Abstract
Arachidonic acid, a dietary cis-polyunsaturated fatty acid, stimulates adhesion and migration of human cancer cells on the extracellular matrix by activation of intracellular signaling pathways. Polyubiquitin chains bearing linkages through different lysine residues convey distinct structural and functional information that is important for signal transduction. We investigated whether ubiquitination was required for arachidonic acid-induced cellular adhesion and migration of MDA-MB-435 cells on collagen type IV. An E1 (ubiquitin-activating enzyme) inhibitor, PYR-431, completely abrogated arachidonic acid-stimulated adhesion. Additionally, expression of a lysine null mutant ubiquitin prevented activation of cellular adhesion. Cells expressing ubiquitin in which lysine 63 (K63) was mutated to arginine (K63R) were unable to adhere to collagen upon exposure to arachidonic acid. When K63 was the only lysine present, the cells retained the ability to adhere, indicating that K63-linked ubiquitin is both necessary and sufficient. Moreover, K63-linked ubiquitin was required for the induction of cell migration by arachidonic acid. The ubiquitin mutants and PYR-431 did not prevent arachidonic acid-induced phosphorylation of TGF-β activated kinase-1 (TAK1) and p38 MAPK, suggesting K63-linked ubiquitination occurs downstream of MAPK. These novel findings are the first to demonstrate a role for K63-linked ubiquitination in promoting cell adhesion and migration.
Keywords: adhesion, ubiquitin, metastasis, signal transduction, arachidonic acid
Introduction
Cellular adhesion and migration are important steps in many biological processes. One such process is cancer metastasis, in which altered cell–cell adhesion and adhesion to extracellular matrix components, such as collagen type IV, allow tumor cells to migrate away from the primary tumor and invade a secondary site (Mack and Marshall 2010). These adhesive events require coordinated signal transduction pathways and can be influenced by factors in the microenvironment of both the primary tumor and the metastatic site (Mack and Marshall 2010; Yilmaz and Christofori 2010).
The n-6 cis-polyunsaturated fatty acid arachidonic acid and its precursor linoleic acid are common dietary fatty acids that, furthermore, may be components of the tumor microenvironment. Arachidonic acid is also incorporated into the cellular membrane in an esterified form and is released by the action of phospholipases, particularly during inflammatory responses (Khanapure et al. 2007). Further metabolism of arachidonic acid by cyclooxygenases and lipoxygenases is a highly regulated process that generates eicosanoids, small bioactive lipids that are involved in a wide range of biological processes and signaling pathways (Khanapure et al. 2007). In animal models, there is substantial evidence linking high fat intake to an increase in tumor cell metastasis (Erickson and Hubbard 1990; Rose et al. 1994; Rose and Connolly 1997). Our laboratory and others have shown that fatty acids and their metabolites can alter adhesion of cancer cells to the extracellular matrix (Jiang et al. 1995; Johanning and Lin 1995; Palmantier et al. 1996). In our previous work, we utilized a highly metastatic human cancer cell line, MDA-MB-435, and demonstrated that arachidonic acid induced cellular adhesion to collagen type IV in an integrin-dependent manner requiring the activation of multiple signal transduction proteins, including a p38 MAPK/RhoA pathway (Palmantier et al. 1996, 2001; Paine et al. 2000; Nony et al. 2005; Garcia et al. 2009).
Ubiquitination is an important cellular process involved in both protein degradation and signal transduction. The ubiquitin monomer is composed of 76 amino acids, 7 of which are lysine residues, and is universally expressed and highly conserved in eukaryotes (Pickart and Eddins 2004). For example, the yeast and human ubiquitin share 96% amino acid sequence identity. The process of ubiquitin conjugation is an ATP-dependent enzymatic cascade that is also highly conserved (Pickart and Eddins 2004). The C-terminal glycine residue of ubiquitin is added to a lysine residue of the target substrate protein or another ubiquitin molecule by the sequential action of 3 enzymes: (i) an activating enzyme (E1), (ii) a conjugating enzyme (E2) that transiently carries the activated ubiquitin molecule, and (iii) a ligase (E3) that transfers the activated ubiquitin from the E2 to the substrate (Hershko et al. 1983; Herrmann et al. 2007). There is a clear hierarchical organization to the ubiquitin enzymatic cascade, as there are only two E1 enzymes known in humans that initiate the reaction (Zacksenhaus and Sheinin 1990; McGrath et al. 1991; Jin et al. 2007). A significant but limited number of E2s have been identified and each E2 serves several E3s, with hundreds of E3 genes existing in the human genome (Hershko and Ciechanover 1998).
Current evidence indicates that ubiquitin chains linked through lysine residue 48 (K48) of ubiquitin target substrates to a multi-subunit proteasome for degradation. A recently discovered and less understood pathway involves the addition of lysine 63 (K63)-linked chains that perform non-proteolytic functions in at least 4 pathways: DNA damage repair, cellular signaling, intercellular trafficking, and ribosomal biogenesis (Welchman et al. 2005). Recent studies have shown that polyubiquitination through K63-linked chains plays an important role in the activation of signal transduction pathways (Wang et al. 2001; Shi and Kehrl 2003; Yamamoto et al. 2006). K63-linked polyubiquitin chains act as scaffolds to assemble protein kinase complexes and mediate their activation by binding to proteins containing ubiquitin binding domains (Kanayama et al. 2004; Kishida et al. 2005). For example, signaling through the IL-1 receptor results in K63 ubiquitination of TNF receptor associated factor 6 (TRAF6), which is important for initiating activation of MAPKs (Kishida et al. 2005).
We have previously shown that arachidonic acid induces cellular adhesion that is dependent on rapid activation of a MAPK cascade (Paine et al. 2000). This activation of p38 MAPK was dependent on phosphorylation of a MAP3K, TGF-β activated kinase-1 (TAK1) (Nony et al. 2005). It is still unclear how arachidonic acid activates the MAPK pathway leading to cellular adhesion. Herein, we investigated the role of ubiquitination in arachidonic acid-induced cellular adhesion and migration on collagen type IV and induction of MAPK signaling.
