Abstract
For over 120 years it has been appreciated that certain salts (kosmotropes) cause the precipitation of proteins, whilst others (chaotropes) increase their solubility. The cause of this, “Hofmeister effect” is still unclear; especially with the original concept that kosmotropic anions “make” water structure and chaotropes “break” it being countered by recent studies suggesting otherwise. Here, we present the first direct evidence that chaotropic anions have an affinity for hydrophobic concavity, and that it is competition between a convex hydrophobe and the anion for a binding site that leads to the apparent weakening of the hydrophobic effect by chaotropes. In combination, these results suggest that chaotropes primarily induce protein solubilization by direct binding to concavity in the molten globule state of a protein.
Since the pioneering work of Hofmeister over 120 years ago,1,2 it has been appreciated that certain salts decrease the solubility of proteins whilst others increase it. This Hofmeister effect is most evident with anions, with studies repeatedly revealing the “Hofmeister series”, typically: F−, SO42−, AcO−, Cl−, Br−, NO3−, ClO3−, I−, ClO4− and SCN−, with highly solvated fluoride and sulfate decreasing the solubility of a protein and increasing the stability of its fold, and weakly solvated anions such as perchlorate and thiocyanate inducing the opposite. Hofmeister linked these phenomena to the already firmly established observation that salts dissolved in water typically result in an increase in viscosity,3 and progress in understanding salt-viscosity relationships4,5 was in part responsible for the idea that salts influence the bulk structure of water. The idea that salting-out anions or kosmotropes (water structure makers), and salting-in anions or chaotropes (water structure breakers) modulate the hydrogen-bonding network of bulk water has been further supported by neutron diffraction experiments.6 However, most recently this idea has been challenged by evidence from femtosecond time-resolved infrared spectroscopy (fs-IR),7–9 dielectric relaxation (DR) spectroscopy,7,10,11 and optical Kerr-effect spectroscopy.10 In combination these studies reveal that salts generally do not exert a significant influence beyond the first or second solvation shell, and that what influence they do have cannot account for all aspects of the Hofmeister effect; and in particular why anionic chaotropes increase the solubility of proteins and unfold their 3° structure to give the molten globule state.12,13 For this reason direct anion-protein interactions have also been investigated. In particular, protein-fold destabilization via specific interactions with peptide groups and other hydrogen bond donor groups of proteins have been closely scrutinized,14–18 with for example a weak association (Ka = 2 M−1) between perchlorate ion and amide groups being determined.19–21 Complementing these studies have been investigations into the possibility that anions interact directly with the hydrophobic surfaces of molecules.22 Thus both in silico studies23,24 and models based on surface tension increments17,25,26 suggest weak interactions between hydrophobic groups and chaotropes. Here, using a model 1:1 host-guest system, we provide the first direct calorimetric and spectroscopic (1H NMR) evidence that chaotropic anions bind to hydrophobic concave surfaces. In combination these studies demonstrate that in the context of a concave surface, these anions effectively compete with hydrophobic guests for hydrophobic concavity, and in doing so modulate the thermodynamics of hydrophobe binding in a way that mirrors the Hofmeister effect. These results suggest that the observed ability of chaotropic anions to weaken the hydrophobic effect and induce salting-in effects arises through binding to the pitted surface of the molten globule state.
Water-soluble host 1 (Figure 1) is a curved amphiphile. It possesses a water-soluble outer coat comprised of eight carboxylic acids, and an 8 Å wide × 8 Å deep hydrophobic pocket that binds guests as small as ethane and as large as adamantanes. The binding pocket of the host also possesses a slightly hydrophobic rim that leads to a predisposition to dimerize in aqueous solution. The resulting supramolecular nanocapsules have been shown to display a wide range of unusual phenomenon,27–30 but when an amphiphilic guest such as adamantane carboxylic acid (AC, Figure 1) binds to the pocket under slightly basic conditions the guest selects a singular binding orientation in which the hydrophobic moiety of the guest occupies the cavity, and the polar carboxylate – located at the entrance to the cavity – is fully solvated by water. As a result, the hydrophobic rim of the host is rendered less so, and only distinct 1:1 host-guest complexation is observed.31
Figure 1.
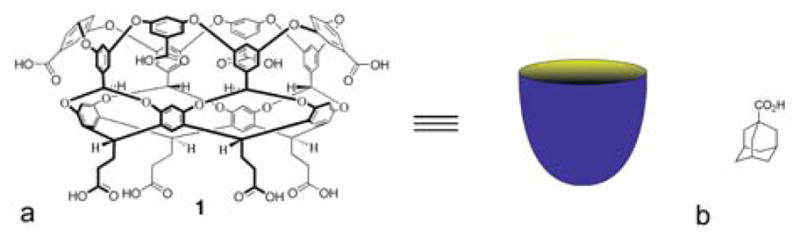
a) Chemical structure of host 1 and a schematic (blue bowl) representation of its overall topology. b) Structure of guest AC.
