Abstract
The dapdiamides are a family of antibiotics that have been presumed to be cleaved in the target cell to enzyme-inhibitory N-acyl-2,3-diaminopropionate (DAP) warheads containing two alternative electrophilic moieties. Our prior biosynthetic studies revealed that an eneamide warhead is made first and converted to an epoxyamide via a three enzyme branch pathway. Here we provide a rationale for this logic. We report that the RR-epoxyamide warhead is a more efficient covalent inactivator of glucosamine-6-phosphate synthase by an order of magnitude over the eneamide, and this difference correlates with a more than ten-fold difference in antibiotic activity for the corresponding acyl-DAP dipeptides.
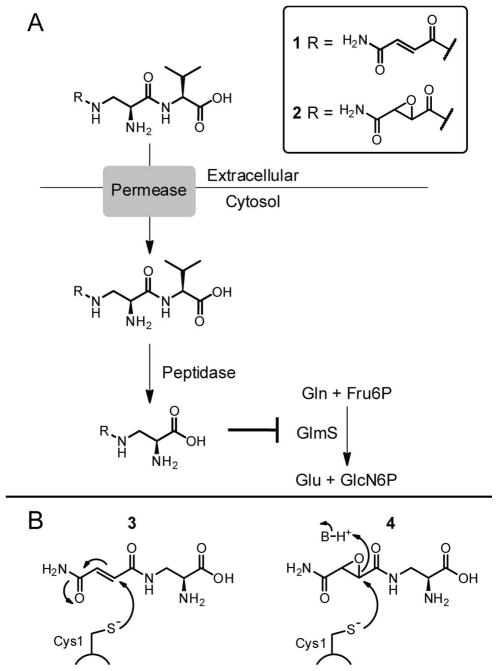
Pantoea agglomerans are epiphytic bacteria which produce a series of peptide-based antimetabolites that suppress the growth of competing microbes (1–4). This species has received attention because it inhibits growth of the economically important plant pathogen Erwinia amylovora, a species that inhabits the same ecological niche and leads to the necrotic plant disease fire blight (5). Among the known P. agglomerans antibiotics are the family of dapdiamides (Scheme S1A), N-acyl-dipeptides where the “dap” refers to the constituent nonproteinogenic L-2,3-diaminopropionyl (DAP) residue (blue in Scheme S1) (6). Antibiotic activity-based screening of a P. agglomerans CU0119 genomic library heterologously expressed in E. coli has recently led to the identification of a biosynthetic gene cluster for the dapdiamides (6). This cluster encodes genes that are necessary and sufficient for production of dapdiamide A (Nβ-fumaramoyl-L-DAP-L-Val) 1 and the corresponding epoxide 2 (Figure 1A) (7, 8).
Figure 1.
Antibiotic activity of the dapdiamides. A) Proposed Trojan horse mechanism of the dapdiamides. B) Role of GlmS glutaminase domain Cys1 in the Gln hydrolysis mechanism. C) Proposed mechanism of GlmS inhibition by dapdiamide N-acyl-DAP warheads.
We have shown previously that at least three different types of ATP-utilizing enzymes participate in this pathway (Scheme S2) (7, 8). DdaG transiently makes fumaroyl-AMP that is captured by the β-NH2 of L-DAP on the way to Nβ-fumaroyl-DAP. After subsequent amidation to Nβ-fumaramoyl-DAP 3 (Figure 1B) by DdaH, DdaF utilizes ATP, this time cleaving it to ADP and Pi and forming an activated Nβ-fumaramoyl-DAP acyl phosphate. This species undergoes nucleophilic attack by the amino group of one of three branched chain aliphatic amino acids, e.g. Val to form 1. We have characterized a third ATP-dependent enzyme, DdaD, which is a nonribosomal peptide synthetase module composed of an adenylation (A) and thiolation (T) domain. DdaD makes Nβ-fumaramoyl-DAP-AMP, and this intermediate is captured by the terminal thiol of the phosphopantetheinyl arm attached to the DdaD T domain. This results in a covalently tethered Nβ-fumaramoyl-DAP-thioester which is the substrate for a nonheme, mononuclear FeII-oxygenase DdaC which epoxidizes the olefin. The tethered epoxysuccinamoyl-DAP-S-DdaD is likely hydrolyzed by the thioesterase DdaE, liberating Nβ-epoxysuccinamoyl-DAP 4 (Figure 1B). This epoxide can be processed by DdaF to yield 2 as an antibiotic endproduct.