Materials and methods
Materials
Arachidonic acid was obtained from Cayman Chemical Company (Ann Arbor, Michigan, USA). Collagen type IV and poly-D-lysine were from BD Biosciences (San Jose, California, USA). Bovine serum albumin (BSA) was obtained from Calbiochem (San Diego, California, USA). FuGENE 6 was from Roche Applied Science (Indianapolis, Indiana, USA). E1 inhibitor (PYR-431) was from BioGenova (Rockville, Maryland, USA). Phospho-TAK1 (Thr187) and phospho-p38 (Thr180/Tyr182) antibodies were from Cell Signaling Technology (Beverly, Massachusetts, USA). A rabbit polyclonal antibody to TAK1, a mouse monoclonal antibody to p38, and a mouse monoclonal antibody to hemagglutinin (HA) tag were from Upstate Cell Signaling Solutions (Lake Placid, New York, USA). The mouse monoclonal GAPDH antibody was from Millipore (Temecula, California, USA). Yeast ubiquitin constructs were a kind gift from Dr. Zhijan Chen at the University of Texas Southwestern Medical Center and were described previously (Wang et al. 2001). Plasmid DNAs were isolated using a Plasmid Maxi Kit (QIAGEN, Valencia, California, USA).
Cell culture and preparation
The highly metastatic human breast cancer cell line MDA-MB-435 was obtained from Dr. Janet Price (M.D. Anderson Cancer Center). Cells were maintained in Eagle’s minimal essential medium (Invitrogen, Carlsbad, California, USA) supplemented with 5% fetal bovine serum (FBS) (Hyclone, Logan, Utah, USA), 1 mmol/L sodium pyruvate, 2 mmol/L L-glutamate, and 2× minimal essential medium vitamin solution (Invitrogen). Subconfluent cells were harvested by a brief incubation with Versene (Invitrogen), washed twice with serum-free medium, and resuspended in serum-free medium at 3 × 105 cells/mL for adhesion assays or 1 × 106 cells/mL for immunoblotting. Before treatments, cells were allowed to equilibrate in serum-free medium for 30 min at 37 °C under 5% CO2. Arachidonic acid (30 μmol/L final concentration; stock diluted in a 328 mmol/L KOH, 0.9% NaCl solution) was added to the cells and incubated for 2 min to 2 h depending on the experiment. Addition of the arachidonic acid solution did not change the pH of the culture medium. When inhibitors were used, they were added to cells in suspension and the cells were incubated for 30 min prior to the addition of arachidonic acid.
Cellular adhesion assays
Adhesion assays were carried out as previously described (Garcia et al. 2009). Briefly, 96-well tissue culture plates were pre-coated with collagen type IV (6.4 μg/mL), poly-D-lysine (32 μg/mL), or BSA (20 mg/mL). The cells were prepared as described and 100 μL of the cell suspension was added to each well and allowed to adhere for 45 min. Adherent cells were fixed with 6% (v/v) glutaraldehyde, stained with 0.05% (w/v) crystal vio let, and solubilized with 1% (w/v) sodium dodecyl sulphate (SDS).
Immunoblotting
After treatment, the cells were washed with PBS and lysed in 50 mmol/L Tris–HCl, pH 7.4, 1% (v/v) Nonidet P-40, 0.25% (w/v) deoxycholate, 150 μmol/L NaCl, 1 μmol/L EDTA, 1 mmol/L phenylmethylsulfonyl fluoride, 1 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin, 1 mmol/L Na3VO4, and 1 mmol/L NaF. Lysates were centrifuged at 16 000g for 10 min at 4 °C and protein concentration was determined using a bicinchoninic acid protein assay kit (Thermo Scientific, Waltham, Massachusetts, USA). The samples were resolved by SDS–PAGE on 4%–12% bis-tris gradient gels and transferred to Invitrolon PVDF membranes (Invitrogen). Membranes were blocked in TBS-T (20 mmol/L Tris, 137 mmol/L NaCl, pH 7.4, plus 0.1% (v/v) Tween 20) with 5% (w/v) non-fat milk, incubated with primary antibody for 1 h to overnight, and incubated with horseradish peroxidase-conjugated secondary antibody for 1 h. Immunoreactivity was detected using ECL Plus chemiluminescent substrate (GE Healthcare, Buckinghamshire, UK). Blots were stripped by incubation in Western Blot Stripping buffer (Thermo Scientific) for 10 min and subsequently rinsed with TBS-T, blocked, and re-blotted as necessary.
Transfection of tumor cells
MDA-MB-435 cells were transfected using FuGENE 6 transfection reagent, as per the manufacturer’s protocol (3:1 ratio of FuGENE (μL): DNA (μg)), with HA-tagged ubiquitin DNA constructs (see Fig. 2A). A transfection efficiency of 70% was achieved based on immunofluorescence staining for the HA tag. Twenty-four hours after transfection, ubiquitin protein expression was assayed by immunoblot using an anti-HA antibody. Cells were then used for adhesion assays as already described.
Fig. 2.
