Abstract
Many recent studies highlight the importance of lipids in membrane proteins, including in the formation of well-ordered crystals. To examine the effect of changes in one lipid, cardiolipin, on the lipid profile and the production, function and crystallization of an intrinsic membrane protein, cytochrome c oxidase, the cardiolipin synthase (cls) gene of Rhodobacter sphaeroides was mutated, causing > 90% reduction in cardiolipin content in vivo and selective changes in the abundances of other lipids. Under these conditions, a fully native cytochrome c oxidase (CcO) was produced, as indicated by its activity, spectral properties and crystal characteristics. Analysis by MALDI tandem mass spectrometry (MS/MS) revealed that the cardiolipin level in CcO crystals, as in the membranes, was greatly decreased. Lipid species present in the crystals were directly analyzed for the first time using MS/MS, documenting their identities and fatty acid chain composition. The fatty acid content of cardiolipin in R. sphaeroides CcO (predominantly 18:1) differs from that in mammalian CcO (18:2). In contrast to the cardiolipin-dependence of mammalian CcO activity, major depletion of cardiolipin in R. sphaeroides did not impact any aspect of CcO structure or behavior, suggesting a greater tolerance of interchange of CL by other lipids in this bacterial system.
Lipids are resolved in the crystal structures of many membrane proteins, implying a significant role in protein structure and function (1, 2). Specific lipids have been shown to be required for correct folding and assembly of several membrane proteins including the plant light harvesting complex (3) and the bacterial lactose permease (4–6). Other studies on bovine and yeast cytochrome bc1, bovine cytochrome c oxidase, and bacterial KcsA K+ channel indicate a requirement for specific lipids for full activity (7–12). In some cases, complete removal of bound lipid in vitro results in abolishment of protein function, an effect that is reversed by adding back certain types of lipids (11–13). Investigating the roles of bound lipids is therefore an important part of understanding the mechanism and regulation of membrane proteins.
Lipids are found to stabilize membrane protein structure (14) and therefore retention of lipid molecules during purification has been emphasized as an important factor in the successful crystallization of membrane proteins (2, 15). The observation that the quality of E. coli lactose permease crystals can be controlled by changing the phospholipid content (16) demonstrates this principle. Similarly, the crystallization of the cytochrome b6f complex was improved by the addition of lipids (14). Efforts to obtain high resolution structures of membrane proteins thus demand a closer scrutiny of the role of protein-associated lipid molecules.
The terminal enzyme of the respiratory electron transport chain, cytochrome c oxidase (CcO) (17, 18), has served as a model system for studying the involvement of lipids in membrane protein structure and function (19, 20) because its spectrally-accessible metal centers provide sensitive intrinsic probes for monitoring structural integrity and activity. A number of lipid molecules co-purify with CcO from bovine heart and R. sphaeroides and the lipid binding sites identified crystallographically in the core subunits of CcO are highly conserved from bacteria to mammals (2, 21, 22), suggesting an important role not only in function but also in achieving high resolution crystal structures of CcO.
Cardiolipin (CL) is one lipid whose requirement has been established by biochemical studies of a number of membrane proteins, particularly respiratory chain complexes from both bacterial and mitochondrial sources (7, 10, 11, 23, 24). Its unique structure, composed of two phosphates, three glycerols and four hydrophobic fatty acid chains, enables cardiolipin to bind to membrane proteins more tenaciously than lipids with only two alkyl chains. Recent evidence suggests that it can act as a glue between proteins that form super-complexes in membranes (25, 26) and that it plays a role in the signaling system that controls apoptosis in eukaryotic cells (27, 28). It has also been reported to be essential for the full activity of mammalian heart CcO (7, 29), but most of these studies removed CL in vitro after the enzyme was purified. In contrast, several in vivo studies in yeast report that a deficiency in cardiolipin did not significantly affect the activity of CcO, nor did it alone cause mitochondrial dysfunction (30–32). However, the deficiency of both phosphatidylglycerol (PG) and CL resulted in very little CcO production and severely dysfunctional mitochondria (30), suggesting CL is functionally replaceable by PG but not other lipids in yeast.
To test if CL is strictly required in CcO from the purple non-sulfur bacterium R. sphaeroides, CcO was expressed in a CL-deficient mutant created by deleting the cls gene encoding CL synthase. The expression, activity, structure, crystallization, and lipid composition of CcO were examined. The results support a flexible lipid requirement in this bacterial enzyme.
Materials and Methods
Media and growth conditions
All R. sphaeroides strains were grown in Sistrom’s succinate-basal salts medium (33, 34). The cells were grown on agar-solidified medium or in liquid cultures at 30 °C under anaerobic photoheterotrophic (35) or aerobic chemoheterotrophic (36) conditions, as previously described. When required, ampicillin (50 μg/mL), kanamycin (25 μg/mL), tetracycline (1 μg/mL), chloramphenicol (25 μg/mL), streptomycin (5 μg/mL) or spectinomycin (5 μg/mL) was added to the media. Escherichia coli cells were grown in LB medium at 37 °C and, when required, antibiotics were added to the solid media or liquid cultures at the concentration of 50 μg/mL of ampicillin, kanamycin or chloramphenicol.
For photosynthetic growth curves, 2.4.1 (wild-type, CL+) (37) and CL3 (cls deletion, CL−) strains were grown in 250 mL sidearm flasks filled completely with Sistrom’s medium under approximately 30 μmol m−2 s−1 light without shaking. For aerobic growth curves, strains 2.4.1 and CL3 were grown at 30 °C in 50 mL of Sistrom’s medium in 250 mL sidearm flasks with vigorous shaking (250 rpm). In each case, cell densities were measured in a Klett photometer with a #66 filter. Note that 1 Klett unit is equivalent to 107 cells per mL (38).
Construction of plasmid pSUP-CLD
Using genomic DNA from the wild-type strain 2.4.1 as a template, a 600 nucleotide region of the cls gene (GenBank YP_353546 coding sequence, nucleotides 1 to 600) was PCR amplified using the primers P1, 5′-GCGGGATCCATGATCGACGACTGGCTGGGCGTCC - 3′, and P3, 5′-GCGGTCGACGTTGCGCCCGCCCACGATGGCC - 3′, SalI. Similarly, a 591 nucleotide region of the gene (GenBank YP_353546 coding sequence, nucleotide 961–1551) was amplified by using another set of primers, P2, 5′-GCGAAGCTTGGATCCTCAGAGGTAGCTCTGGATCG - 3′, HindIII, BamHI and P4, 5′-GCGGTCGACGGCGAGGCGATCACCGCGCTCC - 3′, SalI. The amplified products were cloned into the vectors pPICT-2 and pGEM-T Easy (Promega, Madison, WI), respectively, to generate the plasmids pPICT-2-I and pGEM-T-II. Segment II from the plasmid pGEM-T-II was cut out by double digestion with SalI and HindIII and was ligated into pPICT-2-I which had been digested with the same enzymes to generate the plasmid pPICT-2-I/II. From the pUC4K plasmid (GE Healthcare, Piscataway, NJ), the kanamycin resistance cassette (kanR) was cut out by using SalI sites placed on either side of the gene and was subsequently inserted into the plasmid pPICT-2-I/II which had been digested by the same enzymes. The resulting plasmid pPICT-2-CK contained the kanR cassette inserted between segments I and II. The orientation of kanR was checked by digesting the plasmid pPICT-2-CK with HindIII. Since a HindIII site is placed asymmetrically within kanR, digestion by the enzyme would produce fragments of different sizes based upon the orientation of the gene (39). Using BamHI, the entire gene construct was cut out from the plasmid pPICT2-CK and was ligated into the suicide plasmid pSUP202 (40) by using the BamHI site placed within the tetracycline resistance gene, to obtain the plasmid pSUP-CLD.
Disruption of the cls gene in R. sphaeroides 2.4.1
The plasmid pSUP-CLD was mobilized into the wild-type strain 2.4.1 (39) by using the E. coli strain S17-1 (RP4-2-Tc::Mu-Km::Tn7Tp rSmrPro−; (40)) as a donor. Through selection for kanamycin (100 μg/mL) resistance, the plasmid was forced to recombine with the genome using the regions homologous to cls gene. The exconjugants with double crossovers were selected by ampicillin and chloramphenicol sensitivity (39). The resulting mutant strain of R. sphaeroides was called CL3. The gene disruption in the mutant was verified by PCR.
Quantitative membrane lipid analysis using TLC and radioactive labeling
Cultures of wild-type and mutant strains were grown aerobically overnight. Lipids were then labeled by incubating the cultures with [1-14C]-acetate as described (41). Cells were collected by centrifugation and washed twice and suspended in 50 μL water. From the cells, lipid extracts were prepared and loaded onto activated silica thin-layer chromatography (TLC) plates (Si250; Baker) as previously described (35). The lipids were separated by two-dimensional TLC using chloroform-methanol-water (65:25:4, v/v/v) for the first dimension and chloroform-methanol-acetic acid-water (170:25:25:4 v/v/v/v) for the second dimension. Lipids were visualized by charring with 50% H2SO4 (42). The plates were then exposed to a phosphoimager (GE Healthcare) to obtain an image of the labeled lipids. The lipids on the TLC plates were then visualized by iodine staining and the radioactivity in the individual spots was measured by using a liquid scintillation counter (41). The relative amount of each lipid was calculated as the percentage of total radioactivity present in the lipid sample.
