Abstract
During spermiogenesis in the water fern, Marsilea vestita, basal bodies are synthesized de novo in cells that lack preexisting centrioles, in a particle known as a blepharoplast. We have focused on basal body assembly in this organism, asking what components are required for blepharoplast formation. Spermiogenesis is a rapid process that is activated by placing dry microspores into water. Dry microspores contain large quantities of stored protein and stored mRNA, and inhibitors reveal that certain proteins are translated from stored transcripts at specific times during development. Centrin translation accompanies blepharoplast appearance, while β-tubulin translation occurs later, during axonemal formation. In asking whether centrin is an essential component of the blepharoplast, we used antisense, sense, and double-stranded RNA probes made from the Marsilea centrin cDNA, MvCen1, to block centrin translation. We employed a novel method to introduce these RNAs directly into the cells. Antisense and sense both arrest spermiogenesis when blepharoplasts should appear, and dsRNA made from the same cDNA is an effective inhibitor at concentrations at least 10 times lower than either of the single-stranded RNA used in these experiments. Blepharoplasts are undetectable and basal bodies fail to form. Antisense, sense, and dsRNA probes made from Marsilea β-tubulin permitted normal development until axonemes form. In controls, antisense, sense, and dsRNA, made from a segment of HIV, had no effect on spermiogenesis. Immunoblots suggest that translational blocks induced by centrin-based RNA are gene specific and concentration dependent, since neither β-tubulin- nor HIV-derived RNAs affects centrin translation. The disruption of centrin translation affects microtubule distributions in spermatids, since centrin appears to control formation of the cytoskeleton and motile apparatus. These results show that centrin plays an essential role in the formation of a motile apparatus during spermiogenesis of M. vestita.
INTRODUCTION
Throughout most of the eukaryotic realm, the formation of male gametes involves the synthesis and assembly of a motile apparatus. The motile apparatus comprises flagellar or ciliary axonemes, whose rapid beat frequency and precise beat shape are the consequence of mechanochemical interactions between scores of proteins that are arranged in a remarkably complex cylindrical array (Dutcher, 1995). A controlling template for axonemal formation is the basal body (Mizukami and Gall, 1966), which resides in the cortical cytoplasm at the base of the axoneme (Ringo, 1967; Fulton, 1971; Dibbayawan et al., 1995). Basal bodies closely resemble centrioles (Gould, 1975; Kuriyama and Borisy, 1981; Kochanski and Borisy, 1990), and they are usually formed from or in close association with centrioles (Dirksen, 1991;Marshall and Rosenbaum, 2000), (however, see Fulton [1971]; Fulton and Dingle [1971]; Gould [1975]). The ubiquity of this association has led to the perception that all basal bodies arise from these cytoplasmic organelles. About a century ago, however, lower plants were shown to produce motile male gametes, in a process that involved the formation of basal bodies in cells that lacked preexisting centrioles (Webber, 1897, 1901; Chamberlain, 1898; Sharp, 1914; Duckett, 1973; Hepler, 1976; Vaughn and Harper, 1998).
In the developing spermatids of lower plants, basal bodies form in a particle that was originally called the blepharoplast (Webber, 1897, 1901) because as the spermatids matured, it looked like an eyelash. Ultrastructural analyses reveal that, as a discrete spherical particle in the cytosol, the blepharoplast is the precursor for basal body assembly (Mizukami and Gall, 1966;Hepler, 1976;Doonan et al., 1986;Vaughn et al., 1993), and the elongated “eyelash” is actually a cytoskeletal array now known as a multilayered structure, or MLS (Carothers, 1975; Myles and Hepler, 1977; Duckett, 1973; Marc and Gunning, 1986, 1988; Hoffman and Vaughn, 1995; Wolniak et al., 2000; Klink and Wolniak, 2000), which is common to all plant spermatozoids. The uppermost stratum of the MLS is a ribbon of cross-linked microtubules that appears to control patterns of cell and nuclear elongation (Myles and Hepler, 1977, 1982) in the developing spermatid and is attached to the basal bodies of the motile apparatus (Carothers, 1975; Hoffman and Vaughn, 1995).
Recently, we have focused on blepharoplast formation and de novo basal body assembly in the male gametophytes of the water fern, Marsilea vestita. In this organism, microspores, which are meiotic products, are stored as dry structures that germinate when they are placed into an aqueous medium. The pattern of development in these male gametophytes is precise and synchronous; the entire process reaches completion in an 11-h period at 20°C (Mizukami and Gall, 1966; Hepler, 1976). During the 1st 5.5 h after imbibition, there are 9 mitotic divisions that produce 39 cells, all contained within the original microspore wall (Sharp, 1914). One cell is a prothallial remnant, 6 of the cells are sterile jacket cells, and the remaining 32 cells are spermatids (Sharp, 1914, Hepler, 1976, Pennell et al., 1986, 1988). During the next 5.5 h after imbibition, the spermatids differentiate to become freely swimming, ciliated gametes (Myles and Hepler, 1977).
In our previous studies, we (Hart and Wolniak, 1998, 1999) extended earlier work (Hyams et al., 1983; Pennell et al., 1986, 1988) that focused on patterns of transcription and translation in the gametophyte that are necessary for spermiogenesis. We showed that dry microspores contain substantial quantities of stored proteins, like α-, β- and γ-tubulin, and that spermiogenesis requires the translation of some new proteins from stored mRNA, such as centrin (Hart and Wolniak, 1998). In the male gametophyte of M. vestita, significant quantities of centrin protein first begin to accumulate from the translation of stored mRNA, ∼4 h after the spores are imbibed. In contrast, the translation of additional β-tubulin occurs > 8 h after imbibition, and appears necessary for axonemal formation, late in spermiogenesis (Hart and Wolniak, 1998). Most new centrin protein synthesis coincides with the formation of the blepharoplast, at a stage when the cell division cycles are nearing completion. Centrin abundance reaches maximal levels at ∼6 h after imbibition, as the basal bodies formed from the blepharoplast reach maturity and as the MLS begins to form. The question that remains is whether centrin translation is an essential prerequisite for formation of the blepharoplast.
Centrin is a calcium binding protein (Salisbury, 1995; Schiebel and Bornens, 1995) that resides in and around basal bodies (Baron et al., 1992; Taillon et al., 1992; Levy et al., 1996). It is synthesized during the assembly of a motile apparatus during the amoebo-flagellate transformation in Naegleria (Levy et al., 1998). Centrioles are preexisting structures in the amoebae. Centrins that participate in the organization and function of centrosomes (Middendorp et al., 2000) in higher organisms are also found in the lumenal space of centrioles (Paoletti et al., 1996), but precise roles for centrin in centrosomal and centriolar organization and function remain to be determined. During its ontogeny, the blepharoplast in M. vestita briefly serves as a functional centrosome, and then differentiates as a basal body factory (Hepler, 1976; Wolniak et al., 2000). Centrin proteins are related to basal body-associated proteins in lower eukaryotes (Moudjou et al., 1991; Hart and Wolniak, 1999), and they are also present in or near the blepharoplast and in or near the MLS of fern spermatids (Vaughn et al., 1993; Hoffman et al., 1994). Centrins interact with other proteins (Baron et al., 1991; Klotz et al., 1997), such as pericentrin (Doxsey et al., 1994), γ-tubulin, and cytoplasmic dynein (Purohit et al., 1999; Young et al., 2000).
Is centrin translation in gametophytes of M. vestita an essential and rate-limiting factor for blepharoplast formation? To test for a centrin requirement, we blocked centrin translation in developing male gametophytes of M. vestita in vivo by employing antisense RNA (Holt et al., 1988) that was made from our MvCen1 centrin cDNA clone (Hart and Wolniak, 1999). We devised a novel protocol for the introduction of our constructs into the cytosol of the gametophytes. Our control experiments, using sense constructs, revealed inhibitory effects that resembled those observed in Caenorhabditis elegans and other organisms (Fire et al., 1998; Kennerdell and Carthew, 1998; Montgomery and Fire, 1998; Montgomery et al., 1998; Tabara et al., 1998, 1999; Ngo et al., 1998; Bashirullah et al., 1999; Driver et al., 1999; Fire, 1999; Misquitta and Paterson, 1999; Sanchez-Alvorado and Newmark, 1999; Boscher and Labouesse, 2000; Klink and Wolniak, 2000; Sharp and Zamore, 2000; Wianny and Zernicka-Goetz, 2000), prompting us to generate double-stranded RNA (dsRNA) constructs as a means to assess whether the effects were the result of RNA interference (RNAi). Since no one had heretofore added dsRNA directly to plant cells, a portion of our experimental design required an assessment of the efficacy of the approach; to this end, we performed a variety of additional controls, including treatments of cells with antisense, sense, and dsRNA derived from a β-tubulin cDNA obtained from our library, and a noncoding segment of HIV. Tubulin was selected because its pattern of new translation differs from that of centrin, and HIV was selected because it represents a transcript that should be irrelevant in the fern gametophyte.
In this paper, we show that centrin translation is necessary for the formation of the blepharoplast, and for differentiation of the motile apparatus in the developing spermatid of M. vestita. This is the 1st demonstration of a critical function for centrin protein during the formation of the motile apparatus in a higher eukaryote. To perform RNAi experiments, we developed a novel method of introducing these constructs into the cells, by treating the dry spores with various RNAs at the time of imbibition. We include a large number of control experiments to assess the specificity of the response in the context of the underlying biology. Because this is the 1st demonstration that the direct addition of dsRNA to plant cells results in altered patterns of translation, our experimental design also assesses the efficacy of this approach.
MATERIALS AND METHODS
Sporocarps, the dried structures containing the microspores and megaspores of M. vestita, were originally obtained from Dr. Peter Hepler (University of Massachusetts, Amherst, MA). The original source of sporocarps is a natural population of ferns growing at Lake Laganita, at Stanford, CA (Hepler, 1976). In addition, 10 ponds in the greenhouses at the University of Maryland have been constructed and used to generate additional quantities of sporocarps. Sporocarps were ground with three 1-s bursts in a commercial coffee grinder (Braun, model KSM2, Lynnfield, MA) and sifted through 425 μm and 212 μm wire sieves to separate the microspores from the megaspores. Microspores were cultured in a shaking water bath (New Brunswick Scientific, Gyrorotary, Model G7, New Brunswick, NJ) maintained at 20°C (Brinkmann Lauda, model RM6, Westburg, NY) at a concentration of 10 mg/10 ml of Laetsch's medium (Laetsch, 1967). α-Amanitin and cycloheximide (Sigma, St. Louis, MO) were used at concentrations of 1, 10, and 100 μM.