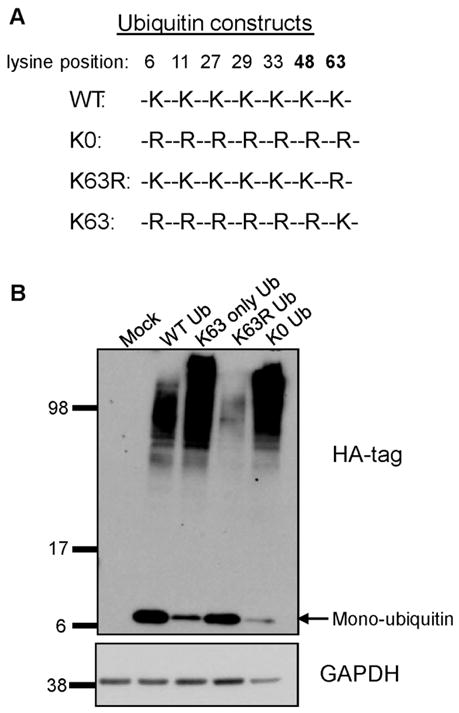
Expression of HA-tagged ubiquitin mutant constructs. (A) Sequences of the ubiquitin constructs depicting the specific mutations of lysine residue(s) to arginine. (B) MDA-MB-435 cells were transfected with 6 μg of DNA and protein expression was visualized after 24 h by immunoblot of whole cell lysates for the HA tag. Mono-ubiquitin appears at a mobility of approximately 8 kDa, whereas high molecular weight bands represent polyubiquitin and ubiquitinated proteins. This membrane was re-probed for GAPDH as a loading control. A similar expression pattern of these ubiquitin mutant constructs has been reported previously (Tan et al. 2008). The labeled molecular weights are in kDa.
Agarose drop migration assay
Cell migration was assayed as previously described (Akiyama et al. 1989; Akiyama 2002) with cells harvested in Versene, washed twice in serum-free medium, incubated in serum-free medium at 37 °C at a concentration of 1.5 × 106 cells/mL for 30 min to recover, collected by centrifugation, and resuspended at a concentration of 1.5 × 107 cells/mL in minimal essential medium containing 10% (v/v) fibronectin-free FBS and 0.2% (w/v) low melting point agarose. Fibronectin-free FBS was prepared using gelatin-Sepharose 4B beads according to the manufacturer’s protocol at a ratio of 4 mL beads: 100 mL FBS (GE Healthcare). Drops (1 μL) of the cell suspension were placed onto substrates prepared with 5 μg/mL collagen type IV and cultured at 37 °C in 5% CO2 for 24 h. Cells were visualized at 0 h and 24 h with a Zeiss Axiovert 35 inverted microscope (Carl Zeiss Microimaging, Inc., Thornwood, New York, USA) using a 2.5× Plan-Neofluar objective lens; images were captured using a Spot Advanced Plus CCD camera (Diagnostic Instruments, Inc., Sterling Heights, Michigan, USA). The surface area of the initial drop and the surface area covered by the drop and the migrated cells after 24 h were determined using MB-Ruler (MB-Software Solutions, Baltimore, Maryland, USA) to analyze the micrographs and Microsoft Excel to calculate the area. Migration was quantified by subtracting the initial area of the drop from the total area covered by the cells and the drop at the end of the experiment.
Statistical analysis
For the adhesion assays, one-way analysis of variance was used to analyze data from 3 to 5 separate experiments. Significant differences were determined with a p value set at < 0.05. Error bars represent one standard deviation above and below the calculated mean. The migration assays were performed 4 times for each condition in each experiment and the experiments were performed 2 times. The error bars represent the standard error. A Student’s t test was performed and a p value < 0.05 was considered statistically significant.
Results
Inhibition of ubiquitin-activating enzyme E1 abrogates arachidonic acid-stimulated adhesion to collagen type IV
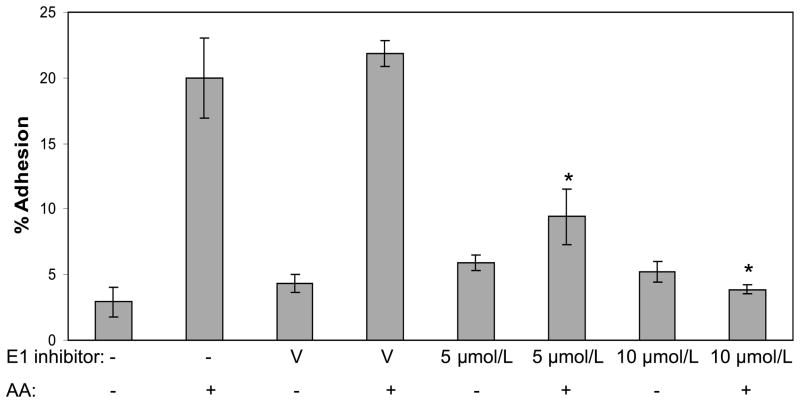
To determine whether ubiquitination played any role in adhesion of MDA-MB-435 cells to collagen type IV, we used a small molecule inhibitor (PYR-431) of the E1 (ubiquitin-activating) enzyme. The E1 inhibitor blocks the loading of ubiquitin into the active site of the E1 enzymes and consequently prevents both degradative (K48) and non-degradative (K63) ubiquitination (Yang et al. 2007). In the absence of inhibitor, arachidonic acid induced a significant increase in adhesion of cells to collagen IV, while the DMSO vehicle had no effect on adhesion (Fig. 1). Pretreatment of the cells with the E1 inhibitor abrogated the arachidonic acid-induced adhesion of the cells to collagen IV at both the 5 μmol/L and 10 μmol/L concentrations of inhibitor. These cells exhibited a basal level of adhesion to collagen IV that was not changed by the E1 inhibitor. Exposure to the E1 inhibitor for the length of the assay did not cause cell death as judged by a trypan blue cell viability assay (data not shown). The ability of the E1 inhibitor to block cell adhesion suggests ubiquitination is required for cell adhesion.
Fig. 1.
Inhibiting ubiquitin-activating enzyme E1 abrogates arachidonic acid-stimulated cell adhesion to collagen IV. MDA-MB-435 cells were pretreated with the E1 inhibitor, PYR-431, for 30 min. The cells were exposed to vehicle (V) or arachidonic acid (AA) and incubated on BSA, poly-D-lysine, or collagen IV-coated wells for 45 min, and then cell adhesion was determined. The results are presented as percent adhesion, with attachment to the poly-D-lysine substrate serving as 100%. An asterisk indicates a significant difference (p < 0.05) when compared with the AA sample (no vehicle or inhibitor). The results are representative of three independent experiments.