Engineering R. sphaeroides 2.4.1 and CL3 strains to overproduce CcO
To overproduce a histidine-tagged version of the CcO enzyme, the pRK-pYJ123H plasmid (36) carrying the genes coding for a wild-type CcO was transferred by conjugation from E. coli S17-1 into R. sphaeroides strains 2.4.1 and CL3 as described (36) to create strains 2.4.1+CcO (CL+) and CL3+CcO (CL−), respectively. These strains were grown aerobically in Sistrom’s media containing 1 μg/mL tetracycline.
To overproduce a version of CcO suitable for crystallization, a histidine tag on subunit II and a shortened version of subunit IV are required to improve molecular homogeneity (2). Therefore, the pCH169 plasmid (2) carrying these versions of the CcO genes was transferred by conjugation into the CL3 mutant strain to create strain 169/CL3 (Supplemental Table S1). The CL(+) crystallography strain used as the control herein is 169, which is derived from a 2.4.1 CcO deletion strain (YZ200, (36)) and carries the pCH169 plasmid (2).
The lipid composition of the membranes from the two different strains without the cls deletion (2.4.1+CcO and 169, both refered to as WT or as CL(+)) were found to be the same, by mass spectrometry lipid analysis. Similarly the two strains carrying the cls deletion (CL3+CcO and 169/CL3, both refered to as CL(−)) also showed the same lipid profile (data not shown), as expected since these strains are derived from the same parent strain, CL3 (43). All strains used for CcO production in this study are listed in Supplemental Materials Table S1.
CcO purification, characterization and crystallization
R. sphaeroides strains overexpressing 6xhistidine-tagged CcO were grown aerobically in Sistrom’s medium with appropriate antibiotics as described (36). CcO protein was purified by using nickel affinity chromatography and monitored by UV-Vis spectroscopy (dithionite-reduced minus ferricyanide-oxidized or dithionite-reduced only) (36). CcO protein for crystallization was purified as described previously (2). Maximal rates of oxygen consumption by purified CcO were measured polarographically in 50 mM KH2PO4 pH 6.5 with 0.05% (m/v) lauryl maltoside, 1 mM N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD), 2.8 mM ascorbate, and 30 μM cytochrome c as the substrate and in the absence or presence of externally-added lipids (a mixture of soybean asolectin at final concentration of 0.44 mg/mL (Avanti Polar Lipids, Alabaster, AL) and cholate at final concentration of 0.02%). The enzymatic activity (turnover number; moles cytochrome c oxidized per mole enzyme per second) was calculated as described (44). Total protein amounts were determined by using the bicinchroninic acid protein assay kit (Pierce, Rockford, IL) including 0.25% deoxycholate in the buffer (45). Four subunit and two subunit CcO protein crystals were obtained using methods described earlier (46) from the crystallization optimized strains.
Direct mass spectrometry analysis of lipids in membranes and protein crystals
Mass spectrometry (MS) and tandem mass spectrometry (MS/MS) analysis of lipids in the isolated membranes and CcO protein crystals were performed using a linear quadrupole ion trap mass spectrometer equipped with a vacuum matrix-assisted laser desorption/ionization ion source (model vMALDI-LTQ, Thermo Scientific, San Jose, CA). Protein crystals were rinsed briefly in water at 4 °C to remove residual protein precipitate and other well-solution components loosely attached to the surface of the crystals before re-dissolving them by extensive vortexing in water. One μL of either resuspended membrane (at a protein concentration of 0.25 mg/mL determined by the BCA assay using bovine serum albumin as the standard) or 1 μL of redissolved CcO crystal solution (at a CcO concentration of 8.3 μM, equivalent to 1.00 mg protein/mL, determined by the BCA assay using purified CcO solution with a known concentration as the standard) were applied to individual spots on the stainless steel sample plate and dried in air. Subsequently, 1 μL of 0.1 M (for membrane samples) or 0.5 M (for crystal samples) 2,5-hydroxy benzoic acid (2,5-DHB, Sigma-Aldrich, Milwaukee, WI) dissolved in acetonitrile (Mallinckrodt Chemicals, Philipsburg, NJ ): water (2:1, v/v) was added. MS and MS/MS spectra were acquired in negative and positive ion modes using enhanced resonance ejection scan conditions. MS/MS spectra were acquired using collision-induced dissociation (CID) typically at a normalized collision energy of 30%, an isolation width of 1.0 or 1.2, and an activation Q value of 0.2. The spectra shown were the average of 250 scans.
Results
Identification of a gene encoding CL synthase in R. sphaeroides
Since the gene encoding CL synthase (cls) had not been previously identified in R. sphaeroides, the candidate cls gene of R. sphaeroides (GenBank Protein ID YP_353546) was selected based on similarity to the amino acid sequence of E. coli CL synthase (27% identity, 44% similarity using BLAST (47)). The candidate gene was cloned into the E.coli expression plasmid pQE-30 (Qiagen, Valencia, CA) and transformed into the E. coli CL-deficient strain SD9 (48). Thin layer chromatograms of extracts from the SD9 transformant harboring the cls expression plasmid showed the presence of CL while no CL was observed for SD9 transformed with empty vector (43). These results suggest that the R. sphaeroides cls gene functionally complements the E. coli CL-deficient mutant and thus encodes a CL synthase of R. sphaeroides.
Construction of R. sphaeroides CL synthase mutant
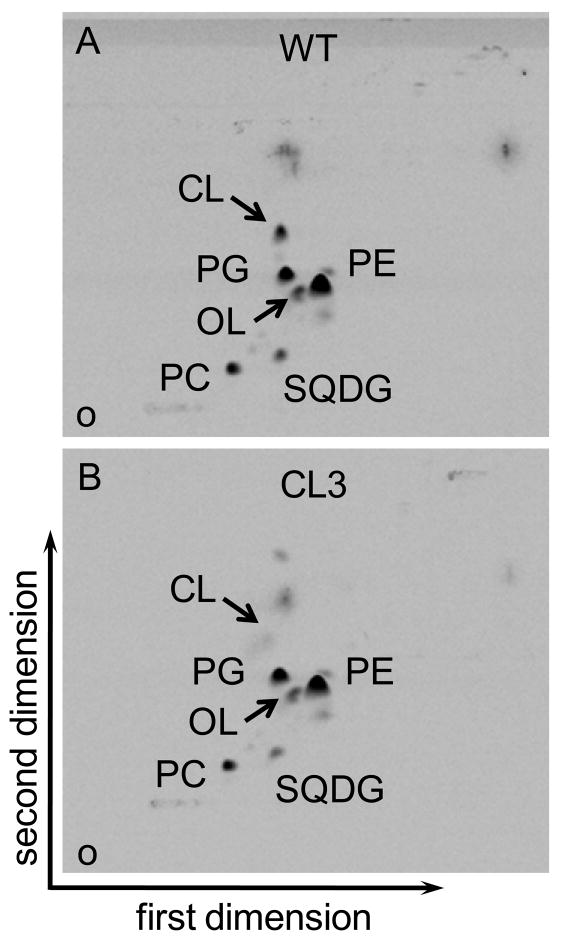
The genomic copy of the cls gene was replaced by a disrupted copy carried by the plasmid pSUP-CLD to obtain a CL synthase-deficient mutant of R. sphaeroides (Figure 1A). Gene disruption was confirmed by PCR analysis of genomic DNA. The PCR amplification using the primers specific for the two ends of the cls gene (P1 and P2) gave a larger amplified product in the mutant compared to the wild type (Figure 1B). The size of the amplified product in the mutant (approximately 2.6 kb) equaled the sum of the sizes of the kanamycin resistant cassette (1.2 kb) plus the 5′ region (0.6 kb) and 3′ region (0.59 kb) of the cls gene that had been amplified and used for obtaining the gene disruption construct. A second PCR using the cls gene specific primer P2 and the kanamycin resistant gene specific primer P5 gave a product of the expected size (approx. 0.96 kb) in the mutant, whereas no amplification was seen in the wild type (Figure 1B). TLC analysis of the lipid extracts from the wild type and the mutant (designated CL3) showed a strong reduction in the level of CL in the mutant (Figure 2). The result clearly indicated that the inactivation of the cls gene impaired CL synthesis in the mutant.
Figure 1.
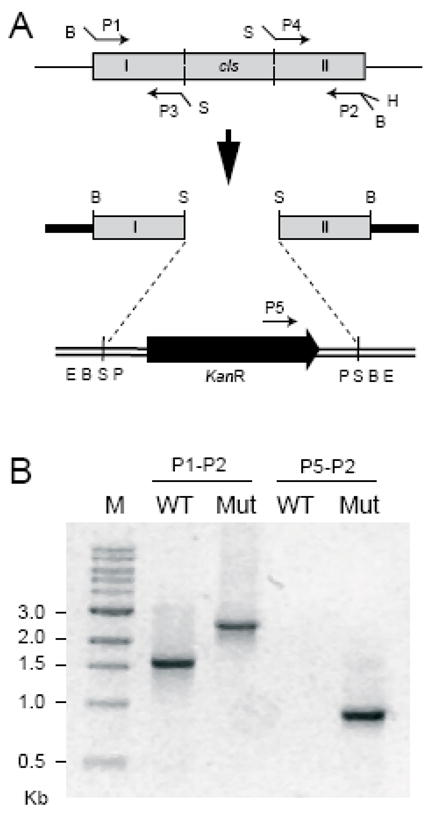
Disruption of the cls gene. (A) Schematic diagram showing the gene disruption strategy. Grey boxes represent the cls gene and amplified segments of the gene. The black block arrow represents the kanamycin resistance cassette and the direction of the arrowhead shows its orientation. Plasmids pPICT-2 and pUC4K are shown as thick solid lines and double lines, respectively. Restriction sites: B, BamHI; S, SalI, H, HindIII; E, EcoRI; P, PstI. (B) PCR verification for the gene disruption. Genomic DNA was isolated from the type (WT) and the cls-deleted mutant (Mut) strains and two PCR reactions were carried out using different sets of primers placed at the positions indicated in Fig. 2A. The sizes of the PCR products were compared by gel electrophoresis. M indicates the marker lane.