RNAi Experiments
A full-length, 750 base pair (bp) centrin cDNA (MvCen1 accession # U92973) was originally isolated from our cDNA library during a heterologous screen, and previously characterized (Hart and Wolniak, 1998). A 3′-truncated centrin cDNA was prepared from MvCen1 by KpnI (Life Technologies, Rockville, MD) cleavage producing a construct ∼250 bp in length. The truncated centrin cDNA clone was transcribed in vitro according to manufacturers instructions (Epicentre Technologies, T3, T7 Ampliscribe transcription kit, Madison, WI). RNA was prepared using protocols adapted from Fire et al. (1998). In parallel, we made comparable 250 bp 5′-truncated sense, antisense and dsRNA probes from a β-tubulin cDNA that we isolated from our M. vestita library. We made 250 bp sense, antisense, and dsRNAs from a cDNA that encodes a random sequence from the HIV genome (kindly provided by Dr. Jeffrey deStefano, University of Maryland, College Park, MD) for use in a series of negative control experiments. Sequence analysis of this HIV-derived probe revealed no homologies with M. vestita (our unpublished results).
Four milligrams of dry microspores were added to 1 ml of sterile water with either sense, antisense, or double-stranded RNA at a concentration of 2000, 200, 20, 2 μg/ml, or untreated in 2-ml centrifuge tubes. Spores were agitated on an Orbitron agitator (model 260200, Boekel Industries, Feasterville, PA) with aeration for a period of 8 h at 20°C. Alternatively, after incubation in the 2-ml microcentrifuge tubes, we transferred the microspores to 50-ml flasks containing 25 ml of medium. For all of these experiments, the cells were harvested 8 h after imbibition and processed for structural observations, or fractionated for biochemical analysis, as described below.
Histology
Excess culture medium was removed from the flask with a pipette, and spores were pooled into a volume of 1 ml and transferred to 2-ml centrifuge tubes. For fixation, 8% paraformaldehyde pH 7.4 in PBS was diluted 1:1 with the culture medium containing spores to bring the final concentration of paraformaldehyde to 4%. The spore walls were fractured in a stainless steel mortar and pestle, and the cells were fixed according to protocols originally described by Hepler (1976). The spores were then transferred to a microcentrifuge tube and allowed to settle without centrifugation. Spores were washed 3 times in PBS (15 min each wash) and dehydrated through 10, 20, 30, 40, 50, 60, and 70% ethanol in PBS (45 min each). Dehydration proceeded through 80% and 90% ethanol in distilled water (45 min each) followed by 3 changes of 100% ethanol. Infiltration and polymerization in polymethacrylate was performed with procedures described by Baskin et al., (1992).
Immunocytochemistry
Male gametophytes from M. vestita exhibit high amounts of autofluorescence from spore wall and cytoplasmic polyphenolics, thereby prompting us to employ immunogold histochemistry. Methacrylate sections (1–2 μm) were placed on cleaned glass microscope slides in drops of water and allowed to adhere to the glass by drying at 40°C on a slide warmer. The plastic was then removed by deep-etching in chloroform for 2 h, followed by a 30-min acetone treatment. The sections were incubated with PBS pH 7.4 (three 5-min incubations) and blocked with a solution containing 1.0% BSA fraction V (Fischer Scientific, Pittsburg, PA), 7.5% glycine (Fischer Scientific), 5.0% Idaho Spuds (Pillsbury, Minneapolis, MN), and 5.0% Carnation nonfat dried milk (Nestle; Solon, OH) in PBS. Slides were transferred to PBS (three 5-min incubations) followed by one 5-min incubation in PBST (PBS with 0.1% Tween 20). Anticentrin (1:50 in PBST) monoclonal antibody 20H5, directed against Chlamydomonas reinhardtii, (a gift of Dr. Jeffery Salisbury, Mayo Clinic, Rochester, MN), anti-α/β-tubulin monoclonal antibodies (Amersham, Buckinghamshire, UK - 1:100 in PBST), and anti-P28 antibody (a gift of Dr. Gianni Piperno, Mt. Sinai School of Medicine, NY), were added to the sections and incubated for 1 h in a humid chamber at room temperature. Slides were transferred to PBST (three 5-min incubations). Gold-conjugated antimouse secondary antibodies (Research Diagnostics, Flanders, NJ; 1:500 in PBST) were incubated on sections in a humid chamber for 1 h at room temperature for detection of anticentrin and anti-β-tubulin antibodies. Gold-conjugated antirabbit IgG secondary antibodies (Research Diagnostics) were used for the detection of P28. Slides were transferred to PBST (three 5-min incubations) followed by immersion in distilled water (three 5-min changes). This transfer was done according to the manufacturer's instructions, since silver precipitation occurs nonspecifically because of salts present in wash solutions. Silver enhancement was performed for 15 min, followed by three 5-min washes in distilled water.
Protein Extraction
Microspores were grown in Laetsch's (1967) medium or in distilled water. Protein extractions were performed on dry spores as previously described (Hart and Wolniak, 1998), and on spores cultured for 30 min at 4°C, and then grown for varying intervals from 1 to 11 h, at 20°C in a shaking water bath. Spores were grown at a density of 1 mg/ml of culture medium. Male gametophytes were collected by centrifugation at 9000 × g for 10 min. Protein extraction, SDS PAGE and protein transfers onto PVDF membranes (Immobilion P, Millipore, Bedford, MA) for immunoblotting were performed according to procedures described earlier (Hart and Wolniak, 1998). Protein concentrations were measured using assays developed by Bradford (1976).
Immunoblotting
PVDF membranes were wetted in methanol for 30 s and transferred to PBS at pH7.4 (three 5-min incubations). Blocking and antibody incubations were the same as those used for immunocytochemistry. Primary antibody concentrations were 1:100 (centrin 20H5 monoclonal antibody), 1:500 (anti-α/β-tubulin antibodies), and 1:250 (anti-P28 antibodies). Antispecies horseradish peroxidase secondary antibodies (Amersham) were diluted 1:1500 in PBST and membranes were incubated in a humid chamber for 1 h at room temperature. Membranes were transferred to PBST (three 10-min incubations and one 20-min incubation). Chemiluminescence detection of binding was performed using a 1:1 dilution of ECL (Amersham) on Kodak X-OMAT AR film (Eastman Kodak, Rochester, NY), or with a STORM 840 PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
RESULTS
Development of the Male Gametophyte
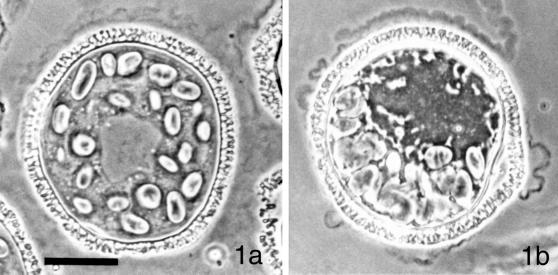
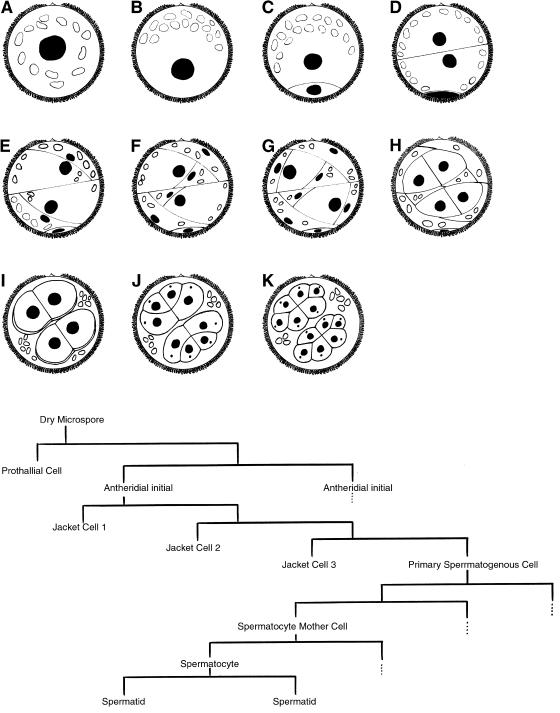
The pattern of development of the male gametophyte of Marsilea vestita, first described by Sharp (1914), consists of a phase of cytoplasmic polarization (Figures 1 and 2) followed by a series of rapid cell divisions (Figure 2), and then, a phase of gamete differentiation (Myles and Hepler, 1977). The entire process lasts ∼11 h and is initiated by placing dry microspores into water or a defined aqueous medium (Laetsch, 1967). At the earliest stage of development, a centrally positioned nucleus is surrounded by dozens of starch-containing plastids (Figures 1a and 2a). Before the 1st mitotic division, the cytoplasm of the hydrated microspore becomes reorganized so that the plastids become situated along 1 side of the cell (Figures 1b and 2b). The nucleus becomes repositioned away from the clustered plastids, and undergoes the 1st mitotic division (Figure 2c), which creates a remnant prothallial cell that does not divide further. The other cell of the gametophyte proceeds to enter a series of 8 additional cell division cycles that are completed by 5.5 h after imbibition and incubation at 20°C (Figure 2, d–k). The 9 division cycles ultimately partition the cytoplasm within the spore wall into a total of 39 cells, 32 of which are spermatids, clustered into 2 groups of 16 cells. These spermatogenous masses are surrounded by 6 sterile jacket cells, which represent the antheridial wall. A fate map (Figure 2, lower panel) shows how further proliferation does not occur in the prothallial and sterile jacket cells, once they are formed. After completion of the cell divisions, the jacket cells become less conspicuous over time as the spermatids enlarge, separate from each other, and mature (Myles and Hepler, 1977). Before the last cell division, each blepharoplast, visible as a discrete particle in the cytosol of each spermatocyte, splits into 2 parts, and functions as centrosomes for the last mitotic spindle (Hepler, 1976). The blepharoplasts disperse and then reform in each of the spermatids, where they become involved in basal body formation (Mizukami and Gall, 1966; Hepler, 1976). During the next 5.5 h after imbibition, each of the spermatids develops a motile apparatus (Hepler, 1976), and undergoes extensive elongation and coiling of the nucleus (Myles and Hepler, 1977, 1982). The elongated nucleus and the motile apparatus ultimately reside at the anterior end of the gamete (Myles and Hepler, 1977). Each mature spermatozoid possesses ∼140 cilia placed along the distal face of its coiled cell body.