Polyubiquitination is critical for arachidonic acid-stimulated adhesion to collagen IV
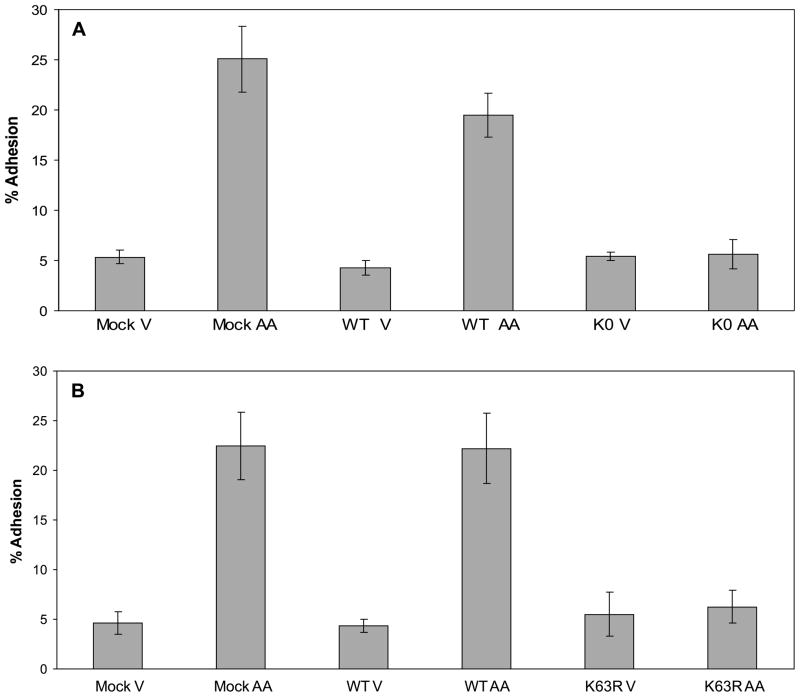
Next, we wanted to confirm the inhibition of adhesion by the E1 inhibitor and begin to characterize the mechanism by which this inhibition occurs by blocking ubiquitination at the level of the ubiquitin protein. This was accomplished by utilizing a series of HA-tagged yeast ubiquitin mutant constructs in which different sets of lysine residues were mutated to arginine residues (see Fig. 2A) (Wang et al. 2001). When the construct in which all of the lysine residues were mutated to arginine residues was expressed, the resulting lysine-null (K0) ubiquitin mutant could attach to a target protein but was not able to form polyubiquitin chains and, therefore, prevented global polyubiquitination. The MDA-MB-435 cells were either mock transfected or transfected with wild-type (WT) ubiquitin or the K0 ubiquitin construct and ubiquitin protein expression was assessed after 24 h by immunoblotting for the HA-tagged ubiquitin (Fig. 2B). Both the WT and K0 ubiquitin were detected as both unincorporated ubiquitin (8 kDa) and polyubiquitin and ubiquitinated proteins (higher molecular weight bands). The transfected cells were then exposed to either vehicle or arachidonic acid and tested in a collagen IV adhesion assay (Fig. 3A). Both the mock transfected and the WT ubiquitin-expressing cells adhered to collagen IV upon exposure to arachidonic acid. However, the cells expressing the K0 ubiquitin showed no induction of adhesion in response to arachidonic acid, confirming that ubiquitination is necessary for cell adhesion.
Fig. 3.
Polyubiquitination through lysine 63 is critical for arachidonic acid-stimulated cell adhesion to collagen IV. (A) Cells were mock transfected (empty vector) or transfected with wild-type (WT) or K0 ubiquitin constructs. Twenty-four hours after transfection, the cells were exposed to vehicle (V) or arachidonic acid (AA) for 45 min in a collagen IV adhesion assay. A representative of three individual adhesion assays is shown. There was no statistically significant difference between the K0 vehicle- and arachidonic acid-treated samples. (B) Cells were mock transfected (empty vector) or transfected with wild-type (WT) or K63R ubiquitin constructs. After 24 h the cells were exposed to vehicle (V) or arachidonic acid (AA) for 45 min in a collagen IV adhesion assay. A representative of three individual adhesion assays is shown. There was no statistically significant difference between the K63R vehicle- and arachidonic acid-treated samples.
Lysine 63 ubiquitination is required for arachidonic acid-stimulated adhesion to collagen IV
Given the prevention of arachidonic acid-induced cell adhesion by inhibiting ubiquitination, we wanted to establish which ubiquitin lysine residue(s) were critical for the fatty acid-stimulated adhesion of the MDA-MB-435 cells. To address this, we used a ubiquitin construct in which the lysine residue at position 63 was mutated to an arginine (K63R) (Fig. 2A). This amino acid change permits ubiquitination through lysine 48 and allows the ubiquitin degradation pathway to function. The K63R ubiquitin was detected both as unincorporated ubiquitin and in ubiquitinated proteins (Fig. 2B). The cells expressing the K63R ubiquitin were unable to adhere to collagen IV in response to arachidonic acid in an adhesion assay (Fig. 3B).