We examined the binding of AC to host 1 ([150 μM]) in 10 mM phosphate buffer at pH 11.3. The basic conditions chosen ensured that the host was sufficiently soluble in water for Isothermal Titration Calorimetry (ITC) experiments, and that the amphiphilic guest remained in its conjugate-base form. ITC was used to determine the association constant (Ka), free energy change (ΔG°), enthalpy change (ΔH°), and entropy change (TΔS°) for binding of AC to 1 both in the absence and presence of 100 mM sodium salts (F−, SO42−, AcO−, Cl−, Br−, NO3−, ClO3−, I−, SCN− and ClO4−). The results are presented in Figure 2 and Supporting Material). In the absence of any added salt, the binding of AC releases 9.1 kcal mol−1 of free energy, the vast majority of which stems from the large enthalpic change upon guest complexation. However, there is also a very small contribution to binding from a net entropy change. We have previously reported an in silico study of the solvation of 1 demonstrating that solvation of its cavity liberates approximately 5 kcal mol−1 of free energy, a result of an enthalpy change of ΔH° = −20 kcal mol−1 which is countered by a comparable entropic penalty of −TΔS° = +15 kcal mol−1.32 Thus, it is the release of ‘ordered’ water molecules of solvation that lie behind the small entropic boost to AC binding. For binding in the presence of kosmotropic salts such as fluoride, as well as mid-Hofmeister series salts such as bromide, the free energy of guest binding is increased by approximately 0.5 kcal mol−1. In other words, as expected, the hydrophobic effect is enhanced by the presence of these salts. Responsible for this small free energy change are a small but significant increase in the exothermicity of complexation and a concomitant increase in the contribution from entropy. In contrast, for the series of chaotropes, chlorate, iodide, thiocyanate and perchlorate there was a monotonic trend of decreasing free energy of complexation such that in the presence of perchlorate the binding of AC was 1.5 kcal mol−1 weaker than fluoride. Hence these salts induce in this complexation process the same salting-in effect observed in proteins and other molecules. More significantly, beneath this weakening of the hydrophobic effect there was a rapid drop in the enthalpic contribution and a corresponding increase in the role of entropy to the overall free energy change. Indeed, with perchlorate, AC binding was actually endothermic and entirely driven by entropy. Hence, relative to the case when fluoride ion was present, there was a drop in enthalpy of approximately 10 kcal mol−1 and an almost compensating change in −TΔS°.
Figure 2.
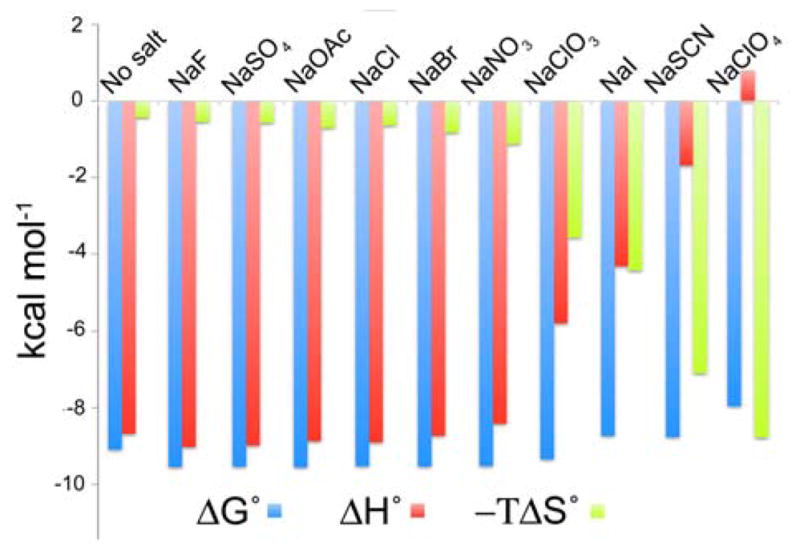
ITC date for the binding of AC to 1 in 10 mM phosphate buffer (pH = 11.3), in the absence and presence of various sodium salts. ([Host] = 150 μM, [salt] = 100 mM, [guest titrant] = 1.5 mM).