Figure 1.
The microgametophyte of M. vestita at and after the time of imbibition. (a) At the time of imbibition, the male gametophyte of M. vestita consists of a single cell that is contained within the microspore wall. A large centrally positioned nucleus is surrounded by dozens of starch-containing plastids. (b) Spermiogenesis is initiated by placing the spore into an aqueous medium, and all of the development occurs within the spore wall. At 30–45 min after imbibition, there is a change in the organization of the cytoplasm: the plastids become aggregated along 1 side of the microspore and the nucleus becomes displaced toward the opposite side of the gametophyte. Images photographed with Phase Contrast optics. Bar = 25 μm.
Figure 2.
Drawings of the cell division patterns during development of the male gametophyte of M. vestita. (a) Microspores at the time of imbibition. The nucleus is depicted as a black circle, and the plastids are depicted as open ellipses. (b) The gametophyte before the 1st division, when cytoplasmic reorganization occurs. (c) After the 1st mitotic division, a prothallial cell (bottom)is cut off from the rest of the gametophyte and no longer divides. (d) The 2nd division gives rise to 2 antheridial initials, essentially of equal size. (e–g) Each antheridial initial undergoes a series of unequal divisions that produce the 3 sterile jacket cells. None of the jacket cells can proliferate further. (h) Each primary spermatogenous cell undergoes a symmetric division. (i) The 2 spermatogenous cells in each antheridium undergoes a symmetric division to produce 4 spermatocyte mother cells. By this stage, the jacket cells become far less conspicuous and they eventually degenerate. (j) The 4 spermatocyte mother cells in each antheridium undergo a division to produce 8 spermatocytes. At this stage, a blepharoplast (dot) appears in each spermatocyte, and then it rapidly disappears (see Hepler [1976]). Later, the blepharoplasts reform, split and function as the centrosomes for the next mitotic cycle, when each spermatocyte undergoes a division that produces 16 spermatids in each antheridium. (Lower panel) A fate map of gametophyte development during spermiogenesis in M. vestita. During the 1st 5.5 h, the gametophytes undergo 9 mitotic division cycles to produce a total of 39 cells, 32 of which are spermatids. The vegetative prothallial cell and the sterile jacket cells lose the capability to proliferate after they are formed. The dotted lines indicate continued proliferation in an identical segment of the gametophyte. The antheridial initials, the primary spermatogenous cells, and their progeny proliferate to produce a total of 32 spermatids.
Translational and Transcriptional Inhibitors Block Spermiogenesis at Different Stages
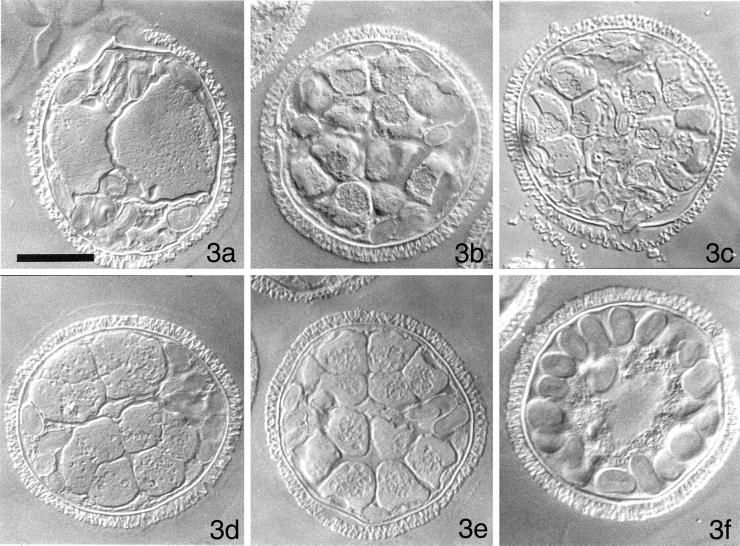
We first tested whether specific stages of spermiogenesis are reliant upon transcription and translation. We compared the pattern of normal gametophyte development in untreated cells fixed at various points after imbibition (Figure 3, a–c) to that of gametophytes treated with α-amanitin, an inhibitor of RNA polymerase II, (Figure 3, d and e), or with the translational inhibitor, cycloheximide (Figure 3f). Each of these general inhibitors was added to the cells at the time of imbibition, and remained present until the time of fixation. The segregation of plastids to the periphery of the gametophyte, a process that precedes the 1st division in untreated microspores (Figure 1b) also occurs in gametophytes treated with α-amanitin (our unpublished results), and by 4 h of incubation in the inhibitor, these cells form blepharoplasts (our unpublished results). Treatment with α-amanitin is followed by a normal pattern of cell division (Figure 3, d and e) and spermatid maturation (our unpublished results), but the developmental process is arrested before spermatozoid release (Hyams et al., 1983; Hart and Wolniak, 1998). In contrast, gametophytes treated with 10 μM cycloheximide fail to exhibit any cytoplasmic changes or cell divisions; even after 11 h, the gametophyte is unicellular, with a large, centrally placed nucleus and with plastids scattered around the periphery of the cell (Figure 3f). Thus, male gametophyte development is blocked at a very early stage in the absence of translation and much of spermatid development is dependent on the translation of stored mRNA.
Figure 3.
Development of the male gametophyte of M. vestita in normal medium, or in medium containing either α-amanitin or cycloheximide. Cells were fixed at various times after imbibition. (a) Untreated gametophyte, fixed after the 3rd cell division, 90 min after imbibition. Each of the antheridial initials has already produced the 1st of 3 sterile jacket cells, which are rich in plastids. (b) Untreated gametophyte, fixed after the 9th cell division, 5 h after imbibition. This 2 μm section depicts 14 of the 32 spermatids. The jacket cells are becoming less conspicuous. (c) Untreated gametophyte, fixed at 8 h after imbibition. The spermatids had initiated nuclear elongation at the time of fixation. (d) A gametophyte incubated with α-amanitin at the time of imbibition and fixed 4 h later. (e) A gametophyte incubated with α-amanitin at the time of imbibition and fixed 8 h later. All of the mitotic division cycles have occurred in these gametophytes. (f) A gametophyte incubated with cycloheximide at the time of imbibition and fixed 10 h later. There have been no cell divisions. Moreover, there have been no apparent changes in cytoplasmic or nuclear organization in the presence of this inhibitor. All images photographed with DIC. Bar = 25 μm.
Centrin, β-Tubulin, and P28 Proteins Accumulate at Different Times and in Different Places
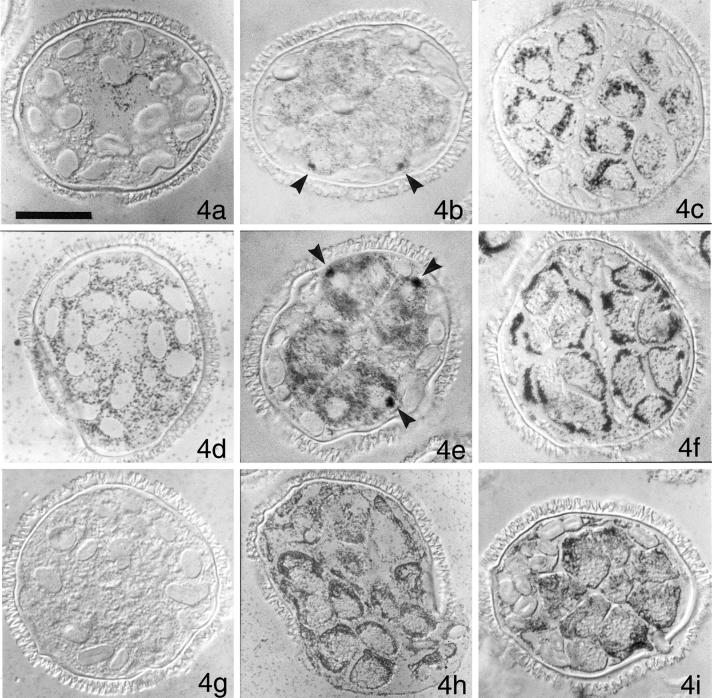
Our inhibitor experiments show that spermiogenesis is dependent on translation of stored mRNA. Our earlier studies (Hart and Wolniak, 1998), show that new translation of centrin and β-tubulin occur at different stages of spermiogenesis and suggest that specific proteins are made at particular times in the developing gametophyte. We screened immunoblots of gametophyte protein isolates with a number of antibodies directed against cytoskeletal, axonemal, and centrosomal antigens, asking if centrin is the only new protein made midway through spermiogenesis, at the time of blepharoplast formation. We found that a ciliary inner-arm component, known as P28 (LeDizet and Piperno, 1995), binds to a single protein band on our blots in the proper molecular weight range, and there is a large increase in the abundance of this antigen ∼5 h after imbibition. Unlike centrin, P28 is undetectable in gametophytes that have been imbibed for < 4 h, and reaches maximal levels of abundance ∼6 h after imbibition (our unpublished results). We performed a series of in situ immunolocalizations on male gametophytes of M. vestita as a means to determine the distribution of these proteins during spermiogenesis. Untreated gametophytes were fixed at various times after imbibition, and then embedded, sectioned, and labeled with antibodies directed against centrin (Figure 4, a–c), β-tubulin (Figure 4, d–f), and P28 (Figure 4, g–i). Anticentrin antibody labeling of gametophytes fixed at the time of imbibition (Figure 4a) is weak but specific, and reveals a perinuclear distribution. Thereafter (Figure 4, b and c), anticentrin antibody labeling is restricted to the spermatogenous mass; the sterile jacket cells, located around and between the spermatogenous cells, are not labeled with the anticentrin antibody (Figure 4b). At 4 h after imbibition, some of the anticentrin antibody is aggregated in the blepharoplasts (Figure 4b, arrows), highlighting its association with, or presence in the blepharoplast (Hoffman et al., 1994). By 8 h after imbibition, the distribution is intense, and again, perinuclear, with most of the label in the anterior region of each spermatid (Figure 4c).
Figure 4.