Figure 2.
Two-dimensional thin-layer chromatogram of 14C-labeled lipids from the aerobically-grown R. sphaeroides (A) wild-type strain 2.4.1 and (B) the cls-deleted mutant CL3. The lipids were visualized by using a phosphoimager. Lipids: PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PC, phosphatidylcholine; SQDG, sulfolipid; OL, ornithine lipid; CL, cardiolipin. O, origin.
Composition of general lipid types in the wild type and the mutant cells
To determine the effect of cls gene inactivation on lipid content, a saturation 14C-acetate labeling approach was used to obtain a quantitative comparison of relative lipid amounts in the wild type and the CL3 mutant. The level of CL in the mutant was strongly reduced, but a trace of CL, less than 10% of wild type, was still present in the mutant (Table 1). The result was consistent with that observed for an E. coli cls knockout mutant, in which a low level of CL was also still present (49). Corresponding to the decrease in CL in the CL3 mutant, an increase in PG, the precursor for CL synthesis, was observed (Table 1). Within statistical limitations, the amounts of phosphatidylcholine (PC), phosphatidylethanolamine (PE), ornithine lipid (OL) and sulfoquinovosyldiacylglycerol (SQDG) remained relatively unchanged. The structures of the molecular species in each type of lipid in the cell membranes were elucidated by MALDI MS and MS/MS as well as ESI MS, MS/MS and multistage tandem mass spectrometry (MSn) in the linear ion trap mass spectrometer (50). The changes in the amounts of the phospholipids revealed by TLC were confirmed by quantitative ESI analysis of the phospholipids using internal standards in both MS and MS/MS (see accompanying paper).
Table 1.
Lipid composition of R. sphaeroides wild-type (2.4.1) and CL-deficient mutant (CL3) determined by thin-layer chromatograpy and radioactive labeling
| Lipid | wild-type | CL3 |
|---|---|---|
| CL | 5.6 ± 1.6 | 0.5 ± 0.1 |
| OL | 3.7 ± 0.2 | 3.5 ± 0.4 |
| PE | 42.4 ± 4.3 | 44.1 ± 2.5 |
| PG | 22.4 ± 0.1 | 26.2 ± 2.2 |
| SQDG | 6.7 ± 0.2 | 6.4 ± 1.3 |
| PC | 19.2 ± 4.4 | 19.2 ± 3.8 |
Keys: The values given represent the mean percentage of radioactivity for individual lipids ± the standard error from 3 independent experiments. CL, cardiolipin; PE, phosphatidylethanolamine; OL, ornithine lipid; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol; PC, phosphatidylcholine.
Effect of CL deficiency on R. sphaeroides cell growth
To analyze the effect of the decreased CL on the overall growth of R. sphaeroides cells, the photosynthetic and respiratory growth of the wild-type and the mutant was compared. Under the conditions used in this study, no significant differences were observed in the photosynthetic growth for both lines (Figure 3A). Respiratory growth was slightly reduced in the mutant, as indicated by the increased lag time (slower approach to final cell density) and lower final cell density (Figure 3B). The results suggest that CL may contribute to optimal respiratory growth, but may be less critical to photosynthetic growth.
Figure 3.
Growth curves of R. sphaeroides wild-type (▲; 2.4.1) and CL-deficient mutant (●; CL3) strains under A) anaerobic photoheterotrophic growth, and B) aerobic chemoheterotrophic growth conditions, both in Sistrom’s media at 30 °C. The data presented are the average of three independent growth cultures and the error bars are the standard deviations. Absorbances are expressed in Klett units, where 1 Klett unit is equivalent to 107 cells per mL (38).
Production of CcO in R. sphaeroides wild-type and CL-deficient cells
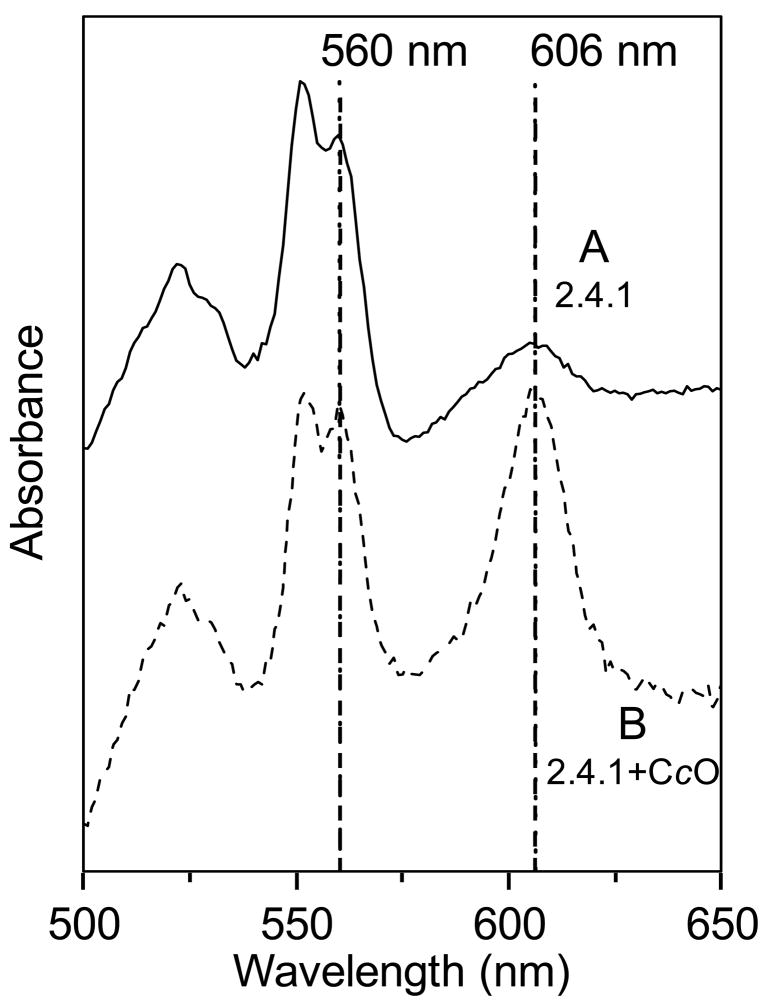
The wild-type 2.4.1 strain of R. sphaeroides naturally produces a low level of CcO in its membranes when grown under aerobic conditions (Figure 4A). Since this strain has been previously used to produce a variety of lipid mutants (39, 42, 51), it was necessary to demonstrate that it is also a suitable host for CcO overproduction to provide the model system for testing the role of lipids in this protein. Optical difference spectra of reduced minus oxidized states provide a facile method to assess the level of CcO in the membrane by the absorbance of its a-type hemes. Difference spectra of the solubilized membrane proteins of the wild-type 2.4.1 strain (Figure 4A) showed that the 606 nm peak, representing a-type hemes of CcO, was much smaller than the 560 nm peak to which the spectra are normalized. (Note that the 560 nm peak represents b-type hemes of the bc1 complex plus a cbb3 oxidase also present in the membranes. In addition, a membrane-bound cytochrome c contributes to the 550 nm region.) After the CcO genes were introduced on the plasmid pRK-pYJ123H to create strain 2.4.1+CcO, the 606 nm peak was strongly increased relative to the 560 nm peak, indicating an overproduction of CcO (Figure 4B). This was confirmed by measuring the concentration of CcO per mg total protein, which showed an equivalent increase. When this plasmid was introduced into the CL-deficient strain CL3, high levels of CcO expression were also seen, but were variable depending on the culturing time and volume of the culture (data not shown). In smaller volume cultures the levels of CcO were high, while they were sometimes lower in larger cultures grown for longer times. This observation indicates a possibly greater sensitivity of the CL3 mutant to the stress experienced during subculturing and dilution into larger flasks (see also accompanying paper).
Figure 4.
Optical difference spectra of dithionite-reduced minus ferricyanide-oxidized solubilized membranes from A) R. sphaeroides strains 2.4.1 (wild-type) and B) 2.4.1+CcO (wild-type with CcO genes added on the expression plasmid PRK- pYJ123H). The relative levels of CcO are indicated by the sizes of the a-type heme absorbance peaks at 606 nm compared to the b-heme at 560 nm. The spectra are normalized to the 560 nm (b-type heme) peak. (Note that these membranes also contain the bc1 complex, a cbb3 type oxidase, and a membrane-bound cytochrome c which all contribute to the 560 nm and 550 nm peaks.)