To investigate the cause of this swing in thermodynamics, we examined the 1H NMR spectrum of host 1 in the presence of different concentrations of each of the Hofmeister salts. For the kosmotropes and mid-Hofmeister series salts only slight signal broadening was observed at higher (600 mM) concentrations. In contrast, with the salts nitrate, chlorate, iodide, thiocyanate and perchlorate a number of spectral changes were observed. Most illustratively, the signal from the four benzal protons that project into the base of the hydrophobic cavity – atoms that are reliable reporters of guest complexation – were noted to shift downfield upon increasing salt concentrations. Figure 3 shows a series of 1H NMR spectra for host 1 in the presence of increasing amounts of NaClO4, and the 1H NMR spectra of the host in the presence of 42 mM salt and one equivalent of organic guest AC. As the NaClO4 concentration is increased there is a downfield shift in the signal from the benzal protons, from ca. 4.38 ppm in the spectrum in the absence of salt to ca. 4.82 ppm in the presence of 42 mM of the salt. Importantly, this downfield shift could be nullified by the addition of one equivalent of strongly binding AC; the resulting NMR spectrum being essentially identical to that obtained for the host-guest complex in the absence of salt. These results demonstrate that the chaotropic anions bind to the hydrophobic pocket of 1, and this was further confirmed by the excellent 1:1 binding isotherms obtained by accurate titrations of the different chaotropic salts into the solution of the host (Figure 1 insert). Fitting of the obtained isotherms for the five chaotrope salts gave association constants of: NaNO3, < 1 M−1, NaClO3 = 3 M−1, NaI = 11 M−1, NaSCN = 33 M−1, and NaClO4 = 95 M−1. Thus, the strongest binding perchlorate anion binds to the host liberating 2.70 kcal mol−1 of free energy; a relatively modest free energy change, but one that is sufficient to allow the salt to effectively compete with the guest when the former is present at much higher concentrations.
Figure 3.

1H NMR spectra of 1 (1 mM) in 10 mM phosphate buffer (pD = 11.3) in the presence of: a) no salt; b) 2 mM NaClO4; c) 8 mM NaClO4; d) 26 mM NaClO4; e) 42 mM NaClO4; f) 42 mM NaClO4 + and 1 mM AC (bound guest signals at highfield). The signal for the benzal hydrogens is labeled (*). The suppressed water signal is at ca. 4.70 ppm. Inset: Binding isotherm based on the benzal hydrogen atoms signal shifts for the complexation of ClO4− to 1. The gap in the data is a result of the signal coalescing with the suppressed water signal.
To our knowledge this is the first observation of anions binding to hydrophobic concavity. Being larger than isoelectric cations, anions have a lower charge to mass ratio and are more challenging targets for recognition because their more diffuse nature results in less effective electrostatic binding interactions.33–35 In addition, anion binding may be pH sensitive because they may become protonated outside a certain pH window, and can form strong hydrogen bonds with hydroxylic solvents such as water. Hence, general practice has been to form receptors that utilize the enthalpically strongest of non-covalent interactions, i.e., those involving ion-ion or ion-dipole forces, in order to compete with the solvation shell and affect recognition. The results here however demonstrate that hydrophobic concavity can bring about the selective recognition of more weakly solvated chaotropic anions.
The dehydration free energy of perchlorate has been accurately determined to be 51 kcal mol−1.36 However, it is difficult to define either an accurate hydration number (the number of moles of slow water dipoles per mole of dissolved salt) or an accurate number of waters in the solvation shell (a thermodynamic ‘average’ picture defined with respect to a distance distribution from the center of the ion). However, computational studies suggest that at least 8 waters of solvation surround the ion.37 The [ClO4(H2O)8]− anion is however too large to fit within the confines of 1 and so must necessarily undergo partial dehydration in order to bind. Indeed, unpublished preliminary in silico studies confirm that perchlorate is solvated within the cavity with 3–4 waters. Hence the observed selectivities in anion binding relate to the adaptability of its solvation shell; the more strongly solvated kosmotropic anions cannot reorganize and/or lose part of their solvation shell so as to be accommodated into the hydrophobic cavity, whereas the chaotropic anions are more flexible in this regard.
To gain further insight into anion binding we first carried out a van’t Hoff plot for the binding of perchlorate to 1 using 1H NMR. The difference between the ΔH° and −TΔS° values for the binding of AC in the absence and presence of perchlorate (Table S1, Supporting Material) suggests that perchlorate binding is strongly exothermic but comes with a large entropic penalty, and the van’t Hoff plot confirmed this: ΔH° = −10.62 kcal mol−1 and −TΔS° = +7.94 kcal mol−1. A previous in silico study of the structure of 1 in pure water revealed that its binding site is solvated with 0–7 water molecules (4.4 on average), and that these waters generally possess fewer hydrogen bonds than the bulk, have slower translational motion and faster orientational motion, and are relatively ordered.32 Hence, in combination with our current studies it is evident that the hydrophobic pocket of 1 is hydrated at an entropic cost, and that these waters are readily displaced by a partially hydrated perchlorate anion but at a further entropic cost. Thus, in the context of concavity it appears that chaotropes (structure breakers) are actually structure makers.