Immunolocalizations reveal differences in the distributions of various antigens in the male gametophytes over time. Gametophytes were fixed immediately after imbibition (a, d, and g), 4 h after imbibition (b, e, and h), and 8 h after imbibition (c, f, and g) in culture medium. After sectioning at a thickness of < 4 μm and acetone-extraction of the plastic embedding medium, the cells were labeled with anticentrin antibody (a–c), anti-β-tubulin antibody (d–f), or anti-P28 antibody (g–i). Secondary gold-conjugated antibodies and silver enhancement were used to image antibody binding. (a) Centrin is detectable at low levels in cells at the time of imbibition and is present near the centrally positioned nucleus. (b) By 4 h after imbibition, dark aggregates of anticentrin antibody can be seen in spermatogenous cells; because of their abundance (1 per cell), their proximity to the unlabeled nucleus, and their size, these aggregates represent blepharoplasts (arrows). (c) By 8 h after imbibition, anticentrin antibody is abundant in the spermatids, in a roughly-perinuclear distribution. Most of the antibody ultimately resides in the apical portion of each forming spermatozoid, as it reaches maturity (our unpublished results). (d) In contrast to centrin, β-tubulin is abundant and scattered through the cytosol from the time that the spores imbibe water. (e) By 4 h after imbibition, the cytoplasm of the spermatogenous cells is heavily labeled with anti-β-tubulin antibody, while the surrounding sterile jacket cells exhibit almost no labeling. The dense labeling in 3 of the spermatids (arrowheads) shows the position of the blepharoplasts, which are sufficiently mature to exhibit anti-β-tubulin labeling. Each of the blepharoplasts is adjacent to a nucleus, represented by a nonlabeled zone in each spermatid. (f) By 8 h after imbibition, the spermatids of the sectioned gametophyte show dense labeling with anti-β-tubulin antibody at their outer edges. In some sections, the pattern resembles a clover leaf, and denotes the formation of the microtubule ribbon that is assembling on dorsal side of the developing cell (see Myles and Hepler, [1977]). (g) P28 is not detectable in newly imbibed microspores. (h) At 4 h after imbibition, the cytoplasm of both spermatogenous and sterile jacket cells is heavily labeled with anti-P28 antibodies, but blepharoplasts are not detectable in these cells. (i) At 8 h after imbibition, anti-P28 antibody is abundant in all cells of the gametophyte. All images photographed with DIC. Bar = 25 μm.
Immunolabeling with anti-β-tubulin antibody reveals a uniform distribution throughout the cytosolic compartment of the single cell gametophyte a few min after imbibition (Figure 4d). The label becomes restricted to the spermatogenous cells of the gametophyte by 4 h after imbibition (Figure 4e), with no label apparent in the surrounding sterile jacket cells (see Figure 2g). Within the spermatogenous cells, aggregates of anti-β-tubulin antibody aggregate at each blepharoplast (Figure 4e, arrows). Blepharoplast staining with anti-β-tubulin antibody reveals that the structure is reaching maturity (Doonan et al., 1986; Pennell et al., 1986; Hoffman and Vaughn, 1995). By 8 h after imbibition, the anti-β-tubulin antibody label becomes localized in the anterior portions of the spermatids (Figure 4f). In 1–3 μm sections of gametophytes labeled with anti-β-tubulin antibody, the most intense labeling often circumscribes the spermatogenous mass. This pattern of labeling is consistent with the positioning of the MLS and the forming microtubule ribbon that ultimately resides on the dorsal face of the cell body of each mature gamete (Myles and Hepler, 1977); at the light microscopic level of resolution, the pattern often resembles a cloverleaf.
The anti-P28 antibody label is not detectable in gametophytes imbibed for < 4 h (Figure 4g). Thereafter, the P28 antigen accumulates both in spermatogenous and sterile cells (Figure 4h), and by 8 h after imbibition, the label is uniformly distributed throughout the gametophyte (Figure 4i) though it seems to be more abundant in the spermatids, especially in the anterior regions of the forming gametes. We found that in gametophytes treated with α-amanitin at the time of imbibition, the distribution and abundance of labeling with anticentrin, anti-β-tubulin, or anti-P28 antibodies was not discernibly different from that observed in untreated gametophytes fixed at the same times during development (our unpublished results).
We observed no labeling with anticentrin or anti-P28 antibodies in gametophytes that had been imbibed with cycloheximide (our unpublished results). Cycloheximide-treated gametophytes exhibit random cytoplasmic labeling with anti-β-tubulin antibodies, but we were unable to detect any aggregation of the anti-β-tubulin antibody label in blepharoplast-like particles, even 10 h after imbibition (our unpublished results).
RNAi Specifically Inhibits Centrin Translation and Disrupts Spermiogenesis
In this study, we were most interested in asking whether centrin translation is necessary for blepharoplast formation and subsequent assembly of the motile apparatus. To address this question, we employed antisense strategies, introducing our RNA constructs into the cells at the time of imbibition by immersing the dry spores into RNA-containing solutions. We found that gametophyte development was arrested as a function of the concentration of centrin antisense RNA added to the microspores. In contrast to untreated gametophytes fixed at 8 h after imbibition, when the divisions are completed and spermatid maturation is well under way (Figure 5a), we found that high concentrations (2 mg/ml) of antisense centrin RNA probes arrested the vast majority of gametophytes in the 1-cell stage of development, with no segregation of plastids to 1 end of the cell (Figure 5b). Most (70%) of the gametophytes present in these large cultures, containing thousands of spores, when treated with the antisense probe, became arrested in prophase of the 1st mitotic division, with condensed chromosomes (Figure 5b), but lacking apparent spindles. For our initial set of controls, we used sense versions of the same RNA, and treated large populations of microspores under identical conditions. Most of the gametophytes (>85%) treated with high concentrations of the sense probe failed to reach prophase of the 1st mitotic division by 8 h (our unpublished results); these cells closely resembled the microspores that had been imbibed in cycloheximide (Figure 3f).
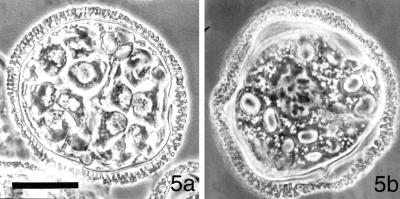
Figure 5.
A high concentration of centrin-based RNA probes arrests development of the male gametophytes of Marsilea vestita. At 8 h after imbibition, untreated gametophytes (a) consist of 39 cells, 32 of which are developing spermatids. Twelve spermatids are visible in this section of a gametophyte. (b) Gametophytes that were imbibed in a solution containing 2 mg/ml antisense centrin RNA and fixed at 8 h were arrested in the 1st mitotic division, with condensed chromosomes (dark structures in the center of the cell), but with no apparent spindles. Images photographed with phase contrast optics. Bar = 25 μm.
In the gametophytes imbibed in the presence of low concentrations (2–20 μg/ml) of centrin antisense or sense probes, we observed normal cytoplasmic partitioning into sterile plastid-containing jacket cells and spermatogenous cells. The early phases of cell division cycles appeared to progress normally. However, 40 to 50% of these gametophytes contained fewer cells than untreated controls, suggesting that they had failed to complete all 9 division cycles. Intermediate concentrations of centrin antisense or sense RNA (200 μg/ml) resulted in a block to development in virtually all of the gametophytes, with > 75% of the cells blocked after the 7th mitotic division. Irrespective of the concentration of probe used, the spermatogenous cells failed to develop further if they were arrested before they completed all of their division cycles (our unpublished results, see below).
The unexpected anomalies observed with our sense probes also led us to suspect that these gametophytes were sensitive to RNA interference (RNAi) effects (Fire et al., 1998) and prompted us to evaluate whether the presence of small quantities of centrin dsRNA might be the factor responsible for the observed inhibition of development. We treated microspores at the time of imbibition with several different concentrations of centrin dsRNA. Microspores were cultured in high (2 mg/ml) concentrations of dsRNA (centrin), similar to those used in sense and antisense experiments. These gametophytes failed to undergo mitosis for > 10 h after imbibition. Arrest of development appeared to coincide with prophase of the 1st mitotic division. Nuclear envelope breakdown did not occur, but chromatin was condensed, in a manner similar to that observed after treatments in antisense centrin RNA. Gametophytes developing in intermediate concentrations of centrin dsRNA (200 μg/ml) undergo 6 or 7 division cycles to produce spermatocyte mother cells or spermatocytes that have plastids localized to the posterior of the cells. These plastids are closely aggregated and the cells have a diffuse region of cytoplasm terminated by an anterior nucleus that fails to undergo coiling. Intermediate concentrations of dsRNA produced arrested development in > 80% of the gametophytes before all of the division cycles were completed. Low concentrations of centrin dsRNA (2 μg/ml) arrested development for ∼50% of the gametophytes after the 6th or 7th division, but in these populations of treated cells, we saw high variation in the stage of arrest (our unpublished results). Whether or not the gametophytes developed beyond the 7th division cycle in the presence of centrin dsRNA, the spermatogenous cells failed to undergo coiling of the cell body or to produce anterior ciliary arrays (our unpublished results). With an apparent concentration-dependent effect on development, it became necessary to assess protein abundance as a function of added-RNA concentration on immunoblots and protein distributions in the treated cells by immunolocalizations.
In order to determine whether the centrin based RNA constructs were affecting translation in different concentration ranges or at different stages during development, we performed a series of dose response studies with centrin RNA probes and populations of cells. We isolated proteins from treated gametophytes 8 h after imbibition for analysis on immunoblots (Figure 6A). We found that centrin dsRNA was at least 10 times more effective in preventing translation of centrin mRNA than centrin sense RNA or centrin antisense RNA in these populations of cells assayed 8 h after imbibition. None of the centrin RNA treatments affected the accumulation of P28 protein in the gametophytes.
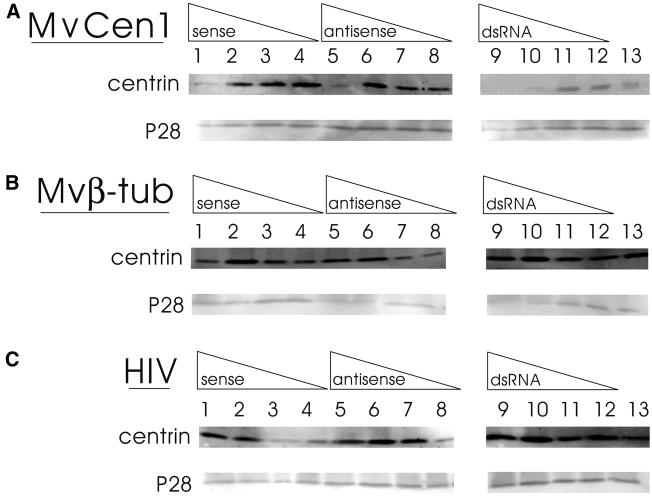
Figure 6.