Properties of CcO purified from wild-type and cls mutant cell lines
Table 2 lists the spectral properties and enzymatic activities of CcO protein purified from R. sphaeroides cell lines overproducing a histidine-tagged version of CcO. The Soret (445 nm) and alpha (606 nm) bands in the optical spectra are diagnostic of the native, reduced heme centers of CcO. No wavelength shifts of peak maxima were observed between the CL(−) mutant cell line (CL3+CcO) and the CL(+) control (2.4.1+CcO), indicating that the absence of the cls gene in the mutant did not impair its ability to correctly assemble and insert the metal centers into CcO. The enzymatic activities of CcO purified from CL(−) and CL(+) strains were also very similar, indicating that the enzyme purified from the CL(−) mutant cells was as active as that purified from the CL(+) cells. The activities of all purified CcOs increased similarly in the presence of externally-added lipids during the assay. This commonly observed phenomenon is likely related to the extent of delipidation of CcO during the purification process (44). When tested for stability at temperatures ranging from 20 °C to 80 °C, C cO purified from both the wild-type and mutant strains behaved similarly (data not shown).
Table 2.
Spectral and enzymatic properties of CcO purified from R. sphaeroides wild-type (2.4.1+CcO) and cardiolipin-depleted (CL3+CcO) cell lines
| Cell Line | Genotype | Spectral Peaks of Purified CcO (nm) | Activity (s−1) no added lipid | Activity (s−1) plus added lipid | Increase in activity with lipid | |
|---|---|---|---|---|---|---|
| Soret | Alpha | |||||
| 2.4.1+CcO | wild-type | 445 | 606 | 1050 ± 100# | 1340 ± 50# | 28 ± 2%# |
| CL3 +CcO | Δcls | 445 | 606 | 1060 ± 109@ | 1410 ± 120@ | 33 ± 8%@ |
Keys: Both strains produced his-tagged CcO from a plasmid (PRK-pYJ123H (36)). The Soret and alpha peaks were from the optical spectra of dithionite-reduced CcO. The molecular activity of purified CcO was measured at pH 6.5 and presented as turnover number (moles cytochrome c oxidized per mole CcO per second). The number of independent measurements was 3# or 5@.
SDS-PAGE analysis of CcO purified from CL(+) and CL(−) mutant strains indicated that the composition and relative levels of all of the subunits were similar (see accompanying paper).
MALDI MS analysis of lipids in the membranes overexpressing CcO
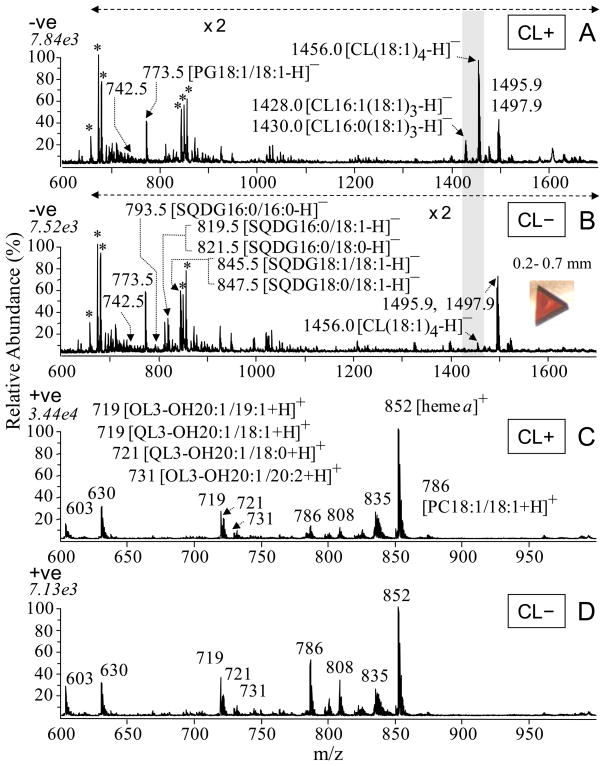
Figure 5(A and B) shows the negative ion mode MALDI MS spectra of the lipids in the isolated membranes of the CL(+) and CL(−) strains of R. sphaeroides. Similar types of lipids were observed in the membranes of both strains, including PE, PG, SQDG, and CL, although with varying abundance. The major molecular species of lipids detected in these strains include the phospholipids PE 18:1/18:1 (m/z 742.5), PG 18:1/18:1 (m/z 773.5), cardiolipins CL(18:1)4 (m/z 1456.0), CL(16:1)1(18:1)3 (m/z 1428.0), CL(16:0)1(18:1)3 (m/z 1430.0) and CL(18:0)1(18:1)3 (m/z 1458.0), as well as sulfolipids SQDG 16:0/16:0 (m/z 793.5), SQDG 16:0/18:1 (m/z 819.5), SQDG 16:0/18:0 (m/z 821.5), SQDG 18:1/18:1 (m/z 845.5) and SQDG 18:0/18:1 (m/z 847.5). The identities of each of the membrane lipid species labeled in these figures were confirmed by MS/MS analysis (50). Note that although MALDI is not generally considered to be a rigorously quantitative method due to issues arising from matrix/sample heterogeneity, these experiments were performed in parallel under identical sample handling and data acquisition conditions and in multiple replicates, giving confidence in a semi-quantitative interpretation of the data, particularly using MS/MS, since the chemical noise observed during MS analysis is mostly eliminated (52). Further, MALDI has the major advantage of being able to analyze samples directly, without chemical solvent extraction, and being more tolerant of the presence of detergent than ESI, thus bypassing problems caused by detergents and detergent removal procedures (50). These facts, along with the good agreement with the radioactive labeling analysis (Table 1) and quantitative ESI measurements described in Figures 2 and 4 of the accompanying paper, lend strength to the quantitative significance of the observation of approximately 5% relative abundance of CL ions in CL(−) membranes compared to the CL(+) membranes. The 5% level is calculated relative to the ion abundances of PE 18:1/18:1 at m/z 742.5 and SQDG 18:1/18:1 and SQDG 18:0/18:1 at m/z 845.5 and 847.5, the most abundant ions within the displayed m/z range (Figures 5A and 5B). In contrast, the abundance of the PG 18:1/18:1 (m/z 773.5), and the SQDG 16:0/18:1 and SQDG 16:0/18:0 lipid species (m/z 819.5 and 821.5, respectively), showed an apparent increase in the CL(−) mutant membrane. (It should be noted that the m/z 1495.9 and m/z 1497.9 ions seen in negative ion mode were determined not to be related to cardiolipin by MS/MS and MS3 analysis.)
Figure 5.
MALDI-MS spectra of membranes isolated from CL(+) (169WT) and CL(−) (169CL3) strains of R. sphaeroides in negative ion mode (A and B) and in positive ion mode (C and D). The highlighted peaks in A) are those of CL at 1456, 1430, and 1428, which are barely discernable in B), the CL(–) sample. A) CL(+) membranes in negative ion mode, B) CL(−) membranes in negative ion mode, C) CL(+) membranes in positive ion mode, D) CL(−) membranes in positive ion mode, each at 0.25 mg/mL membrane protein concentration. The identities of labeled ions in each spectrum were confirmed by MS/MS experiments. Ions labeled with asterisks were identified as 2,5-DHB matrix cluster adducts. The peaks at m/z 1495.9, 1497.9, 1533.8 and 1535.8 were determined by MS/MS and MSn not to be CL or CL derivatives.
The positive ion mode MALDI MS spectra of the membranes of the CL(+) and CL(−) strains are shown in Figures 5C and 5D. In addition to the most abundant PC lipid ions (PC 18:1/18:1 [m/z 786, 808, 824], PC 18:1/19:1 [m/z 800, 838]), PE lipids (mainly PE 18:1/18:1 [m/z 744, 766]) and ornithine lipids (OL 3-OH 20:1/19:1, m/z 719) were also observed in positive ion mode. The recently discovered glutamine lipids (QL) (53) were also identified by ESI analysis of the membrane lipid extract (supplementary of accompanying paper); QL has a relatively lower ionization efficiency in MALDI and overlaps with the m/z 719 of OL. Comparison of Figure 5C with Figure 5D suggests that PC, PE and OL are not significantly affected by the cls mutation, either in molecular structural diversity or in relative quantity.