What if any is the role of the sodium ion? Computational studies suggest that from a thermodynamic viewpoint there is at least one water layer between sodium and perchlorate ions,37 and recent studies that examine water reorientation dynamics around salts using DR spectroscopy38 suggest that even relatively ‘strongly’ associating divalent metal sulfates such as MgSO4 associate only weakly through contact, solvent shared, or double solvent separated ion pairs at the concentrations in question. Furthermore, very recent studies using a combination of fs-IR and DR spectroscopy7 indicate that although combinations of strongly solvated anions and cations can lead to interdependent and non-additive effects upon their solvation shells, the combination of weakly solvated perchlorate ion and moderately solvated sodium ion does not lead to any significant perturbation of the dynamical water structure between them. These results suggest that close association of the sodium ion is not essential for anion binding.
The broader conclusion from these results is that anion-concavity interactions are important in the salting in of macromolecules by chaotropes. Whilst perhaps it is more intuitive that the important anion-protein interactions involve hydrogen bond donor groups, perchlorate ion binding with amide groups of poly(N-isopropylacrylamide) are very weak (ΔG° = 0.41 kcal mol−1),19 much weaker than the association of chaotropic anions to the hydrophobic concavity of octa-anionic 1 (ΔG° = 2.70 kcal mol−;1). Building on this thought, in increasing the solubility of proteins chaotropes generally shift the folding-equilibrium from the fully folded state to the molten globule state. In other words, they break 3° structure by solubilizing the core hydrophobic residues, whilst maintaining 2° structure.12,13 We hypothesize that the molten globule state – which possesses a much larger accessible hydrophobic surface area than a folded protein, and much more defined structure (and concavity) than a fully denatured protein – is the ideal structure for binding chaotropic anions which consequently compete with interactions between hydrophobic surfaces.
In conclusion, chaotropic anions have a considerable affinity for hydrophobic concavity; an affinity that is much larger than that measured between anions and amide groups. This affinity allows chaotropic anions to compete with the interaction of two hydrophobic surfaces, a phenomenon that is manifested in a “weakening” of the hydrophobic effect. These results suggest that an important mechanism by which chaotropic anions destabilize protein folds, and increase their solubility, centers on interactions between these anions and molecular concavity.
Supplementary Material
Acknowledgments
BCG, and CLDG acknowledge the financial support of the National Institutes of Health (GM074031) in carrying out this research.
Footnotes
Supporting Information Available:
ITC and NMR titration data, NMR van’t Hoff plots, and fitting of thermodynamic data. This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Hofmeister F. Arch Exp Pathol Pharmakol. 1888;24:247. [Google Scholar]
- 2.Kunz W, Henle J, Ninham BW. Curr Opin Colloid In. 2004;9:19. [Google Scholar]
- 3.Poiseuille JML. Ann Chim Phys. 1847;21:76. [Google Scholar]
- 4.Jenkins HDB, Marcus Y. Chem Rev. 1995;95:2695. [Google Scholar]
- 5.Jones G, Dole M. J Am Chem Soc. 1929;51:2950. [Google Scholar]
- 6.Mancinelli R, Botti A, Bruni F, Ricci MA, Soper AK. Physical Chemistry Chemical Physics. 2007;9:2959. doi: 10.1039/b701855j. [DOI] [PubMed] [Google Scholar]
- 7.Tielrooij KJ, Garcia-Araez N, Bonn M, Bakker HJ. Science. 2010;328:1006. doi: 10.1126/science.1183512. [DOI] [PubMed] [Google Scholar]
- 8.Omta AW, Kropman MF, Woutersen S, Bakker HJ. Science. 2003;301:347. doi: 10.1126/science.1084801. [DOI] [PubMed] [Google Scholar]