Centrin translation is inhibited specifically and in a concentration-dependent manner by treatment of gametophytes with RNA probes made from MvCen1. Immunoblots of protein isolates obtained at 8 h after imbibition from populations of gametophytes treated with different concentrations of antisense RNA (lanes 1–4), sense RNA (lanes 5–8) or dsRNA (lanes 9–12). RNA probes were made from MvCen1 cDNA (A), which encodes centrin from Marsilea male gametophytes (Hart and Wolniak, 1998), from MvB-Tub cDNA (B), which encodes a β-tubulin from Marsilea male gametophytes, and from a 250 bp random sequence present in the viral genome of HIV (kindly provided by Dr. J. DeStefano, University of Maryland) (C). The concentration of RNA was 1.5 mg/ml was in lanes 1, 5, and 9. The concentration of RNA was 150 μg/ml in lanes 2, 6, and 10. The concentration of RNA was 15 μg/ml in lanes 3, 7, and 1. The concentration of RNA was 1.5 μg/ml in lanes 4, 8, and 12. The immunoblots were probed with anticentrin antibody (upper blot in each pairing) and anti-P28 antibody (lower blot in each pairing). Binding was detected by ECF fluorescence imaging with the STORM PhosphorImager as described in the text. Lane 13 depicts a protein isolate that was obtained from untreated gametophytes, 8 h after imbibition, run in parallel with the RNA-treated spores. (Cells from these populations of control gametophytes revealed normal immunolabeling patterns with anticentrin antibody—see Figure 4). The anticentrin antibody binding in control lanes should be compared separately with binding intensities in the same blots with the same populations of cells treated for MvCen1, Mvβ-tub, and HIV. Though we loaded the same amounts of protein from identical cell populations onto gels, and then labeled and imaged them identically, comparisons of intensities between experiments on different blots is invalid because of nonlinearities in antigen binding and amplified fluorescence detection. (A) MvCen-derived RNA probes reduce the amount of centrin translated by the gametophytes as an inverse function of RNA probe concentration. The use of dsRNA derived from MvCen1 is at least 10 times more effective in blocking centrin translation than either sense or antisense RNA made from the same cDNA. P28 translation is not affected by the presence of MvCen-derived RNA probes. (B and C) RNA probes derived from either Mvβ-tub or HIV are without significant effect on the abundance of either centrin or P28 after 8 h.
The effect of centrin dsRNA is highly specific; we found that antisense, sense and dsRNA derived from M. vestita β-tubulin had no effect on the abundance of centrin protein in the developing gametophytes at 8 h after imbibition (Figure 6B). Moreover, in the presence of our β-tubulin RNA constructs, we found that the pattern of centrin translation/accumulation through gametophyte development was not distinctly different from that described in untreated cells (Hart and Wolniak, 1998). Centrin protein was detectable at very low levels from 0–3 h after imbibition, with an increase in apparent translation at 4 h after imbibition, reaching maximal abundance by ∼6 h after imbibition in the gametophytes, whether they were treated with antisense, sense, or dsRNA made from M. vestita β-tubulin cDNA (Figure 6B). Centrin levels remained high and constant thereafter. As expected, none of the β-tubulin RNA treatments affected the accumulation of P28 protein in the gametophytes. RNA made from a cDNA that encodes a random fragment of the HIV genome were also without effect on the amount of centrin or P28 protein made in the gametophytes (Figure 6C), further attesting to the high specificity of the effects observed with centrin dsRNA.
Patterns of Development Are Altered Specifically by RNAi Treatments
With abnormal cell division patterns resulting from an induced block to centrin translation following the addition of centrin RNA (Figure 6), we assayed for altered abundance and distribution of centrin (Figure 7), β-tubulin (Figure 8), and P28 (Figure 9) by in situ immunolabeling after the gametophytes had been imbibed with 1 of the RNA probes. We assayed fixed and sectioned cells that had been treated with 200 μg/ml sense, or antisense RNA transcribed from centrin or β-tubulin cDNAs obtained from M. vestita. We used 20 μg/ml dsRNA as standard treatments on other populations of cells. In parallel, we fixed and sectioned cells that had been treated with sense, antisense or dsRNA derived from a 250 base segment of the HIV genome, added at the same concentrations. In all cases, 4 mg of cells were treated in 1 ml of RNA-containing solution and the gametophytes were fixed 8 h after imbibition. Intermediate concentrations (200 μg/ml) of centrin sense (Figures 7a and 8a) and antisense (Figures 7b and 8b), enabled gametophytes to undergo some of their mitotic divisions. Because development was uniformly arrested with intermediate concentrations of our sense and antisense RNA probes, it became the focus of our efforts to assess induced changes in development. At these concentrations with our centrin-based RNA probes, we observed arrested development for the vast majority of gametophytes, which, at most, stopped with the 7th cell division cycle. In these gametophytes, the spermatocyte mother cells that were present in the antheridia did not undergo further differentiation. Blepharoplasts, clearly labeled in untreated gametophytes (Figure 4, b and e) were never observed, and in their absence, the subsequent formation of basal bodies (and then, a motile apparatus) could not be expected.
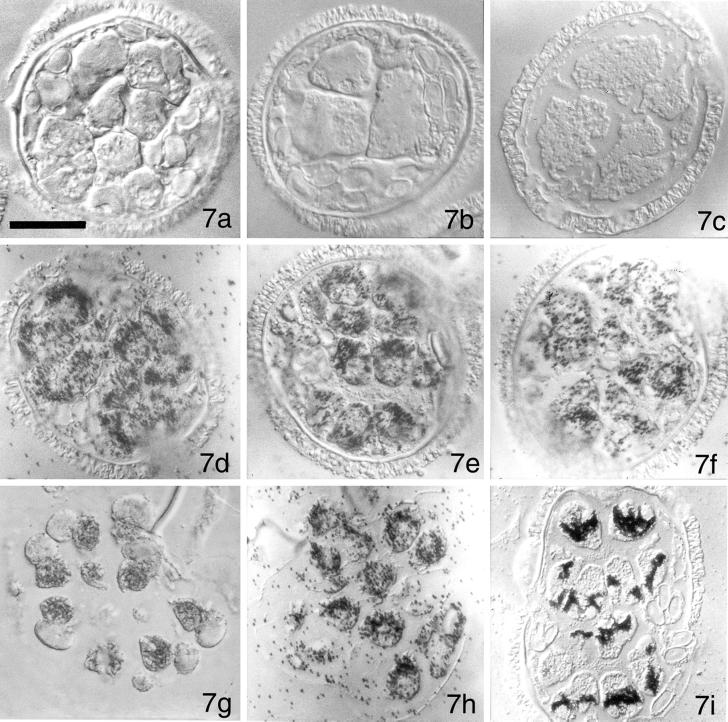
Figure 7.
Immunolocalizations with anticentrin antibody reveal that centrin-derived RNA probes affect development and the accumulation/aggregation patterns of centrin protein. Gametophytes were imbibed with solutions containing MvCen1-derived sense RNA (a), MvCen1-derived antisense RNA (b), or MvCen1-derived dsRNA (c). No centrin protein is detectable in the gametophytes that were treated with centrin RNA probes. In addition, the gametophytes failed to undergo all 9 mitotic division cycles. Gametophytes were imbibed with solutions containing Mvβ-tubulin-derived sense RNA (d), Mvβ-tubulin-derived antisense RNA (e), or Mvβ-tubulin-derived dsRNA (f). Anticentrin antibody labeling reveals that centrin protein accumulation and distribution is not altered by the presence of the Mvβ-tubulin-derived RNA probes. Gametophytes were imbibed with solutions containing HIV-derived sense RNA (g), HIV-derived antisense RNA (h), or HIV-derived dsRNA (i). Anticentrin antibody accumulation and distribution is not altered by the presence of the HIV-derived RNA probes. All images photographed with DIC. Bar = 25 μm.
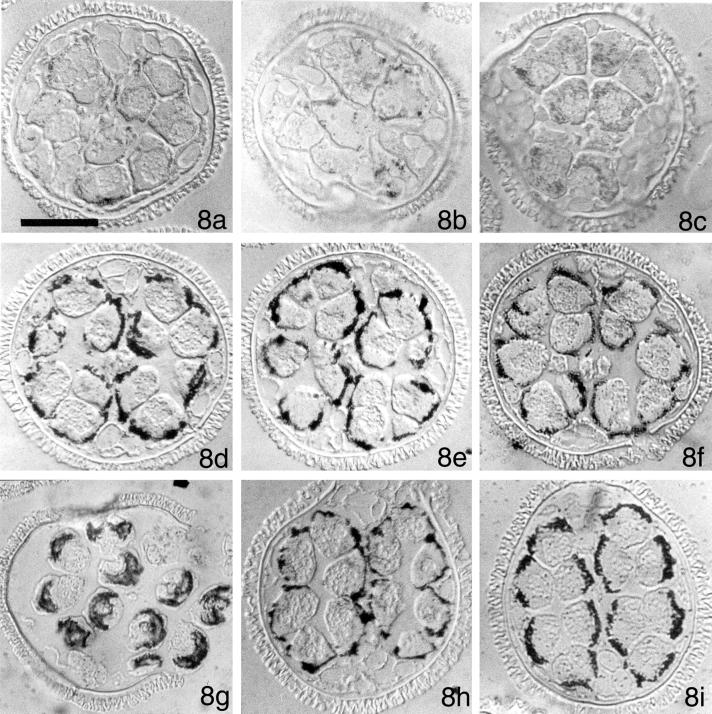
Figure 8.
Immunolocalizations with anti-β-tubulin antibody reveal that centrin-derived RNA probes affect development and the aggregation patterns of β-tubulin protein. Gametophytes were imbibed with solutions containing MvCen1-derived sense RNA (8a), MvCen1-derived antisense RNA (8b), or MvCen1-derived dsRNA (8c). Anti-β-tubulin antibody staining is diffuse in these gametophytes. Moreover, the gametophytes have undergone fewer than the normal 9 division cycles. Gametophytes were imbibed with solutions containing Mvβ-tubulin-derived sense RNA (8d), Mvβ-tubulin-derived antisense RNA (8e), or Mvβ-tubulin-derived dsRNA (8f). Anti-β-tubulin antibody accumulation and distribution is not altered by the presence of the Mvβ-tubulin-derived RNA probes, up through 8 h after imbibition. Gametophytes were imbibed with solutions containing HIV-derived sense RNA (8g), HIV-derived antisense RNA (8h), or HIV-derived dsRNA (8i). Anti-β-tubulin antibody accumulation and distribution is not altered by the presence of the HIV-derived RNA probes. All images were photographed with DIC. Bar = 25 μm.