Purification and crystallization of CcO from wild-type and cls mutant
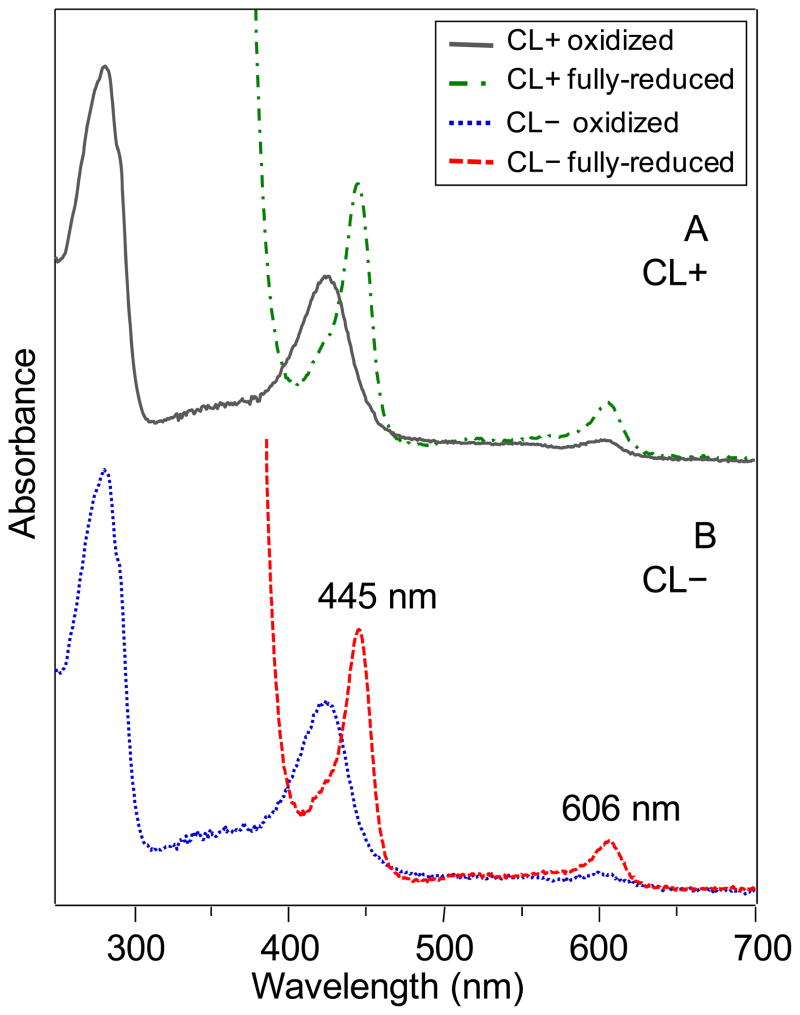
The R. sphaeroides strain containing the cls deletion and a plasmid containing CcO designed for crystallization (169CL3) displayed high expression yield. The UV-Vis spectral characteristics of the CcO purified from this CL(−) strain, in reduced and oxidized forms, were essentially identical in the peak positions of the hemes a and a3 to those of the CcO obtained from the CL(+) strain purified in the same manner (Figure 6).
Figure 6.
Oxidized and reduced spectra of purified CcO samples used to produce protein crystals, purified from A) CL(+) (169WT) and B) CL(−) (169CL3) strains grown aerobically at 30 ºC. The reduced peaks at 445 and 606 nm, which are characteristic of the native heme a and a3 spectra, are seen to be at the same wavelengths in both CcO forms. These peaks have contributions of both hemes at both wavelengths. The spectra were acquired using the same CcO protein concentrations.
The CcO purified from CL(+) and CL(−) strains contained the normal four subunits (I, II III and IV) as revealed by SDS-PAGE analyses (see accompanying paper). Using CcO purified from CL(+) and CL(−) strains, CcO crystals were obtained with four subunits and with two-subunits (subunit I and II only) for both strains, under the same crystallization conditions and yielding the same form (space group P212121 for 2-subunit crystal; R3 for 4-subunit crystal). The resolution obtained for the four-subunit CcO crystals from the CL(−) mutant (3.2 Å) was similar to that of four-subunit crystals from the CL(+) strain (3.3 Å), while the resolution for the 2-subunit crystals from the CL(−) mutant (3.3 Ǻ) was slightly less than that from the CL(+) strain (2.9 Ǻ) based on many repeats with the CL(+) and CL(−) forms, grown, purified and crystallized in parallel over several years (Supplemental Table S2). Thus, the strongly decreased levels of CL observed in the membrane lipid profile did not prevent the crystallization of four-subunit or two-subunit CcO. The four-subunit CcO crystals generated were examined by MALDI MS and MS/MS analysis.
MALDI MS and MS/MS analysis of the lipids in the re-dissolved four-subunit CcO crystals
Figure 7 shows the MALDI MS spectra obtained by direct analysis of the re-dissolved crystals, in negative ion mode (Figures 7A and 7B) and in positive ion mode (7C and 7D).
Figure 7.
MALDI-MS spectra of the four-subunit CcO protein crystals from CL(+) (169WT) and CL(−) (169CL3) strains of R. sphaeroides in negative ion mode (A and B) and positive ion mode (C and D), showing a marked difference in CL content by relative peak height at m/z 1456 (highlighted). A) CcO from CL(+) in negative ion mode, B) CcO from CL(−) in negative ion mode, C) CcO from CL(+) in positive ion mode, D) CcO from CL(−) in positive ion mode. Ions labeled with asterisks were identified as 2,5-DHB matrix cluster adducts. The identities of labeled ions in each spectrum were confirmed by MS/MS experiments. Inset in B: photo of a typical four-subunit CcO crystal used in this analysis.
The data show that CcO crystals from the CL(+) strain retain readily detectable and multiple species of CL and SQDG, as well as PG 18:1/18:1, and OL 3-OH 20:1/19:1 in negative ion mode (Figure 7A), PC18:1/18:1 and PE18:1/18:1 in positive ion mode, as confirmed by MS/MS analysis (Figures 8 and 9). These detailed structures of the lipids in the crystals have not been previously reported. Both cardiolipin and SQDG contained multiple fatty acid chain compositions. In addition to the most abundant CL(18:1)4 (m/z 1456), minor species of CL(16:1)1(18:1)3 (m/z 1428) and CL(16:0)1(18:1)3 (m/z 1430) were also found, in ratios similar to those found in the membrane, indicating no apparent selection for CL on the part of CcO during purification and crystallization. SQDG in the crystals also included a variety of fatty acid chain compositions (Figures 9B and 9C), all at comparable relative abundance. The relative levels of various fatty acids appear to be representative of the membrane background (compare Figure 7 and Figure 5), suggesting that CcO also has little selectivity for fatty acid chain variations in SQDG.
Figure 8.
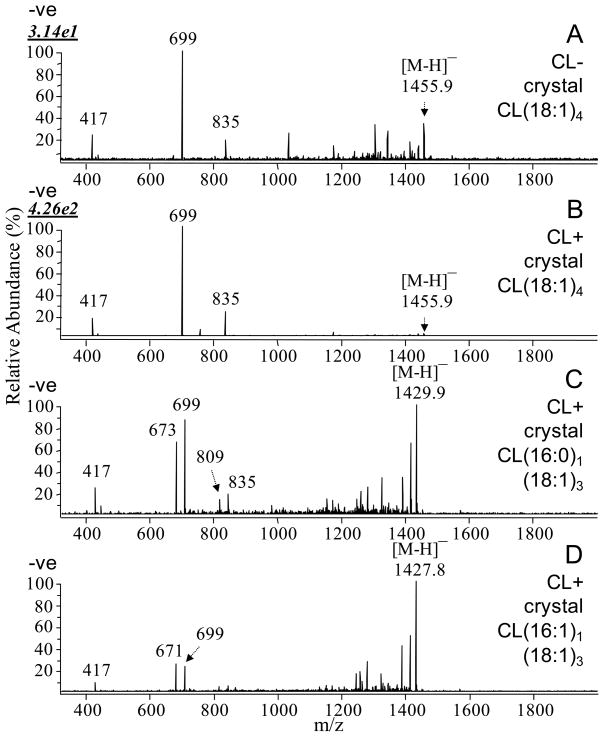
Identification of the cardiolipin molecular species in the four-subunit CcO crystals from CL(+) (169WT) and CL(−) (169CL3) R. sphaeroides using CID MS/MS in negative ion mode on a MALDI-linear ion trap mass spectrometer, showing the marked depletion of CL(18:1)4 in CL(−) crystals compared to CL(+) crystals (A and B) as demonstrated by comparison of the absolute ion abundance of m/z 699 (top left of panels A and B ), the predominant product ion generated by CL(18:1)4 in MS/MS, corresponding to the phosphatidyldioleoylglyceride moiety. The figure identifies multiple CL species in CL(+) CcO crystals (B, C, D) but no CL (18:2)4 at m/z 1448 is seen (see also Figure 7A).
Figure 9.
MS/MS spectra showing the characteristic fragmentation patterns of the precursor ions of the major lipids, providing the identifications of these lipids in the CcO crystals from CL(+) (169WT) and CL(−) (169CL3) R. sphaeroides using CID MS/MS in positive and negative ion modes on a MALDI-linear ion trap mass spectrometer. Representative lipids in the crystals include A) PG 18:1/18:1 at m/z 773 (−ve), B) SQDG 16:0/18:1 at m/z 819 (−ve), C) SQDG 18:0/18:1 at m/z 847 (−ve) and D) OL 3-OH 20:1/19:1 at m/z 719 (+ve). In panels B and C, the fragment ion at m/z 225 is determined by MS3 to represent the sulfoquinovosyl head group of SQDG, and the peaks in the middle of the spectra are generated by the neutral loss of either one of the two fatty acid chains respectively.
In the CcO crystals from the CL(−) mutant, MS clearly reveals significantly decreased levels of CL (Figure 7B) equivalent to those in the CL(−) membrane (Figure 5B), as well as apparently higher abundance of SQDG species (m/z 793, 819, 821, 845, 847) compared to the CL(+) crystal (Figure 7A). MS/MS further demonstrates that the level of CL(18:1)4 (m/z 1456) in the CL-deficient mutant crystal is about ~10% of that seen in the wild type, as indicated by comparison of the absolute abundances of the m/z 699 fragment ion (the 18:1/18:1 phosphatidyldioleoylglyceride ion) (50, 54) in the MS/MS spectra of the m/z 1455.9 precursor ions from CL(−) and CL(+) crystals (Figures 8A and 8B). These results are again consistent with the membrane CL levels and with radioactive labeling (Table 1), suggesting no strong selectivity of CcO for CL in a CL(−) membrane environment. The PG lipid species (m/z 773.5) show increase in abundance to some extent, while PE, PC and OL lipids are observed in MS positive ion mode to be at similar relative levels to those in the crystals of the CL(+) strain (Figures 7C and 7D).