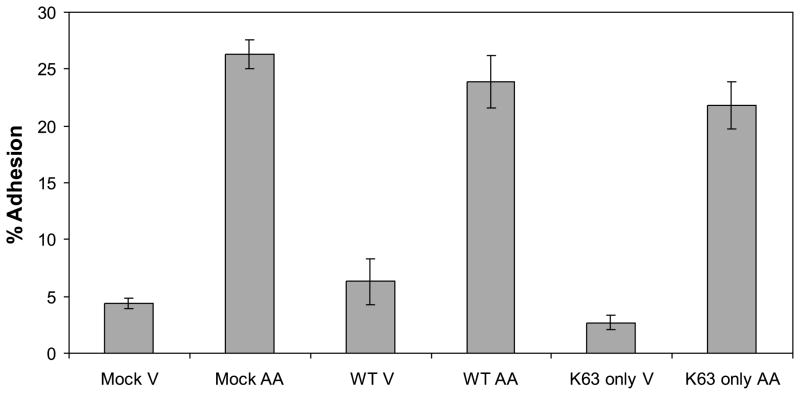
Next we tested whether the ubiquitin K63 residue alone could facilitate arachidonic acid-induced adhesion. We utilized a ubiquitin construct in which all the lysines except K63 were mutated to arginines (K63-only ubiquitin) (Fig. 2A). This mutant ubiquitin is unable to function in the degradation pathway but still functions in the K63-mediated ubiquitin signaling pathway. Expression of the K63-only ubiquitin is shown in the immunoblot for the HA tag in Fig. 2B. Mock transfected cells or cells expressing the WT ubiquitin or the K63-only ubiquitin were exposed to either vehicle or arachidonic acid in a collagen IV adhesion assay (Fig. 4). The K63-only expressing cells displayed the same induction of adhesion as the mock transfected and WT ubiquitin-expressing cells in response to arachidonic acid. None of the ubiquitin constructs had a significant effect on cell viability (data not shown). These findings clearly demonstrate that arachidonic acid induces cell adhesion to collagen IV through a K63-dependent mechanism.
Fig. 4.
Lysine 63 ubiquitination is sufficient to mediate arachidonic acid-stimulated cell adhesion to collagen IV. Cells were mock transfected (empty vector) or transfected for 24 h with wild-type (WT) or K63-only ubiquitin construct. Cells were exposed to vehicle (V) or arachidonic acid (AA) for 45 min in a collagen IV adhesion assay. A representative of three individual adhesion assays is shown. There was no statistical difference between the arachidonic acid-treated samples.
Lysine 63 ubiquitination is required for arachidonic acid-induced cell migration
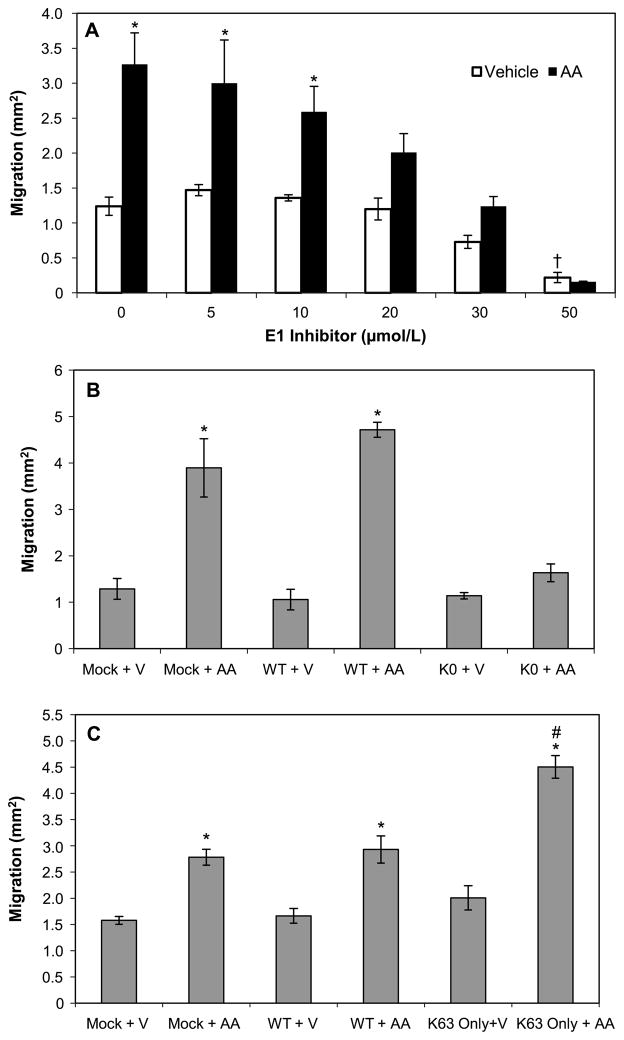
Cell migration is a critical step at several points during tumor cell metastasis (Yilmaz and Christofori 2010). We therefore investigated whether ubiquitination was required for arachidonic acid-induced cell migration out of an agarose droplet containing cells at high density. As depicted in Fig. 5A, at 24 h the cells showed a basal level of migration with vehicle treatment, which significantly increased with arachidonic acid stimulation. The E1 inhibitor blocked cell migration in a concentration-dependent manner, with significant inhibition at 30 μmol/L and complete prevention at 50 μmol/L of E1 inhibitor. At 50 μmol/L, the E1 inhibitor significantly reduced basal migration compared with no inhibitor. However, this decrease in basal migration was not due to loss of cell viability (data not shown). The WT ubiquitin-expressing cells exhibited an increase in cell migration with exposure to arachidonic acid, similar to that of the mock transfected control (Fig. 5B and 5C). However, cells expressing the K0 ubiquitin mutant displayed no statistically significant increase in migration in response to arachidonic acid (Fig. 5B). Most importantly, the cell migration was mediated through K63 ubiquitination. Cells expressing the K63-only ubiquitin not only retained the ability to migrate in response to arachidonic acid but also migrated significantly more than the mock control or WT ubiquitin-expressing cells (Fig. 5C).
Fig. 5.