Compounds closely related to 1 and 2 are known metabolites from other bacteria (Scheme S1B) (4, 9–11) and members of this N-acyl-DAP-aa class likely serve as pro forms of antibiotics. In analogy to the known Trojan horse mechanism of synthetic Nβ-methoxyfumaroyl-DAP-aa compounds (FMDP-aa) (Scheme S3), (12) these natural products are presumably taken up by neighboring microbes through oligopeptide permeases and then undergo cleavage by intracellular peptidases to remove the C-terminal residue (Figure 1A). In the case of 1 and 2, this cleavage would yield the stretched Gln analogues 3 or 4 respectively. Compound 3 has been previously characterized as an irreversible inactivator of bacterial (13, 14) and candidal (15) glucosamine-6-P (GlcN6P) synthase. GlcN6P synthase (termed GlmS in prokaryotes) converts fructose-6-P (Fru6P) to GlcN6P in an isomerization/amination reaction (Scheme S4)(16). Nascent NH3 for the amination step is produced from in situ hydrolysis of L-Gln in the glutaminase active site. The amide of Gln undergoes nucleophilic attack by the side chain thiolate of Cys1 to generate a hemithioaminal which decomposes to a γ-glutamyl-S-enzyme species and nascent NH3. The glutamyl thioester is hydrolyzed to release Glu and the NH3 diffuses down a 20 Å tunnel to the sugar isomerization active site, where Fru6P is converted to GlcN6P. GlcN6P synthase provides the sole route to this aminohexose, which is converted by subsequent enzymatic steps to UDP-N-acetylglucosamine (UDP-GlcNAc), a key precursor for the biosynthesis of both bacterial and fungal cell walls. Interdiction of GlcN6P synthase activity causes cell death and the glutaminase active site has been a target for both natural product antimicrobials (e.g. the dapdiamides and the Bacillus subtilis natural product bacilysin) as well as synthetic electrophilic variants of glutamine (17, 18).
Prior studies in the Badet lab of E. coli GlmS inhibition by the methyl ester analogue of 3, FMDP, suggest that irreversible inactivation with these α,β-unsaturated carbonyl compounds proceeds via Cys1 thiol Michael addition into the fumar(am)oyl moiety (Figure 1B) (19). NMR studies of the reaction of either Cys or of a synthetic CGIVGAIAQR decapeptide that corresponds to the N-terminal sequence of GlmS demonstrated that in both cases FMDP undergoes Michael addition β to the ester by the Cys thiol (19). 4 could in principle similarly covalently modify the glutaminase domain Cys1 via an epoxide ring opening reaction.
Given our recent delineation of the dapdiamide biosynthetic pathway and the utilization of three enzymes dedicated to conversion of 3 to 4, (7, 8) we sought to understand what utility the conversion of one form of electrophilic inhibitor (the fumaramoyl eneamide for conjugate addition) to the second (the epoxide) might offer to the producing microbe. We were also interested in the biological activity of the Nα-fumaramoyl-DAP warhead that is predicted to be formed from peptidease cleavage of dapdiamide D (Nα-fumaramoyl-DAP-Val, Scheme S1A); this moiety is not found in any other known natural products. Finally, we wanted to test the hypothesis that 1 and 2 are Trojan horse antibiotics which require the C-terminal Val for uptake, and then undergo protease cleavage to liberate enzyme-inhibitory acyl-DAP warhead 3 or 4. To these ends we have examined the catalytic efficiency for inactivation of GlcN6P synthase as well as the antibiotic potency of several acyl-DAP compounds.