It is noteworthy that the major molecular species of cardiolipin in the isolated cell membranes and the CcO crystals of R. sphaeroides has four 18:1 fatty acyl chains CL(18:1)4 (m/z 1456) and the three minor species of cardiolipin are CL(16:1)1(18:1)3, CL(16:0)1(18:1)3 and CL(18:0)1(18:1)3. However, CL(18:2)4 (tetralinoleoyl cardiolipin; m/z 1448), the predominant cardiolipin species found in mammalian mitochondria (29, 55), was not observed in any of the samples of R. sphaeroides examined in the current study.
Discussion
Residual CL in the cardiolipin-deficient mutant
The cls-encoded enzyme catalyzes the synthesis of CL from two PG molecules (56). Although lipid analysis showed a significant reduction in the level of CL in the mutant membranes, a low amount of CL (<10% wild-type) still persisted in both the membrane and the CcO crystals of the mutant, as confirmed by saturation radioactive labeling, MALDI MS, MALDI MS/MS, and ESI with internal standards (accompanying paper). At this reduced CL level, no appreciable effects were observed on the protein expression, protein spectroscopic properties, activity, assembly and crystallization.
The source of this residual CL in the mutant still needs to be determined. In the E. coli cls knockout mutant, overexpression of the pss gene led to an increase in the CL level (49), suggesting that phosphatidylserine synthase has CL synthase activity. In a similar experiment, it has been observed that the expression of the pss gene (GenBank protein ID YP_353797) from R. sphaeroides in the E. coli SD9 strain resulted in the synthesis of CL (up to 1%) which showed dependence on the level of expression of the gene (43). Thus, as observed for E. coli, phosphatidylserine synthase may be the source of the residual CL in the CL3 mutant of R. sphaeroides (43).
Difference between molecular components of CL in R. sphaeroides and mammalian cells
Mass spectrometry analysis revealed that the CL molecular species in the membrane, the purified CcO enzymes, and the CcO protein crystals, is mainly CL(18:1)4, while much less abundant CL(16:1)1(18:1)3, CL(16:0)1(18:1)3 and CL(18:0)1(18:1)3 were also present. In contrast to the mammalian mitochondria, where CL(18:2)4 is the predominant and functionally essential species (29, 55, 57), the R. sphaeroides contained no CL(18:2)4 detectable by MS. One important role of cardiolipin in mammalian mitochondria has been proposed to be a part of the signaling system in apoptosis, involving the peroxidizability of the 18:2 unsaturated fatty acyl chains and not the 18:1 acyl chain components (29, 55, 58). The predominant CL species in healthy mammalian mitochondria is CL(18:2)4, and a relative decrease of the 18:2 cardiolipin component has been correlated with diseased mitochondria (29, 55). CL (18:2)4 is also found to activate mammalian CcO more efficiently than other lipids (59). Notably, CL(18:2)4 is not synthesized directly in mammalian cells, but is formed through a modification by acyltransferases of the initial cardiolipin species (58), a process that is absent in the bacterial systems (58, 60). Yeasts also predominantly produce cardiolipin containing 16:1 and 18:1 fatty acyl chains but not 18:2 (61, 62). The observed difference in cardiolipin types may account for fundamental differences in the functional roles that cardiolipin plays in the bacterial, yeast, and mammalian systems.
CL and growth of R. sphaeroides
The CL3 mutant showed some decreased growth under respiratory conditions, suggesting that a normal level of CL is important for robust respiratory growth. A similar effect of CL deficiency on respiratory growth has been observed in a Saccharomyces cerevisiae crd1Δ mutant, in which the loss of CL affects the stability of the respiratory chain complexes (25, 26, 63). The photosynthetic growth of the CL3 mutant was not affected, probably because other lipids such as PG substituted for CL. It has been shown that PG is preferentially increased in purple bacteria during photosynthetic growth (64). It is possible that the level of CL needed for photosynthetic growth is not as high as that needed for respiratory growth, and therefore the low level of CL retained by the CL3 mutant could be sufficient under photosynthetic conditions.
Alternatively, CL may be only required during stress situations. Osmotic stress during growth has been shown to increase CL synthesis in R. sphaeroides (65). Variable, but often lower, levels of CcO production are observed from CL3 cultures, a response which may be due to the added stress of subculturing, dilution and growth at high oxygen levels and high pH (approaching stationary phase). Determining a specific role of CL during respiratory growth in R. sphaeroides will require further studies.
CL in the function and assembly of CcO of R. sphaeroides
Low levels of CL did not appreciably affect the assembly or the activity of the purified enzyme (Table 2). The CcO produced the same type of four-subunit crystals which diffracted to the same level as crystals from the CL(+) strains (Table S2). It was critical to determine whether, despite the low CL levels, CcO was able to preferentially sequester enough CL for its needs. However, the MS and MS/MS analyses reproducibly demonstrated that the purified enzyme and the crystals did not concentrate CL above that found in the membranes. It is reasonable to conclude that CL is not essential for the structure, function or crystallization of Rhodobacter CcO, consistent with several previous studies using bacterial and yeast systems (30–32). This is in contrast to numerous biochemical and biophysical studies which suggest a requirement for CL in mammalian systems (7, 29, 58, 66).
A CL requirement for maintaining supercomplexes between CcO and other membrane proteins of the respiratory chain, and not for the stability or activity of CcO per se, could still be the case in R. sphaeroides, since evidence of such complexes are also found in other bacterial systems (67). Experiments in yeast suggest that complexes III (bc1) and IV (CcO) do not form the normal level of supercomplexes in the mitochondria of CL-deficient mutants, and CL-deficient yeast cultures also grew more slowly and to a lower final density than wild-type yeast cultures (25). Similarly, the CL3 mutant showed slower growth under respiratory conditions in this study (Figure 3) but no direct evidence is available to support a role in supercomplex formation.
The mass spectrometric data on CcO suggests that other lipids may functionally substitute for CL in R. sphaeroides. A relative increase in the levels of SQDG and PG was observed in crystals from the CL(−) mutant strain (Figure 7B). The increase in PG in the CL(−) mutant was also observed in the membrane samples by radioisotope labeling (Table 1). Earlier studies comparing the yeast cls mutant (CL-deficient) and pgs mutant (PG and CL-deficient) suggested that CL is not essential for the functions of CcO or mitochondria in yeast (30–32), but can be functionally replaced by PG (30). The lack of difference in SQDG levels observed in membrane preparations, when analyzed either by thin-layer chromatography or by mass spectrometry, suggests that the altered levels seen in the crystals by MS may reflect some selectivity of CcO, rather than a general metabolic response at the cellular level. Specifically bound SQDG molecules have been previously resolved in the crystals of cyanobacterial photosystem II (68, 69) but not yet in R. sphaeroides CcO. The apparent increased abundance of SQDG in the CcO crystals suggests a possible compensatory role for SQDG during CL deficiency, since both are anionic lipids, and SQDG was found to increase in R. sphaeroides membranes under phosphate-limiting growth conditions (39). Studies in Rhodobacter capsulatus, which has neither CL nor an aa3-type oxidase, have suggested a similar function for ornithine lipid (70). This type of lipid is present in the R. sphaeroides membranes (53) but is not appreciably altered in CL(−) membranes, as seen by thin-layer chromatography and mass spectrometry.
In a previous study (2), high resolution (2.0 Å) two subunit CcO crystals were obtained which contain a potential CL binding site, based on homology with the bovine enzyme, even though the head group is not resolved. In the current study, the 2-subunit crystal form was produced from both CL(+) and CL(−) strains, but in neither case at high enough resolution to visualize the lipids, likely due to the background strain, 2.4.1, not containing the necessary deletion of subunit IV that has been found to be key to giving the highest resolution crystals (2).
Lipid mutants of R. sphaeroides as tools for the study of membrane proteins in vivo
One of the motivations for studying the production, properties, and crystallization of CcO in the CL-deficient mutant of R. sphaeroides was to test the hypotheses that 1) CL is a necessary component of the fully functional enzyme in R. sphaeroides; and 2) the specific lipid composition of a membrane protein sample is an important factor in achieving high quality crystals. The basis for the first hypothesis was a wealth of biochemical evidence supporting a role for CL in CcO function in mammalian systems (7, 12, 20, 66). The basis for the second hypothesis was the observation of specifically bound and conserved lipids in high resolution crystal structures of membrane proteins. A further motivation for these studies was to establish R. sphaeroides as a tool for the overproduction of membrane proteins from many sources for structural studies (71) by providing versions of R. sphaeroides with different and controlled lipid compositions.
The results herein, especially the MS lipid analysis of the CcO crystals, argue against both starting hypotheses and suggest that CcO in R. sphaeroides does not have a strict requirement for CL per se. The results demonstrate that it is possible to overproduce CcO in a CL-deficient mutant, and that the purified enzyme is fully active by available criteria and gives well-resolved crystals, despite a very low CL content. The ability to directly analyze the lipid content of the crystals provides unique insight into the effects of low CL, showing the presence of other bacterial lipids, including SQDG and ornithine lipid, apparently substituting for CL in the crystal. To determine the extent to which the roles of CL can be fully substituted by other lipid species in Rhodobacter sphaeroides, a method to further perturb the lipid profile and produce a more complete CL-deficiency was developed, using a combined metabolic and genetic approach (see accompanying paper).