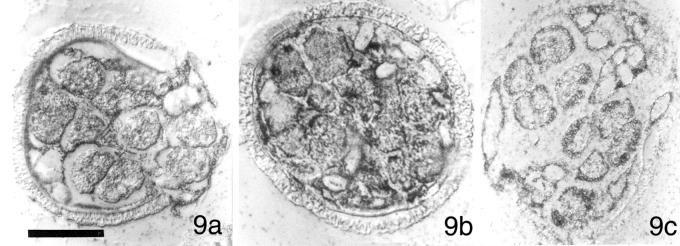
Figure 9.
Immunolocalizations with anti-P28 antibody reveal that our RNA probes exert no effect on development and the aggregation patterns of P28 protein. Gametophytes were imbibed with solutions containing MvCen1-derived dsRNA (9a), Mvβ-tubulin-derived dsRNA (9b), or HIV-derived dsRNA (9c). All images photographed with DIC. Bar = 25 μm.
In contrast to centrin, the abundance of β-tubulin protein does not change substantially until late in spermiogenesis, when it exhibits an increase in abundance that coincides with the formation of ciliary axonemes (Hart and Wolniak 1998). Thus, in the presence of introduced β-tubulin RNA, we anticipated no disruption of spermiogenesis at least up to the stage where substantial levels of newly made β-tubulin were detectable, ∼10 h after imbibition. The addition of β-tubulin sense (Figures 7d and 8d), antisense (Figures 7e and 8e) or dsRNA (Figures 7f, 8f, and 9b) probes to dry microspores at the time of imbibition was followed by a normal pattern of development that included all 9 division cycles. Thereafter, the spermatids appeared to develop normally up to 8 h after imbibition. During this late phase of spermiogenesis, we began to observe subtle anomalies in the spermatozoids that are not the focus of this paper.
Centrin and β-Tubulin Are Affected Differently by RNAi
When immunolabeled with anticentrin antibody, we observed no centrin staining in gametophytes that had been imbibed with sense (Figure 7a), antisense (Figure 7b) or dsRNA (Figure 7c) made from our centrin cDNA. In the absence of labeling with anticentrin antibody in gametophytes treated with centrin sense, antisense, or dsRNA probes, we were unable to detect conspicuous blepharoplast labeling in untreated gametophytes (Figure 4b, arrows). In contrast, when gametophytes were treated with sense (Figure 7d), antisense (Figure 7e), or dsRNA (Figure 7f) that had been made from a M. vestita β-tubulin cDNA, we found intense anticentrin antibody labeling in the spermatids. The distribution of centrin antigen in these cells appeared very similar to that in untreated cells (Figure 4c) that had been fixed at 8 h after imbibition, sectioned, and immunolabeled. Similarly, the presence of sense (Figure 7g), antisense (Figure 7h), or dsRNA (Figure 7i) made from a 250 base segment of the HIV genome had no apparent effects on centrin abundance or on spermatid development.
When gametophytes treated with centrin sense (Figure 8a), antisense (Figure 8b), or dsRNA (Figure 8c) were immunolabeled with anti-β-tubulin antibody, we found diffuse cytoplasmic tubulin staining in the spermatogenous cells. The tubulin aggregation pattern did not resemble the intense antitubulin labeling of blepharoplasts, seen in untreated cells at 4 h after imbibition (Figure 4e), or the anti-β-tubulin antibody labeling that is typical of spermatids observed 8 h after imbibition (Figure 4f), which in some sections, resembles a clover-leaf pattern. Instead, we saw a marked reduction of anti-β-tubulin antibody label intensity in these cells, a result we interpret as indicative of reduced microtubule organization in the gametophyte, rather than lowered β-tubulin abundance. The pattern of anti-β-tubulin antibody labeling in gametophytes treated with sense (Figure 8d), antisense (Figure 8e), or dsRNA (Figure 8f) made from our β-tubulin cDNA all appear markedly different from the gametophytes treated with centrin RNA probes (Figure 8, a–c). In the presence of the β-tubulin RNA probes, there is no apparent disruption of spermiogenesis or β-tubulin aggregation in the spermatids (Figure 8, d–f) through 8 h after imbibition. Gametophytes treated with β-tubulin RNAs begin to display anomalies in spermiogenesis 9–10 h after imbibition, when the cells are actively engaged in axonemal formation (our unpublished results). As expected, treatments of gametophytes with RNA probes made from HIV had no effect on the abundance or distribution of β-tubulin (Figure 8, g–i) in the normally developing spermatids, (where nearly mature gametes are present in Figure 8g, and the clover-leaf pattern of anti-β-tubulin can be seen in Figure 8 (h and i).
Specificity of RNAi
We performed in situ immunolabeling with anti-P28 antibody on fixed, sectioned gametophytes that had been imbibed with the dsRNA probes made from M. vestita centrin cDNA (Figure 9a), M. vestita β-tubulin cDNA (Figure 9b), and HIV cDNA (Figure 9c). The obvious expectation from the addition of irrelevant (noncoding, nonhomologous) RNA probes from HIV to these cells is that we would observe no effect on spermiogenesis. We found that the abundance and the distribution of P28 (Figure 9, a–c) were normal throughout spermiogenesis, a result consistent with our immunoblot studies (Figure 6).
DISCUSSION
We have found that the translation of centrin from stored mRNA is necessary for the formation of the blepharoplast in the male gametophyte of M. vestita. By employing general pharmacological blocks to transcription and translation, we expand upon our earlier observations (Hart and Wolniak, 1998) to show how spermiogenesis in this organism is dependent upon the translation of stored mRNAs. Our RNAi experiments show an essential link between centrin translation and the formation of basal bodies in a developing eukaryotic gamete. Centrin antisense, sense, and dsRNA probes inhibit centrin translation in a gene-specific and concentration-dependent manner. Centrin dsRNA is at least 10 times more effective than either antisense or sense centrin RNA in preventing centrin translation. In the presence of the centrin-derived RNAs, blepharoplasts fail to form, and there is a pronounced disruption in the formation of the motile apparatus. Antisense, sense, or dsRNA made from Marsilea β-tubulin cDNA only affects development late in the process, when new β-tubulin translation appears to occur (Hart and Wolniak, 1998). Blocks to centrin translation affect the distribution of β-tubulin, because these proteins are both present in the blepharoplast and the MLS (Vaughn et al., 1993), and because appear to interact with each other. The addition of HIV RNA exerts no effect on translational patterns in the gametophytes, and is without effect on the spermatozoid development. Thus, our blocks to centrin translation with centrin-derived RNA probes reveal that centrin translation is necessary for blepharoplast formation, and that the centrin protein controls or affects cytoskeletal patterning and formation of the motile apparatus.
Centrin Translation Is Essential for Spermiogenesis
The blepharoplast is a particle unique in the spermatogenous cells of lower plants; it functions briefly as a centrosome, and then it serves as the site for de novo basal body formation. In its capacity as a centrosome for the ultimate division in normally-developing male gametophytes of M. vestita, the blepharoplast resides at spindle poles for just a few minutes (Sharp, 1914; Hepler, 1976). During basal body formation, the particle is recognizable for ∼60 min, a time during which it can be immunolabeled with anticentrin (Figure 4b) and then antitubulin (Figure 4e) antibodies. Centrin has already been shown to associate with the blepharoplast and MLS by EM immunolabeling (Vaughn et al., 1993; Hoffman et al., 1994) and antitubulin antibody labels the blepharoplast only as basal bodies become recognizable (Doonan et al., 1986). The diagnostic pattern of immunolabeling with anti-β-tubulin antibody during later time points in spermiogenesis is in an intense Ag/Au deposit that resembles a cloverleaf pattern in sectioned gametophytes (Figures 4f and 8, d and e, h and i), and signifies the formation of the MLS and its microtubule ribbon along the anterior ends of the spermatids. Deviations from this distinctive labeling pattern (Figure 8, a–c) are indicative of anomalies that occurred earlier in development. In the absence of centrin translation, the gametophytes fail to undergo all 9 mitotic division cycles, blepharoplast formation fails to occur, and spermiogenesis is arrested before the assembly of a motile apparatus. In the absence of centrin translation, there is no aggregation of the anti-β-tubulin antibody label (Figure 4e) that overlies each organized blepharoplast, and the clover-leaf pattern of anti-β-tubulin antibody labeling fails to form.
If cycloheximide is present in the imbibition medium, all gametophyte development is arrested for extended periods (Figure 3f), and no new centrin is translated (Hart and Wolniak, 1998). High concentrations of antisense, sense and dsRNA (centrin) present in the imbibition medium also arrest gametophytes in the single cell stage. High concentrations of antisense centrin RNA blocked the gametophytes at prophase of the 1st division, while sense and dsRNA probes arrested the gametophytes before the 1st division. Perhaps, the probes act through different mechanisms. At high concentrations, centrin antisense RNA appears to act in a gene-specific manner, to lower or block centrin translation necessary for the early division cycles. Small amounts of newly made centrin protein were detectable on immunoblots from untreated gametophytes, shortly after imbibition (Hart and Wolniak, 1998); this centrin protein may be involved in mitotic spindle organization. Centrin appears to function in the organization of mitotic spindles in a variety of organisms (Taillon et al., 1992; Weich et al., 1996; Salisbury, 1995; Middendorp et al., 2000), and plays a prominent role in basal body formation during Naegleria amebo-flagellate transformations (Levy et al., 1996, 1998). Centrin has been immunolocalized in the blepharoplast and the MLS of plant spermatozoids (Vaughn et al., 1993; Hoffman et al., 1994; Hoffman and Vaughn, 1995), but our current work provides the 1st clues about this protein in the formation of these arrays.
Earlier, we (Hart and Wolniak, 1998, 1999) proposed that spermiogenesis in M. vestita relied heavily on the translation of stored mRNAs at specific times during development. Here, our combination of inhibitor and RNAi strategies with immunolabeling reveal that the translation of stored mRNAs occurs at the beginning of development from the time that spores are imbibing culture medium. Spores cultured in the presence of cycloheximide and in high concentrations of either centrin sense RNA or centrin dsRNA all fail to exhibit nuclear migration, plastid reorganization and chromatin condensation, a result suggesting that the initiation of development is controlled by yet-to-be-characterized, untranslated stored mRNAs. It is striking that no development of the gametophyte occurs in the presence of only stored protein (Figure 3f). We suspect that a number of translation products, in addition to centrin, made early in gametophyte development are important components in processes that are manifested morphologically as the segregation of the cytoplasm into spermatogenous and sterile domains that become partitioned later.