The absolute stereochemistry of the oxirane carbons in the N-epoxysuccinamoyl-DAP-Val compounds isolated from P. agglomerans has not been determined, (4, 6) but R,R-epoxide stereochemistry has been found for the related natural product Sch37137 (Scheme S1B) (20). We therefore evaluated both the RR- and SS-4 diastereomers for inhibition of purified GlmS. We chose E. coli GlmS as a model enzyme as it has been well-characterized previously, including a kinact/Kirr (inactivation efficiency) value for 3 (14, 19, 21). We used an established spectro-photometric assay for glutaminase activity to determine a kinact/Kirr for each inhibitor (Figure S1). We found that RR-4 is a potent time-dependent inactivator of GlmS with a kinact/Kirr of 290 M−1s−1 (Table 1). This is approximately seven-fold more potent than 3 which in our hands exhibited a kinact/Kirr of 39 M−1s−1 (Table 1 and Figure S2A).1 The SS-4 diastereomer is a much less efficient GlmS inactivator than either 3 or RR-4; the kinact/Kirr of 5.18 M−1s−1 is approximately 56 fold lower than that for the RR compound. In contrast, Nα-fumaramoyl-DAP exhibited no time-dependent inhibition of GlmS at concentrations up to 600 μM (Figure S2B). We also did not observe any time-dependent GlmS inhibition with 600 μM of the acyl-DAP-Val compounds 1 or RR-2, suggesting that cleavage of the C-terminal Val is required to generate the active enzyme-inhibitory warhead (Figure S2C).
Table 1.
N-acyl-DAP GlmS inactivation efficiencies.
| Inhibitor | kinact/Kirr (M−1s−1) |
|---|---|
| 3 | 39 ± 6 |
| RR-4 | 290 ± 110 |
| SS-4 | 5.18 ± 0.03 |
The order of magnitude increase in inactivation efficiency between 3 and RR-4 offers one rationale for why the producing microbe elaborates the fumaramoyl acyl group to the R,R-epoxide in 4 and 2. Perhaps not coincidentally, DdaF has a kinetic preference for the R,R-diastereomer of 4 over the S,S for ligation to Val, and it is likely RR-2 that is exported by the producer organism (8). In turn GlmS is inactivated much more efficiently by that R,R-diastereomer of 4 than the S,S; this may reflect an evolutionary matching of antibiotic production and target susceptibility.
To determine that the catalytic Cys1 is the nucleophilic target of both 3 and 4, GlmS was treated with each inhibitor and subjected to trypsin digestion and MS. It was possible to isolate the GlmS N-terminal tryptic decapeptide. This peptide underwent the expected mass shift for modification with 3 (Δm =201.1 Da), RR-4 (Δm = 217.1 Da), and SS-4 (Δm = 217.1 Da) (Figures S3–S6, Table S2). Modification of the GlmS N-terminal decapeptide was also seen by MS with Nα-fumaramoyl-DAP (Δm =201.1 Da) (Figure S7, Table S2). The site of modification with these inhibitors was further narrowed by employing tandem MS techniques. MS/MS of the modified decapeptides revealed mass shifts of the b-ion series but not the y-ion series, supporting the hypothesis that the inhibitors were bound at the N-terminus of the peptide (Figure S8–S11). Most conclusively, MS3 fragmentation of the modified CGI b3 ion resulted in the formation of an ion with a mass consistent with the N-terminal CG b2 ion in thioether link-age to the inhibitors (Figure S12–S15). These results validate that, as anticipated, the dapdiamide N-acyl-DAP warheads capture the active site Cys thiolate nucleophile in the glutaminase domain of GlmS.