Lysine 63 ubiquitination is required for arachidonic acid-induced cell migration. Agarose drop migration assays were performed as described in Materials and methods. (A) MDA-MB-435 cells were exposed to vehicle or arachidonic acid (AA) in the presence of E1 inhibitor for 24 h and then the area of cell migration was measured. *, p < 0.05 for AA vs. vehicle; †, p < 0.05 for E1 inhibitor vehicle control vs. no inhibitor vehicle treated control. (B) Cells were either mock transfected with empty vector or transfected with the WT or K0 ubiquitin construct. After 24 h of expression, the cells were harvested and exposed to vehicle (V) or arachidonic acid (AA) in an agarose drop assay for 24 h. *, p < 0.05 for AA vs. vehicle. (C) After 24 h of expression, cells transfected with empty vector (mock) or WT or K63-only ubiquitin were exposed to vehicle (V) or arachidonic acid (AA) for 24 h in a migration assay. *, p < 0.05 for AA vs. vehicle; #, p < 0.05 for K63-only ubiquitin + AA vs. mock + AA sample. Each bar is the mean ± standard error of four separate agarose drop assays.
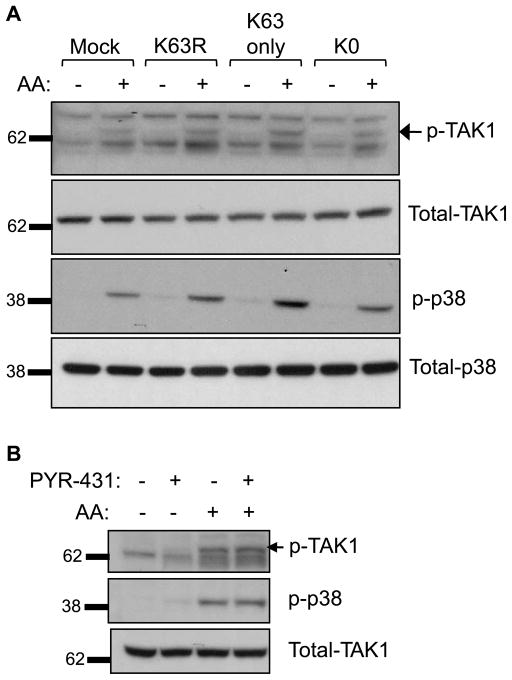
Ubiquitination is not required for phosphorylation of TAK1 and p38 MAPK
From previous work, we know that TAK1 phosphorylation is critical for arachidonic acid-induced p38 phosphorylation, which is required for the induction of cell adhesion (Paine et al. 2000; Nony et al. 2005). We also know that in some signaling pathways, such as in IL-1β receptor signaling, TAK1 phosphorylation is dependent on ubiquitination of upstream activators (Wang et al. 2001). We therefore investigated whether arachidonic acid-induced TAK1 phosphorylation was dependent on ubiquitination. TAK1 phosphorylation was determined by immunoblot using an antibody that specifically recognizes TAK1 phosphorylated at threonine 187. As depicted in Fig. 6A, both TAK1 and p38 MAPK show increased phosphorylation following exposure to arachidonic acid. However, expression of the K0 or K63R ubiquitin mutant did not prevent TAK1 or p38 phosphorylation. (Densitometry analysis of p-p38 and p-TAK1 blots from 3 separate experiments confirmed there was no significant difference between the arachidonic acid-treated samples in Fig. 6A (data not shown).) In addition, the E1 inhibitor, PYR-431, failed to prevent TAK1 and p38 phosphorylation induced by arachidonic acid (Fig. 6B). These data suggest that although ubiquitination appears to play a role in arachidonic acid activation of cell adhesion, it is not required for arachidonic acid activation of at least the upstream kinases in the MAPK signaling pathway.
Fig. 6.
Activation of TAK1 and p38 does not require ubiquitination. (A) MDA-MB-435 cells were mock transfected or transfected with the K63R, K63-only, and K0 ubiquitin mutants for 24 h. The cells were then exposed to arachidonic acid for 2 min and phosphorylation of TAK-1 (p-TAK1) and p38 (p-p38) was assessed by immunoblotting. The immunoblots were re-probed for total TAK1 and total p38 protein as loading controls. The arrow indicates the p-TAK1 band and the other bands represent nonspecific protein reactivity of the antibody. The labeled molecular weights are in kDa. A representative blot of three individual experiments is shown. (B) MDA-MB-435 cells were pretreated with 10 μmol/L of the E1 inhibitor for 30 min, followed by exposure to arachidonic acid (AA) for 2 min. Cell lysates were analyzed by immunoblot for p-TAK1, p-p38, and total TAK1 as a loading control. The experiment shown is representative of three individual experiments.
Discussion
Herein we demonstrate, using a small molecule inhibitor of the E1 ubiquitin-activating enzyme and mutant ubiquitin constructs, that ubiquitination is a necessary component in arachidonic acid-induced cellular adhesion and migration. Through expression of a series of ubiquitin mutants, we found that the dependence of arachidonic acid-induced cellular adhesion and migration on ubiquitination appears to require specifically the lysine 63-mediated pathway. To our knowledge, this is the first report showing that lysine 63-mediated ubiquitination is required for promoting cellular adhesion and migration. These findings could have important potential implications in cancer therapy for targeting steps in tumor cell metastasis.
There are several examples demonstrating the importance of the ubiquitin–proteasome system in cell attachment and migration. Osteoblasts display reduced attachment to fibronectin owing to α5 integrin ubiquitination and degradation in response to fibroblast growth factor receptor signaling (Kaabeche et al. 2005). In another example, talin, a protein important for focal adhesion maintenance, is ubiquitinated by the E3 ligase Smurf1 and degraded, resulting in an increase in focal adhesion turnover and an increase in cell migration (Huang et al. 2009). The mechanism of this increase in cell migration involves K48-linked ubiquitination. Overexpression of a similar E3 ligase, Smurf2, which is involved in the ubiquitin–proteasome system, causes increased invasion and migration of breast cancer cells (Jin et al. 2009).