Supplementary Material
Supplemental Table S1. R. sphaeroides strains and CcO plasmids used in this work.
Supplemental Table S2. X-ray screening results of CcO crystals obtained from CcO expressed in the R. sphaeroides wild type CL(+) and CL(−) mutant at different crystal growth temperatures.
Acknowledgments
We would like to thank Dr. Koichiro Awai for help with the design of the cls gene disruption strategy and Dr. Ling Qin for assistance in crystallizing CcO. We are grateful to Dr. Kouji Matsumato (Saitama University, Japan) for providing the E. coli CL-deficient mutant. Crystal screening was carried out at Argonne National Labs, APS synchrotron, LS CAT beamline.
Abbreviations
- R. sphaeroides
Rhodobacter sphaeroides
- CcO
cytochrome c oxidase
- cls
cardiolipin synthase gene
- CL
cardiolipin
- PG
phosphatidylglycerol
- PE
phosphatidylethanolamine
- PC
phosphatidylcholine
- OL
ornithine lipid
- QL
glutamine lipid
- SQDG
sulfoquinovosyldiacylglyceride
- CL(+)
cardiolipin-proficient strain
- CL(−)
cardiolipin-deficient strain
- MALDI
matrix assisted laser desorption/ionization
- nESI
nanoelectrospray ionization
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- MSn
multistage tandem mass spectrometry
- 2,5-DHB
2, 5-hydroxybenzoic acid
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TLC
thin layer chromatography
- PCR
polymerase chain reactions
- WT
wild-type
Footnotes
The work was supported by NIH R01 GM26916 (SFM) and the Michigan State University Center of Excellence for the Structural Analysis of Membrane Proteins (CB, GER and SFM).
Supporting information can be found at http://pubs.acs.org.
References
- 1.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc Natl Acad Sci U S A. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nussberger S, Doerr K, Wang DN, Kuhlbrandt W. Lipid-protein interactions in crystals of plant light-harvesting complex. J Mol Biol. 1993;234:347–356. doi: 10.1006/jmbi.1993.1591. [DOI] [PubMed] [Google Scholar]
- 4.Bogdanov M, Sun J, Kaback HR, Dowhan W. A phospholipid acts as a chaperone in assembly of a membrane transport protein. J Biol Chem. 1996;271:11615–11618. doi: 10.1074/jbc.271.20.11615. [DOI] [PubMed] [Google Scholar]
- 5.Bogdanov M, Dowhan W. Phospholipid-assisted protein folding: phosphatidylethanolamine is required at a late step of the conformational maturation of the polytopic membrane protein lactose permease. Embo J. 1998;17:5255–5264. doi: 10.1093/emboj/17.18.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogdanov M, Umeda M, Dowhan W. Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J Biol Chem. 1999;274:12339–12345. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- 7.Sedlák E, Robinson NC. Phospholipase A2 digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry. 1999;38:14966–14972. doi: 10.1021/bi9914053. [DOI] [PubMed] [Google Scholar]
- 8.Valiyaveetil FI, Zhou Y, MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 9.Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Awasthi YC, Chuang TF, Keenan TW, Crane FL. Tightly bound cardiolipin in cytochrome oxidase. Biochim Biophys Acta. 1971;226:42–52. doi: 10.1016/0005-2728(71)90176-9. [DOI] [PubMed] [Google Scholar]
- 11.Gomez B, Jr, Robinson NC. Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry. 1999;38:9031–9038. doi: 10.1021/bi990603r. [DOI] [PubMed] [Google Scholar]
- 12.Robinson NC. Specificity and binding affinity of phospholipids to the high affinity cardiolipin sites of beef heart cytochrome c oxidase. Biochemistry. 1982;21:184–188. doi: 10.1021/bi00530a031. [DOI] [PubMed] [Google Scholar]
- 13.Schagger H, Hagen T, Roth B, Brandt U, Link TA, von Jagow G. Phospholipid specificity of bovine heart bc1 complex. Eur J Biochem. 1990;190:123–130. doi: 10.1111/j.1432-1033.1990.tb15554.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Kurisu G, Smith JL, Cramer WA. A defined protein-detergent-lipid complex for crystallization of integral membrane proteins: The cytochrome b6f complex of oxygenic photosynthesis. Proc Natl Acad Sci U S A. 2003;100:5160–5163. doi: 10.1073/pnas.0931431100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garavito RM, Ferguson-Miller S. Detergents as tools in membrane biochemistry. J Biol Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- 16.Guan L, Smirnova IN, Verner G, Nagamori S, Kaback HR. Manipulating phospholipids for crystallization of a membrane transport protein. Proc Natl Acad Sci U S A. 2006;103:1723–1726. doi: 10.1073/pnas.0510922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosler JP, Ferguson-Miller S, Mills DA. Energy transduction: proton transfer through the respiratory complexes. Annu Rev Biochem. 2006;75:165–187. doi: 10.1146/annurev.biochem.75.062003.101730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills DA, Ferguson-Miller S. Understanding the mechanism of proton movement linked to oxygen reduction in cytochrome c oxidase: lessons from other proteins. FEBS Lett. 2003;545:47–51. doi: 10.1016/s0014-5793(03)00392-2. [DOI] [PubMed] [Google Scholar]
- 19.Marsh D, Horvath LI. Structure, dynamics and composition of the lipid- protein interface. Perspectives from spin-labelling. Biochim Biophys Acta. 1998;1376:267–296. doi: 10.1016/s0304-4157(98)00009-4. [DOI] [PubMed] [Google Scholar]
- 20.Powell GL, Knowles PF, Marsh D. Spin-label studies on the specificity of interaction of cardiolipin with beef heart cytochrome oxidase. Biochemistry. 1987;26:8138–8145. doi: 10.1021/bi00399a018. [DOI] [PubMed] [Google Scholar]
- 21.Distler AM, Allison J, Hiser C, Qin L, Hilmi Y, Ferguson-Miller SM. Mass spectrometric detection of protein, lipid and heme components of cytochrome c oxidase from R. sphaeroides and the stabilization of non-covalent complexes from the enzyme. Eur Mass Spectrom. 2004;10:295–308. doi: 10.1255/ejms.594. [DOI] [PubMed] [Google Scholar]
- 22.Svensson-Ek M, Abramson J, Larsson G, Tornroth S, Brzezinski P, Iwata S. The x-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 23.Fyfe PK, Isaacs NW, Cogdell RJ, Jones MR. Disruption of a specific molecular interaction with a bound lipid affects the thermal stability of the purple bacterial reaction centre. Biochim Biophys Acta, Bioenerg. 2004;1608:11–22. doi: 10.1016/j.bbabio.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim Biophys Acta, Biomembr. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 26.Pfeiffer K, Gohil V, Stuart Rosemary A, Hunte C, Brandt U, Greenberg Miriam L, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 27.Sorice M, Manganelli V, Matarrese P, Tinari A, Misasi R, Malorni W, Garofalo T. Cardiolipin-enriched raft-like microdomains are essential activating platforms for apoptotic signals on mitochondria. FEBS Lett. 2009;583:2447–2450. doi: 10.1016/j.febslet.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Tyurin VA, Tyurina YY, Ritov VB, Lysytsya A, Amoscato AA, Kochanek PM, Hamilton R, Dekosky ST, Greenberger JS, Bayir H, Kagan VE. Oxidative lipidomics of apoptosis: quantitative assessment of phospholipid hydroperoxides in cells and tissues. Methods Mol Biol. 2010;610:353–374. doi: 10.1007/978-1-60327-029-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sparagna GC, Chicco AJ, Murphy RC, Bristow MR, Johnson CA, Rees ML, Maxey ML, McCune SA, Moore RL. Loss of cardiac tetralinoleoyl cardiolipin in human and experimental heart failure. J Lipid Res. 2007;48:1559–1570. doi: 10.1194/jlr.M600551-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Chang S-C, Heacock PN, Mileykovskaya E, Voelker DR, Dowhan W. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J Biol Chem. 1998;273:14933–14941. doi: 10.1074/jbc.273.24.14933. [DOI] [PubMed] [Google Scholar]
- 31.Tuller G, Hrastnik C, Achleitner G, Schiefthaler U, Klein F, Daum G. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 1998;421:15–18. doi: 10.1016/s0014-5793(97)01525-1. [DOI] [PubMed] [Google Scholar]
- 32.Jiang F, Rizavi HS, Greenberg ML. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol Microbiol. 1997;26:481–491. doi: 10.1046/j.1365-2958.1997.5841950.x. [DOI] [PubMed] [Google Scholar]
- 33.Sistrom WR. A requirement for sodium in the growth of Rhodopseudomonas spheroides. J Gen Microbiol. 1960;22:778–785. doi: 10.1099/00221287-22-3-778. [DOI] [PubMed] [Google Scholar]
- 34.Sistrom WR. The kinetics of the synthesis of photopigments in Rhodopseudomonas spheroides. J Gen Microbiol. 1962;28:607–616. doi: 10.1099/00221287-28-4-607. [DOI] [PubMed] [Google Scholar]