The Specificity of RNAi and the Requirement of Centrin for Spermiogenesis
Because we had opted to perform direct additions of RNAs to our gametophytes, it was necessary to establish the efficacy of our RNAi approach, by including multiple negative and positive controls in the experimental design. We employed sense RNA and antisense RNA in addition to the dsRNA in parallel dose-response experiments for multiple genes (Figure 6). By assaying for the presence and distribution of centrin, β-tubulin, and P28 in identically treated cells with immunolocalizations (Figures 7–9), we are able to match developmental anomalies with specific changes in antigen distributions. By comparing centrin abundance on immunoblots made from cells treated with each of the RNA probes (Figure 6), we conclude that the inhibitions of centrin-derived RNA probes are almost certainly gene-specific and definitely concentration-dependent.
At low concentrations, our centrin-based RNA probes may prevent centrin translation in sufficient quantities for blepharoplast formation. In the absence of blepharoplasts, we observe no further progression in spermiogenesis; usually we fail to see the final 2 cell division cycles in the gametophyte. We suspect that mitotic arrest after the 7th division cycle provides an indication that the blepharoplast somehow controls mitotic progression in the final stages of proliferation in the gametophyte. The blepharoplast serves as a centrosome for the 9th division in this organism (Hepler, 1976). Centrin has been linked to centrosomal duplication (Jarvik and Suhan, 1991; Middendorp et al., 2000), so perhaps in its absence, duplication of the forming blepharoplast is blocked at this stage, and subsequent formation of the mitotic spindle is prevented. Since mature blepharoplasts produce basal bodies, formation of basal bodies and ciliary axonemes is precluded after these treatments. We see no indication of basal body formation, or ciliogenesis in gametophytes that were blocked with our centrin-based RNAs.
High concentrations of the centrin sense RNA and centrin dsRNA in the gametophytes may be affecting development through an effective, specific block of centrin translation. It was not surprising that the centrin RNA probes are inhibitory, given the fact that spermiogenesis represents a complex developmental program that is reliant on the translation of certain kinds of stored mRNA within a tight time regime. We believe that our centrin RNAs could also alter pool sizes of transcripts in the gametophyte, and affect overall rates of centrin translation through multiple mechanisms. It is reasonable to suspect that the cells possess surveillance mechanisms to detect anomalous quantities or types of RNAs that could be disruptive to the developmental program. The mechanism of inhibition by centrin sense RNA is probably linked to the presence of small amounts of dsRNA present in the transcription mixture, since dsRNA made from the same probe is even more effective at inhibiting development at the same stage. The effects of high doses of both sense and antisense centrin RNA were less dramatic than comparable concentrations of centrin dsRNA (Figure 6A), suggesting that inhibition is the consequence specific RNAi mechanisms affecting translational activities in the cells. Similar multi-tiered effects have been observed in other organisms (Fire et al., 1998). The specificity of the response of this developmental program to the presence of centrin-based RNA probes is highlighted by the inability of our β-tubulin RNA probes to affect development within 8 h of imbibition, or of our HIV-based RNA probes to evoke any anomalies in spermiogenesis.
Intermediate concentrations of antisense centrin RNA usually allow 7 division cycles to reach completion before gametophyte development is arrested. At this stage of development, we would have expected to see blepharoplasts form, but they fail to appear. Since the cells fail to label with anticentrin antibody, we assume that there is insufficient centrin protein present in the gametophyte for detection of the antigen. We performed immunolabeling on these gametophytes with anti-β-tubulin, looking for tubulin aggregations that are diagnostic of blepharoplast maturation (Doonan et al., 1986), but we failed to see any aggregates that hinted at organized blepharoplast assembly. This result was expected because tubulin additions to the blepharoplast occur late in the ontogeny of the organelle (Hepler, 1976; Doonan et al., 1986). Thus, we are led to conclude that at intermediate concentrations, our centrin antisense probes are acting specifically on centrin translation and that centrin synthesis is a necessary prerequisite for blepharoplast formation.
Our centrin dsRNA treatments block the translation of stored centrin mRNA at concentrations ∼10–100 times lower than either sense or antisense RNA (Figure 6). The increased potency of inhibition by dsRNA is a hallmark of RNAi effects in a variety of eukaryotic organisms (Kennerdell and Carthew, 1998; Montgomery et al., 1998; Montgomery and Fire, 1998). After the addition of our centrin-based RNA probes, it is clear that the block to development occurs at or just before the stage of blepharoplast formation, and the further elaboration of the motile apparatus or maturation of the gametes is arrested. We suspect that the anomalies in β-tubulin distribution in cells imbibed with centrin sense (Figure 8d), antisense (Figure 8e) or dsRNA (Figure 8f) is the consequence of the role of centrin protein in controlling microtubule assembly patterns in the developing spermatid (Vaughn et al., 1993; Vaughn and Harper, 1998). In contrast to a block of centrin translation, development is not dramatically altered through 8 h after imbibition by the presence of antisense, sense or dsRNA made from a β-tubulin cDNA isolated from our gametophyte library. Our earlier work showed that new β-tubulin is not translated until late in spermiogenesis, when the spermatids are assembling ciliary axonemes (Hart and Wolniak, 1998). Presumably at that stage of development, this protein becomes limiting for axonemal growth.
Rapid, Efficient, Direct Uptake of Drugs and RNA into Gametophytes during Imbibition
The blocks to spermiogenesis we observe require entry of cycloheximide or these RNAs into the cytosol of the gametophyte. We have found that molecules as large as 450+ bp RNAs will enter the dry microspores rapidly at the time of imbibition. Pettitt (1979) showed that dry megaspores from M. vestita were permeable to large iron particles at the time of imbibition, and that the particles would accumulate in cytosolic space of the gametophyte if they were present in the aqueous medium at the time the dry spores were immersed in the suspension. The drugs or RNAs used in our experiments were present in the aqueous medium at the time the dry microspores were added. Uptake of the probes appears to be rapid and uniform in a population of cells. We can vary RNA concentration in replicate experiments and treat cells directly with RNA, as opposed to being reliant upon expression of an insert in a transformed organism (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000). The level of permeability we observe may be widespread among gametophytes whose germination immediately follows imbibition by dry spores. Thus, in an extension of these particle uptake experiments (Pettitt, 1979), and our previous drug and radiolabel experiments (Hart and Wolniak, 1998), it seems clear that our RNAi probes enter the cells of the gametophytes at the time of imbibition. The utility and efficacy of macromolecular uptake by dry spores at the time of imbibition provides a powerful means with which to perform a variety of experimental treatments.
To ensure specificity with our RNAi treatments, we performed in vitro transcriptions from those portions of the centrin and β-tubulin transcripts that exhibit the lowest similarity to related genes. Centrin appears to be a single-copy or few-copy gene in gametophytes of M. vestita (Hart and Wolniak, 1999). Both centrin and β-tubulin RNAs were made from cDNAs isolated from our M. vestita library. Clearly, these factors contribute substantially to the specificity of the response.
ACKNOWLEDGMENTS
We gratefully acknowledge support for this work from the National Science Foundation (MCB-9809950, MCB-9904435). We are grateful to Dr. Peter Hepler for providing us with our initial supply of sporocarps. In addition, we appreciate the efforts of Drs. Jeff DeStefano, Peter Hart, Jeff Salisbury, Berl Oakley, and Gianni Piperno, who supplied us with probes that proved to be invaluable during various phases of this study. A number of our colleagues at the University of Maryland, including Chia-Wei Tsai, Jeff Molk and Drs. Eric Baehrecke, Margaret De Cuevas, Bill Jeffery, Steve Mount, and Heven Sze have provided us with input, suggestions, and comments at various stages of this project. Their interest, support and encouragement has helped us extend this work in a number of important directions.
Footnotes
*Corresponding author: sw36@umail.umd.edu.