There is one example of K63-mediated ubiquitination of the focal adhesion protein paxillin. However, this resulted in a decrease in cell motility by inducing paxillin to redistribute to the cytoplasm (Didier et al. 2003). Our findings herein suggest that the K48-mediated degradation pathway is not necessary for the induction of cell adhesion or migration, since arachidonic acid was able to stimulate both adhesion and migration in cells expressing the K63-only ubiquitin. In fact, these cells displayed enhanced migration when compared with the mock transfected control cells. However, use of these ubiquitin mutant constructs cannot completely rule out the involvement of the K48-mediated ubiquitination pathway because of the possible contribution of the endogenous wild-type ubiquitin. Overall, these data strongly suggest that the signal transduction pathway activated by arachidonic acid is regulated by K63-linked ubiquitination.
These results led us to test whether ubiquitination was critical for fatty acid-induced MAPK activation, specifically TAK1 and p38 activity. It is known that TAK1 activation can be regulated by K63-linked ubiquitination at 2 levels. First, the TAK1 kinase complex contains adaptor proteins called TABs, of which TAB2 contains an ubiquitin binding domain. In TGF-β signaling, the E3 ligase TRAF6 autoubiquitinates through K63 linkages and binds to the ubiquitin binding domain of TAB2, resulting in TAK1 activation (Sorrentino et al. 2008). Second, TAK1 itself is a target for K63-linked polyubiquitination. This is the case for TNF-α and IL1-β signaling pathways, where K63-linked TAK1 ubiquitination was shown to be essential for TAK1 kinase activity and activation of downstream mediators (Fan et al. 2010). Given this information, we were surprised to find that inhibition of polyubiquitination by the E1 inhibitor or expression of the K0 and K63R ubiquitin mutants did not prevent TAK1 or p38 MAPK phosphorylation. Therefore, the arachidonic acid-induced signal transduction pathway does not utilize K63-linked ubiquitination to activate TAK1 or its downstream mediator p38. We then investigated other signaling molecules that are known to be important for arachidonic acid-mediated cell adhesion. Inhibiting ubiquitination with both the E1 inhibitor and the ubiquitin mutants did not alter arachidonic acid activation of HSP-27, PKD, or RhoA (data not shown). Therefore, we hypothesize that ubiquitination is important for cell adhesion at a step downstream of these functions. Some possible downstream targets of ubiquitination include proteins involved in focal adhesion formation, cytoskeletal rearrangements required for cellular adhesion and migration, or activation of integrins. There are other signaling molecules, such as Akt and β-catenin, for which K63 polyubiquitination has been linked to cancer development (Shekhar et al. 2008; Yang et al. 2009), but we have no evidence for these proteins playing a role in arachidonic acid signaling in our system.
Altogether, the novel findings reported herein suggest that fatty acid induction of cell adhesion and migration utilizes a unique K63 ubiquitin-regulated pathway. Identifying the ubiquitinated proteins in this pathway may lead to new therapeutic targets for preventing metastasis and is the focus of future studies.
Acknowledgments
We thank Drs. Harriet Kinyamu and Thomas Eling for a careful review of the manuscript. This work has been supported in part or in whole by the Intramural Research Program of the NIEHS and the NIH.
Contributor Information
Denise M. Ray, Laboratory of Molecular Carcinogenesis, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA
Brian A. Rogers, Laboratory of Molecular Carcinogenesis, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA; Department of Biology, North Carolina Central University, Durham, NC 27707, USA
Jeffrey A. Sunman, Laboratory of Molecular Carcinogenesis, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA
Steven K. Akiyama, Laboratory of Molecular Carcinogenesis, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA
Kenneth Olden, Laboratory of Molecular Carcinogenesis, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
John D. Roberts, Laboratory of Molecular Carcinogenesis, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
References
- Akiyama SK. Functional analysis of cell adhesion: quantitation of cell-matrix attachment. Methods Cell Biol. 2002;69:281–296. doi: 10.1016/S0091-679X(02)69018-1. [DOI] [PubMed] [Google Scholar]
- Akiyama SK, Yamada SS, Chen WT, Yamada KM. Analysis of fibronectin receptor function with monoclonal antibodies: roles in cell adhesion, migration, matrix assembly, and cytoskeletal organization. J Cell Biol. 1989;109(2):863–875. doi: 10.1083/jcb.109.2.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier C, Broday L, Bhoumik A, Israeli S, Takahashi S, Nakayama K, et al. RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol Cell Biol. 2003;23(15):5331–5345. doi: 10.1128/MCB.23.15.5331-5345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson KL, Hubbard NE. Dietary fat and tumor metastasis. Nutr Rev. 1990;48(1):6–14. doi: 10.1111/j.1753-4887.1990. tb02871.x. [DOI] [PubMed] [Google Scholar]
- Fan Y, Yu Y, Shi Y, Sun W, Xie M, Ge N, et al. Lysine 63-linked polyubiquitination of TAK1 at lysine 158 is required for tumor necrosis factor α- and interleukin-1β-induced IKK/NF-κB and JNK/AP-1 activation. J Biol Chem. 2010;285(8):5347–5360. doi: 10.1074/jbc.M109.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MC, Ray DM, Lackford B, Rubino M, Olden K, Roberts JD. Arachidonic acid stimulates cell adhesion through a novel p38 MAPK-RhoA signaling pathway that involves heat shock protein 27. J Biol Chem. 2009;284(31):20936–20945. doi: 10.1074/jbc.M109.020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J, Lerman LO, Lerman A. Ubiquitin and ubiquitin-like proteins in protein regulation. Circ Res. 2007;100(9):1276–1291. doi: 10.1161/01.RES.0000264500.11888.f0. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67(1):425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258(13):8206–8214. [PubMed] [Google Scholar]