- 9.Moilanen DE, Wong D, Rosenfeld DE, Fenn EE, Fayer MD. Proceedings of the National Academy of Sciences. 2009;106:375. doi: 10.1073/pnas.0811489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turton DA, Hunger J, Hefter G, Buchner R, Wynne K. The Journal of Chemical Physics. 2008;128:161102. doi: 10.1063/1.2906132. [DOI] [PubMed] [Google Scholar]
- 11.Wachter W, Kunz W, Buchner R. J Phys Chem A. 2005;109:8675. doi: 10.1021/jp053299m. [DOI] [PubMed] [Google Scholar]
- 12.Hamada D, Kidokoro SI, Fukada H, Takahashi K, Goto Y. Proceedings of the National Academy of Sciences. 1994;91:10325. doi: 10.1073/pnas.91.22.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynn RM, Konishi Y, Scheraga HA. Biochemistry. 1984;23:2470. doi: 10.1021/bi00306a023. [DOI] [PubMed] [Google Scholar]
- 14.Tadeo X, Lopez-Mendez B, Castano D, Trigueros T, Millet O. Biophys J. 2009;97:2595. doi: 10.1016/j.bpj.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eggers DK, Valentine JS. J Mol Biol. 2001;314:911. doi: 10.1006/jmbi.2001.5166. [DOI] [PubMed] [Google Scholar]
- 16.Lund M, Jungwirth P. J Phys Condens Matter. 2008;20:494218. [Google Scholar]
- 17.Pegram LM, Wendorffa T, Erdmanna R, Shkela I, Bellissimoa D, Felitskya DJ, Record MTJ. Proc Natl Acad Sci U S A. 2010;107:7716. doi: 10.1073/pnas.0913376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timasheff SN. Biochemistry. 2002;41:13473. doi: 10.1021/bi020316e. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Furyk S, Bergbreiter DE, Cremer PS. J Am Chem Soc. 2005;127:14505. doi: 10.1021/ja0546424. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Cremer PS. Curr Opin Chem Biol. 2006;10:658. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 21.Zhang YJ, Cremer PS. Annu Rev Phys Chem. 2010;61:63. doi: 10.1146/annurev.physchem.59.032607.093635. [DOI] [PubMed] [Google Scholar]
- 22.Baldwin RL. Biophys J. 1996;71:2056. doi: 10.1016/S0006-3495(96)79404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zangi R, Hagen M, Berne BJ. J Am Chem Soc. 2007;129:4678. doi: 10.1021/ja068305m. [DOI] [PubMed] [Google Scholar]
- 24.Smith JD, Saykally RJ, Geissler PL. J Am Chem Soc. 2007;129:13847. doi: 10.1021/ja071933z. [DOI] [PubMed] [Google Scholar]
- 25.Pegram LM, Record MTJ. J Phys Chem B. 2007;111:5411. doi: 10.1021/jp070245z. [DOI] [PubMed] [Google Scholar]
- 26.Pegram LM, Record MTJ. Chemical Physics Letters. 2008;467:1. doi: 10.1016/j.cplett.2008.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S, Gan H, Hermann AT, Rick SW, Gibb BC. Nature Chemistry. 2010;2:847. doi: 10.1038/nchem.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Gibb BC. Chem Commun. 2008:3709. doi: 10.1039/b805446k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibb CLD, Sundaresan AK, Ramamurthy V, Gibb BC. J Am Chem Soc. 2008;130:4069. doi: 10.1021/ja7107917. [DOI] [PubMed] [Google Scholar]
- 30.Natarajan A, Kaanumalle LS, Jockusch S, Gibb CLD, Gibb BC, Turro NJ, Ramamurthy V. J Am Chem Soc. 2007;129:4132. doi: 10.1021/ja070086x. [DOI] [PubMed] [Google Scholar]
- 31.Sun H, Gibb CLD, Gibb BC. Supramol Chem. 2008;20:141. [Google Scholar]
- 32.Ewell J, Gibb BC, Rick SW. J Phys Chem B. 2008;112:10272. doi: 10.1021/jp804429n. [DOI] [PubMed] [Google Scholar]
- 33.Sessler JL, Gale PA, Cho W-S. Anion Receptor Chemistry. Royal Society of Chemistry; Cambridge: 2006. [Google Scholar]
- 34.Gale PA. Coord Chem Rev. 2001;213:79. [Google Scholar]
- 35.Caltagirone C, Gale PA. Chem Soc Rev. 2009;38:520. doi: 10.1039/b806422a. [DOI] [PubMed] [Google Scholar]
- 36.Marcus Y. Ion Properties. 1. Marcel Dekker; New York: 1997. [Google Scholar]
- 37.General IJ, Asciutto EK, Madura JD. J Phys Chem B. 2008;12:15417. doi: 10.1021/jp806269w. [DOI] [PubMed] [Google Scholar]
- 38.Buchner R, Chen T, Hefter G. J Phys Chem B. 2004;108:2365. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