Next, we investigated whether the improved enzyme inactivation efficiency of the epoxide-containing acyl-DAP warhead RR-4 would translate to greater antibiotic potency of the epoxyamide dipeptide RR-2 as compared with eneamide 1. Minimum inhibitory concentrations (MICs) for 1 and RR-2 were determined against: an ecologically relevant target, E. amylovora 273; wild type E. coli K12 MG1655; and E. coli NR698, which carries a mutation in the increased membrane permeability (imp) gene that leads to increased outer membrane permeability (Tables 2 and S4)(22). RR-2 was more potent than 1 by 16 fold against E. amylovora, suggesting that the greater in vitro inactivation efficiency against GlmS correlates with in vivo potency against this microbe. In contrast, minimal inhibition of E. coli K12 growth was observed at concentrations of both 1 and RR-2 up to 500 μM (Table S4). In light of our demonstration that 3 and RR-4 inhibit purified E. coli GlmS, we suspected that this absence of antibiotic activity resulted from lack of compound penetration into the target cell cytosol. MICs determined for 1 and RR-2 against E. coli NR698 support this hypothesis; they are similar to the MICs against E. amylovora 273, and here again the epoxyamide is more potent than the eneamide by an order of magnitude. The antibiotic activity of 1 and RR-2 was abrogated in the presence of 167 mM N-acetyl-glucosamine, consistent with the hypothesis that these antibiotics act via blockade of GlcN6P production by GlmS (Table S4). As anticipated based on the hypothesis that the C-terminal Val is required for uptake of acyl-DAP-Val antibiotics by target cell peptide permeases, no antibiotic activity was observed for RR-4 at concentrations up to 417 μM against either E. amylovora or E. coli NR698 (Table S4).
Table 2.
MIC values for 1 and RR-2 against E. amylovora 273 and E. coli NR698.
| Antibiotic | E. amylovora 273 | E. coli NR698 |
|---|---|---|
| 1 | 188 μM | 188 μM |
| RR-2 | 12 μM | 16 μM |
In sum, the dapdiamide antibiotic biosynthetic pathway generates a pair of Nβ-acyl-DAP-Val compounds, exportable pro-toxins containing two distinct inactivating electrophilic moieties. The penultimate incarnation of the acyl group is the fumaramoyl half amide which is known to irreversibly inactivate GlmS (14). Such eneamide functional groups are also found in natural product syringolins and glidobactins which capture the nucleophilic Thr1 of proteasomes (23). Elaboration of the fumaramoyl double bond to the R,R-epoxide by DdaCDE produces the ultimate biosynthetic version of the electrophilic acyl group (8). We have wondered what advantage the epoxide might offer over the eneamide functionality for inactivation of its target enzyme. One possibility could be selectivity/increased stability in biological microenvironments, in that the epoxide opening would typically require protonation of the epoxide oxygen to lower the barrier for C-O cleavage on nucleophilic attack. Consistent with enzymatic assistance for capture of the epoxide is our observation that the SS-4 is approximately 56 fold less efficient a GlmS inhibitor than the R,R-diastereomer, presumably reflecting stereospecific orientation of the epoxide moiety toward a chiral general acid side chain in the active site. Many epoxy metabolites are known in natural products, including the anticapsin moiety of the dipeptide antibiotic bacilysin which targets the same GlmS active site nucleophile (17). It remains to be seen if studies with purified GlmS enzymes from pathogens such as Candida strains and phytopathogenic bacteria such as E. amylovora will show comparable ratios of improved inactivation efficiencies for RR-4 vs. 3, giving insights into the chemical logic used by microbes in design, production, and optimization of mechanism-based enzyme inhibitors with antibiotic activity.
Supplementary Material
Acknowledgments
This work was supported in part by NIH GM 20011 (C.T.W.), GM 07753 (M.A.H.), GM 067725 (N.L.K), and CNRS (B.B.).
Footnotes
This is two-fold lower than the kinact/Kirr for 3 previously reported by Badet and coworkers (13). We tested 3 that was used in that study side-by-side against newly synthesized compound and found a comparable kinact/Kirr of 34 M−1s−1, so we used the more recently determined value as basis for comparison.