- Huang C, Rajfur Z, Yousefi N, Chen Z, Jacobson K, Ginsberg MH. Talin phosphorylation by Cdk5 regulates Smurf1-mediated talin head ubiquitylation and cell migration. Nat Cell Biol. 2009;11(5):624–630. doi: 10.1038/ncb1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WG, Hiscox S, Hallett MB, Scott C, Horrobin DF, Puntis MC. Inhibition of hepatocyte growth factor-induced motility and in vitro invasion of human colon cancer cells by gamma-linolenic acid. Br J Cancer. 1995;71(4):744–752. doi: 10.1038/bjc.1995.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447(7148):1135–1138. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- Jin C, Yang YA, Anver MR, Morris N, Wang X, Zhang YE. Smad ubiquitination regulatory factor 2 promotes metastasis of breast cancer cells by enhancing migration and invasiveness. Cancer Res. 2009;69(3):735–740. doi: 10.1158/0008-5472.CAN-08-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanning GL, Lin TY. Unsaturated fatty acid effects on human breast cancer cell adhesion. Nutr Cancer. 1995;24(1):57–66. doi: 10.1080/01635589509514393. [DOI] [PubMed] [Google Scholar]
- Kaabeche K, Guenou H, Bouvard D, Didelot N, Listrat A, Marie PJ. Cbl-mediated ubiquitination of α5 integrin subunit mediates fibronectin-dependent osteoblast detachment and apoptosis induced by FGFR2 activation. J Cell Sci. 2005;118(6):1223–1232. doi: 10.1242/jcs.01679. [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15(4):535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Khanapure SP, Garvey DS, Janero DR, Letts LG. Eicosanoids in inflammation: biosynthesis, pharmacology, and therapeutic frontiers. Curr Top Med Chem. 2007;7(3):311–340. doi: 10.2174/156802607779941314. [DOI] [PubMed] [Google Scholar]
- Kishida S, Sanjo H, Akira S, Matsumoto K, Ninomiya-Tsuji J. TAK1-binding protein 2 facilitates ubiquitination of TRAF6 and assembly of TRAF6 with IKK in the IL-1 signaling pathway. Genes Cells. 2005;10(5):447–454. doi: 10.1111/j.1365-2443.2005.00852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack GS, Marshall A. Lost in migration. Nat Biotechnol. 2010;28(3):214–229. doi: 10.1038/nbt0310-214. [DOI] [PubMed] [Google Scholar]
- McGrath JP, Jentsch S, Varshavsky A. UBA1: an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991;10(1):227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nony PA, Kennett SB, Glasgow WC, Olden K, Roberts JD. 15(S)-Lipoxygenase-2 mediates arachidonic acid-stimulated adhesion of human breast carcinoma cells through the activation of TAK1, MKK6, and p38 MAPK. J Biol Chem. 2005;280(36):31413–31419. doi: 10.1074/jbc.M500418200. [DOI] [PubMed] [Google Scholar]
- Paine E, Palmantier R, Akiyama SK, Olden K, Roberts JD. Arachidonic acid activates mitogen-activated protein (MAP) kinase-activated protein kinase 2 and mediates adhesion of a human breast carcinoma cell line to collagen type IV through a p38 MAP kinase-dependent pathway. J Biol Chem. 2000;275(15):11284–11290. doi: 10.1074/jbc.275.15.11284. [DOI] [PubMed] [Google Scholar]
- Palmantier R, Roberts JD, Glasgow WC, Eling T, Olden K. Regulation of the adhesion of a human breast carcinoma cell line to type IV collagen and vitronectin: roles for lipoxygenase and protein kinase C. Cancer Res. 1996;56(9):2206–2212. [PubMed] [Google Scholar]
- Palmantier R, George MD, Akiyama SK, Wolber FM, Olden K, Roberts JD. cis-Polyunsaturated fatty acids stimulate β1 integrin-mediated adhesion of human breast carcinoma cells to type IV collagen by activating protein kinases C-ε and -μ. Cancer Res. 2001;61(6):2445–2452. [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Rose DP, Connolly JM. Dietary fat and breast cancer metastasis by human tumor xenografts. Breast Cancer Res Treat. 1997;46(2–3):225–237. doi: 10.1023/A:1005971317978. [DOI] [PubMed] [Google Scholar]
- Rose DP, Connolly JM, Liu XH. Dietary fatty acids and human breast cancer cell growth, invasion, and metastasis. Adv Exp Med Biol. 1994;364:83–91. doi: 10.1007/978-1-4615-2510-3_8. [DOI] [PubMed] [Google Scholar]
- Shekhar MP, Gerard B, Pauley RJ, Williams BO, Tait L. Rad6B is a positive regulator of β-catenin stabilization. Cancer Res. 2008;68(6):1741–1750. doi: 10.1158/0008-5472.CAN-07-2111. [DOI] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. Tumor necrosis factor (TNF)-induced germinal center kinase-related (GCKR) and stress-activated protein kinase (SAPK) activation depends upon the E2/E3 complex Ubc13-Uev1A/TNF receptor-associated factor 2 (TRAF2) J Biol Chem. 2003;278(17):15429–15434. doi: 10.1074/jbc.M211796200. [DOI] [PubMed] [Google Scholar]
- Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, et al. The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10(10):1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17(3):431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6(8):599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Okamoto T, Takeda K, Sato S, Sanjo H, Uematsu S, et al. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol. 2006;7(9):962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67(19):9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325(5944):1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz M, Christofori G. Mechanisms of motility in metastasizing cells. Mol Cancer Res. 2010;8(5):629–642. doi: 10.1158/1541-7786.MCR-10-0139. [DOI] [PubMed] [Google Scholar]
- Zacksenhaus E, Sheinin R. Molecular cloning, primary structure and expression of the human X linked A1S9 gene cDNA which complements the ts A1S9 mouse L cell defect in DNA replication. EMBO J. 1990;9(9):2923–2929. doi: 10.1002/j.1460-2075.1990.tb07483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]