REFERENCES
- Baron AT, Greenwood T, Salisbury JL. Localization of the centrin-related 155,000-Mr protein of PtK2 cells during the cell cycle. Cell Motil Cytoskeleton. 1991;18:1–14. doi: 10.1002/cm.970180102. [DOI] [PubMed] [Google Scholar]
- Baron AT, Greenwood T, Bazinet C, Salisbury JL. Centrin is a component of the pericentriolar lattice. Biol Cell. 1992;76:383–388. doi: 10.1016/0248-4900(92)90442-4. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD. Joint actions of two RNA degradation pathways controls the timing of maternal transcript elimination in the midblastula transition in Drosophila melanogaster. EMBO J. 1999;18:2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T, Busby CH, Fowke LC, Sammut M, Gubler F. Improvements in immunostaining samples embedded in methacrylate: localization of microtubules and other antigens throughout developing organs in plants of diverse taxa. Planta. 1992;187:405–413. doi: 10.1007/BF00195665. [DOI] [PubMed] [Google Scholar]
- Boscher JM, Labouesse M. RNA interference: genetic wand and genetic watchdog. Nat Cell Biol. 2000;2:E31–E36. doi: 10.1038/35000102. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carothers ZB. Comparative studies on spermatogenesis in bryophytes. Biol J Linn Soc. 1975;6(suppl.):71–84. [Google Scholar]
- Chamberlain CJ. The homology of the blepharoplast. Bot Gaz. 1898;26:431–435. [Google Scholar]
- Chuang C-F, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Nat Acad Sci USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbayawan T, Harper JDI, Elliot J, Gunning BES, Marc J. A γ-tubulin that associates specifically with centrioles in HeLa cells and the basal body complex in Chlamydomonas. Cell Biol Int. 1995;19:559–567. doi: 10.1006/cbir.1995.1102. [DOI] [PubMed] [Google Scholar]
- Dirksen ER. Centriole and basal body formation during ciliogenesis revisited. Biol Cell. 1991;72:31–38. doi: 10.1016/0248-4900(91)90075-x. [DOI] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco P, Kirschner M. Pericentrin, a highly conserved protein of centrosomes involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Doonan JH, Lloyd CW, Duckett JG. Anti-tubulin antibodies locate the blepharoplast during spermatogenesis in the fern Platozoma microphyllum R.Br: a correlated immunofluorescence and electron-microscopic study. J Cell Sci. 1986;81:243–265. doi: 10.1242/jcs.81.1.243. [DOI] [PubMed] [Google Scholar]
- Driver SE, Robinson GS, Flanagan J, Shen W, Smith LEH, Thomas DW, Roberts PC. Oligonucleotide-based inhibition of embryonic gene expression. Nat Biotechnol. 1999;17:1184–1187. doi: 10.1038/70724. [DOI] [PubMed] [Google Scholar]
- Duckett JG. An ultrastructural study of the differentiation of the spermatozoid of Equisetum. J Cell Sci. 1973;12:95–120. doi: 10.1242/jcs.12.1.95. [DOI] [PubMed] [Google Scholar]
- Dutcher S. Flagellar assembly in two hundred and fifty easy-to-follow steps. Trends Genet. 1995;11:398–404. doi: 10.1016/s0168-9525(00)89123-4. [DOI] [PubMed] [Google Scholar]
- Fire A. RNA-triggered gene silencing. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fulton C. Centrioles. In: Reinert J, Ursprung H, editors. Origins and Continuity of Cell Organelles. New York: Springer-Verlag; 1971. pp. 170–221. [Google Scholar]
- Fulton C, Dingle AD. Basal bodies, but not centrioles in Naegleria. J Cell Biol. 1971;51:826–836. doi: 10.1083/jcb.51.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould RR. The basal bodies of Chlamydomonas reinhardtii: formation from probasal bodies, isolation, and partial characterization. J Cell Biol. 1975;65:65–74. doi: 10.1083/jcb.65.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PE, Wolniak SM. Spermiogenesis in Marsilea vestita: a temporal correlation between centrin expression and blepharoplast differentiation. Cell Motil Cytoskeleton. 1998;41:39–48. doi: 10.1002/(SICI)1097-0169(1998)41:1<39::AID-CM3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Hart PH, Wolniak SM. Molecular cloning of a centrin homolog from Marsilea vestita and evidence for its translational control during spermiogenesis. Biochem Cell Biol. 1999;77:101–108. doi: 10.1139/o99-013. [DOI] [PubMed] [Google Scholar]
- Hepler PK. The blepharoplast of Marsilea: its de novo formation and spindle association. J Cell Sci. 1976;21:361–390. doi: 10.1242/jcs.21.2.361. [DOI] [PubMed] [Google Scholar]
- Hoffman JC, Vaughn KC, Joshi HC. Structural and immunocytochemical characterization of microtubule organizing centers in pteridophyte spermatogenous cells. Protoplasma. 1994;179:46–60. [Google Scholar]
- Hoffman JC, Vaughn KC. Using developing spermatogenous cells of Ceratopteris to unlock the mysteries of the plant cytoskeleton. Int J Plant Sci. 1995;156:346–358. [Google Scholar]
- Holt JT, Redner RL, Nienhuis AW. An oligomer complementary to c-myc mRNA inhibits proliferation of HL-60 promyelocytic cells and induces differentiation. Mol Cell Biol. 1988;8:963–973. doi: 10.1128/mcb.8.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyams JS, Vondy KP, Luba A, Bell PR. J. Submicrosc. Cytol. 15: 133–138. 1983. Structural and macromolecular events associated with basal body morphogenesis in Marsilea. [Google Scholar]
- Jarvik J, Suhan J. The role of the flagellar transition region: inferences from the analysis of a Chlamydomonas mutant with defective transition region structures. J Cell Sci. 1991;99:731–740. [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Klink, V.P., and Wolniak, S.M. (2000). The utility of RNAi in the study of the plant cytoskeleton. J. Plant Growth Regul. (in press). [DOI] [PubMed]
- Klotz C, deLoubresse NG, Ruiz F, Beisson J. Genetic evidence for a role of centrin-associated proteins in the organization and dynamics of the infraciliary lattice in Paramecium. Cell Motil Cytoskeleton. 1997;38:172–186. doi: 10.1002/(SICI)1097-0169(1997)38:2<172::AID-CM6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Kochanski RS, Borisy GG. Mode of centriole duplication and distribution. J Cell Biol. 1990;110:1599–1605. doi: 10.1083/jcb.110.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Borisy GG. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch WM. Ferns. In: Wilt FH, Wessells NK, editors. Methods in Developmental Biology. New York: Thomas Y. Crowell; 1967. pp. 319–328. [Google Scholar]
- LeDizet M, Piperno G. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol Biol Cell. 1995;6:697–711. doi: 10.1091/mbc.6.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Lai EY, Remillard SP, Heintzelman MB, Fulton C. Centrin is a conserved protein that forms diverse associations with centrioles and MTOCs in Naegleria and other organisms. Cell Motil Cytoskeleton. 1996;33:298–323. doi: 10.1002/(SICI)1097-0169(1996)33:4<298::AID-CM6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Levy YY, Lai EY, Remillard SP, Fulton C. Centrin is synthesized and assembled into basal bodies during Naegleria differentiation. Cell Motil Cytoskeleton. 1998;40:249–260. doi: 10.1002/(SICI)1097-0169(1998)40:3<249::AID-CM4>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Marc J, Gunning BES. Immunofluorescent localization of cytoskeletal tubulin and actin during spermatogenesis in Pteridium aquilinum (L.) Kuhn. Protoplasma. 1986;134:163–177. [Google Scholar]
- Marc J, Gunning BES. Monoclonal antibodies to a fern spermatozoid detect novel components of the mitotic and cytokinetic apparatus in higher plant cells. Protoplasma. 1988;142:15–24. [Google Scholar]
- Marshall WF, Rosenbaum JL. How centrioles work: lessons from green yeast. Curr Opin Cell Biol. 2000;12:119–125. doi: 10.1016/s0955-0674(99)00065-4. [DOI] [PubMed] [Google Scholar]
- Middendorp S, Kuntziger T, Abraham Y, Holmes S, Bordes N, Paintrand M, Paoletti A, Bornens M. A role for centrin 3 in centrosome reproduction. J Cell Biol. 2000;148:405–415. doi: 10.1083/jcb.148.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misquitta L, Paterson BP. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc Nat Acad Sci USA. 1999;96:1451–1456. doi: 10.1073/pnas.96.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami I, Gall J. Centriole replication II. Sperm formation in the fern Marsilea and the cycad. Zamia. J Cell Biol. 1966;29:97–111. doi: 10.1083/jcb.29.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MK, Fire A. Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet. 1998;14:255–258. doi: 10.1016/s0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Nat Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudjou M, Paintrand M, Vigues B, Bornens M. A human centrosomal protein is immunologically related to basal body-associated proteins from lower eukaryotes and is involved in the nucleation of microtubules. J Cell Biol. 1991;115:129–140. doi: 10.1083/jcb.115.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles DG, Hepler PK. Spermiogenesis in the fern Marsilea vestita: microtubules, nuclear shaping, and cytomorphogenesis. J Cell Sci. 1977;23:57–83. doi: 10.1242/jcs.23.1.57. [DOI] [PubMed] [Google Scholar]
- Myles DG, Hepler PK. Shaping of the sperm nucleus in Marsilea: a distinction between factors responsible for shape generation and shape determination. Dev Biol. 1982;90:238–252. doi: 10.1016/0012-1606(82)90373-6. [DOI] [PubMed] [Google Scholar]
- Ngo J, Tshudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Nat Acad Sci USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M, Paintrand M, Salisbury J, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Pennell RI, Hyams JS, Bell PR. The blepharoplast of Marsilea: a structure concerned with basal body assembly lacking tubulin. Eur J Cell Biol. 1986;40:238–241. [Google Scholar]
- Pennell RI, Vondy KP, Bell PR, Hyams JS. Composition and function of the blepharoplast of Marsilea vestita. Eur J Cell Biol. 1988;46:51–60. [Google Scholar]
- Pettitt JM. Ultrastructure and cytochemisty of spore wall morphogenesis. In: Dyer AF, editor. The Experimental Biology of Ferns. New York: Academic Press; 1979. pp. 213–252. [Google Scholar]
- Purohit A, Tynan S, Vallee RV, Doxsey SJ. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J Cell Biol. 1999;147:481–491. doi: 10.1083/jcb.147.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringo DL. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J Cell Biol. 1967;33:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL. Centrin, centrosomes, and mitotic spindle poles. Curr Opin Cell Biol. 1995;7:39–45. doi: 10.1016/0955-0674(95)80043-3. [DOI] [PubMed] [Google Scholar]
- Sanchez-Alvorado A, Newmark P. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci USA. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel E, Bornens M. In search of a function for centrins. Trends Cell Biol. 1995;5:197–201. doi: 10.1016/s0962-8924(00)88999-0. [DOI] [PubMed] [Google Scholar]
- Sharp LW. Spermatogenesis in Marsilea. Bot Gaz. 1914;58:419–432. [Google Scholar]
- Sharp PA, Zamore PD. Molecular biology: RNA interference. Science. 2000;287:2431–2433. doi: 10.1126/science.287.5462.2431. [DOI] [PubMed] [Google Scholar]
- Tabara H, Grishok A, Mello CC. RNAi in C. elegans: soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Taillon BE, Adler SA, Suhan JP, Jarvik JW. Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J Cell Biol. 1992;119:1613–1624. doi: 10.1083/jcb.119.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn KC, Sherman TD, Renzaglia KS. A centrin homolog is a component of the multilayered structure in bryophytes and pteridophytes. Protoplasma. 1993;175:58–66. [Google Scholar]
- Vaughn KC, Harper JDI. Microtubule-organizing centers and nucleating sites in land plants. Int Rev Cytol. 1998;181:75–149. doi: 10.1016/s0074-7696(08)60417-9. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang M-B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber HJ. Notes on the fecundation of Zamia and the pollen tube apparatus of Ginkgo. Bot Gaz. 1897;24:225–235. [Google Scholar]
- Webber, H.J. (1901). Spermatogenesis and fecundation of Zamia. USDA Bur. Plant Ind. Bull. No. 2.
- Weich H, Geir BM, Paschke T, Spang A, Grein K, Steinkotter J, Melkonian M, Schiebel E. Characterization of green alga, yeast, and human centrins. J Biol Chem. 1996;271:22453–22461. doi: 10.1074/jbc.271.37.22453. [DOI] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- Wolniak SM, Klink VP, Hart PE, Tsai C-W. Control of development and motility in the spermatozoids of lower plants. Grav. Sp. Biol Bull. 2000;13:85–93. [PubMed] [Google Scholar]
- Young A, Dictenberg JB, Purohit A, Tuft R, Doxsey SJ. Cytoplasmic dynein-mediated assembly of pericentrin and γ-tubulin onto centrosomes. Mol Biol Cell. 2000;11:2047–2056. doi: 10.1091/mbc.11.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]