- 35.Benning C, Somerville CR. Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1992;174:2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhen Y, Qian J, Follmann K, Hayward T, Nilsson T, Dahn M, Hilmi Y, Hamer AG, Hosler JP, Ferguson-Miller S. Overexpression and purification of cytochrome c oxidase from Rhodobacter sphaeroides. Protein Expression Purif. 1998;13:326–336. doi: 10.1006/prep.1998.0903. [DOI] [PubMed] [Google Scholar]
- 37.van Niel CB. The Culture, General Physiology, Morphology, and Classification of the Non-Sulfur Purple and Brown Bacteria. Bacteriol Rev. 1944;8:1–118. doi: 10.1128/br.8.1.1-118.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeliseev AA, Kaplan S. TspO of rhodobacter sphaeroides. A structural and functional model for the mammalian peripheral benzodiazepine receptor. J Biol Chem. 2000;275:5657–5667. doi: 10.1074/jbc.275.8.5657. [DOI] [PubMed] [Google Scholar]
- 39.Benning C, Beatty JT, Prince RC, Somerville CR. The sulfolipid sulfoquinovosyldiacylglycerol is not required for photosynthetic electron transport in Rhodobacter sphaeroides but enhances growth under phosphate limitation. Proc Natl Acad Sci U S A. 1993;90:1561–1565. doi: 10.1073/pnas.90.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon R, Priefer U, Puehler A. Vector plasmids for in vivo and in vitro manipulations of gram-negative bacteria. Mol Genet Bact-Plant Interact, [Symp] 1983:98–106. [Google Scholar]
- 41.Weissenmayer B, Geiger O, Benning C. Disruption of a gene essential for sulfoquinovosyldiacylglycerol biosynthesis in Sinorhizobium meliloti has no detectable effect on root nodule symbiosis. Mol Plant-Microbe Interact. 2000;13:666–672. doi: 10.1094/MPMI.2000.13.6.666. [DOI] [PubMed] [Google Scholar]
- 42.Arondel V, Benning C, Somerville CR. Isolation and functional expression in Escherichia coli of a gene encoding phosphatidylethanolamine methyltransferase (EC 2.1.1.17) from Rhodobacter sphaeroides. J Biol Chem. 1993;268:16002–16008. [PubMed] [Google Scholar]
- 43.Tamot B. MS thesis. Department of Biochemistry and Molecular Biology, Michigan State University; 2006. Construction and characterization of a cardiolipin-deficient mutant in Rhodobacter sphaeroides. [Google Scholar]
- 44.Hosler JP, Fetter J, Tecklenburg MMJ, Espe M, Lerma C, Ferguson-Miller S. Cytochrome aa3 of Rhodobacter sphaeroides as a model for mitochondrial cytochrome c-oxidase. Purification, kinetics, proton pumping, and spectral analysis. J Biol Chem. 1992;267:24264–24272. [PubMed] [Google Scholar]
- 45.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 46.Qin L, Mills DA, Buhrow L, Hiser C, Ferguson-Miller S. A conserved steroid binding site in cytochrome c oxidase. Biochemistry. 2008;47:9931–9933. doi: 10.1021/bi8013483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibuya I, Miyazaki C, Ohta A. Alteration of phospholipid composition by combined defects in phosphatidylserine and cardiolipin synthases and physiological consequences in Escherichia coli. J Bacteriol. 1985;161:1086–1092. doi: 10.1128/jb.161.3.1086-1092.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishijima S, Asami Y, Uetake N, Yamagoe S, Ohta A, Shibuya I. Disruption of the Escherichia coli cls gene responsible for cardiolipin synthesis. J Bacteriol. 1988;170:775–780. doi: 10.1128/jb.170.2.775-780.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X. PhD thesis. Department of Chemistry and Department of Biochemistry & Molecular Biology, Michigan State University; 2009. Investigating the functional roles of lipids in membrane protein cytochrome c oxidase from Rhodobacter sphaeroides using mass spectrometry and lipid profile modification. [Google Scholar]
- 51.Klug RM, Benning C. Two enzymes of diacylglyceryl-O-4′-(N,N,N,-trimethyl)-homoserine biosynthesis are encoded by btaA and btaB in the purple bacterium Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 2001;98:5910–5915. doi: 10.1073/pnas.101037998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Busch KL. Chemical noise in mass spectrometry. Part III - more mass spectrometry/mass spectrometry. Spectroscopy (Duluth, MN, United States) 2003;18:52–55. [Google Scholar]
- 53.Zhang X, Ferguson-Miller SM, Reid GE. Characterization of ornithine and glutamine lipids extracted from cell membranes of Rhodobacter sphaeroides. J Am Soc Mass Spectrom. 2009;20:198–212. doi: 10.1016/j.jasms.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu F-F, Turk J, Rhoades Elizabeth R, Russell David G, Shi Y, Groisman Eduardo A. Structural characterization of cardiolipin by tandem quadrupole and multiple-stage quadrupole ion-trap mass spectrometry with electrospray ionization. Journal of the American Society for Mass Spectrometry. 2005;16:491–504. doi: 10.1016/j.jasms.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 55.Sparagna GC, Johnson CA, McCune SA, Moore RL, Murphy RC. Quantitation of cardiolipin molecular species in spontaneously hypertensive heart failure rats using electrospray ionization mass spectrometry. J Lipid Res. 2005;46:1196–1204. doi: 10.1194/jlr.M500031-JLR200. [DOI] [PubMed] [Google Scholar]
- 56.Hirschberg CB, Kennedy EP. Mechanism of the enzymatic synthesis of cardiolipin in Escherichia coli. Proc Natl Acad Sci U S A. 1972;69:648–651. doi: 10.1073/pnas.69.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han X, Yang J, Yang K, Zhao Z, Abendschein DR, Gross RW. Alterations in myocardial cardiolipin content and composition occur at the very earliest stages of diabetes: a shotgun lipidomics study. Biochemistry. 2007;46:6417–6428. doi: 10.1021/bi7004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol. 2007;292:C33–C44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- 59.Yamaoka-Koseki S, Urade R, Kito M. Cardiolipins from rats fed different dietary lipids affect bovine heart cytochrome c oxidase activity. J Nutr. 1991;121:956–958. doi: 10.1093/jn/121.7.956. [DOI] [PubMed] [Google Scholar]
- 60.Lopez-Lara IM, Sohlenkamp C, Geiger O. Membrane lipids in plant-associated bacteria: Their biosyntheses and possible functions. Mol Plant-Microbe Interact. 2003;16:567–579. doi: 10.1094/MPMI.2003.16.7.567. [DOI] [PubMed] [Google Scholar]
- 61.Schlame M, Greenberg ML. Cardiolipin synthase from yeast. Biochim Biophys Acta. 1997;1348:201–206. doi: 10.1016/s0005-2760(97)00117-3. [DOI] [PubMed] [Google Scholar]
- 62.Gaspar ML, Aregullin MA, Jesch SA, Nunez LR, Villa-Garcia M, Henry SA. The emergence of yeast lipidomics. Biochim Biophys Acta, Mol Cell Biol Lipids. 2007;1771:241–254. doi: 10.1016/j.bbalip.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 63.Zhong Q, Gohil VM, Ma L, Greenberg ML. Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56. J Biol Chem. 2004;279:32294–32300. doi: 10.1074/jbc.M403275200. [DOI] [PubMed] [Google Scholar]
- 64.Russell NJ, Harwood JL. Changes in the acyl lipid composition of photosynthetic bacteria grown under photosynthetic and non-photosynthetic conditions. Biochem J. 1979;181:339–345. doi: 10.1042/bj1810339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Catucci L, Depalo N, Lattanzio VMT, Agostiano A, Corcelli A. Neosynthesis of cardiolipin in Rhodobacter sphaeroides under osmotic stress. Biochemistry. 2004;43:15066–15072. doi: 10.1021/bi048802k. [DOI] [PubMed] [Google Scholar]
- 66.Robinson NC, Zborowski J, Talbert LH. Cardiolipin-depleted bovine heart cytochrome c oxidase: binding stoichiometry and affinity for cardiolipin derivatives. Biochemistry. 1990;29:8962–8969. doi: 10.1021/bi00490a012. [DOI] [PubMed] [Google Scholar]
- 67.Berry EA, Trumpower BL. Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J Biol Chem. 1985;260:2458–2467. [PubMed] [Google Scholar]
- 68.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 69.Jones MR. Lipids in photosynthetic reaction centres: Structural roles and functional holes. Progress in Lipid Research. 2007;46:56–87. doi: 10.1016/j.plipres.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 70.Aygun-Sunar S, Mandaci S, Koch H-G, Murray IVJ, Goldfine H, Daldal F. Ornithine lipid is required for optimal steady-state amounts of c-type cytochromes in Rhodobacter capsulatus. Mol Microbiol. 2006;61:418–435. doi: 10.1111/j.1365-2958.2006.05253.x. [DOI] [PubMed] [Google Scholar]
- 71.Laible PD, Scott HN, Henry L, Hanson DK. Towards higher-throughput membrane protein production for structural genomics initiatives. J Struct Funct Genomics. 2004;5:167–172. doi: 10.1023/B:JSFG.0000029201.33710.46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1. R. sphaeroides strains and CcO plasmids used in this work.
Supplemental Table S2. X-ray screening results of CcO crystals obtained from CcO expressed in the R. sphaeroides wild type CL(+) and CL(−) mutant at different crystal growth temperatures.