2008. Patterns of genitalic morphology around suture zones in North American Lycaeides (Lepidoptera: Lycaenidae): implications for taxonomy and historical biogeography. Ann. Entomol. Soc. Am. 101, 172–180 10.1603/0013-8746(2008)101[172:POGMAS]2.0.CO;2 (doi:10.1603/0013-8746(2008)101[172:POGMAS]2.0.CO;2) [DOI] [Google Scholar]
- 15.Austin G. T., Leary P. J. 2008. Larval hostplants of butterflies in Nevada. Holarctic Lepid. 12, 1–134 [Google Scholar]
- 16.Knowles L. L., Carstens B. C. 2007. Delimiting species without monophyletic gene trees. Syst. Biol. 56, 887–895 10.1080/10635150701701091 (doi:10.1080/10635150701701091) [DOI] [PubMed] [Google Scholar]
- 17.Seehausen O. 2004. Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207 10.1016/j.tree.2004.01.003 (doi:10.1016/j.tree.2004.01.003) [DOI] [PubMed] [Google Scholar]