SUPPORTING INFORMATION AVAILABLE Supplemental materials and methods, Schemes S1–5, Tables S1–S3, and Figures S1–S15. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Brady SF, Wright SA, Lee JC, Suton AE, Zumoff CH, Wodzinski RS, Beer SV, Clardy J. J Am Chem Soc. 1999:11912–11913. [Google Scholar]
- 2.Jin M, Liu L, Wright SAI, Beer SV, Clardy J. Angew Chem, Int Ed. 2003;42:2898–2901. doi: 10.1002/anie.200351053. [DOI] [PubMed] [Google Scholar]
- 3.Jin M, Fischbach MA, Clardy J. J Am Chem Soc. 2006;128:10660–10661. doi: 10.1021/ja063194c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sammer UF, Volksch B, Mollmann U, Schmidtke M, Spiteller P, Spiteller M, Spiteller D. Appl Environ Microbiol. 2009;75:7710–7717. doi: 10.1128/AEM.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanneste JL. Fire blight: the disease and its causative agent, Erwinia amylovora. xi. CABI Pub; Wallingford, Oxon, UK; New York, NY, USA: 2000. [Google Scholar]
- 6.Dawlaty J, Zhang X, Fischbach MA, Clardy J. J Nat Prod. 2010;73:441–446. doi: 10.1021/np900685z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollenhorst MA, Clardy J, Walsh CT. Biochemistry. 2009;48:10467–10472. doi: 10.1021/bi9013165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollenhorst MA, Bumpus SB, Matthews ML, Bollinger JM, Jr, Kelleher NL, Walsh CT. J Am Chem Soc. 2010:441–446. doi: 10.1021/ja1072367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molloy BB, Lively DH, Gale RM, Forman M, Boeck LD, Higgens CE, Kastner RE, Huckstep LL, Neuss N. J Antibiot. 1972;25:137–140. doi: 10.7164/antibiotics.25.137. [DOI] [PubMed] [Google Scholar]
- 10.Cooper R, Horan AC, Gentile F, Gullo V, Loebenberg D, Marquez J, Patel M, Puar MS, Truumees I. J Antibiot. 1988;41:13–19. doi: 10.7164/antibiotics.41.13. [DOI] [PubMed] [Google Scholar]
- 11.Shoji J, Hinoo H, Sakazaki R, Kato T, Hattori T, Matsumoto K, Tawara K, Kikuchi J, Terui Y. J Antibiot. 1989;42:869–874. doi: 10.7164/antibiotics.42.869. [DOI] [PubMed] [Google Scholar]
- 12.Milewski S, Andruszkiewicz R, Kasprzak L, Mazerski J, Mignini F, Borowski E. Antimicrob Agents Chemother. 1991;35:36–43. doi: 10.1128/aac.35.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chmara H, Andruszkiewicz R, Borowski E. Biochim Biophys Acta. 1985;870:357–366. doi: 10.1016/0167-4838(86)90240-2. [DOI] [PubMed] [Google Scholar]
- 14.Badet B, Vermoote P, Le Goffic F. Biochemistry. 1988;27:2282–2287. doi: 10.1021/bi00407a006. [DOI] [PubMed] [Google Scholar]
- 15.Milewski S, Chmara H, Andruszkiewicz R, Borowski E. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1985;828:247–254. doi: 10.1016/0167-4838(85)90304-8. [DOI] [PubMed] [Google Scholar]
- 16.Mouilleron S, Badet-Denisot M, Badet B, Golinelli-Pimpaneau B. Arch Biochem Biophys. 2010;505:1–12. doi: 10.1016/j.abb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Chmara H. Microbiology. 1985;131:265–271. doi: 10.1099/00221287-131-2-265. [DOI] [PubMed] [Google Scholar]
- 18.Milewski S. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 2002;1597:173–192. doi: 10.1016/s0167-4838(02)00318-7. [DOI] [PubMed] [Google Scholar]
- 19.Kucharczyk N, Denisot MA, Le Goffic F, Badet B. Biochemistry. 1990;29:3668–3676. doi: 10.1021/bi00467a012. [DOI] [PubMed] [Google Scholar]
- 20.Rane DF, Girijavallabhan VM, Ganguly AK, Pike RE, Saksena AK, McPhail AT. Tetrahedron Lett. 1993;34:3201–3204. [Google Scholar]
- 21.Badet B, Vermoote P, Haumont PY, Lederer F, Le Goffic F. Biochemistry. 1987;26:1940–1948. doi: 10.1021/bi00381a023. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Groll M, Schellenberg B, Bachmann AS, Archer CR, Huber R, Powell TK, Lindow S, Kaiser M, Dudler R. Nature. 2008;452:755–758. doi: 10.1038/nature06782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.